Abstract
Nanowire-based detection strategies provide promising new routes to bioanalysis that could one day revolutionize the healthcare industry. This review covers recent developments in nanowire sensors for multiplexed detection of biomolecules such as nucleic acids and proteins. We focus on encoded nanowire suspension arrays and semiconductor nanowire-based field-effect transistors. Nanowire assembly and integration with microchip technology is emphasized as a key step toward the ultimate goal of multiplexed detection at the point of care using portable, low power, electronic biosensor chips.
Introduction
Nanowires are defined here as metallic or semiconducting particles having a high aspect ratio, with cross-sectional diameters «1 µm, and lengths as long as tens of microns. Nanoparticles and nanorods <1 µm in length are also interesting for biomolecular detection, and have been recently reviewed [1]. Nanowires show promise in a number of different sensing strategies, including optical [2], electrical [3], electrochemical [4], and mass-based [5] approaches. They are attractive because of their small size, high surface-to-volume ratios,and/or electronic, optical and magnetic properties, which can differ markedly from those observed for bulk or thin film materials as the nanowire cross-sectional diameter decreases. Recent results suggest the possibility of incorporating large numbers of nanowires into large-scale arrays and complex hierarchical structures for high-density biosensors, electronics, and optoelectronics [6]. Readers are referred to several recent reviews for more general information on nanowire synthesis, characterization, and applications [7,8•,9].
We will focus on the application of nanowires for simultaneous detection of multiple biomolecules (i.e. multiplexing). Simultaneous detection of multiple bio-molecular targets, such as nucleic acids or proteins, is important for medical diagnostics and monitoring the response to treatment. For example, multiplexed tests for respiratory pathogens are urgently needed because they present with similar symptoms; accurate pathogen identification and subtyping are important for patient recovery and public health monitoring [10]. Multiple biomarkers are also desirable for diagnosing cancers [11,12••]. Several nanowire-based detection strategies have shown promise for multiplexed bioanalysis, most of which can be classified as either optical or electrical approaches based on how binding signals are detected. We have chosen to highlight two approaches: first, optically encoded nanowire suspension arrays, with fluorescence microscopy readout, which offer flexible multiplexing using readily available instrumentation and second, semiconductor nanowire field-effect transistors (FETs), which offer ultrasensitive, label-free electrical detection. Both have already been demonstrated for biological multiplexing (as many as 30 and 3 targets, respectively). Challenges and future trends for each of these platforms are explored. We also discuss the emerging techniques for organizing nanowires into arrays to improve the level of multiplexing for electrical detection strategies such as FETs.
Optical detection: barcoded nanowires
Striped nanowires can be employed as encoded supports for fluorescence-based bioassays. Multiplexing is achieved by preparing optically distinguishable nanowires, for example, by altering the sequence of different metal segments in the nanowires [2,13,14], or by changing the diameter along metallic wires [15] or silica tubes [16]. In an alternative process termed On-Wire Lithography, sacrificial Ni segments are etched to leave behind more noble metals [17]. Nanowire patterns encoded in any of these ways can be read out optically, and different assays can be performed simultaneously on the different patterned particles in the same sample. After incubation with analytes and tag molecules, images are collected to determine assay results (e.g. reflectance for particle identification and fluorescence for assay quantification). In comparison with encoded bead-based suspension arrays, which use dyes for both identification and quantification [18], barcoded nanowire platforms carry out multiplexed detection with only one dye, circumventing spectral overlapping. Encoded nanowire suspension arrays provide a flexible, convenient, and powerful platform for multiplexed bioassays. The conventional fluorescence optical microscopes used for quantification are already in widespread clinical use, simplifying the adoption of this technology.
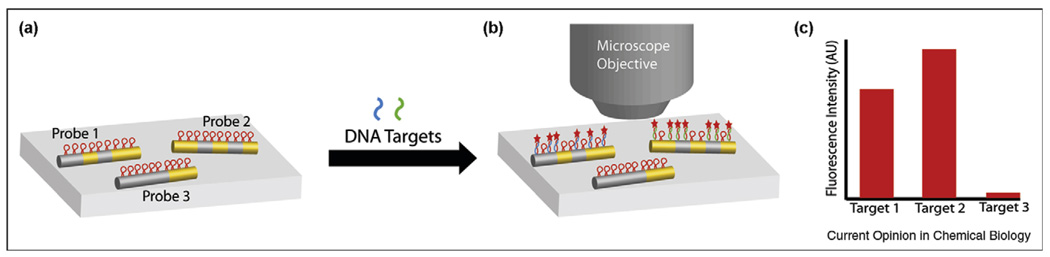
Barcoded metallic nanowires, synthesized by templated electrodeposition of multiple metal segments, have been demonstrated as flexible suspension arrays for multiplexed protein and nucleic acid detection [2,13,14]. These particles are ~6 µm in length and ~300 nm in cross-sectional diameter, with segments generally ≥500 nm. They are stable indefinitely when stored under mildly reducing conditions [19]. Because of their relatively large size, these wires have bulk-like reflectivities for the adjacent metal segments, enabling the barcode striping pattern to be identified via optical microscopy (Figure 1). For example, Ag shows higher reflectivity than Au; therefore, the pattern for a wire composed of Au–Ag–Au–Ag–Au is 0 1 0 1 0. Magnetic segments (Co, Ni) can also be incorporated for the manipulation of the resulting wires [13,20]. As many as tens of thousands of patterns are in principle possible, and a library of 100 patterns has been synthesized and evaluated for software identification; >90% identification accuracy was reported for 74 of the patterns [21]. Target binding is quantified as emission intensity of fluorescent probes associated with each patterned nanowire.
