Abstract
Proteins that are released from cells consist of those in the extracellular matrix, as well as extracellular signaling and adhesion molecules. The majority of these extracellular proteins are, however, unknown. To determine their identity, we have used a proteomics approach to define proteins released from neurons, astrocytes and neural precursor cells. Using 2-dimensional gels and liquid chromatography/mass spectrometry technology, it is shown that while astrocytes release a relatively small number of proteins, neurons and neuronal precursor cells release a larger number of proteins with more functional diversity. Although there is overlap between the different cell types, the exact composition of the extracellular protein pool is unique for each cell population. The various subsets of extracellular neural proteins include those involved in cellular Redox regulation and chaperones. In addition, many proteolytic enzymes are found outside of the cell. These data show that the extracellular space within the nervous system has a more diverse protein composition than previously thought.
Keywords: Neuronal cells, secreted proteins, proteomics, neurons, astrocytes
The extracellular environment of the nervous system is created by molecules that are either actively secreted or shed from the cell surface by proteolytic cleavage. While protein secretion involves the release of proteins via highly specialized vesicular transport systems, the most prevalent mechanism for the generation of extracellular molecules is through a process called shedding in which the ectodomains of transmembrane proteins are released from the cell surface by cleavage from the membrane (Schubert 1973). Examples of shed proteins include growth factors such as proTGF-α (Massague and Pandiella 1993), growth factor receptors (Rose-John and Heinrich 1994), proteoglycans such as Syndecan (Reizes et al. 2006), and adhesion molecules like amyloid precursor protein (Schubert et al. 1988). The rate of release of many of these molecules is regulated by growth factors like insulin (Reizes et al. 2006) and is frequently mediated by metalloproteases (Arribas et al. 1996). Shed membrane ectodomains can act as soluble competitive ligands for their membrane-bound counterpart, both in the context of adhesion and receptor molecules (Hanneken et al. 1995). It follows that secreted and shed molecules play important roles within the nervous system. This class of molecules has not, however, been characterized and the identity of most extracellular molecules is not known. For simplicity, the term “secreted proteins” will be used throughout the text to mean both shed and vesicle-released proteins.
The innate complexity of proteins synthesized by a single cell type makes it impossible to determine the expression of all, or even the majority of its intracellular proteins. In contrast, cells release a much more limited subset of proteins into their extracellular environment. We have previously examined the complexity of proteins secreted from both neural and mesoderm-derived cells using standard 2-dimensional gel electrophoresis (2-D gels) and isotopically labeled proteins (Schubert et al. 1986). It was shown that while the intracellular proteins were very similar between neural and mesodermal cells, the number of extracellular proteins and their diversity was much greater with the neural cells than with mesodermal cells such as muscle and fibroblasts. In addition, unlike nerve cells, the secreted proteins were very similar between the different mesodermal cell types. It was concluded that much of the protein diversity in the nervous system is in proteins that are found in the extracellular space. However, at the time of that publication, the technology was not available to readily determine the identity of this extracellular protein population. Indeed, no studies have previously identified and compared secreted proteins from CNS cell populations. We have therefore used 2-D gel and liquid chromatography/mass spectrometry (LC/MS) technology to identify a subset of extracellular proteins released by cortical nerve and astrocytes as well as by several clonal nerve precursor cell populations isolated from the embryonic and adult rat cortex. Of the 200 proteins identified, it is shown that a large number of proteins with molecular chaperone and antioxidant properties are secreted, and that distinct subsets of proteins are secreted by nerve, astrocytes, and neural precursor cells.
MATERIALS AND METHODS
Cell Lines
Cell lines and preparation of secreted proteins
The nerve-like precursor cells B35, B50, B103 and B104 were cloned from nitrosoethylurea-induced brain tumors (Schubert 1974). The major criterion on which the classification of the nerve cells was made was electrical excitability and veratridine-stimulated sodium flux. A large number of other markers, such as cell-surface antigens, are in agreement with the physiology (Schubert et al. 1986). L6 myoblasts were derived from neonatal skeletal muscle (Yaffe 1968). The clonal adult rat hippocampal cell line, AHP, was obtained from J. Ray (Salk Institute) (Ray et al. 1993; Taupin et al. 2000). Cells were maintained as described in the original texts.
Homogeneous populations of cortical neurons and astrocytes were prepared by the following methods. Cortical neurons were isolated from day 18 Sprague-Dawley rat embryos by tryptic digestion of the tissue. The cells were plated in B27 neurobasal medium (Gibco, Grand Island, NY) in the absence of serum. This medium does not allow the growth of any type of glia (Brewer 1997). The cultures were devoid of astrocytes, 02A oligo dendroglia, progenitor cells and microglia as defined by the absence of immunostaining with glial fibrillary acidic protein (GFAP) (Chemicon, Temecula, CA) monoclonal antibody, VA2B5 (Chemicon, Temecula, CA) and Iba-1 (Wako, Richmond, VA), respectively. Cortical cultures were maintained for 12 to 14 days before the collection of the secreted proteins. Astrocytes were prepared from 2-day-old rats by tryptic digestion of cortical tissue (Liu and Piasecki 2001). The cells were plated in Alpha-Medium (Gibco) containing 10% fetal calf serum. After reaching confluency, the cells were dissociated with trypsin and transferred to 100 mm tissue culture dishes in the same medium. Astrocyte cultures prepared in this manner were devoid of neurons, oligodendrocytes, and microglia as defined by immunostaining with anti-MAP-2 (2at 2b) (Sigma, St. Louis, MO), A2B5 and Iba-1. Serum-free medium was collected 5–7 days after the cultures reached confluency.
