Abstract
We report the effect of surface coverage and sequence on the performance of 5′ thiolated, 3′ fluorophore-labeled DNA hairpin probes bound to Au/Ag striped (“barcoded”) metal nanowires. Coverage was controlled by varying probe concentration, buffer ionic strength, and by addition of short hydroxy-terminated alkanethiol diluent molecules during probe assembly onto the nanowire surface. Surface dilution of the surface-bound probes with aω-hydroxyl alkanethiol, a commonly accepted practice in the surface-bound DNA literature, did not appreciably improve sensor performance as compared to similar probe coverages without hydroxyalkanethiol diluents; this finding underscores the differences between the molecular beacon probes used here and more traditional nonfluorescent, random coil probes. We found that intermediate probe coverage of ~1012 molecules/cm2 gave the best discrimination between presence and absence of a target sequence. Because we are interested in multiplexed assays, we also compared several beacon probe sequences having different stabilities for secondary structure formation in solution; we found that both probe surface coverage and sensor performance varied for different probe sequences. When five different molecular beacon probes, each bound to barcoded nanowires, were used in a multiplexed, wash-free assay for target oligonucleotides corresponding to viral nucleic acid sequences, these differences in probe performance did not prevent accurate target identification. We anticipate that the findings described here will also be relevant to other applications involving molecular beacons or other structured nucleic acid probes immobilized on metal surfaces.
Introduction
Surface films of oligonucleotides can provide selective capture of target sequences for nucleic acid detection. Noble metal surfaces such as Au or Ag are often employed because they are conductive, and can quench or enhance fluorescence, support surface plasmons, and/or provide the heightened electromagnetic fields required for ultrasensitive surface enhanced Raman detection.1–5 Consequently, DNA-coated Au and Ag have been used in a wide variety of sensors based on fluorescence, electrochemistry, surface plasmon resonance, surface-enhanced Raman scattering, and other transduction strategies.6–10 In all of these applications, control of the DNA–surface interface is of critical importance. DNA oligonucleotide probe sequences are commonly attached to noble metal surfaces via a 5′ or 3′ thiol moiety, often with an alkane spacer (e.g., C6H12 or C12H24) and sometimes also additional bases (e.g., poly dT or other sequences). This approach provides single-point attachment to facilitate hybridization to complementary target sequences. Generally probe sequences are selected to avoid the formation of stable secondary structures under experimental conditions (temperature, buffer ionic strength), such that they will be fully available for hybridization with target strands from solution. We will refer to such sequences, which exist as random coils, as “linear probes.”
The DNA–metal interface has been well characterized for thiolated linear probes on noble metals.11–16 At high probe densities, hybridization is hindered by steric and electrostatic repulsions.17–19 Electrostatic repulsions can be manipulated by controlling the ionic strength of the buffers used during probe assembly and/or target hybridization.11,13,20,21 Low ionic strength during probe assembly results in low surface density, which reduces steric hindrance and can result in higher hybridization efficiencies for target binding.11–13,20 Target hybridization is generally performed at relatively high ionic strengths (e.g., 0.3 M) to overcome repulsions between probes and target strands and favor binding. Steric repulsions can also be controlled by surface dilution with thiolated molecules shorter than the probes. Herne and Tarlov pioneered the use of mixed monolayers of thiolated DNA probes and mercaptohexanol on a gold surface.11 The backfilling with mercaptohexanol after initial probe adsorption improved hybridization efficiency by reducing both probe surface density and nonspecific binding events between the nitrogen-containing DNA bases of the probe and the Au surface. This hydroxy-terminated alkanethiol procedure has since been widely accepted.14,21–33
Oligonucleotide probe molecules can also be designed to adopt stable secondary structures under experimental conditions. Molecular beacons (MBs) are oligonucleotide probes designed with self-complementarity at the 3′ and 5′ ends such that they adopt stem-loop or “hairpin” structures.34–37 MBs typically incorporate a quencher and fluorophore at their opposing ends, such that fluorescence is quenched in the hairpin form due to proximity of the quencher to the fluorescent dye.38 Fluorescence can then be detected upon hybridization of a complementary target sequence, which binds to the loop region and separates the fluorophore from the quencher. MBs exhibit an enhanced specificity over linear probes for binding complementary targets.39,40 Additionally, the signaling fluorophore at the beacon 5′ or 3′ end eliminates the need to both label a target sequence and rinse after hybridization. This concept has proven useful in the solution-phase detection of single nucleotide polymorphisms (SNPs) and PCR products.41,42 Another important example of designed secondary structure is aptamers, which are nucleic acids selected for specific binding to any of several classes of targets, including not only nucleic acids, but also small molecules, ions, and proteins. Aptamer conformations, which can include secondary structures such as hairpin loops, bulges, and pseudoknots, are critical to their function.43–47 Aptamer beacons have also been prepared, where a target-induced conformational change leads to the onset of fluorescence in analogy to MBs.46
Surface attachment of MB and aptamer probes is highly attractive as it would enable not only a wide range of surface-based transduction strategies, but also spatial arraying, substantially increasing the degree of multiplexing possible. Thus, interest in the surface attachment of structured probes has been increasing.27,48–53 For example, aptamers have been arrayed on surfaces for multiplexed protein detection.54 MBs have been immobilized on a number of surfaces such as glass,55–60 polystyrene,61 polymethylmethacrylate (PMMA),60 agarose,62 and gold.15,48–50,63–67 MBs attached to metal surfaces are particularly appealing, because the surface itself can take the place of organic quenchers, and can provide lower background fluorescence.48,50,64,65,67,68 While many reports of surface-bound aptamer and beacon probes have appeared, characterization of the probe/surface interface is less common, with probe surface coverages rarely reported, much less varied and correlated to sensor performance. Notable exceptions include electrochemical sensors based on aptamer and beacon probe conformational changes, where similar optimal coverages ~2 × 1012 probes/cm2 were found.49,69
Here, we investigate the effects of probe surface coverage as controlled by ionic strength, time, and dilution with short alkanethiol spacers, and compare several different hairpin probe sequences for use in simultaneous assays on the encoded nanowires. We use barcoded metal nanowires composed of Au and Ag segments as encoded supports and metallic quenchers for bound hairpin probes. This work is significant in that (1) it describes the effects of surface probe density on MB probe performance (intensity in presence and absence of target sequences) for thiolated probes on metal surfaces, and (2) it directly compares surface attachment and probe performance for multiple different probe sequences designed to detect different viral signatures. We previously reported on nucleic acid bioassays employing MB probes immobilized on the surface of barcoded nanowires,15,50,67 which can be performed in a multiplexed, wash-free, closed chamber format.50,70 The studies reported here build on that work to help pave the way for improved multiplexed assays–on barcoded metal nanowires or other encoded supports–in which many different hairpin probe sequences are used to simultaneously report on their targets.
Materials and Methods
Materials
Chemicals and biochemicals were used as received without further purification except where noted. All water used was purified via a Barnstead Nanopure System to a resistivity of 18 MΩ. Barcoded metal nanowires71–75 (NWs, marketed as Nanobarcodes) were obtained from Nanoplex Technologies (now Oxonica, Inc.). The nanowires are approximately 6 µm in length, and 300 nm in cross-sectional diameter, and are patterned by 1-µm segments of Ag and Au metals, which can be distinguished by their optical reflectance. Patterns used here include: 010010, 011000, 000111, 001101, 010101, 100011, and 100101, where 0 = 1 µm of Au, and 1 = 1 µm of Ag. The NWs were received as a suspension of 1 × 109 wires/mL stock concentration in ethanol. Before use, the desired volume of stock NW suspension was rinsed by centrifugation (1 min at 7700g)/resuspension in 2× volume twice in water and once in buffer prior to resuspension in 3× volume buffer. Buffers used in this work were: (1) 0.01 M PBS (0.138 M NaCl; 0.0027 M KCl; 10 mM sodium phosphate, pH 7.4), purchased from Sigma, (2) 0.3 M PBS (0.3 M NaCl; 10 mM sodium phosphate, pH 7.2), and (3) 0.5 M CAC (0.5 M NaCl; 20 mM cacodylic acid; 0.5 mM EDTA, pH 7.0).48
DNA sequences were designed using mfold folding software76 and were complementary to viral sequences as detailed in Table 1. Solution-phase melting transition temperatures were experimentally determined for most of these sequences, and are given in Table 1. We note that mfold predictions of solution secondary structure stability differed from our experimental measurements of solution stability, perhaps due in part to the presence of the dye and thiol moieties in the actual sequences; only experimentally determined Tm’s and ΔG calculated from the melt data are reported and discussed here. All sequences were purchased from Integrated DNA Technologies, Inc. Thiolated DNA probes (received as disulfides) were cleaved before use by reaction with 100 mM dithiothreitol (DTT) for 30 min in 0.1 M phosphate buffer (pH 8.3) and purified by CentriSpin 10 columns (Princeton Separations) according to manufacturer protocols. Attachment of thiolated probe sequences to NWs was performed as described below. Target sequences were synthetic DNA oligonucleotides complementary to the loop regions of the probes.
Table 1.