Figure 1.
Barcoded metallic nanowires for multiplexed biodetection. (a) Nanowires with different patterns of Au and Ag segments are functionalized with different molecular beacon probe sequences, which are nonfluorescent in the absence of target strands. When a mixture of target molecules, in this case complementary to probes 1 and 2, is incubated with the mixture of barcoded nanowires, (b) some nanowire-bound probes become fluorescent because of complementary target binding. Reflectance and fluorescence microscope images are acquired for the identification of nanowires and quantification of fluorescence, respectively, and (c) mean fluorescence intensities are calculated from populations of individual nanowires of each barcode pattern, to quantify the amount of each target present.
Multiplexed detection of biothreat simulants, including protein, spore, and viral targets, has been demonstrated, with sensitivities of 1 × 105 colony-forming or plaque-forming units, and 5 ng/mg for the protein ovalbumin [22]. Averaging signal from tens to thousands of individual nanowires of a given pattern is straightforward, with the order of 100 total wires analyzed per image. Reducing the total number of barcoded nanowires used in an assay can improve detection limits by reducing the number of available binding sites; Tok et al. reported a 100-fold increase in sensitivity for a 100-fold decrease in the number of wires used [22]. As many as 30 different assays have been performed simultaneously on barcoded nanowires to determine the genotypes of single-nucleotide polymorphisms (SNPs) [23•].
The metallic surface of the nanowires can modulate emission intensity of bound fluorescent dyes; this effect can be minimized by coating the wires with SiO2 layers [24] or can take advantage in molecular beacon-style assays in which the wire itself acts as a quencher for fluorescence in the absence of bound target DNA sequences [25•,26••]. These assays have a detection limit of 100 pM DNA target, currently limited by incomplete fluorescence quenching of the probes in the absence of target. These nanowire beacon assays enable multiplexed nucleic acid detection in a single step, no-wash, closed-chamber format that reduces the risk of sample contamination [26••]. RT-PCR products from as many as five viral pathogenic sequences have been monitored in a single sample using a multiplexed nanowire beacon assay [25•].
Looking forward, the development of automated imaging and readout will greatly facilitate the clinical use of barcoded nanowire suspension arrays. Additionally, while high sensitivity is not required for some applications, for example, high viral-load respiratory infection diagnostics, the development of methods for combination with a signal amplification step will be needed for other medical and environmental applications. An additional direction for future work is in the development of barcoded nanowires as tags for biomolecule binding to a planar support. Stoermer and Keating have reported a proof-of-principle competitive binding experiment in which DNA-functionalized, barcoded nanowires were exposed to a surface harboring DNA complementary to the sequence on one of the nanowire patterns. Nanowire binding was selective, with just 5% nonspecific assembly [27]. With further development, this approach could ultimately provide highly sensitive multiplexed detection without the need for fluorescence.
