Abstract
Epigenetic marking on genes can determine whether or not genes are expressed. Epigenetic regulation is mediated by the addition of methyl groups to DNA cytosine bases, of methyl and acetyl groups to proteins (histones) around which DNA is wrapped, and by small interfering RNA molecules. Some components of epigenetic regulation have evolved to permit control of whether maternal or paternal genes are expressed. The epigenetic imprinting of IGF2 expression is an example of maternal and paternal epigenetic marking that modulates fetal growth and fetal size. However, epigenetic regulation also permits the fetus and the infant to adapt gene expression to the environment in which it is growing; sometimes when this adjustment goes awry, the risk of chronic disease is increased. Recent progress in the understanding of nutritional influences on epigenetics suggests that nutrients that are part of methyl-group metabolism can significantly influence epigenetics. During critical periods in development, dietary methyl-group intake (choline, methionine, and folate) can alter DNA and histone methylation, which results in lifelong changes in gene expression. In rodent models, pregnant dams that were fed diets high in methionine, folic acid, and choline produced offspring with different coat colors or with kinked tails. A number of syndromes in humans can be caused by defective epigenetic regulation, including Rett syndrome. There are interesting examples of the effects of nutrition in early life that result in altered health in adults, and some of these could be the result of altered epigenetic regulation of gene expression.
EPIGENETICS
Humans have very similar genetic codes, yet they have great variation in the ultimate result of gene expression, the phenotype. Some of the variation among individuals is due to the ≈50,000 single nucleotide polymorphisms (SNPs) that each of us harbors. However, even monozygous twins are not absolutely identical despite having identical genetic codes. We inherit genes from both our parents, yet, for some of these genes, we express only the gene from one parent. Obviously, phenotype must be determined by more information than that encoded in the DNA sequence alone. In part, it is due to another coding system, the epigenetic code, whereby humans attain flexibility of gene expression, which enables the plasticity of our phenotypes (1–3).
This epigenetic code is a series of marks added to DNA or to the proteins (histones) around which DNA is wrapped. The best-understood marks are DNA methylation, but covalent modifications of histones and chromatin and RNA interference also mediate the epigenetic regulation of gene expression. Some of these epigenetic marks can be inherited by the children, or by the children's children, of the affected individual. Epigenetic mechanisms mediate the size of an infant at birth (4) and likely mediate how cigarette smoking in a grandmother can increase the risk of asthma in her grandchildren (5) or why malnutrition at the time of puberty in a male is associated with a 4-fold lower risk of type 2 diabetes in his grandson (6).
DNA METHYLATION
DNA methylation occurs at cytosine bases that are followed by a guanosine (5′-CpG-3′ sites) (7). In mammals, most CpG sites in DNA are methylated [90–98% (8)], but there are specific CpG-rich areas of DNA where most CpGs are not methylated; these are known as CpG islands (9). CpG islands span the 5′ end of the regulatory region of genes, and when these islands are methylated, gene expression is usually suppressed or silenced (9, 10). The pattern of DNA methylation in CpG islands varies by tissue type and likely accounts for why genes are expressed differentially among tissues (8). As noted earlier, most mammalian DNA is methylated; this includes exons, intergenic DNA, and transposons. Methylation may be the default state for genes, and although the purpose of the methylation of intragenic DNA is unclear at this time, it may protect against expression of unwanted genes (8). Because the CpG islands in the gene promoter region function as the critical switch that regulates gene expression, it is possible for a gene to be hypermethylated overall yet have CpG islands in its promoter that are undermethylated, which results in a gene that is expressed. Conversely, the exons in a gene may be hypomethylated and the CpG islands hypermethylated, which results in a suppressed gene. The inactive X chromosome in women is a good example: the CpG islands in the genes' promoters on this chromosome are hypermethylated whereas the exons are hypomethylated (8).
A family of enzymes known as DNA methyltransferases (DNMT1, DNMT3a, and DNMT3b) catalyze cytosine methylation. DNMT3a and -b can work on unmodified DNA, and these methyltransferases establish DNA methylation patterns, whereas DNMT1 maintains this pattern thereafter (11). DNMT1 has a >10-fold preference for hemi-methylated DNA and transfers the pattern of methylation from the existing to the newly synthesized strand at replication (12). There is also a regulatory factor (Dnmt3L) that is required for DNMT3a or -b function; it stabilizes the active site on these methyltransferases (11). It is apparent that epigenetic marks mediated by DNMTs are critical for embryonic development because Dnmt-null mice die in early gestation (13). Mutations in methyltransferases result in abnormal fetal development and in immunodeficiency as well as in brain abnormalities in humans (14).
