Abstract
The CCAAT/enhancer-binding protein β (C/EBPβ) is required for adipocyte differentiation and maturation. We have studied the role of the transcription factor, C/EBPβ, in the development of diet-induced obesity. Mice with a deletion in the gene for C/EBPβ (C/EBPβ−/−) and wild-type mice were fed a high-fat diet (60% fat) for 12 weeks. The C/EBPβ−/− mice lost body fat, whereas the wild-type mice increased their total body fat on a high-fat diet. The C/EBPβ−/− mice had lower levels of blood triglycerides, free fatty acids, cholesterol, and hepatic triglyceride accumulation compared with the wild-type mice, thus protecting them from diet-induced obesity and fatty liver on a high-fat diet. Deletion of C/EBPβ gene resulted in greatly reducing hepatic lipogenic genes, acetyl CoA carboxylase, and fatty acid synthase and increasing the expression of β-oxidation genes in the brown adipose tissue. CO2 production was significantly higher in the C/EBPβ−/− mice as was the level of uncoupling protein (UCP)-1 and UCP-3 in the muscle. In conclusion, the transcription factor C/EBPβ is an important regulator in controlling lipid metabolism and in the development of diet-induced obesity.
In the last 20 years, the prevalence of obesity has greatly increased by ~10–15% (1). With the increase of obesity, there is an increased risk of certain diseases such as diabetes, hypertension, coronary heart disease, and even some types of cancer (2–4). The development of obesity is a consequence of both environmental and genetic factors. The CCAAT/enhancer-binding protein (C/EBP) family of transcription factors has been shown to coordinate expression of genes involved in adipocyte differentiation (5–8). During adipogenesis, the C/EBP family functions in a transcriptional cascade. C/EBPβ and C/EBPδ are expressed early in development and activate the transcription of peroxisome proliferator–activated receptor γ (PPARγ). This results in induced expression of the C/EBPα gene. C/EBPβ can also activate C/EBPα independently of PPARγ (9,10). These transcription factors activate lipogenic genes necessary for terminal differentiation of the adipocyte.
Much understanding of the roles for the C/EBP family of transcription factors has been elucidated from gene-targeted deletion studies in mice. The mice with a deletion for the C/EBPα gene (C/EBPα−/−) lack the development of adipose tissue and die in the perinatal period with a profoundly fatty liver (7). Mice with a deletion for the C/EBPβ gene (C/EBPβ−/−) have adipocytes but fail to accumulate lipid in the cells even though they have normal levels of PPARγ and C/EBPα (8). Mice with a double gene deletion for C/EBPβ and C/EBPδ have a significant decrease in the weight of epididymal white adipose tissue (WAT), suggesting that both transcription factors have a major role in the development of adipose tissue and thus obesity (8).
In addition to a role in adipose tissue development, the members of the C/EBP family are involved in a broad range of physiological processes. These range from the acute-phase response to regulation of glucose and lipid metabolism (11). C/EBPβ−/− mice have two phenotypes, A and B. Mice with the B phenotype die in the perinatal period because of hypoglycemia and a failure to mobilize their glycogen (12). Mice with the A phenotype survive to adulthood, but exhibit fasting hypoglycemia, with reduced plasma insulin, blood lipids, free fatty acids (FFAs) and impaired hepatic glucose production. The C/EBPβ−/− mice have a blunted response to glucagon and adrenaline primarily due to altered levels of hepatic cAMP production and reduced activity of protein kinase A (6,12). These results suggest that deletion of the C/EBPβ gene affects both glucose homeostasis and lipid homeostasis.
In this study, we wanted to extend the previous studies for the role of C/EBPβ in lipid homeostasis. The main aim of this study was to determine whether the deletion of the gene for C/EBPβ would protect the mice from developing obesity and fatty liver on a high-fat diet.
RESEARCH DESIGN AND METHODS
The C/EBPβ−/− mice were obtained for this study by breeding heterozygous female mice with a targeted deletion in the gene of C/EBPβ (C/EBPβ+/−) with heterozygous male mice. The generation of the C/EBPβ−/− mice and their genetic background has been described previously by Screpanti et al. (13). Briefly, embryonic stem cell clones from the CCE cell line (derived from the 129/Sv/Ev strain) carrying the mutation were injected into C57BL/6J blastocysts and were transplanted into the uteri of F1 (CBA × C57BL/6J) foster mothers. Male chimeras were mated to MF1 females, and offspring heterozygous for the mutant allele were intercrossed to obtain homozygous mice. Adult male and female mice were 8–12 weeks old at the time of their use. Mice containing the gene for C/EBPβ (C/EBPβ+/+) are designated as wild-type animals and are used for controls throughout the study. Screening for the C/EBPβ−/− mice was performed by Southern blot analysis as outlined previously (6). All experimental protocols were approved by the Case Western Reserve University Institutional Animal Care and Use Committee.
Dietary studies
Eight-week-old wild-type and C/EBPβ−/− mice were housed three to five mice per microisolator cage and were maintained on a fixed 12-h light/dark cycle. Animals had free access to water and were fed normal mouse chow (Teklad F6 8664; Harlan Teklad, Madison, WI) or low-fat diet D12450B or high-fat diet D12492 (Research Diets, New Brunswick, NJ). The chow diet contained 27 kcal% protein, 17 kcal% fat, and 57 kcal% carbohydrate. The low-fat diet (D12450B) contained 20 kcal% protein, 10 kcal% fat, and 70 kcal% carbohydrate. The high-fat diet (D12492) contained 20 kcal% protein, 60 kcal% fat, and 20 kcal% carbohydrate. For the high-fat studies, the mice were fed the diet for a total of 12 weeks. The low-fat– and chow-fed animals were fed for 12 weeks and used as controls as indicated. Food intake measurements were measured as grams of food consumed per kilogram body weight over a 24-h period of mice that were individually housed.
Insulin measurements
Insulin was measured in mice that were fed a high-fat diet for 12 weeks. The mice were fasted for 18 h and injected intraperitoneally with 2 g glucose/kg body wt. Blood was sampled from the tail vein and assayed for insulin at 0 and 30 min with an Ultrasensitive Mouse Insulin ELISA enzyme immunoassay from Mercodia (Lincoln Park, MO).
