Summary
A stream of genetic and biochemical information available for the biosynthesis of aminocyclitols over the past few years has provided the foundation to study the modes of formation of this clinically important class of natural products. In addition to work on the identification and functional analysis of aminocyclitol biosynthetic gene clusters, a contingent of recent studies has focused on the detailed analysis of unique enzymatic and catalytic mechanisms inherent to these pathways. The results provide invaluable insights into the biochemical and molecular aspects of aminocyclitol biosynthesis and have revealed diverse and unique features of the pathways.
Introduction
Aminocyclitols are a group of microbially-derived bioactive natural products with important clinical and agricultural applications [1,2]. Members of this class of natural products include the aminoglycoside antibiotics (e.g., streptomycin, hygromycin, butirosin, neomycin), the C7N-aminocyclitols (e.g., acarbose, validamycin, cetoniacytone, salbostatin) and the five-membered ring aminocyclitols (cyclopentitols) (e.g., pactamycin, trehazolin, allosamidin). Aminoglycoside antibiotics were among the first antibiotics used in the clinic and they remain important drugs for the treatment of infections, primarily those caused by aerobic, Gram-negative bacteria, such as Pseudomonas and Enterobacter. In addition, they are occasionally used to treat serious infections caused by some Gram-positive bacteria, e.g., Mycobacterium tuberculosis and methicillin-resistant Staphylococcus aureus [3,4]. The antibacterial activity of this class of compounds is due to their strong affinity for the rRNA of the bacterial ribosome, leading to the disruption of protein synthesis. On the other hand, the C7N-aminocyclitols and the cyclopentitol-derived natural products are structurally more diverse and display a variety of biological activities. Among the valuable members of the C7N-aminocyclitol family are the antidiabetic agent acarbose and the crop protectant validamycin A.
Biosynthetically, aminocyclitols are derived from simple sugar units, formed in most cases by a family of enzymes known as sugar phosphate cyclases (SPCs) [5-7]. These include the 1L-myo-inositol 1-phosphate (MIP) synthases and the 2-deoxy-scyllo-inosose (DOI) synthases involved in the biosynthesis of aminoglycoside antibiotics, as well as the 2-epi-5-epi-valiolone synthases that are involved in the biosynthesis of the C7N-aminocyclitol natural products. The only exceptions are the five-membered ring aminocyclitols, which may be derived from aminosugars, e.g., N-acetylglucosamine, via yet to be determined mechanisms [8-10]. The present review highlights recent reports related to the biosynthesis of aminocyclitols, particularly those derived from sugar phosphate cyclases.
The diversity of sugar phosphate cyclases
Sugar Phosphate Cyclases (SPCs) are a family of enzymes that catalyze the cyclization of sugar phosphates to produce a variety of cyclitol intermediates that serve as the building blocks of many primary metabolites and clinically relevant secondary metabolites (Figure 1) [6]. The two most well-known members of this family of enzymes are the mechanistically distinct MIP synthases and dehydroquinate (DHQ) synthases. The MIP synthases catalyze the conversion of D-glucose 6-phosphate to 1L-myo-inositol 1-phosphate, the first committed step in the production of all inositol-containing compounds including some aminoglycoside antibiotics, and the DHQ synthases catalyze the conversion of the C7-sugar phosphate, 3-deoxy-D-arabino-heptulosonate 7-phosphate (DAHP), to 3-dehydroquinic acid in the shikimate pathway [11,12].
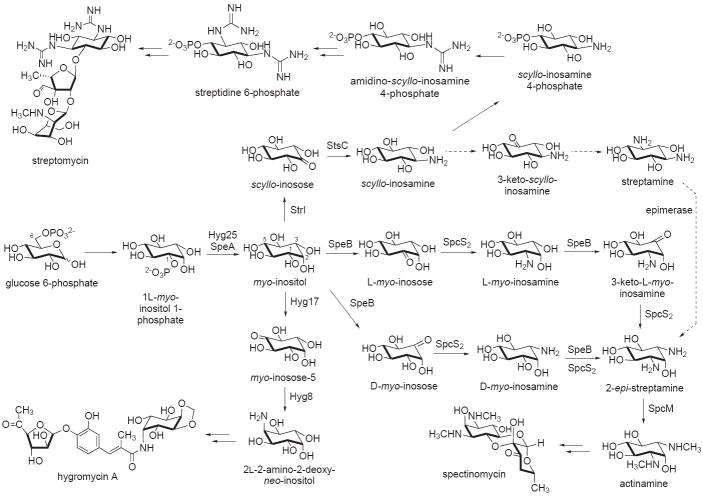
Figure 1.