SIGNAL AMPLIFICATION DOWNSTREAM OF THE EPIGENETIC MARKING
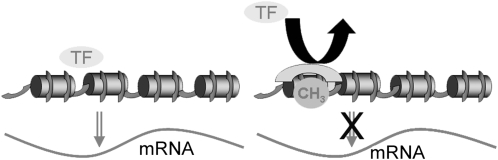
Methylated cytosines serve as docking sites for proteins that prevent transcription factors from accessing their binding sites on the gene promoter (Figure 1). There are 2 families of methyl-CpG-binding proteins (14): the methyl-binding domain proteins that are best exemplified by methyl-CpG-binding protein 2 (MECP2), a 54-kD protein with an N-terminal methyl-binding domain, and the Kaiso family of proteins that contain several zinc fingers that allow them to bind to methylated CpGs. Mutations in the MECP2 gene result in a human developmental delay/autism syndrome (Rett syndrome) that affects mainly females (14). This syndrome is also associated with abnormal organ growth because of the lack of imprinting of IGF2.
FIGURE 1.
Epigenetic marks alter gene expression. Normally, transcription factors bind to promoter regions of DNA and induce gene expression producing mRNA. However, when specific CpG islands in the promoter are methylated, capping proteins that prevent access of the transcription factor to DNA are attracted, and gene expression is suppressed. mRNA, messenger RNA; TF, transcription factor.
HISTONE MODIFICATIONS THAT ARE EPIGENETIC MARKS
Functional interactions between histones and DNA are modulated by their methylation and acetylation status (15–17). Basically, DNA is tightly wound around histones and prevents access to transcription factors. When modified by methylation or acetylation, these proteins loosen up and create gaps through which transcription factors can pass.
Epigenetic marking of histones occurs with the recruiting of G9a methylase, which methylates lysine 9 of histone 3 (K9H3) (18). Mono-methyl or di-methyl K9H3 (1meK9H3 and 2meK9H3) are usually associated with the silencing of genes (19, 20), whereas di-methyl and tri-methyl lysine 4 H3 (2meK4H3 and 3MeK4H3) are enriched in areas with transcriptionally active chromatin. Other posttranslational modifications of histone tails [eg, acetylation of lysines (K) on histone 3 (H3) and on histone 4 (H4)] also alter chromatin architecture at promoter regions (15) and result in a decrease in DNA-histone interaction that results in enhanced gene expression (21).
CROSSTALK
Epigenetic marks on DNA and on histones communicate with each other. Once methyl-CpG-binding proteins attach to methylated cytosines, they attract a variety of other proteins, some of which have enzymatic activity that can further modify neighboring histones by methylating or acetylating specific amino acid residues (14). This reinforces the signals that suppress gene expression. Conversely, histone acetylation inhibits DNA methylation in some promoter areas (21). Thus, epigenetically modified DNA and histones partake in a 2-way conversation that amplifies the desired signals that control gene expression. For example, G9a (ehmt2) histone methylase expression is induced by methylation of a CpG island in its promoter, and this methylase, in turn, methylates lysines 9 (K9) and 27 (K27) on histone H3 (18, 22–25). This increases the binding of the transcriptional repressor restrictive silencing factor (26), which, in turn, recruits a core corepressor complex (which includes histone deacetylase and MeCP2) to repressor element 1 (27, 28).
IMPRINTING
Some components of epigenetic regulation have evolved to permit control of whether maternal or paternal genes are expressed. Most CpG islands in DNA remain unmethylated during development, but a few genes are imprinted genes or genes for which only the paternal or the maternal gene is expressed, and these are specifically methylated (8). The expression of imprinted genes is regulated by methylation of the CpG islands in their promoter, and the markings are stably replicated during cell division but are reversed when inherited through an individual of the opposite sex (4). For example, IGF2, which encodes a growth factor, is maternally suppressed and paternally activated (4): the mother wants a smaller baby whereas the father wants a bigger one.