Electrical detection: field-effect transistors
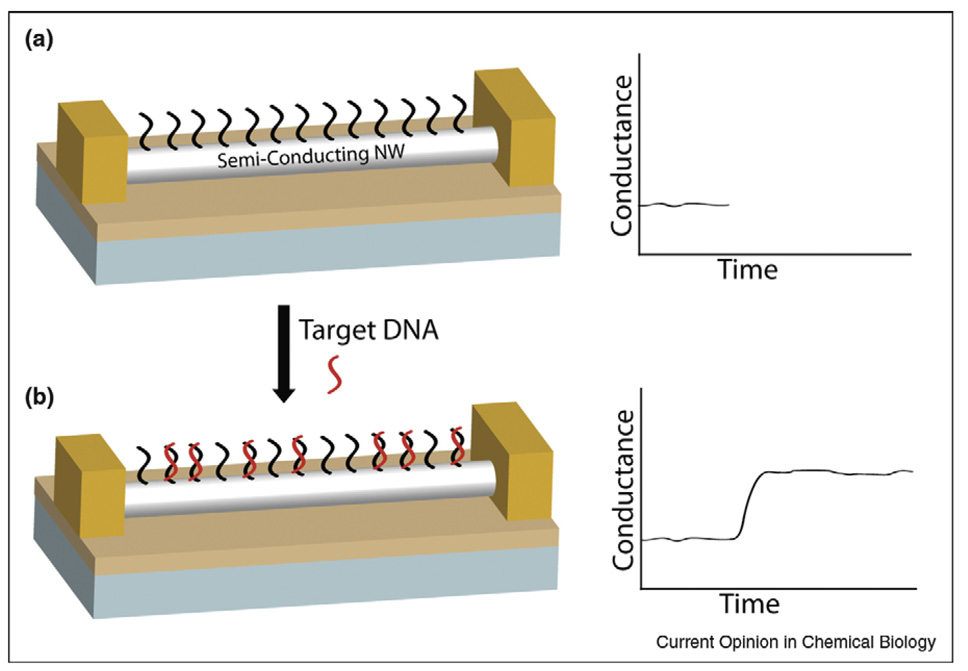
Semiconductor nanowire-based FETs are an exciting new route to ultrasensitive electrical detection of biomolecular interactions [3,28]. In these devices, conductance is monitored to detect binding events occurring on the nanowire surface. Figure 2 shows a peptide nucleic acid (PNA) receptor-modified nanowire FET, in which uncharged PNA probes provide selective binding to detect charged DNA targets [29]. The small diameter of nanowire FETs provides extremely high sensitivity because the binding of target molecules causes accumulation/ depletion of carriers throughout the wire cross-section, and have been used to detect ions, DNA sequences, proteins, and viruses [3,28,30,31•]. Because FETs respond to changes in surface charge, attention must be paid to the buffer solution in which measurements are performed. Physiologically reasonable ionic strength negatively impacts sensitivity by compressing the electrical double layer around the wires. This has been overcome by desalting samples before analysis to increase DeBye length and with it the distance from the wire surface at which changes in charge can be detected [12••]. Recently Heath and coworkers used electrostatically adsorbed DNA probes on functionalized SiNWs to demonstrate DNA detection in 150 mM salt despite the short Debye length at this ionic strength [32•].
Figure 2.
Semiconductor nanowire-based field-effect transistors for biomolecule detection. (a) The surface of a p-type silicon nanowire is modified with peptide nucleic acid (PNA) probe molecules. (b) The binding of DNA target molecules with a net negative charge leads to an accumulation of carriers in the nanowire and a corresponding increase in nanowire conductance.
There are two complementary strategies to fabricate nanowire FETs, top-down and bottom-up. In top-down strategies, the wires themselves, as well as the chip and electronic circuitry, are all fabricated from a bulk silicon wafer using advanced microelectronics technologies (i.e. lithography, etching, and deposition). These technologies are mature and reliable; however, the incorporation of biological probe molecules is limited by harsh conditions (high temperature, solvents, or reactive-ion etching (RIE)) commonly used in fabrication. In bottom-up strategies, nanowire building blocks are synthesized before assembly onto the chip surface, which provides much greater flexibility in nanowire material properties and surface functionalization but does not yet offer the fabrication reliability of top-down approaches.
In top-down strategies, Stern et al. reported a complementary metal oxide semiconductor (CMOS) FET compatible technology to detect antibodies with 100 fM concentration [33•]. An anisotropic wet etch was used instead of RIE, which can degrade device performance [34]. The resulting SiNWs had trapezoidal cross-sections with nm-scale width and height. This approach enables the integration of signals from the nanowire sensors and real-time detection of biomolecules. Gao et al. also used CMOS compatible techniques to fabricate SiNW FET arrays on Silicon-On-Insulator wafers [35]. Thermal oxidation of SiNW and etching off the grown SiO2 layer further decrease the dimension of SiNW. They showed that SiNW arrays with PNA probes could detect complementary target DNA with high sensitivity (10 fM).
The Lieber group has pioneered bottom-up strategies to fabricate SiNW FETs; their contributions to this field have been recently reviewed [3,28]. Notably, they have demonstrated the versatility of SiNW FETs by the detection of several classes of targets including ions, small molecules, proteins, nucleic acids, and viruses with high selectivity and sensitivity [3,12••,28–30,31•]. These authors also demonstrated the highest level of multiplexing to date for nanowire FET sensors in a simultaneous assay for three cancer markers with a detection of 0.9 pg/mL in desalted but undiluted serum samples [12••]. Monoclonal antibodies specific for each of the targets were spotted onto different nanowire FETs; samples solutions were delivered through microfluidic channels and the electrical signal from each FET was monitored in real time during exposure to each of the targets.
Semiconductor nanowire FETs offer ultrasensitive, label-free, real-time electrical detection of biomolecules. To date, most efforts have focused on the fabrication and characterization of the FET devices, with less attention on biological multiplexing, at a current maximum of three targets at once. New methods are needed to enable the controlled placement and integration of large numbers of functional nanowire bioFETs with different probe antibodies or nucleic acid sequences by top-down, bottom-up, or hybrid strategies.