Biochemical analysis
After an overnight fast, mice were killed, and a heart puncture was performed to collect the blood. Plasma was generated using Microtainer plasma separator tubes from Becton Dickinson (Franklin Lakes, NJ). The measurements of cholesterol, β-hydroxybutyrate, FFA, and triglyceride were performed by Veterinary Diagnostic Services (Marshfield Laboratories, Marshfield, WI). The determination of liver triglycerides was performed using the Triglyceride (GPO) reagent set from Pointe Scientific (Lincoln Park, MI). Insulin was determined with an Ultrasensitive Mouse Insulin ELISA enzyme immunoassay from Mercodia.
Body composition
A bolus of labeled water, 2H2O, was administered by intraperitoneal injection to fasted mice. After allowing for isotope equilibration (2 h), a blood sample was collected by the tail vein, and the 2H enrichment of body water was measured using gas chromatography–mass spectrometry (14). Body water was calculated from the isotope dilution, and the fat mass was determined as previously described (15).
CO2 production
The rate of CO2 production was determined using doubly labeled water (16–18). Fed wild-type and C/EBPβ−/− mice were intraperitoneally injected with a bolus of 2H2O and H218O to achieve initial labeling of ~2% 2H and 0.25% 18O. Blood was sampled at 2, 48, and 72 h after injection. CO2 production was determined from the difference between the elimination of 2H and 18O from body water (17). The 2H-labeling was determined after exchange with acetone (14). The 18O enrichment of water was determined after the generation of trimethylphosphate (17). The rate of CO2 production (moles per day) was calculated using the equation: CO2 production rate = 0.5 × N1 (k18O – k2H), where the fractional biological decay constants (k18O and k2H) are in day−1 and N1 is the body water pool (moles) calculated as [(V2H + V18O)/2]. Data are expressed as means ± SE.
Histology of liver
Liver tissues from fasted mice were fixed in 10% formalin (Sigma, St. Louis, MO) at 4°C. The tissues were embedded in paraffin and stained with hematoxylin and eosin by the Pathology Core Facility (Case Western Reserve University, Cleveland, OH). Pictures were taken at ×40 magnifications.
Real-time RT-PCR analysis
Total RNA was isolated with RNeasy Mini Isolation kit from Qiagen (Valencia, CA). The single-strand cDNA was synthesized from 2 µg total RNA with random hexamer primers and MMTV reverse transcriptase (Ambion, Austin, TX). Real-time PCRs were performed as described previously (19). Standard curves were generated by diluting wild-type liver cDNA for Fig. 3 and Fig. 6 and by diluting wild-type muscle cDNA for Fig. 4 (1:4, 1:8, 1:16, 1:32, 1:64, and 1:128 in H2O). One microliter of the dilution was used for PCR. PCR was performed in a 25-µl volume using Syber Green PCR Core reagent mix (Applied Biosystems, Foster City, CA) as a source of Taq, buffer, and dNTPs. Primers were used at a final concentration of 0.2 µmol/l, and the sequence of the primers used for RT-PCR are in Table 1. PCR was performed in the Chromo4 Cycler (MJ Research) with 40 cycles (15 min at 95°C; followed by 40 cycles of 30 s at 95°C, 30 s at 60°C, 45 s at 72°C, and 30 s at 73°C [optimal read]; followed by a 5 min extension at 72°C and a melting curve). The relative amounts of the mRNAs were determined by linear regression from the standard curves derivative maximum method, with Opticon Monitor 3 software (MJ Research), and 18S rRNA was used as a loading control. The RT-PCR reactions were run on RNA isolated from wild-type or C/EBPβ−/− mice that had been fed a high-fat diet for 12 weeks. For mRNA expressed as mRNA fold over wild type, the gene expression from wild-type mice was set arbitrarily at 1 and the gene expression for the C/EBPβ−/− mice is represented as fold difference relative to wild-type mice gene expression. The gene expression was measured for both fed and fasted conditions in separate RT-PCRs with wild-type mice gene expression set at 1 under both conditions. The number of mice for each gene tested is four.
FIG. 3.
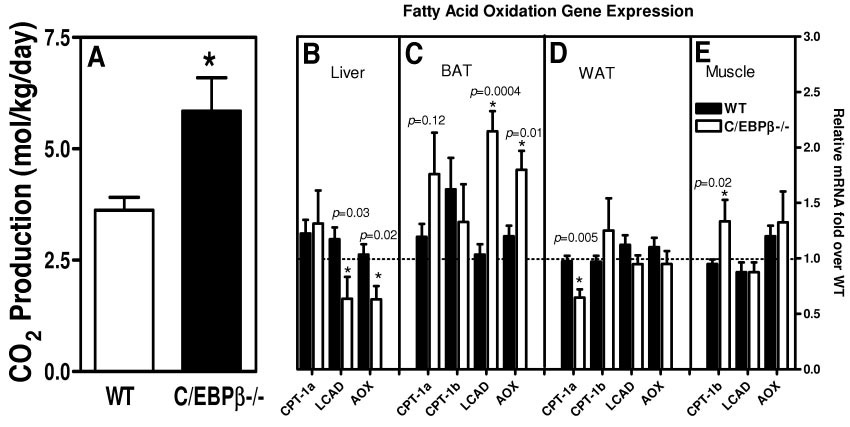
C/EBPβ−/− mice have increased energy expenditure. A: CO2 production for fed wild-type and C/EBPβ−/− mice on a high-fat diet for 12 weeks. The values are the means ± SE for six mice per group for CO2 production. *P < 0.05. Real-time PCR quantification for β-oxidation gene expression in liver (B), BAT (C), WAT (D), and muscle (E) of mice fasted overnight after 12 weeks of high-fat diet. *P < 0.05 compared with wild-type mice. The values are the means ± SE for four mice per group normalized with 18s RNA and expressed as fold difference over wild-type mice in that tissue.
FIG. 6.