NUTRITION AND EPIGENETIC MARKS
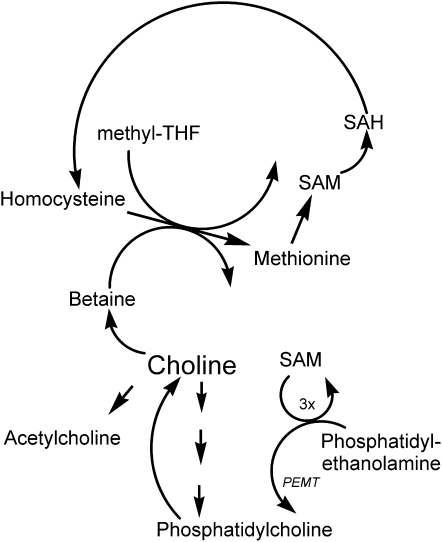
The DNA and histone methyltransferases all use S-adeno-sylmethionine (SAM) as the methyl donor. As will be discussed below, the availability of SAM is directly influenced by diet. Thus, diet can directly influence epigenetic marking. SAM is formed from methyl groups derived from choline, methionine, or methyl-tetrahydrofolate (Figure 2). Choline, methionine, and folate metabolism are metabolically related at the point at which homocysteine is converted to methionine. Thus, the effects of these nutrients on epigenetic marking are interrelated.
FIGURE 2.
Choline-methionine-folate interactions. These 3 nutrients are metabolically interrelated. Methyl-THF, 5-methyl tetrahydrofolate; SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine; PEMT, phosphatidylethanolamine-N-methyltransferase.
Homocysteine can be methylated to form methionine (29) by 2 parallel pathways, both of which lower homocysteine concentrations (30). In the first, vitamin B-12 and folic acid are involved in a reaction catalyzed by methionine synthase (31). A deficiency of these nutrients (32, 33), or SNPs in the genes for the enzymes involved in this pathway (31, 33, 34), can result in elevated plasma homocysteine concentrations. In addition, tetrahydrofolate is needed to scavenge one-carbon groups when betaine is metabolized (35). The alternative pathway for the methylation of homocysteine to form methionine is catalyzed by betaine homocysteine methyltransferase (36), an enzyme whose activity has been reported to increase in rats during methionine excess (37). Betaine, which is derived from dietary choline by the action of choline dehydrogenase, is the methyl group donor in this reaction, and supplemental oral betaine can lower plasma homocysteine concentrations (38, 39).
Any of these pathways can interact to alter epigenetic DNA and histone methylation because perturbing the metabolism of one of the methyl donors results in compensatory changes in the other methyl donors because of the intermingling of these metabolic pathways (40–42). Rats treated with the antifolate methotrexate had diminished pools of choline metabolites in liver (41, 43). Rats ingesting a choline-deficient diet had diminished tissue concentrations of methionine and SAM (44) and doubled plasma homocysteine concentrations (45). Humans who are depleted of choline have a diminished capacity to methylate homocysteine and develop elevated homocysteine concentrations in plasma after a methionine loading test (46).
MATERNAL DIET INFLUENCES FETAL DEVELOPMENT BY CHANGING EPIGENETIC MARKS
Maternal diets high in choline and/or methionine and/or methyl-folate during pregnancy result in epigenetic changes in gene expression in the fetus that have permanent effects on their offspring. Feeding pregnant mice diets low in choline and methionine diminished methylation in CpG islands of genes that controlled brain development in the fetuses (47). This had important function effects. Maternal dietary choline supplementation or choline deficiency during late pregnancy in rodents was associated with significant and irreversible changes in hippocampal function in the adult rodent, including altered long-term potentiation (LTP) (48–50) and altered memory (51–56). Dams fed choline-deficient diets during late pregnancy have offspring with decreased methylation of genes that inhibit cell cycling (47, 57), thereby inducing these genes and causing diminished progenitor-cell proliferation and increased apoptosis in the fetal hippocampus (58, 59), insensitivity to LTP as adult animals (50), and reduced visuospatial and auditory memory (56). More choline (≈4× dietary concentrations) during days 11–17 of gestation in the rodent increased hippocampal progenitor cell proliferation (58, 59), decreased apoptosis in these cells (58, 59), enhanced LTP in the offspring when they were adult animals (48–50), and enhanced visuospatial and auditory memory by as much as 30% in the adult animals throughout their lifetimes (51–53, 55, 56, 60, 61). Indeed, memory in adult rodents decreases as they age, and offspring exposed to extra choline in utero do not show this “senility” (53, 60). Thus, choline supplementation during a critical period in pregnancy causes lifelong changes in brain structure and function by changing epigenetic marking.