Alignment of nanowires for multiplexed sensor arrays
To realize the considerable potential of sensors based on electrical detection, arrays of independent nanodevices must be fully integrated with on-chip computation and wireless communication. Nanowire arrays suitable for electrical detection of large numbers (10s–100s) of different target molecules have not been demonstrated. Challenges include first, positioning addressable nanowire devices individually with correct registry to underlying circuitry; second, the possible incompatibility of biological probe molecules needed for selective recognition with standard lithographic fabrication methodologies; and third, the need for hundreds of different receptor coatings and multiple copies of each type, on nanowires at predetermined chip addresses for multiplexing and statistical significance, respectively. Exciting progress toward these goals has been made in recent years.
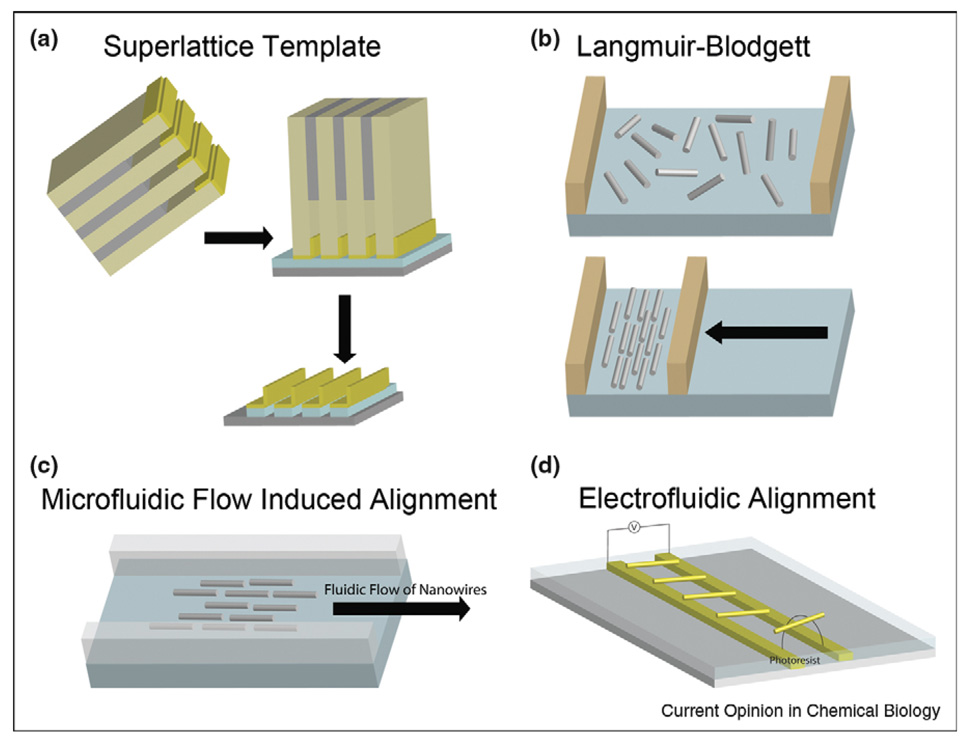
The first of these challenges is easiest to address by fabricating the nanowires in place on the chip using top-down strategies such as that described in the preceding section. Heath and coworkers have developed a superlattice template technique for producing high-density, highly ordered nanowire arrays with wire diameters, and center-to-center distances as small as 8 and 16 nm, respectively, and aspect ratios up to 106 [36]. This technique applies a superlattice template to transfer the thickness of a thin film as the diameter of the nanowires (Figure 3a). The resulting arrays can be transferred to a plastic substrate for flexible chemical sensors [37•]. Although requiring high cost molecular beam epitaxy (MBE) and electron beam lithography, these highly ordered, ultrahigh-density semiconductor nanowire arrays have very low defects and are therefore very promising for large-scale, reproducible, and reliable ultrasensitive biosensors. Multiplexed biosensing will require postfabrication nanowire functionalization with probe molecules, which could potentially be achieved by robotic spotting [38], ink-jet printing [39], selective heating [40], electrochemically [41], or via dip-pen nanolithography [42]. Challenges in postfabrication biofunctionalization include achieving registry between the delivered molecules and the underlying device structure, possible crosscontamination during functionalization of closely spaced features, and the nonplanar device geometries.
Figure 3.
Nanowire alignment techniques. (a) Superlattice template for nanowire patterning: first, a thin layer of metal is deposited on the superlattice while it was tilted at 368; then, the metal layer is brought into contact with an adhesive layer on a semiconductor substrate; finally, metal nanowires are released on the substrate by removing the superlattice and adhesive layer. The thickness of the metal layer is the diameter of the nanowires. (b) Langmuir–Blodgett alignment: nanowires mixed with a surfactant are deposited onto the air/water interface in a Langmuir trough; then, the compression of the nanowires into oriented arrays is achieved by moving a barrier across the air/water interface. (c) Microfluidic flow induced alignment: nanowires are aligned into parallel arrays on a flat substrate by flowing nanowire suspensions in a microfluidic channel. (d) Electrofluidic alignment: nanowires in a dielectric medium are polarized by applying an alternating electric field in the kilohertz range and aligned between two guiding electrodes.