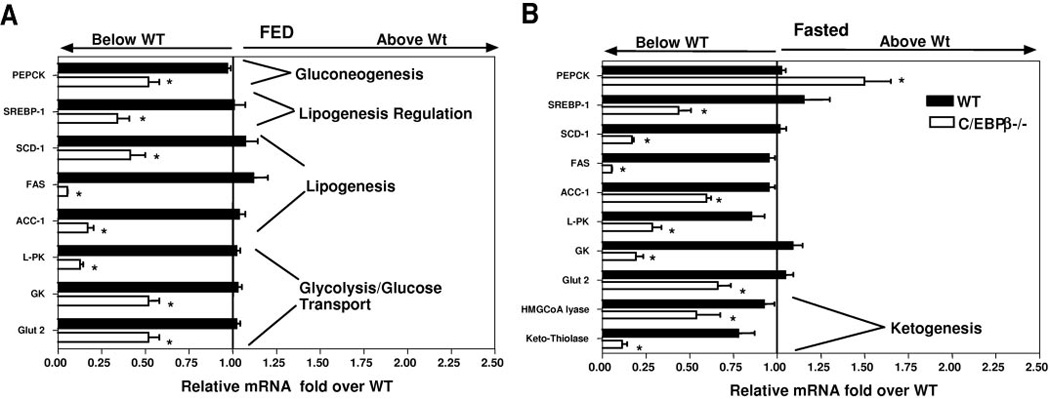
Reduced glycolytic and lipogenic gene expression in C/EBPβ−/− mice. Real-time PCR quantification of liver gene expression for fed (A) and 18-h–fasted (B) wild-type and C/EBPβ−/− mice on a high-fat diet. The values are the means ± SE and expressed as fold over wild type for four mice per group normalized with 18s rRNA (*P < 0.05).
FIG. 4.
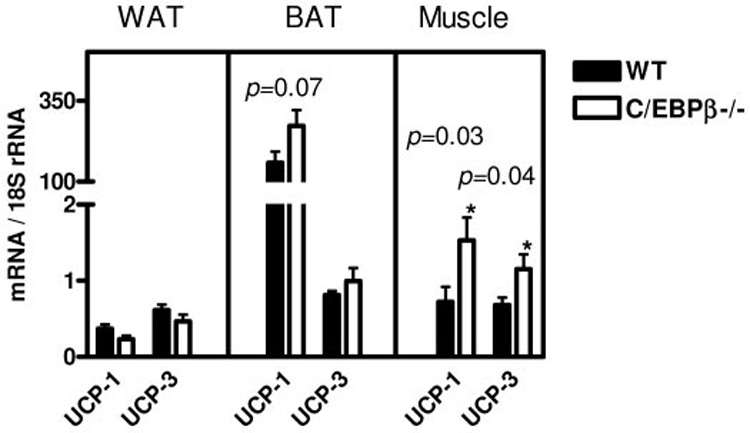
Analysis of UCP gene expression. Real-time PCR quantification in fed mice on a high-fat diet for UCP-1 and UCP-3 gene expression in WAT, BAT, and muscle. The values are the means ± SE for four mice per group normalized with 18s rRNA. *P < 0.05.
TABLE 1.
Primers used for quantitative real-time PCR
| Gene | Primer sequence | Ref. |
|---|---|---|
| GLUT2 | 5′-CCTCAAGAGGTAATAATATCCC | 19 |
| 5′-CCATCAAGAGGGCTCCAGTC | ||
| Glucokinase | 5′-CCCTGAGTGGCTTACAGTTC | 19 |
| 5′-ACGGATGTGGAGTGTTGAAGC | ||
| Liver pyruvate kinase | 5′-CTGGAACACCTCTGCCTTCTG | 19 |
| 5′-CACAATTTCCACCTCCGACTC | ||
| PEPCK | 5′-GTGCTGGAGTGGATGTTCGG | 19 |
| 5′-CTGGCTGATTCTCTGTTTCAGG | ||
| FAS | 5′-AGCGGCCATTTCCATTGCCC | 19 |
| 5′-CCATGCCCAGAGGGTGGTTG | ||
| ACC | 5′-GGGACTTCATGAATTTGCTGATTCTCAGTT | 35 |
| 5′-GTCATTACCATCTTCATTACCTCAATCTC | ||
| SCD-1 | 5′-CCGGAGACCCTTAGATCGA | 19 |
| 5′-TAGCCTGTAAAAGATTTCTGCAAACC | ||
| SREBP-1 | 5′-AACGTCACTTCCAGCTAGAC | 19 |
| 5′-CCACTAAGGTGCCTACAGAGC | ||
| CPT-1a | 5′-AAGCACCAGCACCTGTACCG | 19 |
| 5′-CCTTTACAGTGTCCATCCTCTG | ||
| CPT-1b | 5′-CAAGTTCAGAGACGAACGCC | 37 |
| 5′-TCAAGAGCTGTTCTCCGAACTG | ||
| LCAD | 5′-GCTGCCCTCCTCCCGATGTT | 35 |
| 5′-ATGTTTCTCTGCGATGTTGATG | ||
| AOX | 5′-AAGAGTTCATTCTCAACAGCCC | 19 |
| 5′-CTTGGACAGACTCTGAGCTGC | ||
| Ketothiolase | 5′-TCACGGCAGAAGCAGGATGC | 19 |
| 5′-TGCTCCATCACTCACCTGACTG | ||
| HMG-CoA lyase | 5′-CTGGGCTTAACGGGTCCTC | 19 |
| 5′-TGGCAGTGGACAGCCAATGC | ||
| UCP-1 | 5′-CTGGGCTAGGTAGTGCCAGTG | 36 |
| 5′-CAACCTTGGCTAGACGCACAG | ||
| UCP-3 | 5′-CAACCTTGGCTAGACGCACAG | 36 |
| 5′-TGGAGGTCCGAGGAGAGAGC | ||
| 18S rRNA | 5′-CCATCCAATCGGTAGTAGCG | 19 |
| 5′-GTAACCCGTTGAACCCCATT |
Statistical analysis
Results are expressed as means ± SE. Student’s t tests for paired data were used. Differences were considered significant if P < 0.05.
RESULTS
Reduced accumulation of body fat in C/EBPβ− mice fed a high-fat diet
Both wild-type and C/EBPβ−/− mice were fed either the high-fat diet or low-fat diet for 12 weeks. The animals were weighed weekly, and food intake was measured for a 24-h period weekly (Fig. 1). After 12 weeks on a low-fat diet, wild-type mice had a 17.6% body wt increase, whereas C/EBPβ−/− mice had only 11.1% body wt increase (Fig. 1B and C). However, wild-type mice fed a high-fat diet had a statistically significant increase in body weight of 27%, whereas the C/EBPβ−/− mice had only a 13% increase in body weight (P < 0.03). The wild-type mice consumed less food on the high-fat diet compared with the C/EBPβ−/− mice even though the wild-type mice gained more weight (Fig. 1A) (P < 0.05).
FIG. 1.