There are other good examples of how powerful diet can be in changing epigenetic marks. Feeding pregnant Pseudoagouti Avy/a mouse dams a choline-, methionine-, and folate-supplemented diet altered epigenetic regulation of agouti expression in their offspring, as indicated by increased methylation of the involved gene and by agouti/black mottling of their coats (62, 63). In another example, there was increased DNA methylation of the fetal gene axin fused [Axin(Fu)] after methyl donor supplementation of female mice before and during pregnancy, which reduced by 50% the incidence of tail kinking in Axin(Fu)/+ offspring. It is clear that the dietary manipulation of methyl donors (either deficiency or supplementation) can have a profound and permanent effect on gene expression via changes in epigenetic marks.
SNPs AND DIETARY REQUIREMENT FOR METHYL GROUPS
Epigenetic marks on DNA may be changed by genetic variations in genes of methyl metabolism (methionine, choline, and folate) that alter SAM availability. There are multiple SNPs in genes of methyl-group metabolism that are common in humans and that change human dietary requirements for methyl donors. In folate metabolism, the MTHFR C677T SNP is a well described and common variant that increases the dietary folate requirement (64). Choline metabolism SNPs provide an excellent and well-documented example of how such genetic variation can alter dietary requirements and alter epigenetic methylation. SNPs in folate and choline metabolism genes increase human dietary requirements for choline (65). Although some humans develop fatty liver as well as liver and muscle damage when fed a low-choline diet, others do not. Choline is derived from the diet and by de novo biosynthesis catalyzed by an enzyme coded by the gene PEMT (66). Estrogen causes a marked upregulation in PEMT mRNA expression and enzyme activity (66). Even among premenopausal women who should be resistant to choline deficiency because estrogen induces PEMT, a significant number develop organ dysfunction when deprived of choline (67). Premenopausal women who were carriers of the very common 5,10-methylenetetrahydrofolate dehydrogenase-1958A (MTHFD1; rs2236225) gene allele were >15× as likely as noncarriers to develop signs of choline deficiency on the low-choline diet. Sixty-three percent of the study population had at least one allele for this SNP. Two reactions, mediated by methylenetetrahydrofolate dehydrogenase and methenyltetrahydrofolate cyclohydrolase, can convert 10-formyl tetrahydrofolate to 5,10-methylenetetrahydrofolate. Whereas the formation of 5-methyl tetrahydrofolate is practically irreversible in vivo, the interconversion of 5,10-methylenetetrahydrofolate and 10-formyl tetrahydrofolate is closer to equilibrium (68). This means that 5,10-methylenetetrahydrofolate may be directed either toward homocysteine remethylation or away from it. The MTHFD1 G1958A polymorphism may thus affect the delicately balanced flux between 5,10-methylenetetrahydrofolate and 10-formyl tetrahydrofolate and thereby influence the availability of 5-methyl tetrahydrofolate for homocysteine remethylation. This would increase demand for choline as a methyl-group donor. It is of interest that the risk of having a child with a neural tube defect increases in mothers with the G1958A SNP in MTHFD1 (69).
As noted earlier, PEMT encodes for a protein responsible for endogenous formation of choline. We identified a SNP in the promoter region of the PEMT gene (rs12325817) for which 78% of carriers of the C allele developed organ dysfunction when fed a low-choline diet (odds ratio: 25; 95% CI: 2, 256) (70). Considering the sexual differences in the effect of PEMT rs12325817, it is possible that this SNP alters the estrogen responsiveness of the promoter. The frequency of this variant allele was 0.74. The first of 2 SNPs in the coding region of the choline dehydrogenase gene (CHDH; rs9001) had a protective effect on susceptibility to choline deficiency, whereas a second CHDH variant (rs12676) was associated with increased susceptibility to choline deficiency (70). An SNP in the betaine:homocysteine methyltransferase gene (BHMT; rs3733890) was not associated with susceptibility to choline deficiency (70).
Thus, it appears that genetic variants in methyl-group metabolism are extremely common and that these alter the availability of SAM for epigenetic marking of DNA and histones. When diet is adequate, these marks may be very different from when diet is not, and therefore these marks may reflect the dietary environment of the individual. A critical, but as yet unanswered question is whether this epigenetic responsiveness to diet occurs only during a critical sensitive period of early development or whether responsiveness persists throughout life.