Sequences Used in This Worka
| name | sequence (5′to 3′)b | Hairpin Tm (°C)c | hairpin ΔG (kcal/mol)d | comments |
|---|---|---|---|---|
| DENV-2 | thiol(CH2)6-GCG AGG TGT CTG TTA CCA AGG ATC TGC TCG C-TAMRA | probe specific for a 21-base region of Dengue Virus subtype 2 |
||
| HCV | thiol(CH2)6-GCG AGC ATA GTG GTC TGC GGA ACC GGT GAC TCG C-TAMRA | 62.72 ± 0.06 | −4.8 ± 0.3 | probe specific for a 24-base region of HCV |
| HIV | thiol(CH2)6-GCG AGT GTT AAA AGA GAC CAT CAA TGA GCT CGC-TAMRA | 52.6 ± 0.1 | −2.5 ± 0.5 | probe specific for a 23-base region of HIV |
| INF Be | thiol(CH2)6-GCT GCC AAC GAA GTA GGT GGA GAC GGA GGG GCA GC-TAMRA | 70.40 ± 0.08 | −7.3 ± 0.4 | probe specific for a 23-base region of Influenza B |
| SARS | thiol(CH2)6-GCG AGA GAT GCT GTG GGT ACT AAC CTA CCT CTC GC-TAMRA | 63.83 ± 0.05 | −5.5 ± 0.3 | probe specific for a 25-base region of SARS |
Targets were fully complementary to and the same length as the loop for each probe.
The underlined portions of probe sequences indicate regions of self-complementarity.
Experimentally determined at 1.0 μM in 0.3 M PBS. Please note that these are solution values, and do not represent Tm’s for surface-bound probes.
Calculated from experimental (solution-phase) Tm data.
Sequence for the INF B beacon was taken from ref 77.
SurfaceDilutionofThiolatedMBProbeswithMercaptoethanol
Ninety-six µL of NW stock (pattern 010010) were rinsed and resuspended in 0.01 MPBS as described above, then split into separate 16 µL aliquots for reaction. β-mercaptoethanol (BME, Sigma) and HIV thiolated probe DNA were combined in 0:1, 1:1, 5:1, 10:1, and 20:1 mol ratios. In all samples, the total concentration of adsorbate (DNA + BME) was equal to 1.0 µM. The samples were rotated overnight in the dark at room temperature, after which the volume was doubled by addition of 0.3 M PBS. Rotation continued for another 2 h. Samples were then rinsed and stored in 0.3 M PBS. In the “backfilling” experiment, 1.0 µM BME was added to an aliquot of conjugated 0:1 NWs from above and reacted for 30 min. These wires were then rinsed and stored in 0.3 M PBS.
Effect of Salt Concentration upon Probe Coverage
For these experiments, ionic strength was controlled by KCl concentration in 5 mM phosphate buffer, pH 7.0. Solutions with very low ionic strength (<0.01 M) were prepared by dilution of 5 mM phosphate buffer pH 7.0 with deionized water, the pH of which had been adjusted to 7.2 by addition of very low concentrations of KOH. For each salt concentration, experiments were performed in triplicate to obtain a standard deviation in the measurement of surface coverage. Twenty-five µL of NW stock (pattern 011000) were rinsed by centrifugation as described above and resuspended in a solution of the desired ionic strength (to 83 µL). HIV probe DNA was then added such that its final concentration was equal to 1.0 µM. Each sample was allowed to react for 25 min. Samples were then rinsed by centrifugation (3× volume) and resuspended in the same ionic strength buffer used for attachment (25 µL final volume). Hybridization was performed in 0.5 M CAC buffer as detailed below.
For direct comparison to literature results,11 we also repeated these experiments varying ionic strength by [KH2PO4] alone, without pH adjustment.
Kinetics of Beacon Attachment
Thiolated HIV, HCV, SARS, or INF B MB probes were conjugated to NWs at either 1.0 or 0.25 µM, as noted in the text. For 1.0 µM probe attachment, 60 µL of NW stock solutions for NWs patterned 010010 were rinsed twice with 2×volumes of water and once in 0.3 M PBS before resuspension in 0.3 M PBS to 200 µL. For a DNA concentration of 0.25 µM, 120 µL of 010010 NWs and a final volume of 400 µL were used to facilitate fluorescence quantification, and the remaining procedure was the same for both DNA concentrations. After addition of thiolated MBs to a final concentration of either 1.0 µM or 0.25 µM, samples were rotated at room temperature. At specific time intervals, aliquots containing 5 × 106 NWs for [DNA] = 1 µM and 10 × 106 NWs for [DNA] = 0.25 µM were extracted from the reaction tube and rinsed twice by centrifugation in 3× volume 0.3 M PBS. NWs were then resuspended to their original volumes in 0.3 M PBS and stored at 4 °C.
Determination of Surface Coverage
A modification50 of a procedure first reported by Mirkin and co-workers78 was used to determine the surface coverage of fluorophore-labeled DNA on nanowires. Fluorescent, thiolated probes were removed from the NW surface by exchange with an excess of a β-mercaptoethanol after which NWs were removed and solution fluorescence used to determine probe concentration; surface density was then calculated based on initial NW concentration and dimensions. The detailed procedure was as follows. The solution of NWs bearing the beacons was brought up in volume to 195 µL by the same buffer in which the NWs were previously suspended. Then, 5 µL of β-mercaptoethanol were added to each tube. Samples were vortexed overnight. NWs were then pelleted by centrifugation for 2 min. The fluorescence of the supernatant containing the previously adsorbed DNA was then quantified using a Fluorolog-3 fluorimeter (λex = 558 nm, λem = 580 nm). Coverage was determined by fluorescence calibration of stock fluorescent DNA and based on a concentration of 1 × 109 wires/mL in stock solution.50
Five-Plex Assay
MB probe sequences DENV-2, SARS, INF B, HCV, and HIV (Table 1) were conjugated at a final reaction concentration of 1.0 µM to NWs patterned 000111, 001101, 010101, 100011, and 100101, respectively, through reaction in 0.3 M PBS for 25 min. The conjugates were rinsed twice by centrifugation and resuspension into 0.3 M PBS. Hybridization was performed as described above, with 1 µL of each type of MB probe-coated NWs added to each tube containing target DNA. Different permutations of target sequences (all targets, two targets, or no target) were introduced to the tubes for hybridization.
Hybridization of NW-Bound MB Probes
Aliquots of 3 µL probe-coated NWs, prepared by any of the methods described above, were removed for hybridization to either complementary target (Target sample) or no target (No Target sample). To both Target and No Target tubes were added 37 µL of 0.5 M CAC buffer, and to Target tubes was added 1 µL of target sequence, making these samples 5 µM in final concentration of target sequence. Hybridization was performed while rotating in the dark at room temperature for 1 h. These hybridization conditions apply to all experiments in this work.
Optical Microscopy
Directly after hybridization, and without rinsing,50 a 10 µL aliquot of NW suspension from any of the above experiments was placed onto a glass coverslip. Images were acquired using a Nikon TE-300 inverted microscope with a CFI plan fluor 60x oil immersion lens (N.A. = 1.4) and ImagePro Plus version 4.5 software. Images were captured using a Coolsnap HQ camera (Photometrics). The light source was a 300W ozone-free Xe lamp. A Nikon filter cube specific for TAMRA excitation and emission (excitation: 550/50, emission: 590 long-pass, dichroic: 565 long-pass) was employed to obtain fluorescence images, and reflectance images were obtained at a wavelength of 430 nm (Sutter Instruments filter wheel, Lambda 10-2), as this wavelength provided good contrast between Au and Ag.73,79 NBSee Software (Nanoplex Technologies) was employed to quantify fluorescence intensity.80 Briefly, the reflectance and corresponding fluorescence images were analyzed for several hundred NWs, and a log mean fluorescence intensity was obtained for each NW pattern.50
Results and Discussion
Sensing strategies based on surface-based molecular beacon and aptamer probes are growing in popularity and should offer advantages in selectivity and in the types of targets that can be detected; however, design of these sensors is inhibited by incomplete understanding of how nucleic acid secondary structure affects probe assembly and hybridization on the surface. Here, we used molecular beacon probes as a simple motif to study the effects of structure upon surface coverage and sensor performance, and barcoded metallic nanowires as our solid support.
Probe sequences (Table 1) were designed to adopt stable secondary structures based on inclusion of a 5 or 6 nucleobase “stem” (i.e., the five bases on the 3′and 5′ end of the sequence are complementary). Probe sequences also have a 3′ fluorescent dye and are attached to the nanowires via a 5′ thiol. We demonstrated previously that there is no measurable difference in probe coverage for Au as compared with Ag nanowire segments;15 the effect of these metals on emission intensity is both wavelength and distance-dependent.15,79 Here, we did not vary the nanowire striping (barcode) pattern within any experiment where data were directly compared (see Materials and Methods). Only in the multiplexing experiment are different wire patterns employed simultaneously.
Steric and electrostatic interactions at the surface are expected to play an important role in probe performance, analogous to what is seen for linear probes; surface-bound hairpin probes have additional potential interactions, including changes in the stability and dynamics of secondary structures and the possibility of intermolecular probe-probe binding interactions at the surface due to the complementary regions of adjacent probe molecules. Although MB probe coverage is generally considered important to the performance of surface-based MB sensors, few studies have directly measured coverage or reported performance as a function of coverage.
In contrast, the effect of coverage on surface-bound linear probes has been very well characterized.11–16,18,21,23–26 Several approaches have been used to control surface density of thiolated linear oligonucleotide probes on Au surfaces; these approaches can serve as guides for our studies of the MB-metal interface. They include: (1) surface dilution with another molecule, such as a shorter oligonucleotide or hydroxyalkanethiol, (2) controlling ionic strength during assembly to take advantage of electrostatic repulsions, and (3) including a spacer in the probe molecule itself to increase the separation between the surface and the portion of the probe that will be involved in hybridization reactions. Of these approaches, the latter is incompatible with our surface-quenched beacon probe design, and has shown limited success in alternative MB probe designs based on molecular quenchers. 55,57,62 We discuss our results for approaches (1) and (2) below. In these experiments, a single beacon probe sequence (HIV, see Table 1 for sequences) was used to facilitate comparison. We then go on to compare the HIV probe with other MB probe sequences both alone and combined in a multiplexed assay format.