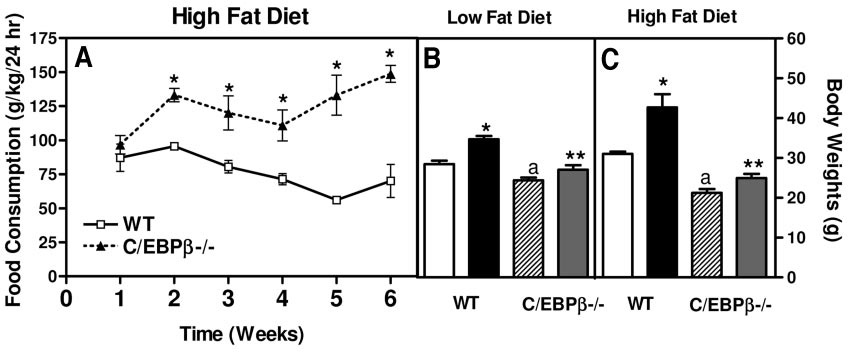
C/EBPβ−/− mice are resistant to increased weight on high-fat diet. A: Food consumed was measured over a 24-h period and normalized by kg body wt for wild-type and C/EBPβ−/− mice. The values are the means ± SE for 11–14 mice per group for food consumption. Body weights were measured for low-fat diets (B) and high-fat diets (C). The wild-type initial (□ 8 weeks old) and wild-type final (■ 20 weeks old) body weights and the C/EBPβ−/− initial (▨ 8 weeks) and C/EBPβ−/− final ( 20 weeks) body weights are shown. The values are the means ± SE for five to eight mice per group. *P < 0.03 compared with wild-type initial body weights; aP < 0.04 compared with wild-type initial body weight; **P < 0.03 compared with wild-type final body weight.
20 weeks) body weights are shown. The values are the means ± SE for five to eight mice per group. *P < 0.03 compared with wild-type initial body weights; aP < 0.04 compared with wild-type initial body weight; **P < 0.03 compared with wild-type final body weight.
To verify whether the increase in body weight was due to an increase in total body lipid, the percentage body fat was measured using the 2H2O method (Fig. 2A). In this experiment, mice were fed a control chow diet, low-fat diet, or high-fat diet for 12 weeks. The wild-type mice had no difference in total body lipid after being fed a low-fat diet. However, after exposure on a high-fat diet for 12 weeks, the wild-type mice had an increase in total body lipid from 27.4 to 36.7% (P < 0.04). The total body lipid for the C/EBPβ−/− mice did not change significantly in response to alteration in diet. The C/EBPβ−/− mice had significantly less total body lipid compared with the wild-type mice on all diets tested (P < 0.04). The C/EBPβ−/− mice dropped total body lipid from 14% on the low-fat diet to 10% on the high-fat diet. Therefore the amount of total body fat was reduced by 29% in the C/EBPβ−/− mice, whereas the wild-type mice increased their total body fat by 34% on a high-fat diet.
FIG. 2.
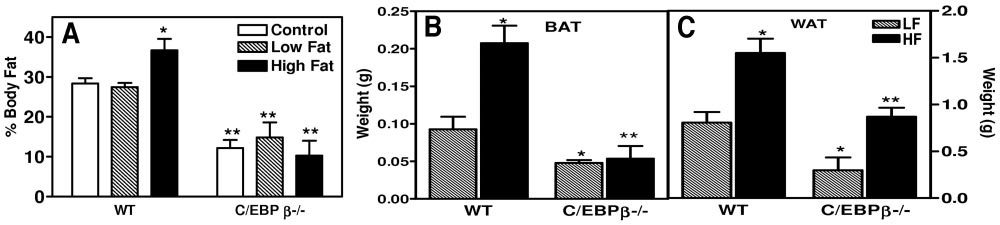
Reduced body fat on high-fat diet. A: The percentage of body fat was measured for wild-type and C/EBPβ−/− mice on control chow, low-fat, or high-fat diets (*P < 0.04 compared with wild-type low-fat and control diet; **P < 0.04 compared with wild-type mice). Fat pads were isolated for BAT (B) and WAT (C) after 12 weeks of either a low-fat or high-fat diet. *P < 0.05 compared with wild-type mice on low-fat diet; **P < 0.05 compared with C/EBPβ−/− mice on low-fat diet. The values are the means ± SE for three to six mice per group.
Brown adipose tissue (BAT) and WAT were isolated and weighed for wild-type and C/EBPβ−/− mice after 12 weeks of either a low-fat or high-fat diet. The wild-type mice had an increased amount of both WAT and BAT on a high-fat diet, whereas the C/EBPβ−/− mice only had a marginal increase in the weight of WAT only. The wild-type mice have a 1.93-fold increase in the weight of the WAT and a 2.2-fold increase in the weight of the BAT on a high-fat diet. The C/EBPβ−/− mice have a 2.9-fold increase of WAT weight on a high-fat diet but no increase in the weight of the BAT on the high-fat diet (Fig. 2B and C).
Increased CO2 production in C/EBPβ−/− mice
Because the C/EBPβ−/− mice consumed more calories than the wild-type mice on a high-fat diet yet remained resistant to diet-induced obesity, we investigated the CO2 production of the mice using the doubly labeled water technique (2H2 18O) (Fig. 3A). We found that the C/EBPβ−/− mice produced 1.6-fold more CO2 compared with the wild-type mice (C/EBPβ−/− 5.8 ± 0.748 and wild type 3.6 ± 0.29 mol CO2 · kg−1 · day−1)(P < 0.04), suggesting increased energy expenditure.
To determine which tissues are contributing to the increased CO2 production, we measured gene expression for the fatty acid oxidation genes and expressed the relative mRNA levels normalized by 18S rRNA as fold over wild-type values for each tissue. The genes tested were the liver-specific form of carnitine palmitoyltransferase (CPT) 1 (CPT-1a), the muscle-specific form of CPT-1 (CPT-1b), long acyl-CoA dehydrogenase (LCAD), and acyl-CoA oxidase (AOX) in fed mice after 12 weeks of a high-fat diet. In the C/EBPβ−/− mice, the liver, WAT, and muscle have similar values of fatty acid oxidation genes for most of the genes tested. In the livers of the C/EBPβ−/− mice, the rate-limiting step of fatty acid oxidation, CPT-1a, is expressed normally. However, there was 0.5- and 0.6-fold less hepatic gene expression for LCAD and AOX, respectively, compared with wild-type mice (Fig. 3B). We tested CPT-1a and CPT-1b in the WAT because both isoforms are expressed in adipose tissues. The level of CPT-1a mRNA expression in WAT is reduced 0.66-fold in the C/EBPβ−/−, whereas the levels of CPT-1b are equally expressed compared with wild-type mice (Fig. 3D). The muscle-specific form of CPT-1, CPT-1b, was elevated by 1.3-fold in the C/EBPβ−/− mice, yet in the muscle of the C/EBPβ−/− mice, we found the same level of LCAD and AOX gene expression compared with wild-type mice.