To assay extracellular proteins, cells were washed twice with serum-free Eagle’s medium (MEM) and incubated in serum-free MEM for 18 hr. The absence of serum for short-term biosynthetic labeling of cells previously grown in serum does not significantly alter the pattern of released extracellular proteins (Truding et al. 1975). The supernatants were filtered through Sephadex G25 to remove salts, and lyophilized. The proteins released into the culture medium were assayed by 2-D gel electrophoresis.
Two-dimensional gel electrophoresis and mass spectrometry analysis
Because the type and amount of proteins vary dramatically with the growth state of cultured cells (Garrels 1979), care was taken to grow and plate cells from the different cell lines under identical conditions. Exponentially dividing cultures were dissociated and replated at 5 × 105 cells per 100 mm tissue culture plate and the following day serum-free growth-conditioned media were prepared. The conditioned medium was desalted and lyophilized. One half ml of 8M urea, 4% (w/v) CHAPS, 40 mM Tris and 0.2% Bio-Lyte 3–10 ampholytes (Biorad, Hercules, CA), and 50 mM DTT were added to the tube. Following centrifugation at 14,000 × g for 10 min, protein amount was determined by the Lowry assay and 200 μg of supernatant was loaded onto pH 3–10 isoelectric focusing strips (Biorad) and electrophoresed to 60,000 volt-hrs on a Biorad Protean IEF machine. The strip was then applied to the top of an 18 cm Biorad precast 12% acrylamide gel and electrophoresed at 25 milliamps per gel until the dye front was at the bottom. Gels were fixed in 50% methanol overnight and silver stained according to the method of Shevchenko (Shevchenko et al. 1996).
Gel spots from 2-D gels were excised and in-gel digested with trypsin (Shevchenko et al. 1996) and analyzed by liquid chromatography electrospray ionization mass spectrometry (LC-MS). Briefly, samples were loaded onto a fused silica capillary column (PicoFrit Column, New Objective, Woburn, MA) packed with reversed phase material (Zorbax C-18, 5 μm particle size, Agilent, Santa Clara, CA). The mobile phase consisted of aqueous 0.1% formic acid (A Buffer) and 0.1% formic acid in 80% acetonitrile/water (B buffer). Elution was achieved by a gradient of 5% to 70% B buffer in 65 min at a flow rate of 150 nl/min. The eluant was electrosprayed into a Bruker Esquire 3000 Plus quadrupole ion trap mass spectrometer (Bruker Daltonics, Billerica, MA). Spectra were measured for the 3 most intense species in each time window. For each mass, MS and MS/MS spectra were recorded. After two sets of spectra, the parent mass was excluded for 1 minute. The complete data set for each gel spot was searched using the Mascot algorithm (Matrix Science, London, UK) against a recent release of the nonredundant NCBI database. Only results that gave significant Mascot scores greater than 40 were reported.
RESULTS
The following experiments examine the proteins that are found in the growth-conditioned medium of cultured nerve, astrocytes, and neural precursor cells. 2-D gels were used with a broad 3–10 pH isoelectric focusing range in the first dimension and 12% acrylamide gels in the second dimension. These conditions were chosen to cover the broadest range possible of isoelectric points as well as to have access to low molecular weight proteins. It is recognized that the more basic proteins are missing, that the resolution of the high molecular weight proteins is less than ideal and that very low abundance proteins will not be detected. In addition, there are several possibilities for the origin of the proteins in this experimental paradigm. These include the secretion or shedding from cells, cell lysis, or residual serum protein. Since the identity of the majority of the proteins in the 2-D gels described here is known, as are the most abundant serum proteins, a comparison between the two groups of proteins indicates that with the rare exception of albumin, no serum protein is present. Data were searched against the NCBI rodent protein sequence database and against the entire mammalian database. Sequence differences were sufficient to distinguish between rodent and bovine sequences, for example, in the case of bovine serum albumin originating from culture medium (Mizejewski 1995).
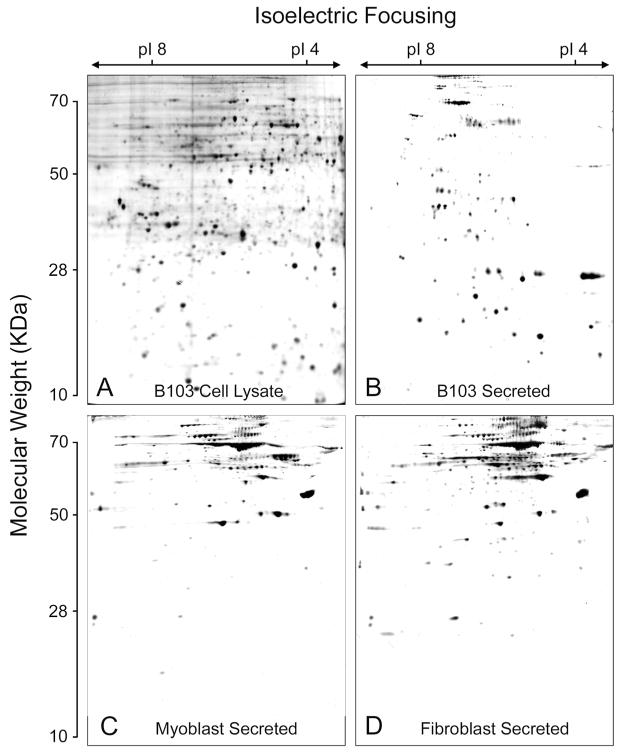
The other undesirable alternative is that the extracellular proteins are the result of cell lysis. Two sets of data make this alternative unlikely. First, when the profile of extracellular proteins is compared to that of a cell lysate on 2-D gels, the protein patterns are very distinct. Figure 1 shows 2-D gels of a silver stained cell lysate (A) and secreted proteins (B) from a CNS nerve precursor cell line called B103. Clearly, the gel patterns are very different. Second, if the 2-D gel of a cell lysate is stained with silver and allowed to develop for a long period of time, it becomes black because the gel contains thousands of proteins of lower abundance that constitute the background of the most abundant 500–1000 proteins that are visible in the more lightly stained gel. The background proteins become evident over longer developing periods. In contrast, 2-D gels of secreted proteins can be developed for an extended time without the appearance of the background stain. Figures 1B, C, and D and Fig. 2 show silver stained gels of secreted proteins from B103 (Fig. 1B), myoblasts (Fig. 1C) and fibroblasts (Fig. 1D) and from primary cultures of cortical astrocytes (Fig. 2A) and nerve (Fig. 2B), with minimal background. While it is unlikely that the extracellular proteins assayed in this study are derived from cell lysis, we cannot rule out the possibility that some lysis occurs that is detected by the highly sensitive LC/MS technology. Secreted proteins can therefore be used as a relatively limited data set to identify physiologically relevant molecules that are associated with the extracellular space.
