Summary
The eukaryotic nucleus is a highly compartmentalized and dynamic environment. Chromosome territories are arranged non-randomly within the nucleus and numerous studies have indicated that a gene’s position in the nucleus can impact its transcriptional activity. Here, we focus on recent advances in our understanding of the influence of specific nuclear neighborhoods on gene expression or repression. Nuclear neighborhoods associated with transcriptional repression include the inner nuclear membrane/nuclear lamina and peri-nucleolar chromatin, whereas neighborhoods surrounding the nuclear pore complex, PML nuclear bodies, and nuclear speckles seem to be transcriptionally permissive. While nuclear position appears to play an important role in gene expression, it is likely to be only one piece of a flexible puzzle that incorporates numerous parameters. We are still at a very early, yet exciting stage in our journey toward deciphering the mechanism(s) that govern the permissiveness of gene expression/repression within different nuclear neighborhoods.
INTRODUCTION
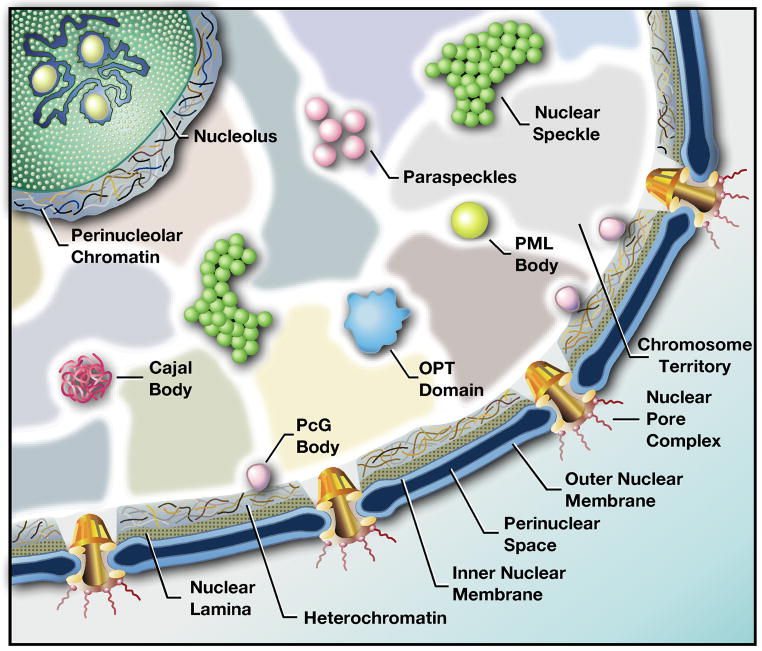
The interphase nucleus of higher eukaryotes is a well organized, compartmentalized, and dynamic organelle [for review, see 1,2]. The entire genome is packaged within the confines of the nucleus, such that genes are able to dynamically interact with the nuclear neighborhood that surrounds them and regulatory proteins can access genes via a diffusion-based mechanism. Individual chromosomes occupy distinct and limited regions, called chromosome territories (Figure 1), which are arranged non-randomly in the nucleus [for review, see 3]. In addition, various specialized nuclear compartments exist (Figure1), including nuclear pore complexes, the nuclear lamina, and the inner nuclear membrane, as well as more than ten nuclear bodies, including nucleoli, nuclear speckles, Cajal bodies, and PML bodies [for review, see 4]. Evidence has accumulated supporting the notion that gene positioning may have a functional impact on gene regulation [for reviews, see 1,2,5]. This stems from the observation that chromosome positioning is non-random, and that each nuclear compartment, or neighborhood, is composed of dynamically exchanging protein constituents with distinct roles in gene expression. If a gene resides in or moves to a particular nuclear neighborhood, could other residents of that neighborhood (chromatin, ribonucleoprotein (RNP) complex, or protein) modulate its expression?
Figure 1. Nuclear Neighborhoods.
A cartoon of the mammalian cell nucleus showing some of the numerous nuclear domains, or neighborhoods, that have been identified. Several nuclear neighborhoods that have been implicated in gene activation or repression are discussed in the text.
In order to address the impact of nuclear position on gene expression, recent studies have examined whether a gene’s activity may correlate with its position relative to the center of the nucleus (radial position) [6,7,8*], relative to other genes/chromosomes [7,9*,10,11,12**,13–15], and/or relative to various nuclear neighborhoods [16*,17**,18*,19*,20,21**,22*,23**,24*,25*,26*]. While it is clear that a gene’s expression can be affected by its nuclear location, the specific mechanisms by which a particular nuclear neighborhood affects gene expression remain unclear. In this review, we focus on specific nuclear neighborhoods, and recent studies that implicate these nuclear compartments in transcriptional regulation. In the process, we will discuss the direct or indirect effects that specialized nuclear compartments could have on the transcription of genes in close proximity. We will not focus on the effects of radial positioning, inter-/intra-chromosomal interactions, and DNA looping on gene expression; nor will we discuss the potential factors that determine a gene’s nuclear location, such as local gene density or chromatin dynamics, as these subjects are reviewed in other articles in this issue and elsewhere [1,2,5,27,28].