DIET AND EPIGENETICS AS DETERMINANTS OF LATER HEALTH OUTCOMES
As noted earlier, epigenetic marks can be inherited by the children or by the children's children of the affected individual. Epigenetic mechanisms likely mediate how cigarette smoking in a grandmother can increase the risk of asthma in her grandchildren (5) or why malnutrition at the time of puberty in a male is associated with a 4-fold lower risk of type 2 diabetes in his grandson (6). In rats, nurturing behaviors of pup licking and grooming and arch-back nursing are inherited from parents via an epigenetic signal involving DNA methylation and histone alterations in the hippocampus (71). Behavior in humans may also be modulated via epigenetic marks. In brains from suicide victims, methylation of the 5′ regulatory region of genes encoding ribosomal RNA was significantly increased and was associated with a history of early childhood neglect/abuse (72). DNA methylation patterns are significantly different in humans with psychotic disorders (73).
How do epigenetic marks become heritable? Although most DNA methylation is erased in the germ line, some methylated sites survive and then are replicated by DNMT1 every time a cell divides. When genes are passed on in the germ line, this DNA is passed along with associated histones. If these histones are epigenetically marked, these markings can then be copied every time the cell divides, influencing gene expression throughout life. Thus, epigenetic mechanisms provide very reasonable explanations for how the dietary exposures of grandparents or how the dietary exposures early in life can be determinants of later health outcomes. (Other articles in this supplement to the Journal include references 74–81.)
Acknowledgments
The author received grant support from the NIH, the USDA, Mead Johnson Nutritionals, and the Egg Nutrition Research Center for studies other than those described in this article and serves on advisory boards for the Solae Company, the Dupont Company, Metabolon, and Pogo Health. The author had no financial conflict of interest in relation to this study.
REFERENCES
- 1.Jaenisch R. DNA methylation and imprinting: why bother? Trends Genet 1997;13:323–9 [DOI] [PubMed] [Google Scholar]
- 2.Jones PA, Gonzalgo ML. Altered DNA methylation and genome instability: a new pathway to cancer? Proc Natl Acad Sci USA 1997;94:2103–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robertson KD, Wolffe AP. DNA methylation in health and disease. Nat Rev Genet 2000;1:11–9 [DOI] [PubMed] [Google Scholar]
- 4.Killian JK, Byrd JC, Jirtle JV, et al. M6P/IGF2R imprinting evolution in mammals. Mol Cell 2000;5:707–16 [DOI] [PubMed] [Google Scholar]
- 5.Li YF, Langholz B, Salam MT, Gilliland FD. Maternal and grandmaternal smoking patterns are associated with early childhood asthma. Chest 2005;127:1232–41 [DOI] [PubMed] [Google Scholar]
- 6.Pembrey ME. Time to take epigenetic inheritance seriously. Eur J Hum Genet 2002;10:669–71 [DOI] [PubMed] [Google Scholar]
- 7.Holliday R, Grigg GW. DNA methylation and mutation. Mutat Res 1993;285:61–7 [DOI] [PubMed] [Google Scholar]
- 8.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet 2008;9:465–76 [DOI] [PubMed] [Google Scholar]
- 9.Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. ChemBioChem 2002;3:274–93 [DOI] [PubMed] [Google Scholar]
- 10.Bird AP. CpG-rich islands and the function of DNA methylation. Nature 1986;321:209–13 [DOI] [PubMed] [Google Scholar]
- 11.Cheng X, Blumenthal RM. Mammalian DNA methyltransferases: a structural perspective. Structure 2008;16:341–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatchwell E, Greally JM. The potential role of epigenomic dysregulation in complex human disease. Trends Genet 2007;23:588–95 [DOI] [PubMed] [Google Scholar]
- 13.Li E, Bestor T, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 1992;69:915–26 [DOI] [PubMed] [Google Scholar]
- 14.Clouaire T, Stancheva I. Methyl-CpG binding proteins: specialized transcriptional repressors or structural components of chromatin? Cell Mol Life Sci 2008;65:1509–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felsenfeld G, Groudine M. Controlling the double helix. Nature 2003;421:448–53 [DOI] [PubMed] [Google Scholar]