Fig. 1.
Total cell lysate (A) and secreted proteins (B–D) from three cell lines. Two hundred micrograms of protein were run on 2-D gels according to procedures in Materials and Methods stained with silver, and developed for the same length of time. A) B103 nerve-like cell line, cell lysate. B) B103 secreted. C) L6 myoblasts secreted. D) RAT 1 fibroblasts secreted.
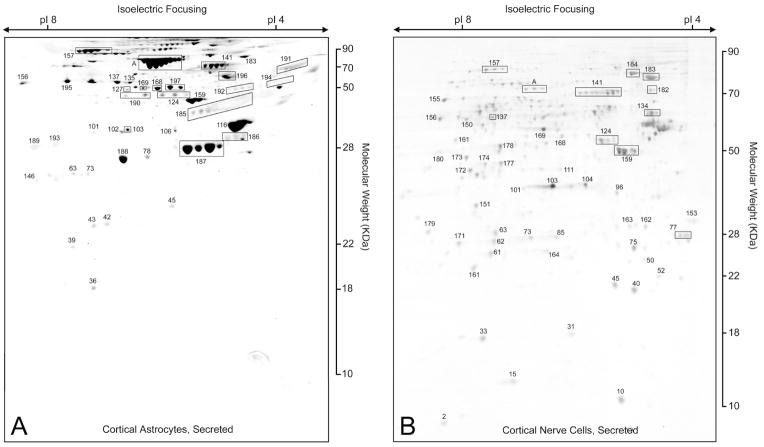
Fig. 2.
Secreted proteins from primary cultured cortical astrocytes (A) and nerve (B). Two hundred micrograms of protein was electrophoresed on 2-D gels and stained with silver. The numbered proteins are identified and defined in the text (Fig. 3 and Supplementary Table S1). Boxed A is bovine serum albumin.
Cortical Nerve and Astrocytes
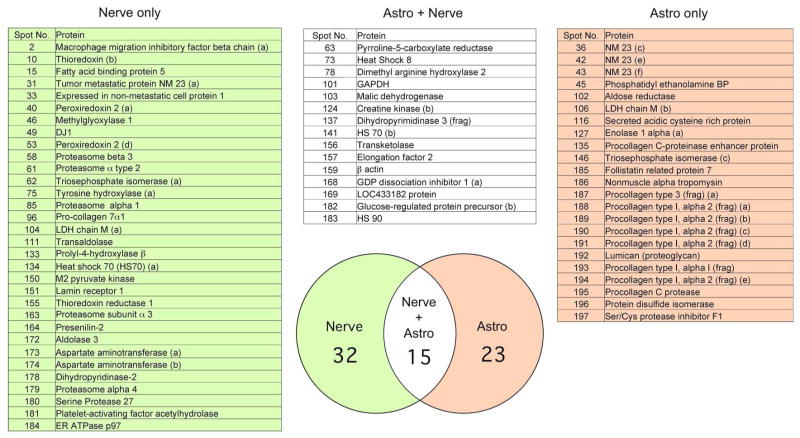
Figure 2 shows silver stained 2-D gels of proteins secreted from astrocytes (A) prepared from newborn rats after 12 days in culture and from E18 cortical neurons (B) after 12 days in culture. Proteins were excised, digested with trypsin, and identified by LC/MS. Proteins are recorded as a solid identification only after three independent LC/MS hits with probability-base Mowse scores of greater than 40 as determined using the Mascot search algorithm (Pappin et al. 1993; Perkins et al. 1999). Only rarely did the second identification not confirm the first. All of the proteins discussed in this text are sequentially numbered and identified in supplementary Table S1. The boxed proteins in the figures are either enclosed for clarity or all of the spots are the same protein as identified by MS but differ systematically in size due to unknown secondary modifications, such as glycosylation or phosphorylation. It must also be noted that many proteins exist in multiple forms, with different molecular weights and isoelectric points. For example, tumor metastatic protein NM23 is found in six different locations; these are indicated as NM23a, b, or c, etc. At present, it is not clear if this heterogeneity is due to proteolysis, secondary modifications, or splice variation. The identification of secreted proteins from primary cultures of nerve or astrocytes is found in Fig. 3 and supplemental material (Table S1). Of the 47 nerve and 38 astrocytes secreted proteins that were identified, only 15 are shared while 23 are unique to astrocytes and over 32 are unique to nerve (Fig. 3).