INNER NUCLEAR MEMBRANE AND NUCLEAR LAMINA
Early electron microscopy studies revealed a close relationship between perinuclear heterochromatin, the inner nuclear membrane (INM) and the nuclear lamina (NL) [for review, see 29]. The NL, a mesh-like structure directly beneath the inner nuclear membrane (INM), is a unique characteristic of metazoan cells [for review, see 30]. The major constituents of the NL are Lamin A/C and Lamin B, which are type V intermediate filament (IF) proteins [for review, see 30]. Integral membrane proteins that reside in the INM include lamin B receptor (LBR), Lap2-emerin-MAN1 (LEM) domain containing proteins and Sad1-UNC homology domain (SUN) proteins, which are collectively referred to as lamin-associated polypeptides (LAPs) [for review, see 31]. There is growing evidence that both INM and NL proteins can affect gene regulation through direct chromatin binding [for review, see 30,32] or via interactions with transcriptional regulators [for review, see 31].
Two recent genome-wide mapping studies, investigating in vivo genome-binding sites of chromatin associated proteins, were carried out using the DNA adenine methyltransferase identification (DamID) technique. Results of these studies in Drosophila melanogaster cells [33*] and human lung fibroblasts [34*] support the view of the NL as a transcriptionally repressive compartment. In Drosophila, Lamin B bound regions of the genome are characterized by low levels of transcription, absence of active histone marks, mid to late replication timing, and low gene density [33*]. In human lung fibroblasts, the nearly identical DamID profiles of Lamin B1 and the INM protein emerin suggest that gene expression levels are also generally low within lamina-associated domains (LADs) [34*]. In addition, lamin B1 and emerin were shown to colocalize with a large transcriptionally inactive region of the IgH locus in mouse fibroblasts, however these interactions are lost in pro-B cells when the locus is transcriptionally active [23**]. These results show a developmental specificity of nuclear position that correlates with gene expression.
In yeast, an integral LEM containing INM protein, Src1/Heh1, binds directly to specific DNA sequences or nucleosomes in telomeric and subtelomeric chromatin resulting in clustering of these genomic regions at the nuclear periphery [18*]. Subsequently, silencing factors and transcriptional activators enriched at the nuclear periphery cooperate to regulate gene expression of the associated subtelomeric and telomeric chromatin [18*]. INM proteins are also involved in tethering rDNA repeats to the nuclear periphery in S. cerevisiae [35] and centromeres to cytoplasmic microtubles in S. pombe [36]. These results suggest that the interactions between INM proteins and chromosomal proteins may be a conserved mechanism by which organisms can ensure genome stability and maintain normal gene expression/repression [35]. However, it is not yet clear whether such transcriptional repression is the cause or effect of a gene’s position at the INM/NL.
INM/NL proteins may create a transcriptionally repressive nuclear compartment via multiple mechanisms. In HeLa cells, emerin and Lap2β have been reported to interact with histone deacetylase 3 (HDAC3), and over-expression of Lap2β induces a global deacetylation of histone H4 [37]. Also, the lamin B receptor, another INM protein, can bind DNA, histone H3-H4 tetramers, mitotic chromosomes, and heterochromatin protein 1 (HP1) in vitro [33*]. It has been suggested that such interactions may result in an increase in the local concentration of HP1, histone deacetylases, and transcriptional repressors, as well as a decrease in the availability of transcription factors at the INM, thus contributing to the repressive nature of this nuclear neighborhood [for review, see 31]. These results are consistent with recent studies, which demonstrate that tethering of a genomic region to the INM/NL by Lap2β [17**] or a truncated form of emerin [23**] can result in some level of transcriptional repression of the associated genes.
Although the nuclear periphery is generally considered a transcriptionally repressive environment, micro-domains of the INM/NL that are transcriptionally permissive may exist. For example, full-length lamin B1 has been implicated in both transcriptional activation [38] and repression [34*]. In addition, repositioning an inducible transgene array to the nuclear lamina with a LacI-LaminB1 fusion protein was not sufficient to inhibit inducible gene activation [21**]. Indeed, the activation kinetics of this locus were unaffected by its nuclear location. It is possible that a large transgene array of this kind, driven by a strong transcriptional activator, could create a transcriptional microenvironment that counteracts the otherwise repressive effects of the nuclear periphery neighborhood [21**]. Alternatively, this array may have incorporated into an existing microenvironment that is permissive for transcriptional activation. Thus, lamina associated gene expression may vary depending on the gene evaluated, the strength of its activator, and its localized microenvironment.