Effect of Surface Dilution with Mercaptoethanol on MB–Nanowire Assay
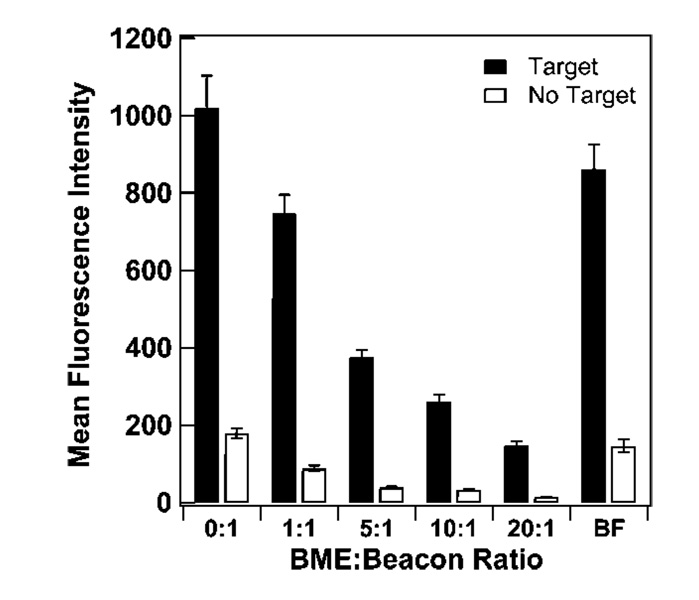
The addition of diluent short-chain hydroxy-terminated alkanethiols on arrays of linear DNA probes has been shown to increase hybridization efficiency by reducing probe coverage and removing nonspecific adsorption via the DNA bases, such that remaining oligonucleotides are attached at a single point (the terminal thiol).11,14,22–28,30–33,53,81 It is not obvious that surface dilution will have the same benefits for MB probes, which may have less propensity to bind the metal surface through their nucleobases as compared with similar length linear (i.e., random coil) probes. Additionally, nonspecific adsorption of target strands to the metal surface during hybridization should not impact MB probe performance to the same extent as for linear probes, since for MBs no signal will arise from binding unless it results in unfolding of the MB secondary structure. To test the effects of short alkanethiol spacers on a system with a probe containing secondary structure, we employed β-mercaptoethanol (BME) as both a coadsorbent and as a postassembly “backfilling” diluent, following literature protocols.11,48 Figure 1 shows fluorescence intensity in the presence and in the absence of target strands, as a function of BME to HIV beacon probe mole ratio during probe assembly onto the nanowires. By increasing the BME:beacon ratio, probe surface coverage was decreased more than 10-fold, from 6.4 × 1012 (0:1 ratio) to 4.8 × 1011 probes/cm2 (20:1 ratio) (Table 2), coverages that largely encapsulate the range of 1.2 × 1011−4 × 1012 probes/cm2 we have reported in previous works.15,50,67 Fluorescence signal decreased with decreasing probe coverage, consistent with less dye moieties; this was true both when complementary target was added and when it was not (Figure 1). In addition to fluorescence intensity, a common figure of merit for beacon performance is the quenching efficiency, defined as [1-(no target signal/target signal)] × 100%. For these data, intensity in the absence of target initially decreased more than that in the presence of target, yielding an increase in quenching efficiency from 82% when no BME coadsorbate was present to 88–90% at all BME:probe ratios tested (Table 2).
Figure 1.
Effect of β-mercaptoethanol (BME) spacers on the fluorescence hybridization signal of HIV probes bound to Au/Ag nanowires. Ratios are mole ratios of BME to beacon. BF sample represents beacon-coated nanowires that were backfilled with BME. Error bars represent the 95% confidence interval.
Table 2.
Quenching Efficiencies and Coverages of Different BME/Beacon Ratios
| BME:beacon ratio | surface coverage (probes/cm2) | Quenching efficiency (%) |
|---|---|---|
| 0:1 | 6.4 × 1012 | 82 ± 2 |
| 1:1 | 2.4 × 1012 | 88 ± 1 |
| 5:1 | 1.6 × 1012 | 90 ± 1 |
| 10:1 | 9.9 × 1011 | 88 ± 1 |
| 20:1 | 4.8 × 1011 | 90 ± 1 |
| BF (backfilled) | 2.0 × 1012 | 83 ± 2 |
Krauss and co-workers have reported coadsorption data for ratios of mercaptopropanol (MP) to beacon probes on a planar gold surface and found fluorescence in the presence of target initially increased with increasing MP:probe ratio, before decreasing at the highest MP:probe ratios. A 4-fold increase in fluorescence was obtained at 10:1 vs 0:1 MP:probe ratio.48 This differs from our finding that hybridization signal decreased steadily with an increasing BME:DNA ratio (Figure 1). Krauss and co-workers did not directly measure surface probe densities, but estimated based on hybridization ~1012 probes/cm2 for their 10:1 mercaptopropanol:probe ratio; no coverage data was given for other ratios. We hypothesize that the initial probe coverage at 0:1 may have been considerably higher in the work by Krauss and co-workers, such that improvements in hybridization efficiency upon dilution outweighed the concomitant reduction in total available fluorophores. Our coverage at 0:1 ratio was 6.4 ×1012 probes/cm2; any improvements in hybridization efficiency upon dilution from this initial coverage were less important than losses in the number of fluorophores available (Figure 1). Alternatively, the curvature of the nanowires used here may decrease the impact of steric hindrance somewhat, such that hybridization efficiencies for identical coverages may differ on wires vs planar surfaces. However, we do not expect this to be a large factor, due to the large size of the nanowires (~300 nm in cross-sectional diameter).
Exposure to hydroxyalkanethiol after initial probe assembly (backfilling) is also a commonly used technique for improving hybridization efficiency of linear probes on Au. Upon the exposure of BME to the 0:1 ratio wires for 30 min (BF sample), surface coverage was reduced by more than half due to thiol-thiol exchange resulting in desorption of beacon probes from the surface. Quenching efficiency was not significantly improved as compared to the 0:1 sample, despite a surface coverage more comparable to the 1:1 and 5:1 coadsorbed samples (Table 2). These data could indicate differences in probe distribution on the surface for coadsorbed vs backfilled routes to this probe surface density. If coadsorption vs backfilling resulted in different initial probe distributions, these could be expected to persist over the time scale of our experiments, given reports of slow surface diffusion of alkanethiols on Au.82,83
In general, surface dilution of MB probes with BME reduced fluorescence signal intensity but provided some improvement in quenching efficiency. For linear probes, performance is often reported as hybridization efficiency, but can also be evaluated in terms of the number of targets bound per unit area. For linear probes on Au, Levicky et al. demonstrated a target oligonucleotide coverage of 3 × 1012/cm2 (100% hybridization efficiency) after exposure of the probe-coated gold surface to mercaptohexanol, a 5-fold improvement compared to a target coverage of 6 × 1011/cm2 (10% hybridization efficiency) beforeMCHexposure.23 Gong et al. observed a near doubling in target coverage, from 4.7×1012 to 8.9×1012/cm2 after exposure to mercaptoundecanol for 1 h.14 While we were not able to directly monitor bound target density, it should be proportional to fluorescence intensity in presence of target. Figure 1 shows no improvement–rather, a decrease–in fluorescence intensity with surface dilution, suggesting that the effect of reduced binding sites (i.e., probe coverage) was greater than any increase in hybridization efficiency resulting from improved MB probe accessibility.
Our findings on surface dilution with BME indicate much less benefit for MB probes as compared with more traditional linear probes. This can be explained by several important differences between hairpin and linear oligonucleotide probes that may impact their response to hydroxyalkanethiol diluents. First, since they are partially double-stranded, hairpins may be less likely than linear probes to nonspecifically adsorb to the metal through the nitrogen atoms of the nucleobases. Diluent thiols are therefore less likely to exhibit an effect on MB orientation. Second, MBs bind target strands differently than do linear probes. MB probes interact with targets primarily through their loops,40 which, if a beacon is oriented normal to the gold surface, are more exposed to the solution-phase target strands. To bind linear probes, assuming full complementarity, targets must penetrate deeper into the probe layer, potentially encountering greater steric hindrance.11,23,84 Third, the signal in molecular beacon assays originates from the probe molecule itself. This is in contrast to signal originating from the target molecule during a binding event, as is generally the case with hybridization efficiency determinations for surface-bound linear probes.11,14,15,22,27,78,84,85 As such, we note that signal in molecular beacon assays will only be observed upon binding of the loop region to a complementary target; nonspecific binding of a target strand directly to a gold surface will not directly impact signal. Thus, any surface protection role played by the hydroxyalkanethiol in linear probe systems will be less noticeable for beacon probes.