An advantage of bottom-up nanowire assembly methods is increased control during nanowire synthesis and functionalization. For example, multisegmented nanowires with metallic, semiconducting, and/or polymeric segments can be prepared by templated electrodeposition [8•]. Similarly, doping levels can be varied during nanowire growth by the vapor–liquid–solid method, to prepare wires with highly and lightly doped regions [43]. It is desirable to include nanowires with different materials properties to, for example, increase dynamic range. Off-chip functionalization of the nanowires with biorecognition probe chemistry provides greater flexibility and enables independent optimization of attachment chemistries for the different probe molecules before assembly onto the chip. Routes to bottom-up nanowire assembly include methods based on electrical fields [44••,45,46], magnetic fields [9], microfluidic flow [47], Langmuir–Blodgett (LB) [48] alignment, selectively grown on device [49], programmed dip-coating [50], growth substrate contact printing [51], blown-bubble film [52], and electroplating in nanochannel [53]. Fluidic and electrofluidic methods have dominated sensor-driven nanowire assembly efforts to date.
LB techniques have been used to align nanowires on centimeter length scales [48]. Nanowires are suspended at the air/water interface and compressed using a LB trough, resulting in ‘rafts’ of nanowires oriented parallel to the barrier of the instrument (Figure 3b). These nanowire rafts are then transferred from the air/water interface onto a planar substrate to produce nanowire arrays.
Flow induced alignment has been used to produce parallel arrays and complex hierarchical structures [47,54]. Nanowires are aligned on a flat substrate by flowing nanowire suspensions in microfluidic channels, as illustrated in Figure 3c. The substrate can be functionalized or patterned by electron beam lithography to further assist nanowire alignment [47]. The flow rate and the direction control the degree of alignment and the orientation of the aligned nanowires, respectively, while the flow duration controls the nanowire surface coverage on the substrate. Although LB techniques and microfluidic flow alignment have attractive features, they are not commonly used in SiNW FET biosensors because they require greater effort and much larger amounts of nanowires as compared to simple pipette deposition [31•]. Furthermore, they cannot accurately control individual nanowire positioning into specific chip address, and they have some problems while aligning Si nanowires with the diameter less than 15 nm [51].
Electric-field-assisted alignment is a promising tool that results in high yields of nanowire arrays in controlled positions. Nanowires in a dielectric medium can be polarized by applying an alternating current (AC) electric field between two guiding electrodes [45,46,55]. This phenomenon, termed dielectrophoresis, results in nanowires moving to the region of highest field and bridging the electrodes. The process is self-limiting: once a wire has aligned between the electrodes the driving force for a second wire to align there is greatly reduced [45]. Mayer and coworkers recently reported an electrofluidic method that provides large-area alignment in registry on-chip lithographic features for nanowires which were grown and functionalized off-chip [44••]. Three mechanisms were combined: first, the AC electric fields assembled nanowires into photoresist wells that spanned pairs of electrodes; second, the capillary forces held the nanowires in these wells during the evaporation of the nanowire suspension solvent; and third, the misaligned nanowires outside the wells were removed during photoresist liftoff. More than 2000 nanowires were assembled into well-defined chip addresses with high yields (~80%) (Figure 3d). PNA probe molecules attached to the nanowires before alignment maintained their selectivity for binding complementary DNA targets after the array was formed. This approach provides a powerful platform for chip-scale nanowire alignment to well-controlled positions for biosensing.
Combinations of top-down and bottom-up methods are likely to yield functional, chip-scale sensor devices in the near future. Methods have not yet been reported to allow bottom-up assembly of nanowires functionalized with different biorecognition coatings at distinct, predetermined chip locations. Thus, at present both top-down and bottom-up nanowire devices must rely on postassembly/fabrication functionalization if they are to simultaneously detect multiple different target biomolecules.
Conclusions
Nanowire-based detection strategies offer several promising routes to multiplexed bioanalysis. Barcoded nanowires provide flexible multiplexing as suspension arrays using conventional fluorescence optical microscopes for readout, and are promising for application in point-of-care clinical settings. Semiconducting nanowire FETs offer ultrasensitive electrical detection in a label-free format that is in principle compatible with portable, low-power microchip-based devices that could process multiplexed data from many individual sensors. Such devices could be used outside of clinical environments, and would be extraordinarily attractive for the future of personalized medicine. These devices have not yet been demonstrated, and routes to multiplexed nanowire FETs are at present limited; however, new approaches to combining bottom-up nanowire assembly with top-down chip fabrication are under development to overcome this challenge.