Fig. 3.
Venn diagram and list of proteins secreted from primary cultures of rat cortical nerve and astrocytes. The identified proteins shown in Fig. 2 were compared between nerve and glia. Of the 47 identified secreted nerve and 38 secreted glial proteins (85 total), only 15 overlapped in expression.
While the mix of secreted protein from rat astrocytes is both limited in number and quite clean in the sense that further development of the gel does not reveal additional proteins, the proteins secreted by cortical neurons are more numerous and varied. Many of the astrocyte secreted proteins are proteolytic fragments of pro-collagen, with a large amount of secreted acidic cystein-rich protein (spot 116, Fig. 2A). A significant amount of bovine serum albumin is also detected (Fig. 2A, boxed A) in the astrocyte preparations and much less from nerve (Figure 2B, boxed A), perhaps because astrocytes are binding and recycling albumin from the culture medium. In contrast to astrocytes, nerve cells secrete a much more varied set of proteins, but only about half of the proteins are very reproducible between preparations. Since the proteins secreted from primary nerve cultures are more variable in appearance and amount, only the consistently reproducible proteins from over a dozen independent sets of 2-D gels done with this cell type are listed in supplementary Table S1 and Fig. 3. These include several heat shock proteins and a number of enzymes that have historically been considered cytoplasmic proteins. The possible reason for the appearance of the latter in the extracellular space will be presented in the Discussion.
In contrast to the very distinct populations of secreted proteins from neuroectoderm-derived nerve and glia (Figs. 2A and B), the secreted proteins from two mesodermally-derived cell types, skeletal muscle myoblasts and fibroblasts, are very similar to each other, quite distinct from nerve and glia, and more limited in number than those from nerve (Figs. 1C and D) (Schubert et al. 1986). They will not be discussed further.
Protein Secretion from Precursor Populations
Our laboratory has created a set of cell lines that are derived from dividing neural precursor cells of E16 rats (Schubert et al. 1974). Of the approximately 100 cell lines, five have a phenotype that is more nerve-like than the others, with the major criteria being that they are capable of making neurotransmitters and generating action potentials (Schubert et al. 1974). They also express several nerve-specific cell surface markers (Stallcup and Cohn 1976a, b; Schubert et al. 1986). To gain some insight into the nature of the extracellular proteins derived from nerve-like precursor cells, the extracellular protein profile of four of these cell lines was examined along with that of a rat adult hippocampal nerve-glial precursor cell line called AHP (Ray et al. 1993). The AHP cells are capable of differentiating into both nerve and glial cells, while the other clonal cell lines have a more limited differentiation potential toward nerve. It should be pointed out, however, that these cells are distinct from embryonic stem cells and are limited in their differentiation pathways to nerve and glial populations.
To examine the proteins from the E16-derived CNS cells, we ran 2-D gels of the four nerve-like cell lines individually, and also mixed the supernatants from the four lines together to create the master gel shown in Supplemental Fig. S1. All but a few of the proteins in this master mix were identified by LC/MS and are listed in supplementary Table S1. Within the pH and molecular weight range on the 2-D gels that we are using, there are approximately 150 proteins in the combined CNS cell line supernatants with each cell line secreting around 100 proteins (Tables 1 and S1).
Table 1.
Shared Proteins
| B35 | B50 | B103 | B104 | AHP | Nerve | Astro | |
|---|---|---|---|---|---|---|---|
| Total | [104] | [119] | [102] | [125] | [65] | [47] | [38] |
| B35 | 104 (100) | 88 (74) | 84 (82) | 90 (72) | 37 (59) | 28 (62) | 16 (42) |
| B50 | 88 (84) | 88 (100) | 96 (94) | 100 (80) | 43 (67) | 27 (60) | 18 (47) |
| B103 | 84 (81) | 96 (81) | 84 (100) | 96 (77) | 33 (52) | 25 (56) | 19 (50) |
| B104 | 90 (87) | 100 (84) | 96 (94) | 125 (100) | 48 (75) | 33 (73) | 19 (50) |
| AHP | 37 (36) | 43 (36) | 34 (34) | 48 (38) | 60 (100) | 32 (71) | 16 (42) |
| Nerve | 28 (27) | 27 (23) | 25 (25) | 33 (26) | 32 (50) | 45 (100) | 14 (36) |
| Astro | 13 (12) | 18 (15) | 18 (18) | 19 (15) | 16 (25) | 14 (31) | 38 (100) |
The number of individual proteins shared between the different groups of cells. For example, B103 had 102 identified protein spots on 2D gels, of which 84 were in common with B35 and 25 in common with primary nerve. The percentage of the shared proteins of the proteins from the cells on the left and the cells at the top compared to the number of proteins in the cells at the top (for example 88% of the B35 proteins (89/104 were shared with B104) are indicated in parentheses.