Effect of Ionic Strength on Surface Coverage
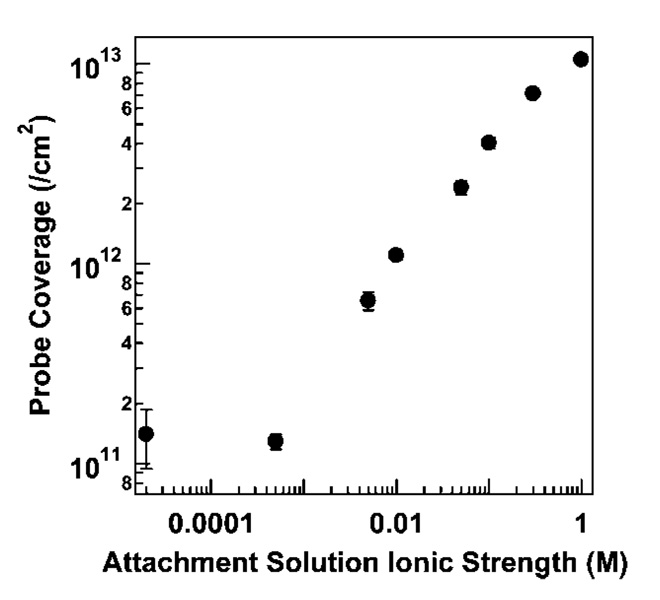
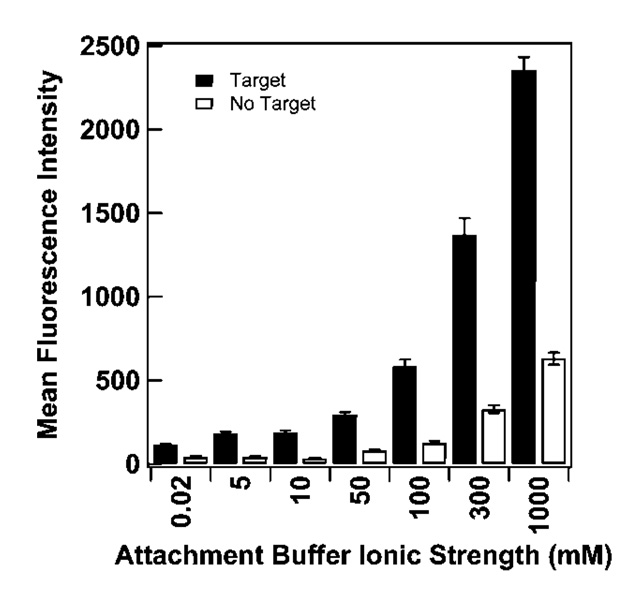
Surface probe density can also be controlled by the ionic strength of the attachment buffer, which determines the length scale of electrostatic repulsions between negatively charged DNA probes; lower coverages are achieved at lower salt concentrations. Because ionic strength also impacts probe secondary structure, which might be expected to influence probe adsorption behavior, we were interested in determining the effect of ionic strength during adsorption on MB probe coverage and performance. Figure 2 shows the dependence of HIV probe surface density on solution ionic strength, which varies over 2 orders of magnitude from 1011 to 1013 probes/cm2 for the range of ionic strengths evaluated.
Figure 2.
Effect of attachment buffer ionic strength on surface coverage for HIV probes attached to Au/Ag nanowires. Ionic strength was adjusted at pH 7.2 using KCl and phosphate buffer. Error bars represent the standard deviation of the surface coverage measurement.
Herne and Tarlov11 performed an analogous experiment using linear probes, in which they varied KH2PO4 concentration during assembly of thiolated 25-base oligonucleotides onto planar gold surfaces. They observed a strong dependence of coverage on ionic strength at low salt concentrations, with a maximum coverage reached by ~0.4 M KH2PO4.11 Coverage was already ~75% of the maximum by 0.1 M KH2PO4. In contrast, in our data, HIV MB probe coverage has not leveled off by 1.0 M ionic strength. At an ionic strength of 0.1 M, HIV coverage was <50% of that obtained at 1.0 M. The fact that beacon probes required a higher ionic strength for surface adsorption supports the hypothesis that electrostatic repulsions would impact MBs differently than linear probes. In low salt, beacon probes experience strong electrostatic repulsions, limiting surface densities and destabilizing the hairpin conformation. We note that varying [KH2PO4] alone results in some difference of pH over the concentration range used here (pH at 1.0 M = 4.0; pH of DI water = 5.3). DNA assembly is generally governed more by ionic strength than pH;12,86,87 nonetheless, to eliminate the possibility that differences between the results for linear probes in reference 16 and those for hairpin probes reported here arose from differences in pH, we repeated the experiment in Figure 2 varying [KH2PO4] alone. The results were comparable to that in Figure 2 (Supporting Information Figures 1–3).
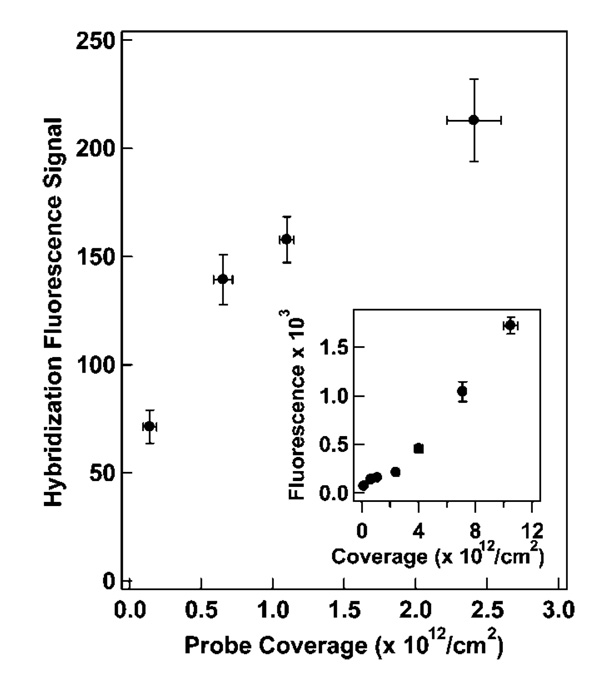
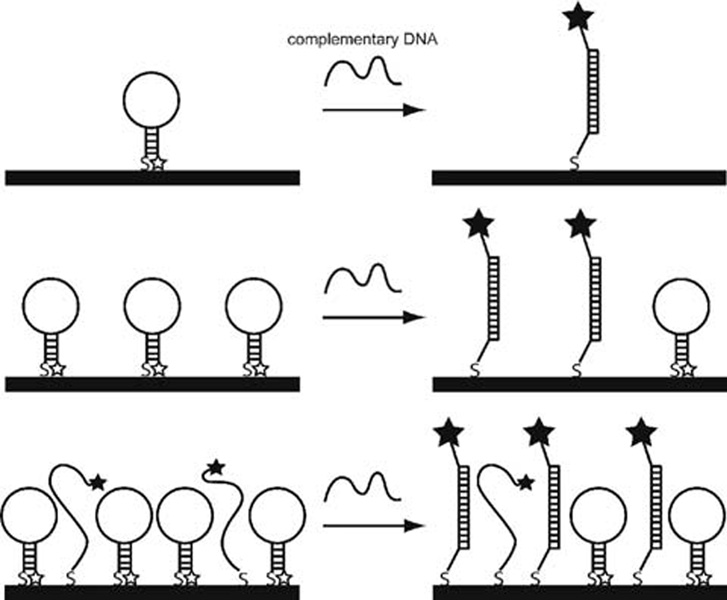
To test the effects of surface coverage on hybridization, we exposed the probe-coated NW samples prepared at different ionic strengths (HIV probe surface densities 1011 to 1013/cm2), to an oligonucleotide target strand complementary to the loop region. All hybridizations were performed in 0.5 M NaCl CAC buffer. Target binding led to increased fluorescence due to unquenching of the MB 3′ fluorophore. Figure 3 shows posthybridization fluorescence intensity minus no target signal as a function of probe coverage. Prehybridization background signal, which was not constant for the different probe densities, was subtracted in order to focus on signal obtained from hybridization events. At the lowest coverages, fluorescence intensity after hybridization increased with coverage. As probe density increased, however, past 6 × 1011 probes/cm2, hybridization fluorescence intensity began to level off before increasing with a higher slope above 4 × 1012 probes/cm2. This response was less straightforward than that observed for linear probes,19,78 and presumably reflects the role of beacon probe secondary structure. We postulate that the initial increase in fluorescence (from ~70 to ~140 counts) was due to the increase in probe density, with minimal interaction between individual beacon probes (Scheme 1, top). At these low coverages, hybridization efficiency would be high, with maximum fluorescence intensity limited by the number of probes on the NW surface. At intermediate coverages, despite the greater number of fluorophores present, steric and electrostatic repulsions would begin to interfere with target binding, resulting in lower hybridization efficiency than at lower coverages (Scheme 1, middle). We interpret the increased fluorescence seen in the highest coverage regime as resulting from greater probe accessibility due to steric inhibition of hairpin formation (Scheme 1, bottom), in addition to the still increasing number of fluorophores present. When a significant fraction of the probes fail to adopt the stem-loop conformation, more target binding events may occur due to increased accessibility of the bases that in the hairpin conformation make up the loop. It should be noted that at this coverage, any unfolded probes could be seen as linear probes, which have been shown to have a lower number of binding events at high probe densities.12 These MB probe sequences, however, contain a stem sequence of 5–6 bases at the 5′ and 3′ ends; the extra 5′ bases separate the hybridization region from the nanowire surface, improving steric accessibility as compared to traditional linear probes lacking a nonhybridizing spacer sequence. Addition of nonhybridizing nucleobases between the sequence of interest and the surface is known to increase hybridization efficiency.16,20
Figure 3.
Hybridization signal (background subtracted) as a function of surface coverage for HIV probes attached to nanowires in solutions of differing ionic strengths and hybridized to their complementary target. All hybridizations were performed at 0.5 M ionic strength. Inset: Expanded version of signal versus coverage, which encompasses the entire range of surface coverages examined. X-axis error bars represent the standard deviation in surface coverage measurement, and y-axis error bars represent the 95% confidence interval in hybridization fluorescence.
Scheme 1.
Illustration of Hybridization of Surface-Immobilized MB Probes at Low (Top), Intermediate (Middle), and High (Bottom) Coverages
Unfolded or improperly folded beacon probes (e.g., suboptimal intramolecular folds and/or intermolecular folds between adjacent probes) exhibit reduced quenching due to the increased separation between the fluorophore and the metal surface.15 Incomplete quenching at higher probe surface densities is apparent in Figure 4, which plots fluorescence intensity in the presence/absence of complementary target as a function of attachment buffer ionic strength (coverages for the data exhibited in Figure 4 can be read from Figure 2). Thus, higher fluorescence intensities in the presence of target for higher coverages result from a combination of a greater number of target binding events and reduced quenching.