The increased CO2 production in the C/EBPβ−/− mice may be explained by increased fatty acid oxidation gene expression in the BAT (Fig. 3C). The amount of CPT-1a in the BAT is elevated by 1.5-fold but is not statistically significant (P = 0.12). The expression of CPT-1b in the BAT of C/EBPβ−/− mice is equal to the gene expression in wild-type mice. However, in the BAT of the C/EBPβ−/− mice, there was an increase of 2.0-fold more LCAD gene expression and 1.4-fold more AOX expression compared with wild-type mice. Therefore the BAT tissue has increased fatty acid oxidation gene expression that may contribute to the resistance of obesity on high-fat diet.
Increased expression of uncoupling protein-1 and −3 in the muscle of C/EBPβ−/− mice
To conclude whether energy was lost through uncoupling proteins (UCPs), we measured the levels of UCP-1 and UCP-3 gene expression in the BAT, WAT, and muscle of fed wild-type and C/EBPβ−/− mice after 12 weeks of a high-fat diet. We found that the C/EBPβ−/− mice had a significant increase of UCP-1 gene expression in muscle tissue compared with wild-type mice (Fig. 4) (P < 0.03). The level of UCP-3 gene expression was significantly increased in muscle of C/EBPβ−/− mice as well (P = 0.04). However, the contribution of the increased gene expression is minimal compared with the primary site of UCP-1 mRNA expression found in the BAT. The C/EBPβ−/− mice have increased levels of UCP-1 mRNA, but the level is not statistically significant (P = 0.07). The WAT has very little mRNA expressed for UCP-1 and UCP-3.
Metabolic consequences of the high-fat diet
An increase in adiposity often results in an increase of triglyceride accumulation in the liver. To test the role of C/EBPβ in the accumulation of hepatic triglycerides, we isolated livers from wild-type and C/EBPβ−/− mice after consuming the high-fat diet for 12 weeks and fasted overnight. Hematoxylin and eosin staining was done on paraffin-embedded sections of the liver. The wild-type mice had an accumulation of macrovesicular and microvesicular fat droplets, whereas the C/EBPβ−/− mice had no visible lipid droplets (Fig. 5).
FIG. 5.
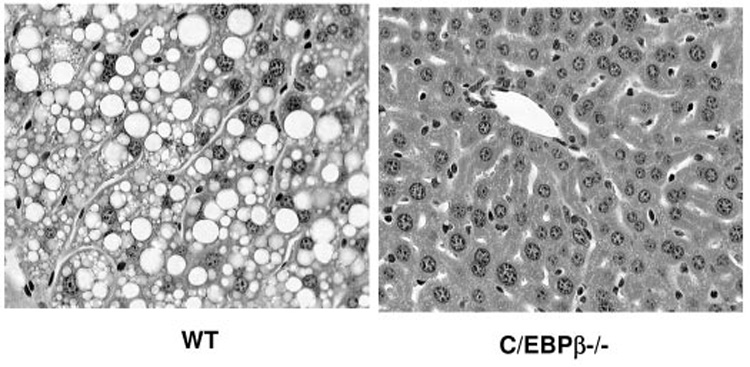
The C/EBPβ−/− mice are resistant to fatty liver. Hematoxylin and eosin–stained sections of wild-type and C/EBPβ−/− livers after 12 weeks on high-fat diet.
To confirm that the droplets seen in the livers from wild-type mice were due to an accumulation of triglyceride, we measured the triglyceride content in the livers. The wild-type mice had 107 ± 17.85 mg/g liver triglycerides, whereas the C/EBPβ−/− mice had 27.57 ± 9.11 mg/g liver triglycerides (Table 2) (P < 0.05).
TABLE 2.
Metabolic analysis of mice on a high-fat diet
| High-fat diet fasted | Wild type | C/EBPβ−/− |
|---|---|---|
| Blood glucose (mg/dl) | 74.75 ± 8.9 | 41.75 ± 5.6* |
| Triglyceride (mg/dl) | 63.75 ± 5.3 | 37.9 ± 2.1* |
| FFA (mmol/l) | 1.53 ± 0.088 | 0.815 ± 0.039* |
| β-Hydroxybutyrate (mmol/l) | 2.15 ± 0.229 | 1.7 ± 0.231* |
| Cholesterol (mg/dl) | 151 ± 2.49 | 94.33 ± 0.667* |
| Triglyceride (mg/g liver) | 107.9 ± 17.8 | 27.57 ± 9.11* |
| Insulin (ng/ml) | 0.581 ± 0.07 | 0.598 ± 0.161 |
Data are means ± SE for five to six mice per group. All mice were fed a high-fat diet for 12 weeks. Blood was sampled after an overnight fast.
P < 0.05.
We were also interested in whether the C/EBPβ−/− mice had lipid accumulating in the blood. We isolated plasma after an overnight from mice fed the high-fat diet for 12 weeks. The C/EBPβ−/− mice had lower levels of blood glucose, triglyceride, FFAs, and cholesterol. All values were statistically significant (P < 0.05). Therefore the C/EBPβ−/− mice are resistant to the deleterious effects of a high-fat diet. They have lower blood triglycerides (C/EBPβ 37.33 ± 2.8 mg/dl and wild type 63.75 ± 5.94) and lower liver triglycerides, even though the C/EBPβ−/− mice have consumed more of the high-fat diet.
The levels of β-hydroxybutyrate were also greatly reduced in the C/EBPβ−/− mice. In Table 2, the C/EBPβ−/− mice have 1.7 ± 0.23 mmol/l β-hydroxybutyrate compared with 2.1 ± 0.229 mmol/l in wild-type mice (P < 0.05). We measured hepatic gene expression of the ketogenic genes, ketothiolase, and hydroxymethylglutaryl (HMG)-CoA lyase in fasted mice (Fig. 6B). The C/EBPβ−/− mice have less ketothiolase gene expression (0.2-fold of wild type) and less HMG-CoA lyase gene expression (0.6-fold of wild type) (P < 0.05).