NUCLEAR PORE COMPLEX
The nuclear membrane is interrupted in places by nuclear pore complexes (NPC) that allow for the exchange of materials between the nucleus and cytoplasm. Each NPC is composed of approximately 30 different proteins, each in multiple copies [for review, see 39]. Although the NPC’s function in nucleo-cytoplasmic exchange is well established [for review, see 39], recent studies also suggest that the NPC functions as a transcriptionally permissive nuclear neighborhood to facilitate gene expression [24*,40*,41, for a review of earlier studies, see 42].
Protein components of the NPC may affect transcription in several ways. For example, genes may interact with NPCs and contribute to gene activation directly via specific promoter sequences, such as the nucleopore-promoter interaction (Nup-PI) in yeast [41]. Protein components of the NPC may also mediate efficient transcription, messenger RNA processing, and export through interactions with the 3′ untranslated region (UTR) of nascent transcripts [24*]. This is illustrated by the subtelomeric gene, hexokinase isoenzyme 1 (HXK1), which requires NPC association at it’s 3′ UTR for efficient transcription [24*]. Alternatively, the NPC may interact with 3′UTR bound proteins such as THO/sub2, to mediate persistent association of transcribed DNA with the nuclear pore complex, allowing for efficient transcription and mRNP biogenesis [40*]. Chromatin bound to the NPC may also acquire transcriptionally permissive histone modifications due to the direct association of NPC components with the SAGA histone acetyltransferase complex [43] or CREB-binding protein (CBP) [44]. Consistent with this hypothesis, a global increase of acetylation induces extensive genomic reorganization, including recruitment of euchromatin-rich promoter regions to the NPC [45**]. In yeast, DNA boundary element binding proteins, such as CTCF, may also function in NPC-mediated gene activation by blocking the spread of telomeric heterochromatin to surrounding active NPC-associated genes [for review, see 42]. Since NPC anchorage can be re-established after mitosis, NPC components may also contribute to transcriptional regulation in a heritable manner through recognition and binding of the histone variant H2A.Z to promote reactivation of genes after cell division [46*].
All of these interactions, mediated by DNA, RNA and/or proteins, suggest that the NPC can act as a mediator of active gene expression at the nuclear periphery. The mechanisms by which this is achieved remain unclear. A better understanding of the dynamics and the mechanisms governing gene positioning and gene expression at the NPC will provide insight into factors that influence transcription on a genome-wide scale.
PERI-NUCLEOLAR CHROMATIN
The structure of nucleoli and the integrity of tandem rDNA repeats within it are maintained in part by the shell of perinucleolar heterochromatin that surrounds the nucleolus [for review, see 47]. Perinucleolar heterochromatin contains satellite DNA that surrounds nucleolar organizer regions (NORs), and silent rDNA clusters [for review, see 47]. Recent reports implicate peri-nucleolar heterochromatin in the establishment and maintenance of silencing of non-rDNA-related genomic regions.
For example, early studies indicated that the inactive X chromosome (Xi) in females is sometimes localized to the nucleolar region in certain cell types [48]. Interestingly, in a recent report utilizing synchronized female mouse fibroblasts, an Xist ncRNA dependent S-phase association of Xi with the peri-nucleolar region was observed in 80–90% of nuclei examined [26*]. A separate study found Kcnqlot1 ncRNA to be necessary for localizing an imprinted region of paternal chromosome 7 to the peri-nucleolar neighborhood, and subsequent bidirectional repression of eight genes spread over a megabase region, including H19/Igf2 [22*]. Fifty-five percent of the cells that exhibited peri-nucleolar localization were in mid-late S-phase. Based on this evidence, it is tempting to speculate that transcriptional repression of these chromosomal regions could be maintained during DNA replication by virtue of their proximity to the peri-nucleolar region, and its associated chromatin and DNA modifying enzymes. However, the precise role that ncRNAs such as Xist and Kcnqlot1 play in peri-nucleolar localization and gene repression remains unclear. In addition, the mechanisms that drive this repression and whether other chromosome territories are stably or transiently associated with the peri-nucleolar neighborhood remains to be determined.
PML BODIES
Promyelocytic leukemia nuclear bodies (PML-NBs) have been implicated in a wide variety of cellular processes, including tumor suppression, viral defense, DNA repair, and/or transcriptional regulation [for review, see 49]. In several cases, PML-NBs have been shown to localize to particularly gene-rich and transcriptionally active regions of chromatin, including the major histocompatability complex (MHC) class I gene cluster region and the p53 gene locus [for review, see 49]. Such findings have led to speculation that PML-NBs may be able to modulate transcription at specific genomic loci.