Figure 4.
Effect of attachment buffer ionic strength on hybridization of complementary oligonucleotides to nanowire-bound HIV probes. Filled and open bars represent hybridization in the presence or absence of complementary target, respectively. Error bars represent the 95% confidence level.
Table 3 summarizes the quenching efficiency for the samples described in Figure 2–Figure 4. Quenching efficiency was lowest for the lowest probe density, 1.4 × 1011/cm2. The highest quenching efficiencies observed were for the 10 and 100 mM ionic strength adsorptions (82 and 78%, at probe densities of 1 and 4 × 1012/cm2), with intermediate, higher and lower densities giving slightly lower quenching efficiency. We attribute this behavior to the interplay between maximizing hybridization signal, which is best at higher probe densities, and minimizing signal in the absence of target, which is best at the lowest probe densities (Figure 3 and Figure 4). The low quenching efficiency at 50 mM adsorption (72%, 2.4 × 1012 probes/cm2) is anomalous, and may be a spurious data point (small changes in “no target” signal can disproportionally impact quenching efficiency) or may point to a difference in the probe densities required to optimize hybridization vs quenching. We note that this anomaly at ~2 × 1012 probes/cm2 was also observed in supporting Table 1. The effects of coverage on overall signal intensity and quenching have recently been evaluated by Plaxco and co-workers for a gold surface-immobilized molecular beacon incorporating a redox-active tag in place of the fluorophore.49 These authors found that electrochemical signal suppression (conceptually similar to fluorescence quenching efficiency here) was best (71%) at 2.1 × 1012 probes/cm2, the highest coverage tested, and dropped by about a factor of 2 below 1 × 1012 probes/cm2 (<40%). In our experiments, quenching efficiency was less sensitive to probe density, presumably due to differences in probe design or signal transduction mechanism (electrochemical vs optical).
Table 3.
Quenching Efficiencies for HIV Beacons Attached in Different Salt Concentrations
| Ionic strength (mM) | Surface coverage (/cm2) | Quenching efficiency (%) |
|---|---|---|
| 0.02 | 1.4 (±0.5) × 1011 | 63 ± 5 |
| 5 | 6.5 (±0.7) × 1011 | 76 ± 3 |
| 10 | 1.10 (±0.05) × 1012 | 82 ± 2 |
| 50 | 2.4 (±0.2) × 1012 | 72 ± 3 |
| 100 | 4.0 (±0.3) × 1012 | 78 ± 2 |
| 300 | 7.11 (±0.04) × 1012 | 76 ± 2 |
| 1000 | 1.05 (±0.05) × 1013 | 73 ± 2 |
Effect of Probe Solution Structure
Hairpin probes do not all have identical secondary structures. We previously investigated the effect of varying stem and loop length on surface-immobilized beacon probe performance for a given target sequence.50 Even for probes of more similar overall lengths, however, differences in secondary structures might be expected to impact surface adsorption and sensor performance; such differences could be relevant to multiplexed experiments in which several different probes (on different patterned NWs, or arrayed on a surface) would be used simultaneously. To test this, we compared four MB probe sequences specific for detection of partial regions of the viruses HIV, SARS, Influenza B, and HCV. We began by monitoring the attachment of these thiolated probes to NWs as a function of assembly time.
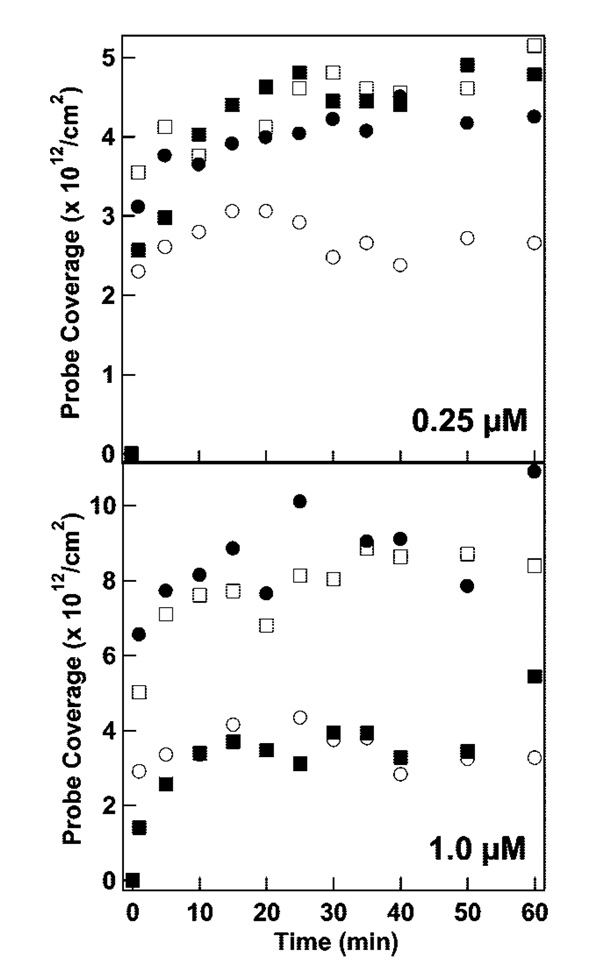
Figure 5 shows surface coverage for the four different thiolated MB probes as a function of adsorption time at two probe concentrations. In all cases, the assembly was complete by 25 min, which is consistent with rapid mixing due to sample agitation during the assembly process and electrostatic and steric repulsions by already-immobilized probes.11,24,86,88,89 Coverages ranged from 3 × 1012 to 9 × 1012 probes/cm2, corresponding to 11–33 nm2/probe. At the lower probe assembly concentration (0.25 µM), similar final coverages of 4–5 × 1012/cm2 were achieved for HIV, HCV, and SARS probes, with lower coverage ~3 × 1012/cm2 for INF B. When probe concentration during assembly was increased to 1 µM, surface coverage for the HIV and HCV probes increased approximately 2-fold to ~8 × 1012/cm2. Final densities of SARS and INF B probes were much lower, 3–4 × 1012/cm2.
Figure 5.
Beacon probe attachment to Au/Ag nanowires as a function of time in 0.3 M NaCl at initial probe concentrations of 0.25 µM (top) and 1.0 µM (bottom). Symbols represent different probes sequences: HIV (filled circles), SARS (filled squares), INF B (open circles), and HCV (open squares) probes.
HIV and HCV probes were able to pack more densely when larger probe concentrations were available. In contrast, INF B, which has the most stable secondary structure in solution (followed by SARS, Table 1), and SARS had final surface coverages that appear to be dictated to a greater extent by their secondary structure than by the solution probe concentration. Probe length,13,24 and to a lesser extent sequence,20,87 have been shown to greatly impact surface coverage on gold for linear probes; however, our beacon probes exhibit very different coverages even though they are approximately the same length (33–35 bases), and have similar stem sequences (i.e., the region closest to the metal). These results underscore the importance of probe sequence and conformation in the surface assembly of structured probes, and suggest a connection between solution phase thermodynamics and surface coverage.
Increased solution concentration can lead to probe homodimer formation, which could influence surface assembly, as homodimers may assemble at a different coverage than hairpins. To account for this possibility, we have modeled homodimer formation using the DINAMelt server (Supporting Information Figures 4–5).90 The results indicate a negligible population of dimers at room temperature. To supplement these findings, we have performed melting experiments of each beacon probe at concentrations of 3, 1, and 0.5 µM in 0.3 M PBS buffer (Supporting Information Figure 6); a substantial change in the population of hairpins would impact Tm. Fitting of the melt curves yielded Tm values that were not statistically significant from each other and gave no discernible trend (Supporting Information Table 2). Therefore, while some dimer structures may exist in solution at probe concentrations of 0.25 and 1 µM, we do not expect a significant contribution from dimers between these two concentrations with respect to surface coverage.
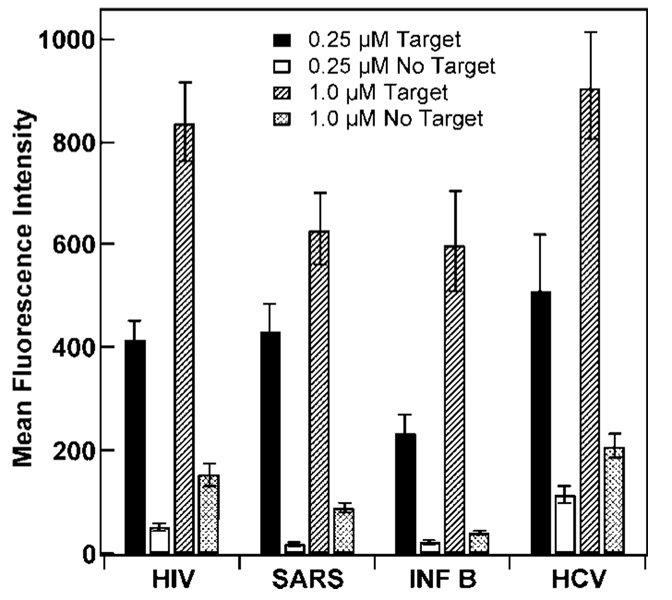
Hybridization data were obtained for probe-coated wires attached in the same fashion as in Figure 5 for both concentrations of DNA. Signals obtained upon hybridization with complementary target (Figure 6, black and hatched bars) for both DNA reaction concentrations were in very good agreement with coverages in Figure 5. For the 0.25 µM assembly condition, the fluorescence intensity in the presence of target was higher for HIV, HCV, and SARS, which had reached higher surface densities than INF B (Figure 5), however we note that the fluorescence intensities differed by nearly 2x, which is larger than the difference in probe coverage. For 1.0 µM probe assembly, similar results are seen, with greater fluorescence response upon hybridization for the HIV and HCV, which assembled at higher surface densities than SARS and INFB. Both absolute quenching (signal in absence of target) and quenching efficiency were better for SARS and INFB, consistent with their more stable solution-phase secondary structures (Table 4). For HIV and SARS probes, the 0.25 µM (lower coverage) samples gave improved quenching efficiencies over the 1.0 µM samples, while for the other two probes little change in quenching efficiency was observed (Table 4). Background signal decreased at lower HIV and SARS probe densities more than anticipated based on the reduction in number of fluorescent molecules, presumably due to reduced steric and electrostatic repulsions between adjacent probes hindering hairpin formation and therefore quenching.