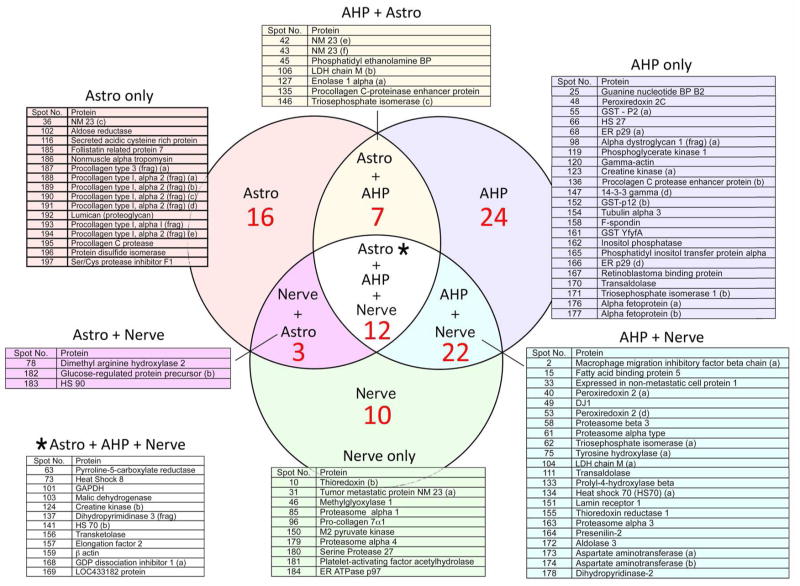
Figure S2 shows a 2-D gel of the proteins secreted from the AHP precursor cell line, and the overlap in proteins between AHP and nerve and glia is shown in Fig. 4. The Venn diagram indicates that AHP is more related to nerve than to astrocytes. Table 1 is a matrix in which the number and percent of secreted proteins in common from each cell population is compared against all others. The total number of identified proteins is shown horizontally across the top, and the number of shared proteins indicated in the columns. In parentheses is the percent of total proteins in the cell line at the top that is shared with each other cell type. These data show that the proteins secreted by the CNS cell lines are more similar to each other than to cortical nerve, astrocytes or AHP cells. They are, however, more similar to the well characterized CNS precursor cell line AHP and primary nerve than to astrocytes. It should also be noted that the total number of proteins secreted by astrocytes (less than 50) is lower than that secreted by any of the neuronal precursor lines. The specificity of the LC/MS data was confirmed by western blotting of the secreted proteins from four different cells with antibodies to four different proteins (Fig. S3).
Fig. 4.
Venn diagram showing the overlap of secreted proteins between cortical neurons and astrocytes and the AHP neural precursor cell line. This figure shows that in the context of extracellular proteins, the AHP cell line is more closely related to nerve than astrocyte.
Functional Analysis of Secreted Proteins
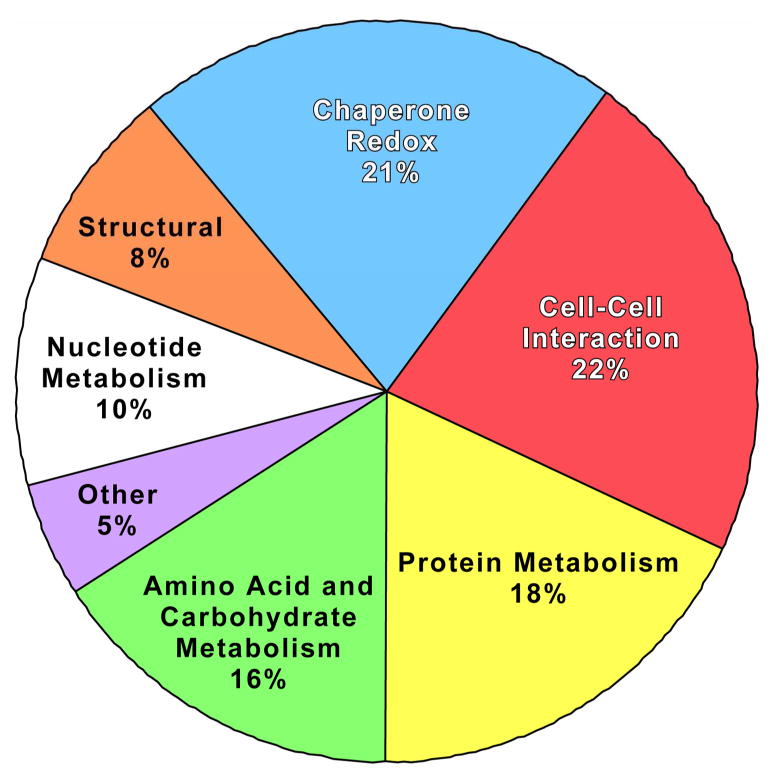
The above data show that among a collection of proteins secreted by primary nerve and glial cells, four CNS nerve-like precursor cell lines, and a nerve-glial precursor cell line, over 250 unique protein spots are resolved on 2-D acrylamide gels. Of these, about 200 have been identified by mass spectroscopy. However, since many of the extracellular proteins exist as multiple molecular weights and isoelectric points, there are only 109 distinct proteins within the larger group. To clarify the potential functional role of these molecules, the 109 proteins were separated according to their known biological activities. The listing of all of the secreted proteins is shown in Supplemental Table S2, along with a description of their known function. The functional grouping is summarized in Fig. 5.
Fig. 5.
Pie chart of secreted proteins according to function.
The major group of extracellular proteins contains, as expected, proteins involved in cell-cell interactions, many likely derived from membrane shedding. Molecular chaperone proteins and Redox proteins were also abundant and are grouped together because they frequently have similar biological roles. The most surprising groups of proteins found outside the cell are those involved in carbohydrate metabolism and the proteasome subunits. This observation has been made on a limited scale before, and its implications will be addressed in the Discussion. There are smaller subsets of secreted proteins involved in lipid, nucleotide and neurotransmitter metabolism. One of the smallest groups of extracellular proteins is that of structural proteins, which are the most abundant in amount within the cell. The lack of significant amounts and spot intensity for these proteins in the 2-D gels is a further argument that cell lysis does not contribute significantly to our analysis.