Biosynthetic pathways to various aminocyclitol natural products. MIPS, 1L-myo-inositol 1-phosphate synthase; DOIS, 2-deoxy-scyllo-inosose synthase; EVS, 2-epi-5-epi-valiolone synthase; DHQS, dehydroquinate synthase; aDHQS, aminodehydroquinate synthase.
A comparative analysis of a number of other sugar phosphate cyclases involved in the biosynthesis of secondary metabolites, e.g., the aminoDHQ synthases (in rifamycin, ansamitocin, geldanamycin, and mitomycin biosyntheses), the DOI-synthases (in butirosin, neomycin, kanamycin, and gentamicin biosyntheses), and the 2-epi-5-epi-valiolone (EV) synthases (in acarbose, validamycin, cetoniacytone, salbostatin pathways), revealed their close relationship to DHQ synthases [6]. However, among members of the SPC superfamily, only the MIP synthases, the DOI synthases, and the EV synthases are actually involved in the biosynthesis of aminocyclitol natural products. The DHQ synthases and the aminoDHQ synthases mostly produce aromatic C7N units, e.g., 3-aminobenzoic acid and 3-amino-5-hydroxybenzoic acid (AHBA), that are precursors of many important microbial secondary metabolites [13].
Biosynthesis of the myo-inositol-derived aminocyclitol units of aminoglycoside antibiotics
In addition to its enormous role in many cellular and physiological processes in living organisms, myo-inositol serves as the precursor of several aminoglycoside antibiotics. This ubiquitous compound is typically modified to more complex aminocyclitols by a series of tailoring reactions, although direct attachment of myo-inositol to various aminosugar moieties is also evident in some aminoglycoside antibiotics such as kasugamycin [14].
In streptomycin and bluensomycin biosyntheses, myo-inositol is oxidized to scyllo-inosose by myo-inositol dehydrogenase (StrI), and subsequently transaminated to scyllo-inosamine by scyllo-inosose aminotransferase (StsC) [15]. While a similar pathway has been proposed for spectinomycin biosynthesis [16,17], more recent evidence seems to suggest that an alternative pathway may be involved. It is possible that the pathway to the aminocyclitol moiety of spectinomycin involves either C-1 oxidation of myo-inositol to L-myo-inosose or C-3 oxidation to D-myo-inosose (Figure 2). The product is then transaminated to L- or D-myo-inosamine. A subsequent second round of oxidation and transamination of either one of them would give 2-epi-streptamine. While this alternative pathway contradicts the earlier report by Jo et al. [17], it is more consistent with heterologous expression results, in which only three genes, speA (a monophosphatase), speB (a dehydrogenase), and spcS2 (an aminotransferase), are sufficient for the production of a bis-demethylated form of actinamine (2-epi-streptamine) in Streptomyces venezuelae YJ003 [16]. This finding also suggests that both SpeB and SpcS2 are used twice in the pathway. Therefore, the involvement of scyllo-inosose and scyllo-inosamine in 2-epi-streptamine formation seems to be less likely, as it would requires an additional epimerase enzyme that was not present in the heterologous system [16]. Alternatively, if scyllo-inosose and scyllo-inosamine are intermediates, this would imply the presence of an endogenous epimerase activity within the heterologous host, S. venezuelae YJ003, or that the reported product was streptamine instead of 2-epi-streptamine. Further and careful characterization of this pathway is crucial, as the lack of adequate data (e.g., NMR and [α]D) for the proper identification of enzyme products reported in some of the recent literature may undermine the accurate discrimination of these highly polar stereoisomers.
Figure 2.
Proposed pathways to various myo-inositol-derived aminoglycosides. The Hyg proteins are from the hygromycin pathway, the Spe and Spc proteins are from the spectinomycin pathway, and the Str and Sts proteins are from the streptomycin pathway.