Figure 6.
Effect of beacon secondary structure and DNA attachment concentration on hybridization of in the presence or absence of complementary oligonucleotides to nanowire-bound HIV, SARS, INF B, and HCV probes. Error bars represent the 95% confidence level.
Table 4.
Quenching Efficiencies for Beacons Attached at Concentrations of 1.0 or 0.25 µM
| beacon | 1.0 µM quenching efficiency (%) | 0.25 µM quenching efficiency (%) |
|---|---|---|
| HIV | 82 ± 3 | 88 ± 2 |
| SARS | 86 ± 2 | 96 ± 1 |
| INF B | 93 ± 2 | 91 ± 2 |
| HCV | 77 ± 4 | 78 ± 6 |
Probe Comparisons and Effect of Postassembly Salt Treatment
In addition to the factors discussed above, treatment of the system during the probe assembly process may play a role in probe arrangement and assay performance. We previously reported a two-step assembly process in which DNA is first incubated with nanowires overnight in 0.01 M PBS and then aged in 0.3 M PBS for two hours as a post-treatment step.15,50,67 For the experiments described above, kinetics and salt studies warranted attachment of probe molecules in one buffer and for a set amount of time. Table 5 displays the maximum quenching efficiencies obtained for several MB probes assembled either with 0.3 M PBS post-treatment (after initial reaction overnight in 0.01 M PBS) or with 0.3 M PBS for 25 min without previous treatment in 0.01 M PBS. These results show no strong correlation between probe surface coverage and quenching efficiency, and no uniformly positive effect of post-treatment (Table 5). Though the DENV-2 probe was insensitive to the two assembly techniques employed and packed with the same density in both cases, the HIV probe exhibited a higher quenching efficiency when assembled onto the surface of the nanowire in 0.3 M PBS and for 25 min, even though the surface coverages in these instances were similar. Our data suggest that behavior on the NW surface is probe-specific and that each probe may need to be optimized individually via assembly salt/DNA concentration or time. Because our ultimate goal is to mix beacon-coated nanowires for bioassays and employ the metal barcodes for multiplexed detection of pathogens, we realize it may not be possible to use fully optimized conditions for each MB simultaneously. Therefore, beacon performance should be optimized for the set of nanowire probes to be used together in a multiplexed experiment (through salt concentration, temperature, etc.).
Table 5.
Quenching Efficiencies and Coverages for Beacons Undergoing Post-Assembly Treatmenta or No Post-Assembly Treatment b
| post-treatment | no post-treatment | |||
|---|---|---|---|---|
| beacons | Quenching efficiency (%) | coverage (/cm2) | quenching Efficiency (%) | coverage (/cm2) |
| DENV-2 | 95 ± 1 | 6.3 × 1012 | 95 ± 1 | 6.9 × 1012 |
| HIV | 82 ± 2 | 6.4 × 1012 | 87 ± 2 | 5.8 × 1012 |
| INF B | 91 ± 1 | 4.8 × 1012 | 93 ± 2 | 5.8 × 1012 |
| HCV | − | − | 77 ± 6 | 1.2 × 1013 |
| SARS | − | − | 86 ± 2 | 4.2 × 1012 |
Postassembly treatment procedure entails nanowire-DNA reaction overnight in 0.01 M PBS with a 2 h aging step in 0.3 M PBS.
No post-treatment assembly entails reaction in only 0.3 M PBS for 25 min.
Five-Plex Assay
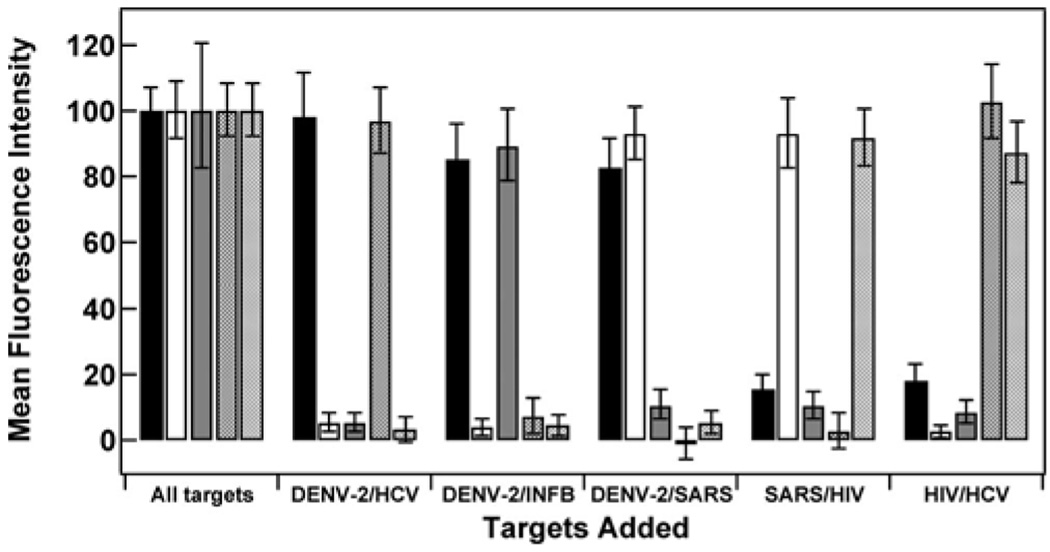
To evaluate the performance of the immobilized MB probes studied in this work in a simultaneous assay, we employed five different barcoded nanowire patterns, each conjugated to a different MB probe sequence. Probes were assembled similarly as for the data shown in Figure 5 (bottom). The MB-conjugated NWs were then mixed together and exposed to different combinations of targets. We used a high target concentration (5 µM) here for ease of comparison with the other experiments presented in this manuscript, in which high concentrations were used to drive hybridization in order to evaluate the behavior of surface-immobilized hairpins. Sensitivity measurements are reported elsewhere.50 The background-subtracted, normalized quantification of the resulting fluorescence signal for experiments where all five targets or different combinations of just two targets were added is presented in Figure 7. Additional target combinations, raw data, and sample microscope images are reported as Supporting Information (supporting Figures 7–9). Quenching efficiencies for each probe (Table 6) were calculated by taking ratios of fluorescence signal when no targets were present (supporting Figure 8) to signal when all targets were present.
Figure 7.
Hybridization fluorescence signal for a 5-plex MB assay on barcoded nanowires. The background fluorescence (no targets added) has been subtracted, and the resulting signal has been normalized to the signal obtained from the sample containing all five target sequences, which are identified as DENV-2 (black bars), SARS (open bars), INF B (gray bars), HCV (hatched bars), and HIV (cross-hatched bars).
Table 6.
Quenching Efficiencies of Beacons Employed in a Five-Plex Assay
| beacon | quenching efficiency (%) |
|---|---|
| DENV-2 | 90 ± 2 |
| SARS | 92 ± 2 |
| INF B | 91 ± 2 |
| HCV | 73 ± 4 |
| HIV | 85 ± 2 |
Quenching efficiencies for each probe (Table 6) were consistent with those reported above under nonmultiplex conditions (Table 4 and Table 5). Remaining fluorescence signal after background subtraction and in the absence of the correct target sequence (Figure 7) can be attributed to either nonspecific binding of targets to the incorrect probe or nanowire pattern misidentification by the analysis software.80 As anticipated from the discussion above, the five probes exhibited different behavior, presumably due to their different sequences and conformations on the nanowire surface. For example, the HCV probe was consistently brighter and exhibited a worse quenching efficiency; this can be understood as less stable formation of the desired hairpin secondary structure at the surface, which facilitates hybridization while limiting quenching. This is despite its predicted solution folding thermodynamics (Table 1), which is intermediate among the probes used here. Surface attachment clearly impacts the thermodynamics of the probe secondary structure. We postulate that the high surface density of probes leads to not only steric and electrostatic repulsions to forming secondary structure (which should be similar for HCV and HIV based on Figure 5 coverage data), but also facilitates interstrand hybridization, leading to misfolded probes for which the TAMRA dye is not close enough to the surface to be well quenched. SARS performed well in the multiplexed experiment, but in general gives more variable (and coverage-dependent) quenching efficiency than other probes. INF B, was darker in the presence of, but also quenched better in the absence, of complementary targets. This is consistent with the solution stability of the hairpin (Table 1). We note that mfold predictions of solution secondary structure stability differed from our experimental measurements of solution stability, perhaps due in part to the presence of the dye moieties in the actual sequences. These results underscore the importance of the surface-probe interface and the difficulties that remain in predicting the performance of any particular probe sequence. Nonetheless, despite the complication of probe-specific responses, MB probes immobilized on metal nanowires were able to readily discriminate between complementary and noncomplementary target nucleic acid sequences in the five-plex experiment shown here and show promise for future multiplexed biosensing applications.