Reduced hepatic lipogenic gene expression in C/EBPβ−/− mice on high-fat diet
The reduced levels of triglycerides in the liver and FFAs in the blood of the C/EBPβ−/− mice suggest that the mice may have reduced fatty acid synthesis. To establish whether the lipogenic and glycolytic genes are affected, we performed RT-PCR. We measured the key genes for glucose uptake, lipogenesis, and gluconeogenesis in livers of C/EBPβ−/− and wild-type mice in both the fasted and fed states after 12 weeks of a high-fat diet (Fig. 6A and B). The gene expression for glucose uptake, GLUT2, and glucokinase was greatly reduced. The gene expression for the regulator of lipogenesis, sterol regulatory element binding protein-1 (SREBP-1), was also greatly reduced in both the fed and fasted states. The target genes for the SREBP-1 transcription factor, acetyl CoA carboxylase-1 (ACC-1), fatty acid synthase (FAS), and sterol CoA desaturase-1 (SCD-1) were significantly reduced. We measured the gene expression of the rate-limiting step in the gluconeogenic pathway, the cytosolic form of PEPCK-C, and determined that the mRNA levels of PEPCK-C were slightly elevated in fed mice and significantly reduced in the fasted C/EBPβ−/− mice.
Insulin regulates glucose uptake and fatty acid synthesis. To ascertain whether the C/EBPβ−/− mice had decreased insulin concentration on a high-fat diet, we measured their insulin levels after an intraperitoneal injection of glucose (2 g/kg body wt). The levels of insulin at 0 and 30 min are 0.5 and 1.1 ng/ml, respectively, for C/EBPβ−/− and wild-type mice. The concentration of insulin was induced equally in response to an injection of glucose for the C/EBPβ−/− and wild-type mice (data not shown).
DISCUSSION
The balance of energy intake and expenditure has been well documented in studies on obesity. In genetic models of mice susceptible to obesity, such as db/db mice, the lack of leptin production results in hyperphagia and obesity (20). Environmental factors such as a high-fat diet in the C57BL/6J inbred mouse strain have focused on the mechanism of the environmental influence for obesity (21,22). To better understand the mechanism for the development of obesity, we focused on the role of a transcription factor C/EBPβ. We studied the effect of deleting C/EBPβ on energy storage and the development of obesity from a high-fat diet. We found the C/EBPβ−/− mice consumed more food than wild-type mice, yet were protected against increased total body lipid and against lipid accumulation in the liver when fed a high-fat diet. This decreased energy storage indicates an increase of energy metabolism. The C/EBPβ−/− mice had higher CO2 production, suggesting that the C/EBPβ−/− mice have increased flux through the tricarboxylic acid cycle and increased energy expenditure. The BAT in the C/EBPβ−/− mice had significantly increased levels of fatty acid oxidation genes, LCAD and AOX. The β-oxidation of very long–chain fatty acids occurs in peroxisomes, and the rate-limiting enzyme of peroxisomal fatty acid β-oxidation is AOX (23,24). In the C/EPBβ−/− mice, the BAT has 1.4-fold increased gene expression of AOX and 2.0-fold increase of LCAD gene expression over wild-type gene expression. Peroxisomal β-oxidation produces an increase of short- and medium-chain fatty acids that can now then enter the mitochondria for β-oxidation (25). The increase of LCAD gene expression would also contribute to the increased overall oxidation of fatty acids on a high-fat diet. The size of the BAT in the C/EBPβ−/− mice remains unchanged after high-fat feeding. These data suggest that increased metabolic activity in the BAT protects the C/EBPβ−/− mice against obesity. The excess energy in the triglycerides from the high-fat diet is lost in a futile cycle in the BAT of the C/EBPβ−/− mice. Future studies will focus on the role of C/EBPβ in oxidation of fatty acids.
Other mechanisms for increased energy expenditure are UCPs. UCP-1 is well accepted as the mechanism for nonshivering thermogenesis in BAT, and its expression is induced by cold exposure and fatty acid administration (26). In most tissues, oxidation of fuels results in ATP production by oxidative phosphorylation in the mitochondria (27). The presence of UCP-1 in the mitochondrial membrane produces a leak in the proton gradient, which results in a loss of ATP synthesis as heat (28). Transgenic mice that overexpress UCP-1 in the skeletal muscle have 98% higher oxygen consumption and were resistant to diet-induced obesity on a high-fat diet similar to the phenotype in the C/EBPβ−/− mice (29). We have shown that the C/EBPβ−/− mice had significantly higher levels of UCP-1 and UCP-3 gene expression in the skeletal muscle and an increase of UCP-1 in the BAT that was approaching significance. This increased UCP-1 gene expression may contribute to the overall increased energy expenditure and resistance to obesity on a high-fat diet in the C/EBPβ−/− mice.
Alterations in food intake and overall metabolism require peripheral metabolic signals to the brain such as leptin and cytokines from the adipose tissue and peptide YY and ghrelin from the gastrointestinal tract. Signaling in the brain by metabolic sensors such as neuropeptide Y and melanocortin peptides are also required for brain-mediated responses to nutritional challenges (30). These may be altered in the C/EBPβ−/− mice but have not yet been tested.
Possible regulation by the transcription factor C/EBPβ for the reduced triglyceride formation in the blood and liver may be the reduced gene expression of key genes in the fatty acid synthesis pathway in both the liver and adipose tissues. Carmona et al. (31) have reported reduced gene expression of FAS in BAT of C/EBPβ−/− mice at 21 and 4°C during a cold challenge. We have shown that ACC, FAS, and SREPB gene expression is greatly reduced in the livers of C/EBPβ−/− mice on a high-fat diet. C/EBPβ can bind directly to the cAMP responsive element–binding protein regulatory region (CRE) in promoters of various genes. Tae et al. (32) have reported that C/EBPβ is required for the full induction of ACC gene expression by cAMP through the CRE region in the promoter. SREBP is a key regulator of hepatic lipogenesis. Overexpression studies of SREBP-1a in cultured cells or animal livers have resulted in activation of lipogenic genes (33). Selective overexpression of SREBP-1c in adipose tissue resulted in animals that developed insulin-resistant hypoglycemia and fatty livers (34). In the C/EBPβ−/− mice, the lower gene expression of SREBP and the deletion of C/EBPβ contribute to the reduced ACC and FAS gene expression.