DISCUSSION
Despite the fact that extracellular proteins mediate cell migration and differentiation within the developing nervous system, there has been no systematic study of the proteins found in the extracellular space, thus limiting our understanding of both the identity and function of this important class of molecules. From this initial study of identifying a subset of secreted proteins made by CNS astrocytes, nerve and nerve precursor cells, we can make the following conclusions about both their composition and biological function: 1) Cortical nerve and glial cells each secrete a unique set of proteins as well as a more limited subset that are shared between the two cell types. 2) The number and variability of the secreted proteins from nerve and neural precursor cells is greater than glia. 3) The number of proteins secreted from a pluripotent CNS precursor cell line are more similar to cortical nerve than glia. 4) The composition of the secreted proteins contains some unexpected molecules, such as protein molecular chaperones, proteasome subunits, and metabolic enzymes. The biochemical function of the classes of secreted proteins allows us to identify new roles for the extracellular space within the nervous system. It should be pointed out, however, that the technology used in this report only identified a subset of secreted proteins. Clearly, proteins such as growth factors have been missed because of low abundance and their basic isoelectric points.
The only systematic studies of extracellular proteins among the cell types of the brain are our work published over 20 years ago before the advent of MS technology to identify proteins, and a more recent identification of some proteins released by cultured astrocytes (Schubert et al. 1986; Lafon-Cazal et al. 2003; Kim et al. 2007). While we could not identify proteins in our initial study, we were able to demonstrate that cells from the CNS secreted a greater number of proteins than mesoderm derived cells and that among the CNS cell lines examined there was extensive overlap between the sets of proteins secreted, with about 15% of the proteins being distinct between pairs of cell lines. Similar results were obtained in the present study with the E-16 nerve-like precursor cells. However, when the secreted proteins from primary cultures of astrocytes, nerve, and CNS precursor cells are included in the data set, the data are more interesting. Assuming the AHP cells are bona fide nerve/glia precursors, then it is quite clear that the CNS cell lines that were derived from transformed E16 rat embryos are more similar to the AHP precursor cells than to astrocytes (Table 1). The secreted proteins from both E16 and adult hippocampus precursor cells are more similar to nerve than to glia, and have a greater number of unique proteins than glia. It is therefore likely that both the E16 cortical cell lines and AHP cells are both more related to nerve than glia.
Many of the secreted proteins fall into the family of multifunctional stress proteins that assist in the assembly and folding of other proteins, the molecular chaperones. These proteins are classically thought to mediate peptide folding, signaling, chaperoning and cytoprotection inside cells. However, recently within the immune system, classical chaperones such as HSP70 were established as extracellular immunomodulators and have been linked to inflammation, antigen presentation and the stimulation of cell division (Asea 2005). It is shown here that these proteins are found outside the cells of the nervous system. A less classical group of chaperones that are abundant in the secreted milieu of nerve and glia are members of the thioredoxin family. These include thioredoxin 1, protein disulfide isomerase, and macrophage migration inhibiting factor (MIF), a classical pro-inflammatory cytokine (Nakamura 2008). In contrast to MIF, thioredoxin is anti-inflammatory, an antioxidant, and reduces disulfides in oxidized proteins. Thioredoxin (Trx) is also released from glial cells during ischemia (Tomimoto et al. 1993) and exogenous Trx has been shown to be neuroprotective in both in vitro and in vivo nerve cell models (Hori et al. 1994; Andoh et al. 2002; Inomata et al. 2006). Peroxiredoxins (Prx) are peroxidases that reduce H2O2 and organic hydroperoxides using reducing equivalents supplied by Trx, and systemically administered recombinant Prx5 can protect against excitotoxic brain lesions in newborn mice (Plaisant et al. 2003). Thus the coordinated secretion of thioredoxin and related family members from both neurons and astrocytes may play a key role in detoxifying extracellular oxidants and regulating Redox-sensitive proteins that are also secreted or reside on the cell surface. For example, most cell surface receptors and many growth factors have extensive intramolecular disulfide bonding (Sun and Davies 1995), and the presence of this reductases in the extracellular space may be required to maintain the function of these proteins.
The study with cultured rat astrocytes identified over 30 secreted proteins using an approach similar to ours; many of these proteins were also found in our study (Lafon-Cazal et al. 2003). A curious observation is that both studies identified a number of “housekeeping” proteins, such as lactate dehydrogenase, creatine kinase, and malate dehydrogenase among the milieu of secreted proteins. This observation and the fact that proteins like the intermediate filament protein vimentin, are also actively secreted (Mor-Vaknin et al. 2003), may be a reflection of the selective release of a subset of internal cellular contents via exosomes, which are vesicles containing a variety of cytoplasmic proteins that are deposited outside the cell (Tytell 2005). As with secreted proteins in general, the role of exosomes in the release of proteins from cells of the nervous system has not been studied.
In addition to proteins involved in energy metabolism, another subset of extracellular proteins is proteasome subunits. Proteasomes may be removed by the cell along with other damaged proteins via exosomes. It should be noted that proteasomes have been found in the blood and elevated levels are associated with several types of cancers (Stoebner et al. 2005). In some cases, serum proteasome concentration varies with the disease state and can be used as a prognostic factor for survival (Jakob et al. 2007). Proteasomes secreted from cells may have some physiological function, for it has been observed that during the differentiation of leukocytes there is a decrease in cytoplasmic and nuclear proteasomes and an increase in plasma membrane-associated proteasomes along with the secretion of proteasome subunits (Lavabre-Bertrand et al. 2000). These observations support the notion that the secreted proteins identified in cultured cells reflect those that are released from cells in vivo. However, except by studying the distribution of each protein in the animal, one cannot state with certainty that there is a one to one correlation of protein secretion in vitro and in vivo. Indeed, this may not be expected because much of the secretion must be physiologically regulated.