Acknowledgements
We thank the National Institutes of Health (R01 EB00268 and R33 CA118591) and the National Science Foundation (NIRT CCR-0303976 and NER CHE 0304575) for financial support, and Prof Theresa Mayer for stimulating discussions on nanowire assembly and integration.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Rosi NL, Mirkin CA. Nanostructures in biodiagnostics. Chem Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 2.Brunker SE, Cederquist KB, Keating CD. Metallic barcodes for multiplexed bioassays. Nanomedicine. 2007;2:695–710. doi: 10.2217/17435889.2.5.695. [DOI] [PubMed] [Google Scholar]
- 3.Patolsky F, Timko BP, Zheng G, Lieber CM. Nanowire-based nanoelectric devices in the life sciences. MRS Bull. 2007;32:142–149. [Google Scholar]
- 4.Wang J. Nanomaterial-based electrochemical biosensors. Analyst. 2005;130:421–426. doi: 10.1039/b414248a. [DOI] [PubMed] [Google Scholar]
- 5.Carrascosa LG, Moreno M, Alvarez M, Lechuga LM. Nanomechanical biosensors: a new sensing tool. Trends Anal Chem. 2006;25:196–206. [Google Scholar]
- 6.Lieber CM, Wang ZL. Functional nanowires. MRS Bull. 2007;32:99–108. [Google Scholar]
- 7.Xia Y, Yang P, Sun Y, Wu Y, Mayers B, Gates B, Yin Y, Kim F, Yan H. One-dimensional nanostructures: synthesis, characterization, and applications. Adv Mater. 2003;15:353–389. [Google Scholar]
- 8. Hurst SJ, Payne EK, Qin L, Mirkin CA. Multisegmented one-dimensional nanorods prepared by hard-template synthetic methods. Angew Chem Int Ed. 2006;45:2672–2692. doi: 10.1002/anie.200504025.. Excellent review of properties and applications of nanowires fabricated by template synthesis methods.
- 9.Wanekaya AK, Chen W, Myung NV, Mulchandani A. Nanowire-based electrochemical biosensors. Electroanalysis. 2006;18:533–550. [Google Scholar]
- 10.Hinman AR. Global progress in infectious disease control. Vaccine. 1998;16:1116–1121. doi: 10.1016/s0264-410x(98)80107-2. [DOI] [PubMed] [Google Scholar]
- 11.Wulfkuhle JD, Liotta LA, Petricoin EF. Early detection: proteomic applications for the early detection of cancer. Nat Rev Cancer. 2003;3:267–275. doi: 10.1038/nrc1043. [DOI] [PubMed] [Google Scholar]
- 12. Zheng G, Patolsky F, Cui Y, Wang WU, Lieber CM. Multiplexed electrical detection of cancer markers with nanowire sensor arrays. Nat Biotechnol. 2005;23:1294–1301. doi: 10.1038/nbt1138.. Highest level of biological multiplexing for nanowire FETs till date, demonstrating label-free, ultrasensitive electrical detection of cancer markers.
- 13.Nicewarner-Pena SR, Griffith Freeman R, Reiss BD, He L, Pena DJ, Walton ID, Cromer R, Keating CD, Natan MJ. Submicrometer metallic barcodes. Science. 2001;294:137–141. doi: 10.1126/science.294.5540.137. [DOI] [PubMed] [Google Scholar]
- 14.Keating CD, Natan MJ. Striped metal nanowires as building blocks and optical tags. Adv Mater. 2003;15:451–454. [Google Scholar]
- 15.Matthias S, Schilling J, Nielsch K, Muller F, Wehrspohn RB, Gosele U. Monodisperse diameter-modulated gold microwires. Adv Mater. 2002;14:1618–1621. [Google Scholar]
- 16.He B, Son SJ, Lee SB. Suspension array with shape-coded silica nanotubes for multiplexed immunoassays. Anal Chem. 2007;79:5257–5263. doi: 10.1021/ac0704964. [DOI] [PubMed] [Google Scholar]
- 17.Qin L, Banholzer MJ, Millstone JE, Mirkin CA. Nanodisk codes. Nano Lett. 2007;7:3849–3853. doi: 10.1021/nl072606s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LaFratta CN, Walt DR. Very high density sensing arrays. Chem Rev. 2008;108:614–637. doi: 10.1021/cr0681142. [DOI] [PubMed] [Google Scholar]
- 19.Stoermer RL, Sioss JA, Keating CD. Stabilization of silver metal in citrate buffer: barcoded nanowires and their bioconjugates. Chem Mater. 2005;17:4356–4361. [Google Scholar]
- 20.Lee K-B, Park S, Mirkin Chad A. Multicomponent magnetic nanorods for biomolecular separations. Angew Chem Int Ed (in English) 2004;43:3048–3050. doi: 10.1002/anie.200454088. [DOI] [PubMed] [Google Scholar]
- 21.Walton ID, Norton SM, Balasingham A, He L, Oviso DF, Jr, Gupta D, Raju PA, Natan MJ, Freeman RG. Particles for multiplexed analysis in solution: detection and identification of striped metallic particles using optical microscopy. Anal Chem. 2002;74:2240–2247. doi: 10.1021/ac020073w. [DOI] [PubMed] [Google Scholar]
- 22.Tok JBH, Chuang FYS, Kao MC, Rose KA, Pannu SS, Sha MY, Chakarova G, Penn SG, Dougherty GM. Metallic striped nanowires as multiplexed immunoassay platforms for pathogen detection. Angew Chem Int Ed. 2006;45:6900–6904. doi: 10.1002/anie.200601104. [DOI] [PubMed] [Google Scholar]
- 23. Sha MY, Walton ID, Norton SM, Taylor M, Yamanaka M, Natan MJ, Xu C, Drmanac S, Huang S, Borcherding A, et al. Multiplexed SNP genotyping using nanobarcode particle technology. Anal Bioanal Chem. 2006;384:658–666. doi: 10.1007/s00216-005-0225-0.. Highest level of multiplexing for barcoded nanowires till date.