Conclusions
In this work, we have investigated the factors that influence molecular beacon surface coverage on metal nanowires and the impact of coverage on beacon performance in hybridization assays. Short hydroxy-terminated alkanethiol spacers were found to have only minimal positive effect on probe performance. While quenching efficiency was increased by coadsorbing at mercaptoethanol: probe ratios between 1:1 and 20:1, fluorescence intensities in the presence of target strands were substantially decreased. Backfilling with the thiol, a technique commonly employed for linear DNA on gold surface to increase hybridization efficiency, did not significantly improve quenching efficiency. We found that the ionic strength of the buffer during probe assembly greatly impacted both probe density and beacon performance. At low coverages, binding events could not be detected, whereas at high coverages, the beacons were unable to quench efficiently. As is the case with linear DNA probes bound on a metal surface, there is a density between the two extremes that produces optimal performance. However, unlike linear probes, the behavior of beacons is more complicated due to their secondary structure, and this optimal density may vary as a function of probe sequence and secondary structure shape and stability. Assembly of thiolated MB probes onto the nanowires was also dictated by probe reaction concentration and sequence. Probe sequences with higher solution hairpin stabilities assembled at lower surface coverages.
These findings were applied to a multiplexed bioassay, in which MB probes immobilized on metal nanowires exhibited excellent selectivity for their complementary nucleic acid targets. Because of their hairpin structure, molecular beacons introduce complexity into the more traditional system of single-stranded DNA monolayers immobilized onto metal surfaces. Thus, while general statements can be made concerning the performance of beacon probes (lower coverage should improve quenching efficiency, increased thermodynamic stability for the folded structure improves quenching, etc.), our incomplete understanding of the role of surface-probe and probe-probe interactions limits our ability to rationally design probes for optimal performance in surface-based assays. The results reported here, in particular comparisons of different methods of achieving desired probe coverages, and of the effect of changing hairpin probe sequence, help to shed light on the MB–metal surface interaction and its role in biodetection.
Acknowledgment
This work was funded by the National Institutes of Health (R01 EB000268) and Pennsylvania State University. We thank Dr. Philip Bevilacqua and Nate Siegfried for their help with DNA hairpin melting and Tm determinations. C.D.K. also acknowledges support from a Beckman Foundation Young Investigator Award, a Sloan Fellowship, and a Dreyfus Teacher-Scholar Award.
Footnotes
Supporting Information Available; Evaluation of HIV probe coverage and hybridization behavior as a function of NaH2PO4 concentration during the assembly process, hairpin melting curves and Tm determinations, 5-plex additional normalized/background subtracted data, raw data, and microscopy pictures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Murphy L. Curr. Opin. Chem. Biol. 2006;10:177–184. doi: 10.1016/j.cbpa.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 2.(a) Moskovits M. Rev. Mod. Phys. 1985;57:783–826. [Google Scholar]; (b) Raether H. Surface Plasmons on Smooth and Rough Surfaces and on Gratings. Berlin: Springer-Verlag; 1988. [Google Scholar]; (c) Meitiu H. Prog. Surf. Sci. 1984;17:153–320. [Google Scholar]
- 3.(a) Lakowicz JR. Anal. Biochem. 2001;298:1–24. doi: 10.1006/abio.2001.5377. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lakowicz JR. Anal. Biochem. 2005;337:171–194. doi: 10.1016/j.ab.2004.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Haynes CL, McFarland AD, Van Duyne RP. Anal. Chem. 2005;77:338A–346A. [Google Scholar]; (b) Zhang X, Shah NC, Van Duyne RP. Vibr. Spectrosc. 2006;42:2–8. [Google Scholar]
- 5.Neumann T, Johansson M-L, Kambhampati D, Knoll W. Adv. Funct. Mater. 2002;12:575–586. [Google Scholar]
- 6.Rosi NL, Mirkin CA. Chem. Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 7.Bally M, Halter M, Voros J, Grandin HM. Surf. Interfac. Anal. 2006;38:1442–1458. [Google Scholar]
- 8.(a) Vo-Dinh T, Yan F, Wabuyele MB. Topics Appl. Physics. 2006;103:409–427. [Google Scholar]; (b) Ataka K, Heberle J. Anal. Bioanal. Chem. 2007;388:47–54. doi: 10.1007/s00216-006-1071-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Aslan K, Gryczynski I, Malicka J, Matveeva E, Lakowicz JR, Geddes CD. Curr. Opin. Biotechnol. 2005;15:55–62. doi: 10.1016/j.copbio.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J. Small. 2005;1:1036–1043. doi: 10.1002/smll.200500214. [DOI] [PubMed] [Google Scholar]
- 10.(a) Wegner GJ, Lee HJ, Corn RM. In: Surface Plasmon Resonance Imaging Measurements of DNA, RNA, and Protein Interactions to Biomolecular Arrays. In Protein Microarray Technology. Kambhampati D, editor. Weinheim: Wiley-VCH; 2004. pp. 107–129. [Google Scholar]; (b) Hutter E, Fendler JH. Adv. Mater. 2004;16:1685–1706. [Google Scholar]
- 11.Herne TM, Tarlov MJ. J. Am. Chem. Soc. 1997;119:8916–8920. [Google Scholar]
- 12.Peterson AW, Heaton RJ, Georgiadis RM. Nucleic Acids Res. 2001;29:5163–5168. doi: 10.1093/nar/29.24.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castelino K, Kannan B, Majumdar A. Langmuir. 2005;21:1956–1961. doi: 10.1021/la047943k. [DOI] [PubMed] [Google Scholar]
- 14.Gong P, Lee C-Y, Gamble LJ, Castner DG, Grainger DW. Anal. Chem. 2006;78:3326–3334. doi: 10.1021/ac052137j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoermer RL, Keating CD. J. Am. Chem. Soc. 2006;128:13243–13254. doi: 10.1021/ja0637200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Opdahl A, Petrovych DY, Kimura-Suda H, Tarlov MJ, Whitman LJ. Proc. Natl. Acad. Sci. U.S.A. 2007;104:9–14. doi: 10.1073/pnas.0608568103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halperin A, Buhot A, Zhulina EB. Biophys. J. 2004;86:718–730. doi: 10.1016/S0006-3495(04)74150-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shchepinov MS, Case-Green SC, Southern EM. Nucleic Acids Res. 1997;25:1155–1161. doi: 10.1093/nar/25.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicewarner-Peña SR, Raina S, Goodrich GP, Fedoroff NV, Keating CD. J. Am. Chem. Soc. 2002;124:7314–7323. doi: 10.1021/ja0177915. [DOI] [PubMed] [Google Scholar]
- 20.Hurst SJ, Lytton-Jean AKR, Mirkin CA. Anal. Chem. 2006;78:8313–8318. doi: 10.1021/ac0613582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee C-Y, Nguyen P-CT, Grainger DW, Gamble LJ, Castner DG. Anal. Chem. 2007;79:4390–4400. doi: 10.1021/ac0703395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peterlinz KA, Georgiadis RM. J. Am. Chem. Soc. 1997;119:3401–3402. [Google Scholar]
- 23.Levicky R, Herne TM, Tarlov MJ, Satija SK. J. Am. Chem. Soc. 1998;120:9787–9792. [Google Scholar]
- 24.Steel AB, Levicky RL, Herne TM, Tarlov MJ. Biophys. J. 2000;79:975–981. doi: 10.1016/S0006-3495(00)76351-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aqua T, Naaman R, Daube SS. Langmuir. 2003;19:10573–10580. [Google Scholar]
- 26.Lee C-Y, Gong P, Harbers GM, Grainger DW, Castner DG, Gamble LJ. Anal. Chem. 2006;78:3316–3325. doi: 10.1021/ac052137j. [DOI] [PubMed] [Google Scholar]
- 27.Gao Y, Wolf LK, Georgiadis RM. Nucleic Acids Res. 2006;34:3370–3377. doi: 10.1093/nar/gkl422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moses S, Brewer SH, Lowe LB, Lappi SE, Gilvey LBG, Sauthier M, Tenent RC, Feldheim DL, Franzen S. Langmuir. 2004;20:11134–11140. doi: 10.1021/la0492815. [DOI] [PubMed] [Google Scholar]
- 29.Mbindyo JKN, Reiss BD, Martin BR, Keating CD, Natan MJ, Mallouk TE. Adv. Mater. 2001;13:249–254. [Google Scholar]
- 30.Arinaga K, Rant U, Tornow M, Fujita S, Abstreiter G, Yokoyama N. Langmuir. 2006;22:5560–5562. doi: 10.1021/la060324m. [DOI] [PubMed] [Google Scholar]