New insight into the mechanism by which PML-NB association may affect MHC gene transcription comes from the discovery of protein:protein interaction between the PML protein and matrix attachment region (MAR)-binding protein, special AT-rich sequence binding protein 1 (SATB1) [20]. This study shows that SATB1, which functions as both a chromatin organizer and transcription factor, is able to bind to multiple DNA regions throughout the MHC class I gene cluster [20]. Together with various isoforms of PML, SATB1 mediates the formation of a complex and dynamic DNA loop structure at this locus. Thus, PML and SATB1 may orchestrate changes transcription through dynamic alterations to chromatin-loop architecture [20]. It is still unclear, however, whether the formation of PML-NBs at the MHC class I loci precedes transcriptional activation, or if PML-NBs form de novo at gene loci that are already active. Alternatively, it is possible that the formation of PML-NBs at these loci is not required for gene activation. Rather, soluble isoforms of PML protein diffusing away from PML-NBs may be able to mediate DNA looping at the MHC class I locus. Future experiments must address the formation of PML-NBs at specific gene loci with high spatial and temporal resolution, in order to answer these questions.
NUCLEAR SPECKLES
Nuclear speckles, also known as interchromatin granule clusters (IGCs), are enriched in splicing related factors, such as small nuclear ribonucleoprotein complexes (snRNPs), and SR proteins [for review, see 50]. Most mammalian nuclei contain between 30–50 nuclear speckles, which are not transcriptionally active regions [for review, see 50]. However, several recent studies have shown that transcriptionally active genes preferentially associate with nuclear speckles [16*,19*,25*]. For example, the active allele of the mono-allelically expressed glial fibrillary acidic protein (GFAP) gene was shown to associate with speckles in 70% of cells, while the inactive allele did not [25*]. In another study, co-expressed erythroid genes were also shown to cluster around nuclear speckles [16*]. However, the basis of non-random active gene positioning at nuclear speckles remains unclear. This study suggests that nuclear speckles form de novo after cell division by the recruitment of splicing factors to active genes, and subsequent association of genes at a common nuclear speckle is facilitated by chromatin dynamics [16*]. Therefore, the clustering of active genes around nuclear speckles may depend on gene position during the G1 phase of the cell cycle, when nuclear speckles begin to re-form after mitosis. Thus, in one view, the association of certain genes with nuclear speckles is a probabilistic event that follows cell division and transcriptional activation.
An alternative possibility is that the integration of RNA polymerase II and splicing factors at speckle-associated loci facilitates transcription and processing of active genes [for review, see 51]. In a recent report, estrogen receptor alpha (ERalpha) bound genes were shown to be dynamically repositioned adjacent to nuclear speckles upon ligand activation [19*]. The authors propose that this actin/myosin dependent, rapid, hormone responsive nuclear rearrangement allows the enhanced, coordinated transcription of nuclear receptor target genes. One could envisage a model where certain genes that are rapidly activated in response to a cellular signal could achieve efficient co-transcriptional RNA processing by being in proximity to the nuclear speckles. Further research, including live-cell imaging of chromatin dynamics, will be required to determine the biological significance of nuclear speckle gene associations, and whether such associations occur in a regulated or probabilistic fashion.
CONCLUSION
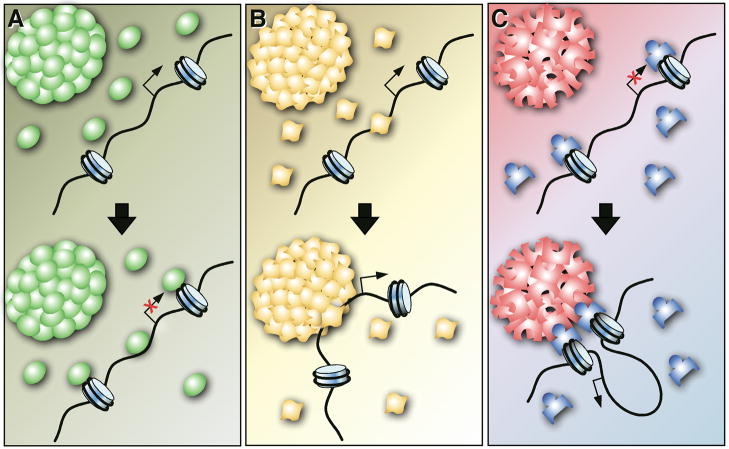
Much progress has recently been made to improve our understanding of the relationship between genome organization and specific nuclear neighborhoods. A gene in close proximity to a particular nuclear neighborhood may interact with one or more diffusible constituents of the neighborhood (e.g. chromatin remodeling, histone modification or DNA methylation factors etc.), thereby directly or indirectly influencing its transcription (Figure 2A). Alternatively, the association of a particular gene locus with a nuclear neighborhood may be a consequence of gene activation/repression, rather than the cause (Figure 2B). In this instance, a freely diffusible protein constituent may bind and activate/repress genes (Figure 2B) and subsequently become associated with a particular nuclear compartment through either dynamic localization of the bound gene to an existing nuclear compartment, or through the nucleation and de novo formation of a nuclear compartment at the bound gene. In addition, proteins associated with a nuclear neighborhood may indirectly affect transcription by stabilizing inter-/intra-chromosomal interactions, or shielding associated genes from the effect of nearby heterochromatin (e.g. SATB1/PML and CTCF/NPC associations) (Figure 2C). Regardless, a gene’s activity is likely not influenced simply by its nuclear neighborhood, but rather by dynamic processes that involve a multitude of factors, that may include cellular signaling events, chromosomal interactions, microenvironment, and/or stochastic gene activation. To fully understand the relationship between nuclear position and gene expression, one must consider the effect of multiple parameters on the position and activity of a single gene.