In summary, these data show that cells from the central nervous system secrete a mixture of proteins that is both diverse in structure and function and different in composition than many would have been predicted. In addition to adhesion and antioxidant proteins such as glutathione S-transferases and peroxiredoxins, cells release enzymes involved in intermediary metabolism and proteolysis. However, the repertoire of secreted proteins is unique for each cell type and represents a distinct set of molecular identifiers for individual phenotypes within the nervous system.
Supplementary Material
Acknowledgments
The project described was supported by Award Number 5R01 AG025337 from the National Institute on Aging. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute on Aging or the National Institutes of Health. The Mass Spectrometry Facility at the Salk Institute is supported by a grant from NCRR (S10RR022988) and by the Vincent J. Coates Foundation.
Footnotes
Publisher's Disclaimer: This is an Accepted Article that has been peer-reviewed and approved for publication in the Journal of Neurochemistry, but has yet to undergo copy-editing and proof correction. Please cite this article as an "Accepted Article"; doi: 10.1111/j.1471-4159.2009.05968.x
References
- Andoh T, Chock PB, Chiueh CC. The roles of thioredoxin in protection against oxidative stress-induced apoptosis in SH-SY5Y cells. J Biol Chem. 2002;277:9655–9660. doi: 10.1074/jbc.M110701200. [DOI] [PubMed] [Google Scholar]
- Arribas J, Coodly L, Vollmer P, Kishimoto TK, Rose-John S, Massague J. Diverse cell surface protein ectodomains are shed by a system sensitive to metalloprotease inhibitors. J Biol Chem. 1996;271:11376–11382. doi: 10.1074/jbc.271.19.11376. [DOI] [PubMed] [Google Scholar]
- Asea A. Stress proteins and initiation of immune response: chaperokine activity of hsp72. Exerc Immunol Rev. 2005;11:34–45. [PMC free article] [PubMed] [Google Scholar]
- Brewer GJ. Isolation and culture of adult rat hippocampal neurons. J Neurosci Methods. 1997;71:143–155. doi: 10.1016/s0165-0270(96)00136-7. [DOI] [PubMed] [Google Scholar]
- Garrels JI. Two dimensional gel electrophoresis and computer analysis of proteins synthesized by clonal cell lines. J Biol Chem. 1979;254:7961–7977. [PubMed] [Google Scholar]
- Hanneken A, Maher PA, Baird A. High affinity immunoreactive FGF receptors in the extracellular matrix of vascular endothelial cells--implications for the modulation of FGF-2. J Cell Biol. 1995;128:1221–1228. doi: 10.1083/jcb.128.6.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Katayama M, Sato N, Ishii K, Waga S, Yodoi J. Neuroprotection by glial cells through adult T cell leukemia-derived factor/human thioredoxin (ADF/TRX) Brain Res. 1994;652:304–310. doi: 10.1016/0006-8993(94)90241-0. [DOI] [PubMed] [Google Scholar]
- Inomata Y, Nakamura H, Tanito M, Teratani A, Kawaji T, Kondo N, Yodoi J, Tanihara H. Thioredoxin inhibits NMDA-induced neurotoxicity in the rat retina. J Neurochem. 2006;98:372–385. doi: 10.1111/j.1471-4159.2006.03871.x. [DOI] [PubMed] [Google Scholar]
- Jakob C, Egerer K, Liebisch P, Turkmen S, Zavrski I, Kuckelkorn U, Heider U, Kaiser M, Fleissner C, Sterz J, Kleeberg L, Feist E, Burmester GR, Kloetzel PM, Sezer O. Circulating proteasome levels are an independent prognostic factor for survival in multiple myeloma. Blood. 2007;109:2100–2105. doi: 10.1182/blood-2006-04-016360. [DOI] [PubMed] [Google Scholar]
- Kim S, Ock J, Kim AK, Lee HW, Cho JY, Kim DR, Park JY, Suk K. Neurotoxicity of microglial cathepsin D revealed by secretome analysis. J Neurochem. 2007 Oct 22;103:2640–2650. doi: 10.1111/j.1471-4159.2007.04995.x. [DOI] [PubMed] [Google Scholar]
- Lafon-Cazal M, Adjali O, Galeotti N, Poncet J, Jouin P, Homburger V, Bockaert J, Marin P. Proteomic analysis of astrocytic secretion in the mouse. J Biol Chem. 2003;278:24438–24448. doi: 10.1074/jbc.M211980200. [DOI] [PubMed] [Google Scholar]
- Lavabre-Bertrand T, Henry L, Guiraud I, Carillo S, Bureau JP. The proteasome and malignant hemopathies. Morphologie. 2000;84:39–43. [PubMed] [Google Scholar]
- Liu Y, Piasecki D. A cell-based method for the detection of nanomolar concentrations of bioactive amyloid. Anal Biochem. 2001;289:130–136. doi: 10.1006/abio.2000.4928. [DOI] [PubMed] [Google Scholar]
- Massague J, Pandiella A. Membrane-anchored growth factors. Annu Rev Biochem. 1993;62:515–541. doi: 10.1146/annurev.bi.62.070193.002503. [DOI] [PubMed] [Google Scholar]
- Mizejewski GJ. The phylogeny of alpha-fetoprotein in vertebrates: survey of biochemical and physiological data. Crit Rev Eukaryot Gene Expr. 1995;5:281–316. doi: 10.1615/critreveukargeneexpr.v5.i3-4.40. [DOI] [PubMed] [Google Scholar]