- 16.Esteve PO, Chin HG, Smallwood A, et al. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev 2006;20:3089–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quina AS, Buschbeck M, Di Croce L. Chromatin structure and epigenetics. Biochem Pharmacol 2006;72:1563–9 [DOI] [PubMed] [Google Scholar]
- 18.Roopra A, Qazi R, Schoenike B, Daley TJ, Morrison JF. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol Cell 2004;14:727–38 [DOI] [PubMed] [Google Scholar]
- 19.Peters AH, Kubicek S, Mechtler K, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell 2003;12:1577–89 [DOI] [PubMed] [Google Scholar]
- 20.Rice JC, Briggs SD, Ueberheide B, et al. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell 2003;12:1591–8 [DOI] [PubMed] [Google Scholar]
- 21.Mutskov VJ, Farrell CM, Wade PA, Wolffe AP, Felsenfeld G. The barrier function of an insulator couples high histone acetylation levels with specific protection of promoter DNA from methylation. Genes Dev 2002;16:1540–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem 2001;276:25309–17 [DOI] [PubMed] [Google Scholar]
- 23.Tachibana M, Sugimoto K, Nozaki M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 2002;16:1779–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tachibana M, Ueda J, Fukuda M, et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev 2005;19:815–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao B, Jing C, Wilson JR, et al. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature 2003;421:652–6 [DOI] [PubMed] [Google Scholar]
- 26.Chong JA, Tapia-Ramirez J, Kim S, et al. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell 1995;80:949–57 [DOI] [PubMed] [Google Scholar]
- 27.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 2005;121:645–57 [DOI] [PubMed] [Google Scholar]
- 28.Ballas N, Mandel G. The many faces of REST oversee epigenetic programming of neuronal genes. Curr Opin Neurobiol 2005;15:500–6 [DOI] [PubMed] [Google Scholar]
- 29.Finkelstein JD. Pathways and regulation of homocysteine metabolism in mammals. Semin Thromb Hemost 2000;26:219–25 [DOI] [PubMed] [Google Scholar]
- 30.Olthof MR, van Vliet T, Boelsma E, Verhoef P. Low dose betaine supplementation leads to immediate and long term lowering of plasma homocysteine in healthy men and women. J Nutr 2003;133:4135–8 [DOI] [PubMed] [Google Scholar]
- 31.Weisberg IS, Jacques PF, Selhub J, et al. The 1298A→C polymorphism in methylenetetrahydrofolate reductase (MTHFR): in vitro expression and association with homocysteine. Atherosclerosis 2001;156:409–15 [DOI] [PubMed] [Google Scholar]
- 32.Shelnutt KP, Kauwell GP, Chapman CM, et al. Folate status response to controlled folate intake is affected by the methylenetetrahydrofolate reductase 677C→T polymorphism in young women. J Nutr 2003;133:4107–11 [DOI] [PubMed] [Google Scholar]
- 33.Jacques PF, Bostom AG, Wilson PW, Rich S, Rosenberg IH, Selhub J. Determinants of plasma total homocysteine concentration in the Framingham Offspring cohort. Am J Clin Nutr 2001;73:613–21 [DOI] [PubMed] [Google Scholar]
- 34.Watkins D, Ru M, Hwang HY, et al. Hyperhomocysteinemia due to methionine synthase deficiency, cblG: structure of the MTR gene, genotype diversity, and recognition of a common mutation, P1173L. Am J Hum Genet 2002;71:143–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mudd SH, Ebert MH, Scriver CR. Labile methyl group balances in the human: the role of sarcosine. Metabolism 1980;29:707–20 [DOI] [PubMed] [Google Scholar]
- 36.Sunden SL, Renduchintala MS, Park EI, Miklasz SD, Garrow TA. Betaine-homocysteine methyltransferase expression in porcine and human tissues and chromosomal localization of the human gene. Arch Biochem Biophys 1997;345:171–4 [DOI] [PubMed] [Google Scholar]
- 37.Holm PI, Bleie O, Ueland PM, et al. Betaine as a determinant of postmethionine load total plasma homocysteine before and after B-vitamin supplementation. Arterioscler Thromb Vasc Biol 2004;24:301–7 [DOI] [PubMed] [Google Scholar]
- 38.Steenge GR, Verhoef P, Katan MB. Betaine supplementation lowers plasma homocysteine in healthy men and women. J Nutr 2003;133:1291–5 [DOI] [PubMed] [Google Scholar]
- 39.Wendel U, Bremer H. Betaine in the treatment of homocystinuria due to 5,10-methylenetetrahydrofolate reductase deficiency. Eur J Pediatr 1984;142:147–50 [DOI] [PubMed] [Google Scholar]
- 40.Kim Y-I, Miller JW, da Costa K-A, et al. Severe folate deficiency causes secondary depletion of choline and phosphocholine in rat liver. J Nutr 1994;124:2197–203 [DOI] [PubMed] [Google Scholar]