Figure 2. The Impact of Nuclear Neighborhoods on Gene Expression.
(A) Protein factors (green) that are enriched in a particular nuclear neighborhood could diffuse to nearby gene loci, thereby negatively (or positively) affecting the expression of genes. (B) The association of a particular gene locus with a nuclear neighborhood may be a consequence of gene activation (or repression), rather than its cause. For example, a gene could be bound and activated by a transcription factor (yellow). Subsequently, a nuclear body may form de novo at the site where the transcription factor is already bound. Alternatively, DNA bound by a protein constituent of a nuclear body (yellow) may dynamically relocate to an already existing nuclear body. (C) Nuclear neighborhood associated proteins (red), may indirectly affect transcription in combination with other protein factors (blue) by stabilizing inter-/intra-chromosomal interactions, or by shielding associated genes from the effect of nearby heterochromatin.
Whether the nuclear position of a gene is part of the cause or consequence of gene activation remains an open question. In the future, it will be important to test the transcriptional consequence of manipulating nuclear gene position in developmental contexts [for review, see 52]. In addition, global studies of genome dynamics upon induced gene activation combined with dynamic imaging of nuclear domains will provide significant insight into the regulation of gene activity. Multi-dimensional live-cell imaging and genome-wide application of the chromosome conformation capture assay (4C) [12**], will be powerful tools to help decipher the transcriptional causes and effects of genome positioning events. Furthermore, additional studies focused on de novo formation of nuclear compartments [53] and tethered gene repositioning experiments [17**,21**,23**] will help to further address the impact of nuclear organization on gene regulation.
Acknowledgments
We thank members of the Spector lab for helpful discussions and R. Ileng Kumaran for comments on the manuscript. Research in the Spector lab is supported by NIH/GM42694, NIH/GM71407 and NIH/EY18244. M.S.B. is supported by a Starr Centennial Pre-Doctoral Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- 2.Kumaran RI, Thakar R, Spector DL. Chromatin dynamics and gene positioning. Cell. 2008;132:929–934. doi: 10.1016/j.cell.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cremer T, Cremer M, Dietzel S, Müller S, Solovei I, Fakan S. Chromosome territories--a functional nuclear landscape. Curr Opin Cell Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Spector DL. SnapShot: Cellular bodies. Cell. 2006;127:1071. doi: 10.1016/j.cell.2006.11.026. [DOI] [PubMed] [Google Scholar]
- 5.Takizawa T, Meaburn KJ, Misteli T. The meaning of gene positioning. Cell. 2008;135:9–13. doi: 10.1016/j.cell.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Küpper K, Kölbl A, Biener D, Dittrich S, von Hase J, Thormeyer T, Fiegler H, Carter NP, Speicher MR, Cremer T, et al. Radial chromatin positioning is shaped by local gene density, not by gene expression. Chromosoma. 2007;116:285–306. doi: 10.1007/s00412-007-0098-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lanctôt C, Kaspar C, Cremer T. Positioning of the mouse Hox gene clusters in the nuclei of developing embryos and differentiating embryoid bodies. Exp Cell Res. 2007;313:1449–1459. doi: 10.1016/j.yexcr.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 8•.Meaburn KJ, Misteli T. Locus-specific and activity-independent gene repositioning during early tumorigenesis . J Cell Biol. 2008;180:39–50. doi: 10.1083/jcb.200708204. A number of non-random gene repositioning events were identified during differentiation in a 3-D mammary epithelial cell culture model, and during tumorigenesis, the alterations of spatial positioning patterns were found to be unrelated to gene activity. This study presents an example of activity-independent genome repositioning events that occur in the early stages of tumor formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9•.Augui S, Filion GJ, Huart S, Nora E, Guggiari M, Maresca M, Stewart AF, Heard E. Sensing X chromosome pairs before X inactivation via a novel X-pairing region of the Xic. Science. 2007;318:1632–1636. doi: 10.1126/science.1149420. This study utilizes 3-D RNA and DNA FISH to identify a novel X-pairing region (Xpr) of the X inactivation center (Xic) that drives Xic trans-associations. Importantly, the authors propose that the Xpr may coordinate reciprocal Xist/Tsix expression during X chromosome inactivation. [DOI] [PubMed] [Google Scholar]