Another example of unusual modification of myo-inositol is observed in the biosynthesis of hygromycin A, a traditional aminoglycoside antibiotic that has received new interest due to its effective use in the treatment of swine dysentery caused by the anaerobic spirochaet Serpulina hyodysenteriae. Formation of the aminocyclitol unit of this antibiotic involves a proposed oxidation of C-5 of myo-inositol to give myo-inosose-5, catalyzed by a putative myo-inositol dehydrogenase Hyg17 (Figure 2) [18]. The product is then converted to 2L-2-amino-2-deoxy-neo-inositol by the PLP-dependent aminotransferase Hyg8 [19]. One of the fascinating biosynthetic features of the aminocyclitol portion of hygromycin A is the formation of the C-4 and C-5 methylene bridge. The origin of this methylene carbon has been established to be methionine, but its mode of formation has yet to be determined. While biochemical characterization of enzymes directly related to hygromycin A biosynthesis is limited, an O-phosphotransferase (Hyg21) that catalyzes phosphorylation of hygromycin A has been identified recently in the antibiotic-producing organism S. hygroscopicus NRRL 2388 [20]. The enzyme catalyzes the transfer of a γ-phosphoryl group of ATP to the 2″-hydroxyl group of hygromycin A, abolishing its antibacterial properties and suggesting its role in a self-resistance mechanism.
Biosynthesis of 2-deoxy-scyllo-inosose-derived aminocyclitol units of aminoglycoside antibiotics
In recent years, a number of biosynthetic pathways to the DOI-derived aminocyclitols have been extensively investigated, with pathways leading to butirosin [21-25], neomycin [25,26], kanamycin [27], and gentamicin [28,29] being the most well-studied. Despite the fact that they are produced by different genera of bacteria, they appear to share early biosynthetic steps utilizing the amino-pseudodisaccharide paromamine as the common intermediate (Figure 3). The core set of genes required for paromamine biosynthesis is highly conserved within these related biosynthetic gene clusters, which facilitates the initial prediction of their biosynthetic pathways. However, more recent characterization of various enzymes involved in the pathways and the successful expressions of the minimal sets of genes necessary for the production of the antibiotics in heterologous hosts have provided invaluable insights into the assembly process of this class of antibiotics [27,28].
Figure 3.
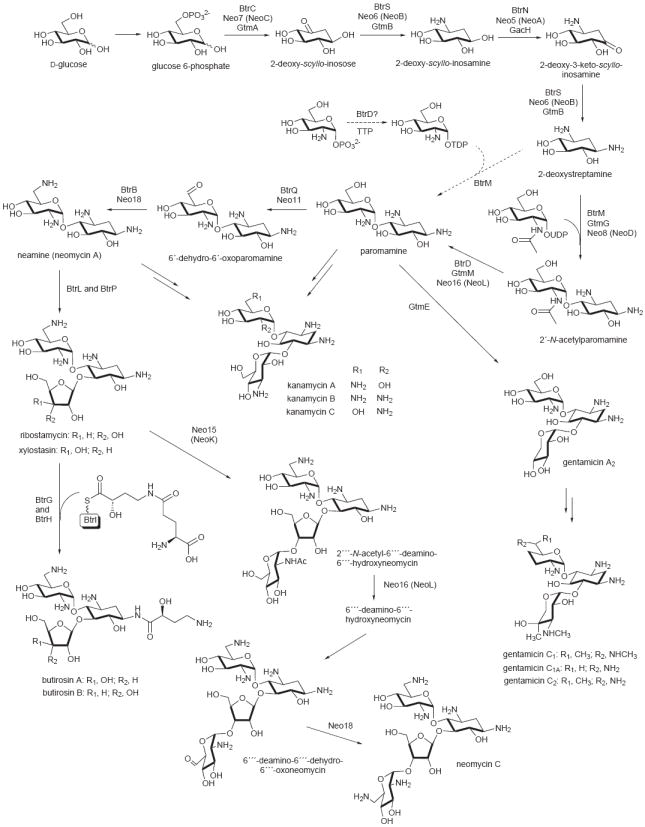
Proposed biosynthetic pathway for the production of DOI-derived aminoglycosides. Multiple protein names above the arrows correspond to homologous enzymes from different pathways (Btr, from the butirosin pathway; Neo, from the neomycin pathway; and Gtm and Gac, from the gentamicin pathway) that catalyze the same reaction. Note that a number of these enzymes are used repetitively in the pathways.
A key metabolic enzyme in the formation of paromamine is the sugar phosphate cyclase, DOI synthase, which catalyzes the conversion of glucose 6-phosphate to DOI. Analysis of the crystal structure reported recently for the butirosin DOI synthase (BtrC) from Bacillus circulans SANK 72073, and that of a DHQ synthase, revealed the structural similarities and differences of these two proteins [30]. However, the mechanism of catalysis of the DOI synthases was proposed to be different from that of DHQ synthases and proceeds through an extraordinary multi-step process including alcohol oxidation, phosphate syn-elimination, carbonyl reduction, ring opening and intramolecular aldol condensation through a boat conformation [24,30,31].