- 41.Selhub J, Seyoum E, Pomfret EA, Zeisel SH. Effects of choline deficiency and methotrexate treatment upon liver folate content and distribution. Cancer Res 1991;51:16–21 [PubMed] [Google Scholar]
- 42.Varela-Moreiras G, Selhub J, da Costa K, Zeisel SH. Effect of chronic choline deficiency in rats on liver folate content and distribution. J Nutr Biochem 1992;3:519–22 [Google Scholar]
- 43.Pomfret EA, da Costa K, Zeisel SH. Effects of choline deficiency and methotrexate treatment upon rat liver. J Nutr Biochem 1990;1:533–41 [DOI] [PubMed] [Google Scholar]
- 44.Zeisel SH, Zola T, da Costa K, Pomfret EA. Effect of choline deficiency on S-adenosylmethionine and methionine concentrations in rat liver. Biochem J 1989;259:725–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Varela-Moreiras G, Ragel C, Perez de Miguelsanz J. Choline deficiency and methotrexate treatment induces marked but reversible changes in hepatic folate concentrations, serum homocysteine and DNA methylation rates in rats. J Am Coll Nutr 1995;14:480–5 [DOI] [PubMed] [Google Scholar]
- 46.da Costa KA, Gaffney CE, Fischer LM, Zeisel SH. Choline deficiency in mice and humans is associated with increased plasma homocysteine concentration after a methionine load. Am J Clin Nutr 2005;81:440–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niculescu MD, Craciunescu CN, Zeisel SH. Dietary choline deficiency alters global and gene-specific DNA methylation in the developing hippocampus of mouse fetal brains. FASEB J 2006;20:43–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pyapali GK, Turner DA, Williams CL, Meck WH, Swartzwelder HS. Prenatal choline supplementation decreases the threshold for induction of long-term potentiation in young adult rats. J Neurophysiol 1998;79:1790–6 [DOI] [PubMed] [Google Scholar]
- 49.Montoya DA, White AM, Williams CL, Blusztajn JK, Meck WH, Swartzwelder HS. Prenatal choline exposure alters hippocampal responsiveness to cholinergic stimulation in adulthood. Brain Res Dev Brain Res 2000;123:25–32 [DOI] [PubMed] [Google Scholar]
- 50.Jones JP, Meck W, Williams CL, Wilson WA, Swartzwelder HS. Choline availability to the developing rat fetus alters adult hippocampal long-term potentiation. Brain Res Dev Brain Res 1999;118:159–67 [DOI] [PubMed] [Google Scholar]
- 51.Meck WH, Williams CL. Perinatal choline supplementation increases the threshold for chunking in spatial memory. Neuroreport 1997;8:3053–9 [DOI] [PubMed] [Google Scholar]
- 52.Meck WH, Williams CL. Characterization of the facilitative effects of perinatal choline supplementation on timing and temporal memory. Neuroreport 1997;8:2831–5 [DOI] [PubMed] [Google Scholar]
- 53.Meck WH, Williams CL. Simultaneous temporal processing is sensitive to prenatal choline availability in mature and aged rats. Neuroreport 1997;8:3045–51 [DOI] [PubMed] [Google Scholar]
- 54.Meck WH, Smith RA, Williams CL. Organizational changes in cholinergic activity and enhanced visuospatial memory as a function of choline administered prenatally or postnatally or both. Behav Neurosci 1989;103:1234–41 [DOI] [PubMed] [Google Scholar]
- 55.Meck WH, Smith RA, Williams CL. Pre- and postnatal choline supplementation produces long-term facilitation of spatial memory. Dev Psychobiol 1988;21:339–53 [DOI] [PubMed] [Google Scholar]
- 56.Meck WH, Williams CL. Choline supplementation during prenatal development reduces proactive interference in spatial memory. Brain Res Dev Brain Res 1999;118:51–9 [DOI] [PubMed] [Google Scholar]
- 57.Niculescu MD, Yamamuro Y, Zeisel SH. Choline availability modulates human neuroblastoma cell proliferation and alters the methylation of the promoter region of the cyclin-dependent kinase inhibitor 3 gene. J Neurochem 2004;89:1252–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Albright CD, Tsai AY, Friedrich CB, Mar MH, Zeisel SH. Choline availability alters embryonic development of the hippocampus and septum in the rat. Brain Res Dev Brain Res 1999;113:13–20 [DOI] [PubMed] [Google Scholar]
- 59.Albright CD, Friedrich CB, Brown EC, Mar MH, Zeisel SH. Maternal dietary choline availability alters mitosis, apoptosis and the localization of TOAD-64 protein in the developing fetal rat septum. Brain Res Dev Brain Res 1999;115:123–9 [DOI] [PubMed] [Google Scholar]