In conclusion, the data presented in this study indicate that the C/EBPβ−/− mice are protected from obesity and fatty liver on a high-fat diet because of reduced hepatic expression of lipogenic genes, ACC, and FAS and increased CO2 production from increased metabolism in the BAT and muscle. The experiments presented indicate that C/EBPβ plays a highly integrative role in the regulation of whole-body energy metabolism and is required for the development of fatty liver during obesity. The C/EBPβ gene may be a potential target for drug therapy in the prevention of fatty liver due to obesity.
ACKNOWLEDGMENTS
We are grateful to Dr. Valeria Poli and Dr. Richard Hanson for the generous gift of the C/EBPβ−/− mice.
Glossary
- ACC
acetyl CoA carboxylase
- AOX
acyl-CoA oxidase
- BAT
brown adipose tissue
- C/EBPβ
CCAAT/enhancer-binding protein β
- CPT
carnitine palmitoyltransferase
- CRE
cAMP responsive element–binding protein regulatory region
- FAS
fatty acid synthase
- FFA
free fatty acid
- HMG
hydroxy-methylglutaryl
- LCAD
long acyl-CoA dehydrogenase
- PRARγ
peroxisome proliferator–activated receptor γ
- SCD-1
sterol CoA desaturase-1
- SREBP-1
sterol regulatory element–binding protein-1
- UCP
uncoupling protein
REFERENCES
- 1.Flegal KM, Carroll MD, Ogden CL, Johnson CL. Prevalence and trends in obesity among US adults, 1999–2000. JAMA. 2002;288:1723–1727. doi: 10.1001/jama.288.14.1723. [DOI] [PubMed] [Google Scholar]
- 2.Willett WC, Dietz WH, Colditz GA. Guidelines for healthy weight. New Engl J Med. 1999;341:427–434. doi: 10.1056/NEJM199908053410607. [DOI] [PubMed] [Google Scholar]
- 3.Kriska AM, Saremi A, Hanson RL, Bennett PH, Kobes S, Williams DE, Knowler WC. Physical activity, obesity, and the incidence of type 2 diabetes in a high-risk population. Am J Epidemiol. 2003;158:669–675. doi: 10.1093/aje/kwg191. [DOI] [PubMed] [Google Scholar]
- 4.Bray GA. Medical consequences of obesity. J Clin Endocrinol Metab. 2004;89:2583–2589. doi: 10.1210/jc.2004-0535. [DOI] [PubMed] [Google Scholar]
- 5.Croniger C, Trus M, Lysek-Stupp K, Cohen H, Liu Y, Darlington GJ, Poli V, Hanson RW, Reshef L. Role of the isoforms of CCAAT/enhancer-binding protein in the initiation of phosphoenolpyruvate carboxykinase (GTP) gene transcription at birth. J Biol Chem. 1997;272:26306–26312. doi: 10.1074/jbc.272.42.26306. [DOI] [PubMed] [Google Scholar]
- 6.Liu S, Croniger C, Arizmendi C, Harada-Shiba M, Ren J, Poli V, Hanson RW, Friedman JE. Hypoglycemia and impaired hepatic glucose production in mice with a deletion of the C/EBPbeta gene. J Clin Invest. 1999;103:207–213. doi: 10.1172/JCI4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang ND, Finegold MJ, Bradley A, Ou CN, Abdelsayed SV, Wilde MD, Taylor LR, Wilson DR, Darlington GJ. Impaired energy homeostasis in C/EBP alpha knockout mice. Science. 1995;269:1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka T, Yoshida N, Kishimoto T, Akira S. Defective adipocyte differentiation in mice lacking the C/EBPbeta and/or C/EBPdelta gene. EMBO J. 1997;16:7432–7443. doi: 10.1093/emboj/16.24.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang QQ, Zhang JW, Daniel Lane M. Sequential gene promoter interactions of C/EBPbeta, C/EBPalpha, and PPARgamma during adipogenesis. Biochem Biophys Res Commun. 2004;319:235–239. doi: 10.1016/j.bbrc.2004.04.176. [DOI] [PubMed] [Google Scholar]
- 10.Rosen ED. The transcriptional basis of adipocyte development. Prostaglandins Leukot Essent Fatty Acids. 2005;73:31–34. doi: 10.1016/j.plefa.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365:561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Croniger CM, Millward C, Yang J, Kawai Y, Arinze IJ, Liu S, Harada-Shiba M, Chakravarty K, Friedman JE, Poli V, Hanson RW. Mice with a deletion in the gene for CCAAT/enhancer-binding protein beta have an attenuated response to cAMP and impaired carbohydrate metabolism. J Biol Chem. 2001;276:629–638. doi: 10.1074/jbc.M007576200. [DOI] [PubMed] [Google Scholar]
- 13.Screpanti I, Musiani P, Bellavia D, Cappelletti M, Aiello FB, Maroder M, Frati L, Modesti A, Gulino A, Poli V. Inactivation of the IL-6 gene prevents development of multicentric Castleman’s disease in C/EBP beta-deficient mice. J Exp Med. 1996;184:1561–1566. doi: 10.1084/jem.184.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCabe BJ, Previs SF. Using isotope tracers to study metabolism: application in mouse models. Metab Eng. 2004;6:25–35. doi: 10.1016/j.ymben.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 15.McCabe BJ, Bederman IR, Croniger C, Millward C, Norment C, Previs SF. Reproducibility of gas chromatography-mass spectrometry measurements of (2)H labeling of water: application for measuring body composition in mice. Anal Biochem. 2006;350:171–176. doi: 10.1016/j.ab.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 16.Brunengraber DZ, McCabe BJ, Kasumov T, Alexancer JC, Chandramouli V, Previs SF. Influence of diet on the modeling of adipose tissue triglycerides during growth. Am J Physiol Endocrinol Metab. 2003;285:E917–E925. doi: 10.1152/ajpendo.00128.2003. [DOI] [PubMed] [Google Scholar]
- 17.Brunengraber DZ, McCabe BJ, Katanick J, Previs S. Gas chromatography-mass spectrometry assay of 18O enrichment of water. Anal Biochem. 2002;306:278–282. doi: 10.1006/abio.2002.5720. [DOI] [PubMed] [Google Scholar]