- 10.Bacher CP, Guggiari M, Brors B, Augui S, Clerc P, Avner P, Eils R, Heard E. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol. 2006;8:293–299. doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- 11.Kosak ST, Scalzo D, Alworth SV, Li F, Palmer S, Enver T, Lee JS, Groudine M. Coordinate gene regulation during hematopoiesis is related to genomic organization. PLoS Biology. 2007;5:e309. doi: 10.1371/journal.pbio.0050309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12••.Noordermeer D, Branco MR, Splinter E, Klous P, van Ijcken W, Swagemakers S, Koutsourakis M, van der Spek P, Pombo A, De Laat W. Transcription and chromatin organization of a housekeeping gene cluster containing an integrated beta-globin locus control region. PLoS Genet. 2008;4:e1000016. doi: 10.1371/journal.pgen.1000016. The human beta-globin locus control region (LCR) was targeted in two opposite orientations to a gene-dense region in the mouse genome containing mostly housekeeping genes. Analysis of the dynamics of the integrated LCR revealed that although it was able to influence the expression of surrounding genes, transcriptional enhancement was not due to repositioning of the locus outside of its chromosome territory (CT), as previously reported. Rather, the authors conclude that LCR makes gene contacts via chromatin looping, leading to enhanced transcription of LCR-associated genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osborne CS, Chakalova L, Mitchell JA, Horton A, Wood AL, Bolland DJ, Corcoran AE, Fraser P. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 2007;5:e192. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu M, Cook PR. The role of specialized transcription factories in chromosome pairing. Biochim Biophys Acta. 2008;1783:2155–2160. doi: 10.1016/j.bbamcr.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 15.Xu N, Tsai CL, Lee JT. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 2006;311:1149–1152. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- 16•.Brown JM, Green J, das Neves RP, Wallace HA, Smith AJ, Hughes J, Gray N, Taylor S, Wood WG, Higgs DR, et al. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J Cell Biol. 2008;182:1083–1097. doi: 10.1083/jcb.200803174. Using human erythropoiesis as a model, the authors report that five co-transcribed genes spatially associate with each other in the nucleus at significant but variable frequencies. Interestingly, the co-transcribed genes were found to cluster around nuclear speckles, contradicting earlier studies that implicate transcription foci in the clustering of co-transcribed genes. This study suggests that nuclear speckles form de novo after cell division by the recruitment of splicing factors to active genes, and subsequent association of genes at a common nuclear speckle is facilitated by chromatin dynamics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17••.Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. Relocation of specific human chromosomes to the nuclear periphery by tethering them to a inner nuclear membrane protein Lap2-beta can reversibly suppress the expression of some endogenous human genes, meanwhile the expression of many other genes is not detectably reduced. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18•.Grund SE, Fischer T, Cabal GG, Antúnez O, Pérez-Ortín JE, Hurt E. The inner nuclear membrane protein Src1 associates with subtelomeric genes and alters their regulated gene expression. J Cell Biol. 2008;182:897–910. doi: 10.1083/jcb.200803098. This study reveals that the integral inner nuclear membrane protein Src1 is associated with subtelomeric chromatin. This may either help cluster genes for cooperative gene regulation at the nuclear periphery or facilitate the TREX-dependent messenger RNA export though the nuclear pore complexes. Src1 provides a good example for understanding the complex role of inner nuclear membrane proteins in gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19•.Hu Q, Kwon YS, Nunez E, Cardamone MD, Hutt KR, Ohgi KA, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG, et al. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci USA. 2008 doi: 10.1073/pnas.0810634105. The authors report rapid, dynamic, hormone-induced and actin-dependent reorganization of estrogen receptor alpha (ERα) responsive genes close to nuclear speckles. Re-positioning of genes to nuclear speckles was associated with enhanced transcription. Evidence is presented that suggests nuclear speckles could act as “hubs” of inter-chromosomal interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar PP, Bischof O, Purbey PK, Notani D, Urlaub H, Dejean A, Galande S. Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat Cell Biol. 2007;9:45–56. doi: 10.1038/ncb1516. [DOI] [PubMed] [Google Scholar]