- 60.Meck WH, Williams CL. Metabolic imprinting of choline by its availability during gestation: implications for memory and attentional processing across the lifespan. Neurosci Biobehav Rev 2003;27:385–99 [DOI] [PubMed] [Google Scholar]
- 61.Williams CL, Meck WH, Heyer DD, Loy R. Hypertrophy of basal forebrain neurons and enhanced visuospatial memory in perinatally choline-supplemented rats. Brain Res 1998;794:225–38 [DOI] [PubMed] [Google Scholar]
- 62.Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J 1998;12:949–57 [PubMed] [Google Scholar]
- 63.Cooney CA, Dave AA, Wolff GL. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J Nutr 2002;132:2393S–400S [DOI] [PubMed] [Google Scholar]
- 64.Davis SR, Quinlivan EP, Shelnutt KP, et al. The methylenetetrahydrofolate reductase 677C->T polymorphism and dietary folate restriction affect plasma one-carbon metabolites and red blood cell folate concentrations and distribution in women. J Nutr 2005;135:1040–4 [DOI] [PubMed] [Google Scholar]
- 65.Kohlmeier M, da Costa KA, Fischer LM, Zeisel SH. Genetic variation of folate-mediated one-carbon transfer pathway predicts susceptibility to choline deficiency in humans. Proc Natl Acad Sci USA 2005;102:16025–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Resseguie M, Song J, Niculescu MD, da Costa KA, Randall TA, Zeisel SH. Phosphatidylethanolamine N-methyltransferase (PEMT) gene expression is induced by estrogen in human and mouse primary hepatocytes. FASEB J 2007;21:2622–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fischer LM, daCosta K, Kwock L, et al. Sex and menopausal status influence human dietary requirements for the nutrient choline. Am J Clin Nutr 2007;85:1275–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Horne DW. Neither methionine nor nitrous oxide inactivation of methionine synthase affect the concentration of 5,10-methylenetetrahydrofolate in rat liver. J Nutr 2003;133:476–8 [DOI] [PubMed] [Google Scholar]
- 69.Brody LC, Conley M, Cox C, et al. A polymorphism, R653Q, in the trifunctional enzyme methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase/formyltetrahydrofolate synthetase is a maternal genetic risk factor for neural tube defects: report of the Birth Defects Research Group. Am J Hum Genet 2002;71:1207–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.da Costa KA, Kozyreva OG, Song J, Galanko JA, Fischer LM, Zeisel SH. Common genetic polymorphisms affect the human requirement for the nutrient choline. FASEB J 2006;20:1336–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weaver IC, Cervoni N, Champagne FA, et al. Epigenetic programming by maternal behavior. Nat Neurosci 2004;7:847–54 [DOI] [PubMed] [Google Scholar]
- 72.McGowan PO, Sasaki A, Huang TC, et al. Promoter-wide hypermethylation of the ribosomal RNA gene promoter in the suicide brain. PLoS One 2008;3:e2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mill J, Tang T, Kaminsky Z, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet 2008;82:696–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Atkinson SA. Introduction to the workshop. Am J Clin Nutr 2009;89(suppl):1485S–7S [DOI] [PubMed] [Google Scholar]
- 75.Bouchard C. Childhood obesity: are genetic differences involved? Am J Clin Nutr 2009;89(suppl):1494S–501S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koletzko B, von Kries R, Monasterolo RC, et al. Can infant feeding choices modulate later obesity risk? Am J Clin Nutr 2009;89(suppl):1502S–8S [DOI] [PubMed] [Google Scholar]
- 77.Ordovas JM. Genetic influences on blood lipids and cardiovascular disease risk: tools for primary prevention. Am J Clin Nutr 2009;89(suppl):1509S–17S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Symonds ME, Stephenson T, Budge H. Early determinants of cardiovascular disease: the role of early diet in later blood pressure control. Am J Clin Nutr 2009;89(suppl):1518S–22S [DOI] [PubMed] [Google Scholar]
- 79.Carlson SE. Early determinants of development: a lipid perspective. Am J Clin Nutr 2009;89(suppl):1523S–9S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Field CJ. Summary of the workshop. Am J Clin Nutr 2009;89(suppl):1533S–9S [DOI] [PubMed] [Google Scholar]
- 81.Rai D, Larson B. Driving research in infant and children's nutrition: a perspective on industry. Am J Clin Nutr 2009;89(suppl):1530S–2S [DOI] [PubMed] [Google Scholar]