- Mor-Vaknin N, Punturieri A, Sitwala K, Markovitz DM. Vimentin is secreted by activated macrophages. Nat Cell Biol. 2003;5:59–63. doi: 10.1038/ncb898. [DOI] [PubMed] [Google Scholar]
- Nakamura H. Extracellular functions of thioredoxin. Novartis Found Symp. 2008;291:184–192. doi: 10.1002/9780470754030.ch14. discussion 192–195, 221–224. [DOI] [PubMed] [Google Scholar]
- Pappin DJ, Hojrup P, Bleasby AJ. Rapid identification of proteins by peptide-mass fingerprinting. Curr Biol. 1993;3:327–332. doi: 10.1016/0960-9822(93)90195-t. [DOI] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Plaisant F, Clippe A, Vander Stricht D, Knoops B, Gressens P. Recombinant peroxiredoxin 5 protects against excitotoxic brain lesions in newborn mice. Free Radic Biol Med. 2003;34:862–872. doi: 10.1016/s0891-5849(02)01440-5. [DOI] [PubMed] [Google Scholar]
- Ray J, Peterson DA, Schinstine M, Gage FH. Proliferation, differentiation, and long-term culture of primary hippocampal neurons. Proc Natl Acad Sci U S A. 1993;90:3602–3606. doi: 10.1073/pnas.90.8.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizes O, Goldberger O, Smith AC, Xu Z, Bernfield M, Bickel PE. Insulin promotes shedding of syndecan ectodomains from 3T3-L1 adipocytes: a proposed mechanism for stabilization of extracellular lipoprotein lipase. Biochemistry. 2006;45:5703–5711. doi: 10.1021/bi052263h. [DOI] [PubMed] [Google Scholar]
- Rose-John S, Heinrich PC. Soluble receptors for cytokines and growth factors: generation and biological function. Biochem J. 1994;300:281–290. doi: 10.1042/bj3000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert D. Protein secretion by clonal glial and neuronal cell lines. Brain Res. 1973;56:387–391. doi: 10.1016/0006-8993(73)90358-2. [DOI] [PubMed] [Google Scholar]
- Schubert D. Induced differentiation of clonal rat nerve and glia cells. Neurobiology. 1974;4:376–387. [PubMed] [Google Scholar]
- Schubert D, Brass B, Dumas JP. Protein complexity of central nervous system cell lines. J Neurosci. 1986;6:2829–2836. doi: 10.1523/JNEUROSCI.06-10-02829.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert D, Schroeder R, LaCorbiere M, Saitoh T, Cole G. Amyloid beta protein precursor is possibly a heparan sulfate proteoglycan core protein. Science. 1988;241:223–226. doi: 10.1126/science.2968652. [DOI] [PubMed] [Google Scholar]
- Schubert D, Heinemann S, Carlisle W, Tarikas H, Kimes B, Steinbach JH, Culp W, Brandt BL. Clonal cell lines from the rat central nervous system. Nature. 1974;249:224–227. doi: 10.1038/249224a0. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Stallcup WB, Cohn M. Correlation of surface antigens and cell type in cloned cell lines from the rat central nervous system. Exp Cell Res. 1976a;98:285–297. doi: 10.1016/0014-4827(76)90440-7. [DOI] [PubMed] [Google Scholar]
- Stallcup WB, Cohn M. Electrical propertiers of a clonal cell line as determined by measurement of ion fluxes. Exp Cell Res. 1976b;98:277–284. doi: 10.1016/0014-4827(76)90439-0. [DOI] [PubMed] [Google Scholar]
- Stoebner PE, Lavabre-Bertrand T, Henry L, Guiraud I, Carillo S, Dandurand M, Joujoux JM, Bureau JP, Meunier L. High plasma proteasome levels are detected in patients with metastatic malignant melanoma. Br J Dermatol. 2005;152:948–953. doi: 10.1111/j.1365-2133.2005.06487.x. [DOI] [PubMed] [Google Scholar]
- Sun PD, Davies DR. The cystine-knot growth-factor superfamily. Annu Rev Biophys Biomol Struct. 1995;24:269–291. doi: 10.1146/annurev.bb.24.060195.001413. [DOI] [PubMed] [Google Scholar]
- Taupin P, Ray J, Fischer WH, Suhr ST, Hakansson K, Grubb A, Gage FH. FGF-2-responsive neural stem cell proliferation requires CCg, a novel autocrine/paracrine cofactor. Neuron. 2000;28:385–397. doi: 10.1016/s0896-6273(00)00119-7. [DOI] [PubMed] [Google Scholar]
- Tomimoto H, Akiguchi I, Wakita H, Kimura J, Hori K, Yodoi J. Astroglial expression of ATL-derived factor, a human thioredoxin homologue, in the gerbil brain after transient global ischemia. Brain Res. 1993;625:1–8. doi: 10.1016/0006-8993(93)90130-f. [DOI] [PubMed] [Google Scholar]
- Truding R, Shelanski ML, Morell P. Glycoproteins released into the culture medium of differentiating murine neuroblastoma cells. J Biol Chem. 1975;250:9348–9354. [PubMed] [Google Scholar]
- Tytell M. Release of heat shock proteins (Hsps) and the effects of extracellular Hsps on neural cells and tissues. Int J Hyperthermia. 2005;21:445–455. doi: 10.1080/02656730500041921. [DOI] [PubMed] [Google Scholar]
- Yaffe D. Retention of differentiation potentialities during prolonged cultivation of myogenic cells. Proc Natl Acad Sci U S A. 1968;61:477–483. doi: 10.1073/pnas.61.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.