- 21••.Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol. 2008;180:51–65. doi: 10.1083/jcb.200706060. Repositioning an inducible gene locus to the nuclear lamina by LacI-LaminB1 was not sufficient to inhibit inducible gene activation. Interestingly, repositioning required passage through mitosis. Components of the gene expression machinery are recruited to the targeted locus with kinetics similar to the non-targeted locus. This 200-copy transgene array driven by a strong activator may create a microenviroment to counteract the repressive effects of the nuclear periphery or it may have incorporated into an existing microenvironment that is permissive to transcriptional activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22•.Mohammad F, Pandey RR, Nagano T, Chakalova L, Mondal T, Fraser P, Kanduri C. Kcnq1ot1/Lit1 noncoding RNA mediates transcriptional silencing by targeting to the perinucleolar region. Mol Cell Biol. 2008;28:3713–3728. doi: 10.1128/MCB.02263-07. Using an episomal vector system, this study characterizes a silencing domain (SD) at the 5′ end of the Kcnq1ot1 anti-sense non-coding RNA (ncRNA), an ncRNA that has been implicated in transcriptional silencing of linked genes. The SD region of the Kcnqlot1 transcript was found to be necessary for the bidirectional spread of H3K9 trimethylation to neighboring chromosomal regions. Intriguingly, the authors also find that the SD domain is also required to target the episomal vector to the perinucleolar chromatin compartment. Collectively, these data suggest a model where an antisense ncRNA mediates transcriptional gene silencing by targeting genes to a heterochromatic nuclear compartment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23••.Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. Repositioning of a specific genetic locus to the inner nuclear membrane (INM) using an LacI-emerin fusion protein resulted in transcriptional repression of many genes on the associated chromosome. Interestingly, repositioning of the gene locus required passage through mitosis. [DOI] [PubMed] [Google Scholar]
- 24•.Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. doi: 10.1038/nature04845. The authors report that a subtelomeric gene, HXK1 (hexokinase isoenzyme 1), associated with nuclear pores through its 3′ untranslated region (UTR) in an transcription dependent manner. This finding suggests that nuclear position is actively involved in optimizing gene expression levels. [DOI] [PubMed] [Google Scholar]
- 25•.Takizawa T, Gudla PR, Guo L, Lockett S, Misteli T. Allele-specific nuclear positioning of the monoallelically expressed astrocyte marker GFAP. Genes Dev. 2008;22:489–498. doi: 10.1101/gad.1634608. This study examines the nuclear positioning of the mono-allelically expressed astrocyte-specific glial fibrillary acidic protein (GFAP) gene. The authors note that GFAP alleles are differentially positioned within the nucleus, and that an allele’s radial position is dependent on gene activity. Interestingly, this report also provides evidence that the active allele of GFAP frequently associates with nuclear speckles. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26•.Zhang LF, Huynh KD, Lee JT. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell. 2007;129:693–706. doi: 10.1016/j.cell.2007.03.036. This study addresses the question of how mammalian female cells maintain silencing of the inactive X chromosome (Xi) over successive cell division cycles. The authors present evidence that the Xi associates with perinucleolar heterochromatin, while the active X chromosome does not. Curiously, the Xi-perinucleolar associations occurred largely during mid-to-late S-phase. Thus, these data suggest a role for the perinucleolar heterochromatic region in the maintenance of Xi silencing during S-phase. [DOI] [PubMed] [Google Scholar]
- 27.Lanctôt C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 28.Chuang CH, Belmont AS. Moving chromatin within the interphase nucleus-controlled transitions? Semin Cell Dev Biol. 2007;18:698–706. doi: 10.1016/j.semcdb.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fawcett DW. The Cell. 2. W.B. Saunders Company; 1981. Nucleus; pp. 195–302. [Google Scholar]
- 30.Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, Goldman RD. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008;22:832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heessen S, Fornerod M. The inner nuclear envelope as a transcription factor resting place. EMBO Reports. 2007;8:914–919. doi: 10.1038/sj.embor.7401075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mattout A, Goldberg M, Tzur Y, Margalit A, Gruenbaum Y. Specific and conserved sequences in D. melanogaster and C. elegans lamins and histone H2A mediate the attachment of lamins to chromosomes. J Cell Sci. 2007;120:77–85. doi: 10.1242/jcs.03325. [DOI] [PubMed] [Google Scholar]
- 33.Pickersgill H, Kalverda B, De Wit E, Talhout W, Fornerod M, van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nature Genetics. 2006;38:1005–1014. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- 34•.Pickersgill H, Kalverda B, De Wit E, Talhout W, Fornerod M, van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nature Genetics. 2006;38:1005–1014. doi: 10.1038/ng1852. Using the DamID method, the authors identified ~500 Drosophila melanogaster genes that interact with B-type lamin (Lam), all of which exhibited repressed gene expression. This Lam-association can be interrupted by enhanced acetylation. This study shows that the nuclear lamina can function in both chromatin positioning and gene regulation. [DOI] [PubMed] [Google Scholar]