- 24.Sioss JA, Stoermer RL, Sha MY, Keating CD. Silica-coated, Au/Ag striped nanowires for bioanalysis. Langmuir. 2007;23:11334–11341. doi: 10.1021/la7019846. [DOI] [PubMed] [Google Scholar]
- 25. Sha MY, Yamanaka M, Walton ID, Norton SM, Stoermer RL, Keating CD, Natan MJ, Penn SG. Encoded metal nanoparticle-based molecular beacons for multiplexed detection of DNA. Nanobiotechnology. 2005;1:327–335. doi: 10.1385/NBT:1:4:327.. Demonstrates the use of nanowire beacons for simultaneous detection of five different pathogen-related RT-PCR products.
- 26. Stoermer RL, Cederquist KB, McFarland SK, Sha MY, Penn SG, Keating CD. Coupling molecular beacons to barcoded metal nanowires for multiplexed, sealed chamber DNA bioassays. J Am Chem Soc. 2006;128:16892–16903. doi: 10.1021/ja0658261.. Demonstration of multiplexed, wash-free nucleic acid detection using barcoded nanowires with molecular beacon probes.
- 27.Stoermer RL, Keating CD. DNA-directed assembly of barcoded nanowires onto glass slides for biosensing applications. Proc SPIE Int Soc Opt Eng. 2004;5588:51–58. [Google Scholar]
- 28.Patolsky F, Zheng G, Lieber CM. Nanowire-based biosensors. Anal Chem. 2006;78:4260–4269. doi: 10.1021/ac069419j. [DOI] [PubMed] [Google Scholar]
- 29.Hahm J-I, Lieber CM. Direct ultrasensitive electrical detection of DNA and DNA sequence variations using nanowire nanosensors. Nano Lett. 2004;4:51–54. [Google Scholar]
- 30.Cui Y, Wei Q, Park H, Lieber CM. Nanowire nanosensors for highly sensitive and selective detection of biological and chemical species. Science. 2001;293:1289–1292. doi: 10.1126/science.1062711. [DOI] [PubMed] [Google Scholar]
- 31. Patolsky F, Zheng G, Lieber CM. Fabrication of silicon nanowire devices for ultrasensitive, label-free, real-time detection of biological and chemical species. Nat Protoc. 2006;1:1711–1724. doi: 10.1038/nprot.2006.227.. Detailed experimental description of bottom-up SiNW FET device fabrication and use.
- 32. Bunimovich YL, Shin YS, Yeo W-S, Amori M, Kwong G, Heath JR. Quantitative real-time measurements of DNA hybridization with alkylated nonoxidized silicon nanowires in electrolyte solution. J Am Chem Soc. 2006;128:16323–16331. doi: 10.1021/ja065923u.. Real-time detection of DNA by top-down fabricated SiNW FETs in physiologically relevant salt concentrations.
- 33. Stern E, Klemic JF, Routenberg DA, Wyrembak PN, Turner-Evans DB, Hamilton AD, LaVan DA, Fahmy TM, Reed MA. Label-free immunodetection with CMOS-compatible semiconducting nanowires. Nature (London, United Kingdom) 2007;445:519–522. doi: 10.1038/nature05498.. Excellent performance for top-down fabricated SiNW FETs with sub-100 fM detection limits for antibodies.
- 34.Li Z, Chen Y, Li X, Kamins TI, Nauka K, Williams RS. Sequence-specific label-free DNA sensors based on silicon nanowires. Nano Lett. 2004;4:245–247. [Google Scholar]
- 35.Gao Z, Agarwal A, Trigg AD, Singh N, Fang C, Tung C-H, Fan Y, Buddharaju KD, Kong J. Silicon nanowire arrays for label-free detection of DNA. Anal Chem. 2007;79:3291–3297. doi: 10.1021/ac061808q. [DOI] [PubMed] [Google Scholar]
- 36.Melosh NA, Boukai A, Diana F, Gerardot B, Badolato A, Petroff PM, Heath JR. Ultrahigh-density nanowire lattices and circuits. Science. 2003;300:112–115. doi: 10.1126/science.1081940. [DOI] [PubMed] [Google Scholar]
- 37. McAlpine MC, Ahmad H, Wang D, Heath JR. Highly ordered nanowire arrays on plastic substrates for ultrasensitive flexible chemical sensors. Nat Mater. 2007;6:379–384. doi: 10.1038/nmat1891.. Transfer printing of SiNW arrays onto flexible substrates.