- 31.Georgiadis R, Peterlinz KP, Peterson AW. J. Am. Chem. Soc. 2000;122:3166–3173. [Google Scholar]
- 32.Rant U, Arinaga K, Fujita S, Yokoyama N, Abstreiter G, Tornow M. Langmuir. 2004;20:10086–10092. doi: 10.1021/la0492963. [DOI] [PubMed] [Google Scholar]
- 33.Park S, Brown KA, Hamad-Schifferli K. Nano Lett. 2004;4:1925–1929. [Google Scholar]
- 34.Tyagi S, Kramer FR. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 35.Broude NE. TRENDS Biotechnol. 2002;20:249–256. doi: 10.1016/s0167-7799(02)01942-x. [DOI] [PubMed] [Google Scholar]
- 36.Fang X, Li JJ, Perlette J, Tan W, Wang K. Anal. Chem. 2000:747A–753A. doi: 10.1021/ac003001i. [DOI] [PubMed] [Google Scholar]
- 37.Tan W, Wang K, Drake TJ. Curr. Opin. Chem. Biol. 2004;8:547–553. doi: 10.1016/j.cbpa.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 38.Marras SAE, Kramer FR, Tyagi S. Nucleic Acids Res. 2002;30:e122. doi: 10.1093/nar/gnf121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonnet G, Tyagi S, Libchaber A, Kramer FR. Proc. Natl. Acad. Sci. U.S.A. 1999;96:6171–6176. doi: 10.1073/pnas.96.11.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsourkas A, Behlke MA, Rose SD, Bao G. Nucleic Acids Res. 2003;31:1319–1330. doi: 10.1093/nar/gkg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marras SAE, Kramer FR, Tyagi S. Genet. Anal. Biomol. Eng. 1999;14:151–156. doi: 10.1016/s1050-3862(98)00018-7. [DOI] [PubMed] [Google Scholar]
- 42.Vet JAM, Majithia AR, Marras SAE, Tyagi S, Dube S, Poiesz BJ, Kramer FR. Proc. Natl. Acad. Sci. U.S.A. 1999;96:6394–6399. doi: 10.1073/pnas.96.11.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Famulok M, Hartig JS, Mayer G. Chem. Rev. 2007;107:3715–3743. doi: 10.1021/cr0306743. [DOI] [PubMed] [Google Scholar]
- 44.Baker BR, Lai RY, Wood MS, Doctor EH, Heeger AJ, Plaxco KW. J. Am. Chem. Soc. 2006;128:3138–3139. doi: 10.1021/ja056957p. [DOI] [PubMed] [Google Scholar]
- 45.Bang GS, Cho S, Kim B-G. Biosens. Bioelectron. 2005;21:863–870. doi: 10.1016/j.bios.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 46.Hamaguchi N, Ellington A, Stanton M. Anal. Biochem. 2001;294:126–131. doi: 10.1006/abio.2001.5169. [DOI] [PubMed] [Google Scholar]
- 47.Radi A-E, Sanchez JLA, Baldrich E, O’Sullivan CK. J. Am. Chem. Soc. 2006;128:117–124. doi: 10.1021/ja053121d. [DOI] [PubMed] [Google Scholar]
- 48.Du H, Strohsahl CM, Camera J, Miller BL, Krauss TD. J. Am. Chem. Soc. 2005;127:7932–7940. doi: 10.1021/ja042482a. [DOI] [PubMed] [Google Scholar]
- 49.Ricci F, Lai RY, Heeger AJ, Plaxco KW, Sumner JJ. Langmuir. 2007;23:6827–6834. doi: 10.1021/la700328r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stoermer RL, Cederquist KB, McFarland SK, Sha MY, Penn SG, Keating CD. J. Am. Chem. Soc. 2006;128:16892–16903. doi: 10.1021/ja0658261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Riccelli PV, Merante F, Leung KT, Bortolin S, Zastawny RL, Janeczko R, Benight AS. Nucleic Acids Res. 2001;29:996–1004. doi: 10.1093/nar/29.4.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Balamurugan S, Obubuafo A, Soper SA, McCarley RL, Spivak DA. Langmuir. 2006;22:6446–6453. doi: 10.1021/la060222w. [DOI] [PubMed] [Google Scholar]
- 53.Steichen M, Buess-Herman C. Electrochem. Commun. 2005;7:416–420. [Google Scholar]
- 54.Cho EJ, Collett JR, Szafranska AE, Ellington AD. Anal. Chim. Acta. 2006;564:82–90. doi: 10.1016/j.aca.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 55.Brown LJ, Cummins J, Brown T. Chem. Commun. 2000:621–622. [Google Scholar]
- 56.Liu X, Tan W. Anal. Chem. 1999;71:5054–5059. doi: 10.1021/ac990561c. [DOI] [PubMed] [Google Scholar]
- 57.Yao G, Tan W. Anal. Biochem. 2004;331:216–223. doi: 10.1016/j.ab.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 58.Yao G, Fang X, Yokota H, Yanagida T, Tan W. Chem. Eur. J. 2003;9:5686–5692. doi: 10.1002/chem.200304977. [DOI] [PubMed] [Google Scholar]
- 59.Liu X, Farmerie W, Schuster S, Tan W. Anal. Biochem. 2000;283:56–63. doi: 10.1006/abio.2000.4656. [DOI] [PubMed] [Google Scholar]
- 60.Situma C, Moehring AJ, Noor MAF, Soper SA. Anal. Biochem. 2007;363:35–45. doi: 10.1016/j.ab.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Steemers FJ, Ferguson JA, Walt DR. Nat. Biotechnol. 2000;18:91–94. doi: 10.1038/72006. [DOI] [PubMed] [Google Scholar]
- 62.Wang H, Li J, Liu H, Liu Q, Mei Q, Wang Y, Zhu J, He N, Lu Z. Nucleic Acids Res. 2002;30:e61. doi: 10.1093/nar/gnf061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fan C, Plaxco KW, Heeger AJ. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9134–9137. doi: 10.1073/pnas.1633515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dubertret B, Calame M, Libchaber AJ. Nat. Biotechnol. 2001;19:365–370. doi: 10.1038/86762. [DOI] [PubMed] [Google Scholar]
- 65.Du H, Disney MD, Miller BL, Krauss TD. J. Am. Chem. Soc. 2003;125:4012–4013. doi: 10.1021/ja0290781. [DOI] [PubMed] [Google Scholar]
- 66.Lubin AA, Lai RY, Baker BR, Heeger AJ, Plaxco KW. Anal. Chem. 2006;78:5671–5677. doi: 10.1021/ac0601819. [DOI] [PubMed] [Google Scholar]
- 67.Sha MY, Yamanaka M, Walton ID, Norton SM, Stoermer RL, Keating CD, Natan MJ, Penn SG. Nanobiotechnol. 2005;1:327–335. doi: 10.1385/NBT:1:4:327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maxwell DJ, Taylor JR, Nie S. J. Am. Chem. Soc. 2002;124:9606–9612. doi: 10.1021/ja025814p. [DOI] [PubMed] [Google Scholar]
- 69.Cheng AKH, Ge B, Yu H-Z. Anal. Chem. 2007;79:5158–5164. doi: 10.1021/ac062214q. [DOI] [PubMed] [Google Scholar]
- 70.Brunker SE, Cederquist KB, Keating CD. Nanomedicine. 2007;2:695–710. doi: 10.2217/17435889.2.5.695. [DOI] [PubMed] [Google Scholar]
- 71.Al-Mawlawi D, Liu CZ, Moskovits J. J. Mater. Res. 1994;9:1014–1018. [Google Scholar]
- 72.Martin CR. Science. 1994;266:1961–1966. doi: 10.1126/science.266.5193.1961. [DOI] [PubMed] [Google Scholar]
- 73.Nicewarner-Pena SR, Freeman RG, Reiss BD, He L, Pena DJ, Walton ID, Cromer R, Keating CD, Natan MJ. Science. 2001;294:137–141. doi: 10.1126/science.294.5540.137. [DOI] [PubMed] [Google Scholar]
- 74.Reiss BD, Freeman RG, Walton ID, Norton SM, Smith PC, Stonas WG, Keating CD, Natan MJ. J. Electroanal. Chem. 2002;522:95–103. [Google Scholar]
- 75.Keating CD, Natan MJ. Adv. Mater. 2003;15:451–454. [Google Scholar]
- 76.Zuker M. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Templeton KE, Scheltinga SA, Beersma MFC, Kroes ACM, Claas ECJ. J. Clin. Microbiol. 2004;42:1564–1569. doi: 10.1128/JCM.42.4.1564-1569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Demers LH, Mirkin CA, Mucic RC, Reynolds RA, III, Letsinger RL, Elghanian R, Viswanadham G. Anal. Chem. 2000;72:5535–5541. doi: 10.1021/ac0006627. [DOI] [PubMed] [Google Scholar]
- 79.Nicewarner-Pena SR, Carado AJ, Shale KE, Keating CD. J. Phys. Chem. B. 2003;107:7360–7367. [Google Scholar]
- 80.Walton ID, Norton SM, Balasingham A, He L, Oviso DF, Jr, Gupta D, Raju PA, Natan MJ, Freeman RG. Anal. Chem. 2002;74:2240–2247. doi: 10.1021/ac020073w. [DOI] [PubMed] [Google Scholar]
- 81.Wackerbarth H, Grubb M, Zhang J, Hansen AG, Ulstrup J. Langmuir. 2004;20:1647–1655. doi: 10.1021/la035547g. [DOI] [PubMed] [Google Scholar]
- 82.Ionita P, Volkov A, Jeschke G, Chechik V. Anal. Chem. 2008;80:95–106. doi: 10.1021/ac071266s. [DOI] [PubMed] [Google Scholar]
- 83.Imabayashi S, Hobara D, Kakiuchi T. Langmuir. 2001;17:2560–2563. [Google Scholar]
- 84.Peterson AW, Heaton RJ, Georgiadis R. J. Am. Chem. Soc. 2000;122:7837–7838. [Google Scholar]
- 85.Casero E, Darder M, Diaz DJ, Pariente F, Martin-Gago JA, Abruna H, Lorenzo E. Langmuir. 2003;19:6230–6235. [Google Scholar]
- 86.Petrovych DY, Kimura-Suda H, Whitman LJ, Tarlov MJ. J. Am. Chem. Soc. 2003;125:5219–5226. doi: 10.1021/ja029450c. [DOI] [PubMed] [Google Scholar]
- 87.Wolf LK, Gao Y, Georgiadis RM. Langmuir. 2004;20:3357–3361. doi: 10.1021/la036125+. [DOI] [PubMed] [Google Scholar]
- 88.Huang E, Satjapipat M, Han S, Zhou F. Langmuir. 2001;17:1215–1224. [Google Scholar]
- 89.Mourougou-Candoni N, Naud C, Thibaudau F. Langmuir. 2003;19:682–686. [Google Scholar]
- 90.Markham NR, Zuker M. Nucleic Acids Res. 2005;33:W577–W581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]