- 35•.Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, De Laat W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. In this study, the authors generate a high-resolution DamID map of Nuclear Lamina (NL) interactions in human lung fibroblasts. The map shows more than 1,300 sharply defined large, discrete lamina-associated domains (LADs) characterized by low gene expression levels. This result suggests the NL not only serves as a transcriptionally inactive region but also plays an important role in genome organization. [DOI] [PubMed] [Google Scholar]
- 36.King MC, Drivas TG, Blobel G. A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell. 2008;134:427–438. doi: 10.1016/j.cell.2008.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Somech R, Shaklai S, Geller O, Amariglio N, Simon AJ, Rechavi G, Gal-Yam EN. The nuclear-envelope protein and transcriptional repressor LAP2beta interacts with HDAC3 at the nuclear periphery, and induces histone H4 deacetylation. J Cell Sci. 2005;118:4017–4025. doi: 10.1242/jcs.02521. [DOI] [PubMed] [Google Scholar]
- 38.Tang CW, Maya-Mendoza A, Martin C, Zeng K, Chen S, Feret D, Wilson SA, Jackson DA. The integrity of a lamin-B1-dependent nucleoskeleton is a fundamental determinant of RNA synthesis in human cells. J Cell Sci. 2008;121:1014–1024. doi: 10.1242/jcs.020982. [DOI] [PubMed] [Google Scholar]
- 39.Tran EJ, Wente SR. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125:1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 40.Rougemaille M, Dieppois G, Kisseleva-Romanova E, Gudipati RK, Lemoine S, Blugeon C, Boulay J, Jensen TH, Stutz F, Devaux F, et al. THO/Sub2p functions to coordinate 3′-end processing with gene-nuclear pore association. Cell. 2008;135:308–321. doi: 10.1016/j.cell.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 41.Schmid M, Arib G, Laemmli C, Nishikawa J, Durussel T, Laemmli UK. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Molecular Cell. 2006;21:379–391. doi: 10.1016/j.molcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 42•.Rougemaille M, Dieppois G, Kisseleva-Romanova E, Gudipati RK, Lemoine S, Blugeon C, Boulay J, Jensen TH, Stutz F, Devaux F, et al. THO/Sub2p functions to coordinate 3′-end processing with gene-nuclear pore association. Cell. 2008;135:308–321. doi: 10.1016/j.cell.2008.08.005. A mutation in THO/sub2p was shown to result in the accumulation of a stalled intermediate in mRNP synthesis, which contained nuclear pore components and polyadenylation factors associated with chromatin. These findings suggest that the THO/sub2p complex functions after the commitment to 3′ end processing, and plays a role in coordinating mRNP export. [DOI] [PubMed] [Google Scholar]
- 43.Luthra R, Kerr SC, Harreman MT, Apponi LH, Fasken MB, Ramineni S, Chaurasia S, Valentini SR, Corbett AH. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. Journal of Biological Chemistry. 2007;282:3042–3049. doi: 10.1074/jbc.M608741200. [DOI] [PubMed] [Google Scholar]
- 44.Ryan CM, Harries JC, Kindle KB, Collins HM, Heery DM. Functional interaction of CREB binding protein (CBP) with nuclear transport proteins and modulation by HDAC inhibitors. Cell Cycle. 2006;5:2146–2152. doi: 10.4161/cc.5.18.3207. [DOI] [PubMed] [Google Scholar]
- 45.Brown CR, Kennedy CJ, Delmar VA, Forbes DJ, Silver PA. Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev. 2008;22:627–639. doi: 10.1101/gad.1632708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH. H2A. Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McStay B, Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu Rev Cell Dev Biol. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- 48.Bourgeois CA, Laquerriere F, Hemon D, Hubert J, Bouteille M. New data on the in-situ position of the inactive X chromosome in the interphase nucleus of human fibroblasts. Human Genetics. 1985;69:122–129. doi: 10.1007/BF00293281. [DOI] [PubMed] [Google Scholar]
- 49.Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8(12):1006–16 . doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]; Nature Reviews Molecular Cell Biology. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 50.Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 51.Pandit S, Wang D, Fu XD. Functional integration of transcriptional and RNA processing machineries. Current Opinion in Cell Biology. 2008;20:260–265. doi: 10.1016/j.ceb.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dillon N. The impact of gene location in the nucleus on transcriptional regulation. Dev Cell. 2008;15:182–186. doi: 10.1016/j.devcel.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 53.Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science. 2008;322:1713–1717. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]