- 38.Auburn RP, Kreil DP, Meadows LA, Fischer B, Matilla SS, Russell S. Robotic spotting of cDNA and oligonucleotide microarrays. Trends Biotechnol. 2005;23:374–379. doi: 10.1016/j.tibtech.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Bietsch A, Zhang J, Hegner M, Lang HP, Gerber C. Rapid functionalization of cantilever array sensors by inkjet printing. Nanotechnology. 2004;15:873–880. [Google Scholar]
- 40.Park I, Li Z, Pisano AP, Williams RS. Selective surface functionalization of silicon nanowires via nanoscale joule heating. Nano Lett. 2007;7:3106–3111. doi: 10.1021/nl071637k. [DOI] [PubMed] [Google Scholar]
- 41.Bunimovich YL, Ge G, Beverly KC, Ries RS, Hood L, Heath JR. Electrochemically programmed, spatially selective biofunctionalization of silicon wires. Langmuir. 2004;20:10630–10638. doi: 10.1021/la047913h. [DOI] [PubMed] [Google Scholar]
- 42.Salaita K, Wang Y, Mirkin CA. Applications of dip-pen nanolithography. Nat Nanotechnol. 2007;2:145–155. doi: 10.1038/nnano.2007.39. [DOI] [PubMed] [Google Scholar]
- 43.Gudiksen MS, Lauhon LJ, Wang J, Smith DC, Lieber CM. Growth of nanowire superlattice structures for nanoscale photonics and electronics. Nature. 2002;415:617–620. doi: 10.1038/415617a. [DOI] [PubMed] [Google Scholar]
- 44. Li M, Bhiladvala RB, Morrow TJ, Sioss JA, Lew K-K, Redwing JM, Keating CD, Mayer TS. Bottom-up assembly of large-area nanowire resonator arrays. Nat Nanotechnol. 2008;3:88–92. doi: 10.1038/nnano.2008.26.. Probe chemistry-compatible, bottom-up assembly, and integration of large-area nanowire array with high yields and low defect densities.
- 45.Smith PA, Nordquist CD, Jackson TN, Mayer TS, Martin BR, Mbindyo J, Mallouk TE. Electric-field assisted assembly and alignment of metallic nanowires. Appl Phys Lett. 2000;77:1399–1401. [Google Scholar]
- 46.Duan X, Huang Y, Cui Y, Wang J, Lieber CM. Indium phosphide nanowires as building blocks for nanoscale electronic and optoelectronic devices. Nature. 2001;409:66–69. doi: 10.1038/35051047. [DOI] [PubMed] [Google Scholar]
- 47.Huang Y, Duan X, Wei Q, Lieber CM. Directed assembly of one-dimensional nanostructures into functional networks. Science. 2001;291:630–633. doi: 10.1126/science.291.5504.630. [DOI] [PubMed] [Google Scholar]
- 48.Tao AR, Huang J, Yang P. Langmuir-Blodgettry of nanocrystals and nanowires. Acc Chem Res. 2008 doi: 10.1021/ar8000525. in press. [DOI] [PubMed] [Google Scholar]
- 49.He R, Gao D, Fan R, Hochbaum AI, Carraro C, Maboudian R, Yang P. Si nanowire bridges in microtrenches: integration of growth into device fabrication. Adv Mater. 2005;17:2098–2102. [Google Scholar]
- 50.Huang J, Fan R, Connor S, Yang P. One-step patterning of aligned nanowire arrays by programmed dip coating. Angew Chem Int Ed. 2007;46:2414–2417. doi: 10.1002/anie.200604789. [DOI] [PubMed] [Google Scholar]
- 51.Javey A, Nam S, Friedman RS, Yan H, Lieber CM. Layer-by-layer assembly of nanowires for three-dimensional, multifunctional electronics. Nano Lett. 2007;7:773–777. doi: 10.1021/nl063056l. [DOI] [PubMed] [Google Scholar]
- 52.Yu G, Cao A, Lieber CM. Large-area blown bubble films of aligned nanowires and carbon nanotubes. Nat Nanotechnol. 2007;2:372–377. doi: 10.1038/nnano.2007.150. [DOI] [PubMed] [Google Scholar]
- 53.Yun M, Myung NV, Vasquez RP, Lee C, Menke E, Penner RM. Electrochemically grown wires for individually addressable sensor arrays. Nano Lett. 2004;4:419–422. [Google Scholar]
- 54.Lu W, Lieber CM. Nanoelectronics from the bottom up. Nat Mater. 2007;6:841–850. doi: 10.1038/nmat2028. [DOI] [PubMed] [Google Scholar]
- 55.Hamers RJ, Beck JD, Eriksson MA, Li B, Marcus MS, Shang L, Simmons J, Streifer JA. Electrically directed assembly and detection of nanowire bridges in aqueous media. Nanotechnology. 2006;17:S280–S286. [Google Scholar]