- 18.Yu L, Sinha AK, Previs SF. Effect of sampling interval on the use of “doubly labeled” water for measuring CO2 production. Anal Biochem. 2005;337:343–346. doi: 10.1016/j.ab.2004.09.042. [DOI] [PubMed] [Google Scholar]
- 19.Foretz M, Ancellin N, Andreelli F, Saintillan Y, Grondin P, Kahn A, Thorens B, Vaulont S, Viollet B. Short-term overexpression of a constitutively active form of AMP-activated protein kinase in the liver leads to mild hypoglycemia and fatty liver. Diabetes. 2005;54:1331–1339. doi: 10.2337/diabetes.54.5.1331. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman JM. Positional cloning of the mouse obese gene and its human homologue. Nature. 1994;372:425–432. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]
- 21.Black BL, Croom J, Eisen EJ, Petro AE, Edwards CL, Surwit RS. Differential effects of fat and sucrose on body composition in A/J and C57BL/6 mice. Metabolism. 1998;47:1354–1359. doi: 10.1016/s0026-0495(98)90304-3. [DOI] [PubMed] [Google Scholar]
- 22.Surwit RS, Feinglos MN, Rodin J, Sutherland A, Petro AE, Opara EC, Kuhn CM, Rebuffe-Scrive M. Differential effects of fat and sucrose on the development of obesity and diabetes in C57BL/6J and A/J mice. Metabolism. 1995;44:645–651. doi: 10.1016/0026-0495(95)90123-x. [DOI] [PubMed] [Google Scholar]
- 23.Rao MS, Reddy JK. Peroxisomal beta-oxidation and steatohepatitis. Semin Liver Dis. 2001;21:43–55. doi: 10.1055/s-2001-12928. [DOI] [PubMed] [Google Scholar]
- 24.Reddy JK, Mannaerts GP. Peroxisomal lipid metabolism. Annu Rev Nutr. 1994;14:343–370. doi: 10.1146/annurev.nu.14.070194.002015. [DOI] [PubMed] [Google Scholar]
- 25.Yanagitani A, Yamada S, Yasui S, Shimomura T, Murai R, Murawaki Y, Hashiguchi K, Kanbe T, Saeki T, Ichiba M, Tanabe Y, Yoshida Y, Morino S, Kurimasa A, Usuda N, Yamazaki H, Kunisada T, Ito H, Shiota G. Retinoic acid receptor alpha dominant negative form causes steatohepatitis and liver tumors in transgenic mice. Hepatology. 2004;40:366–375. doi: 10.1002/hep.20335. [DOI] [PubMed] [Google Scholar]
- 26.Argyropoulos G, Harper ME. Uncoupling proteins and thermoregulation. J Appl Physiol. 2002;92:2187–2198. doi: 10.1152/japplphysiol.00994.2001. [DOI] [PubMed] [Google Scholar]
- 27.Rolfe DF, Brown GC. Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiol Rev. 1997;77:731–758. doi: 10.1152/physrev.1997.77.3.731. [DOI] [PubMed] [Google Scholar]
- 28.Nicholls DG. The thermogenic mechanism of brown adipose tissue (Review) Biosci Rep. 1983;3:431–441. doi: 10.1007/BF01121954. [DOI] [PubMed] [Google Scholar]
- 29.Li B, Nolte LA, Ju JS, Han DH, Coleman T, Holloszy JO, Semenkovich CF. Skeletal muscle respiratory uncoupling prevents diet-induced obesity and insulin resistance in mice. Nat Med. 2000;6:1115–1120. doi: 10.1038/80450. [DOI] [PubMed] [Google Scholar]
- 30.Broberger C. Brain regulation of food intake and appetite: molecules and networks. J Intern Med. 2005;258:301–327. doi: 10.1111/j.1365-2796.2005.01553.x. [DOI] [PubMed] [Google Scholar]
- 31.Carmona MC, Hondares E, Rodriguez de la Concepcion ML, Rodriguez-Sureda V, Peinado-Onsurbe J, Poli V, Iglesias R, Villarroya F, Giralt M. Defective thermoregulation, impaired lipid metabolism, but preserved adrenergic induction of gene expression in brown fat of mice lacking C/EBPbeta. Biochem J. 2005;389:47–56. doi: 10.1042/BJ20050009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tae HJ, Zhang S, Kim KH. cAMP activation of CAAT enhancer-binding protein-beta gene expression and promoter I of acetyl-CoA carboxylase. J Biol Chem. 1995;270:21487–21494. doi: 10.1074/jbc.270.37.21487. [DOI] [PubMed] [Google Scholar]
- 33.Shimano H, Horton JD, Hammer RE, Shimomura I, Brown MS, Goldstein JL. Overproduction of cholesterol and fatty acids causes massive liver enlargement in transgenic mice expressing truncated SREBP-1a. J Clin Invest. 1996;98:1575–1584. doi: 10.1172/JCI118951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimomura I, Hammer RE, Richardson JA, Ikemoto S, Bashmakov Y, Goldstein JL, Brown MS. Insulin resistance and diabetes mellitus in transgenic mice expressing nuclear SREBP-1c in adipose tissue: model for congenital generalized lipodystrophy. Genes Dev. 1998;12:3182–3194. doi: 10.1101/gad.12.20.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan W, Yanase T, Nomura M, Okabe T, Goto K, Sato T, Kawano H, Kato S, Nawata H. Androgen receptor null male mice develop late-onset obesity caused by decreased energy expenditure and lipolytic activity but show normal insulin sensitivity with high adiponectin secretion. Diabetes. 2005;54:1000–1008. doi: 10.2337/diabetes.54.4.1000. [DOI] [PubMed] [Google Scholar]
- 36.Clapham JC, Coulthard VH, Moore GB. Concordant mRNA expression of UCP-3, but not UCP-2, with mitochondrial thioesterase-1 in brown adipose tissue and skeletal muscle in db/db diabetic mice. Biochem Biophys Res Commun. 2001;287:1058–1062. doi: 10.1006/bbrc.2001.5698. [DOI] [PubMed] [Google Scholar]
- 37.Gentile L, Monti M, Sebastiano V, Merico V, Nicolai R, Calvani M, Garagna S, Redi CA, Zuccotti M. Single-cell quantitative RT-PCR analysis of Cpt1b and Cpt2 gene expression in mouse antral oocytes and in preimplantation embryos. Cytogenet Genome Res. 2004;105:215–221. doi: 10.1159/000078191. [DOI] [PubMed] [Google Scholar]