DOI is converted to 2-deoxy-scyllo-inosamine by a PLP-dependent aminotransferase. The high-resolution crystal structures of the PLP- and PMP-bound forms of the aminotransferase (BtrR, also called BtrS) from the butirosin pathway has been reported rather recently, revealing its structural similarity to the aspartate aminotransferase family [22]. In addition, this enzyme class also shares a relationship to the SMAT (secondary metabolite aminotransferases) subfamily of aminotransferases [22]. Interestingly, BtrR was found to catalyze not only the conversion of 2-deoxy-scyllo-inosose to 2-deoxy-scyllo-inosamine but also that of 2-deoxy-3-keto-scyllo-inosamine to 2-deoxystreptamine (Figure 3) [32]. This interesting phenomenon is apparently not unique to the butirosin pathway, as it was also observed in other aminoglycoside biosyntheses, including neomycin and gentamicin (Figure 3), and those of the myo-inositol-derived aminoglycosides [15,16,25,28]. These dual functional aminotransferases may share the same ancestral protein and the encoding genes were distributed amongst myo-inositol- and DOI-derived aminoglycoside producing bacteria via lateral gene transfer. Therefore, it is tempting to assume that these aminotransferases would recognize both inosose and deoxyinosose substrates. In fact, such activity has previously been observed in fresh cell-free extracts of spectinomycin producing bacteria [33].
The repetitive use of enzymes in aminoglycoside biosynthesis is not limited to the aminotransferases alone but also occurs in other key catalytic enzymes. As described earlier, the dehydrogenase (SpeB) from the spectinomycin pathway is also used twice in the pathway, as is the deacetylase NeoL (also called Neo16) from the neomycin pathway. NeoL catalyzes the conversion of 2′-N-acetylparomamine to paromamine and that of 2′″-N-acetyl-6′″-deamino-6′″-hydroxyneomycin to 6′″-deamino-6′″-hydroxyneomycin (Figure 3) [26]. The repetitive use of these enzymes is remarkably unique and highlights the efficient nature of aminoglycoside biosynthetic machineries. Interestingly, a NeoL-homologous protein from the butirosin pathway, BtrD, was initially observed to catalyze the synthesis of thymidine 5′-diphosphoglucosamine from thymidine 5′-triphosphate (TPP) and glucosamine 1-phosphate, and thus was identified as a nucleotidyltransferase [34]. However, this result appeared to be irreproducible and the enzyme was later identified as a 2′-N-acetylparomamine deacetylase [21].
Despite the fact that the butirosin and neomycin pathways share similar reaction steps to the paromamine moiety, the oxidation of 2-deoxy-scyllo-inosamine to 2-deoxy-3-keto-scyllo-inosamine involves two mechanistically different enzymes. In the neomycin pathway, the reaction is catalyzed by a Zn-dependent dehydrogenase (NeoA) [35], whereas in the butirosin pathway, the conversion is catalyzed by an unusual radical SAM dehydrogenase (BtrN) [23,36]. Interestingly, genes encoding proteins homologous to NeoA are consistently found in other DOI-derived aminoglycoside biosynthetic gene clusters [15], which suggests that the majority of the DOI-derived aminoglycoside pathways utilize a Zn-dependent dehydrogenase. Although there is a NeoA-homologous protein present in the butirosin pathway (BtrE), it appeared to be incapable of catalyzing the oxidation reaction, as it lacks two zinc-binding motifs important for activity [23].
With many highly homologous enzymes being shared across the aminoglycoside pathways, it is somewhat surprising to realize the remarkable structural diversity of this class of natural products. One key class of enzymes that contributes to this diversity are the glycosyltransferases that control the attachments of various sugar moieties to specific positions within the molecules. Recently, two distinct N-acetylglucosaminyltransferases have been identified in neomycin biosynthesis; NeoD (Neo8) catalyzes the conversion of 2-deoxystreptamine to 2′-N-acetylparomamine, and NeoK (Neo15) involved in the transfer of N-acetylglucosamine to ribostamycin [26]. The latter compound is derived from neamine through BtrL-catalyzed ribosylation followed by a BtrP-catalyzed dephosphorylation process [37]. Ribostamycin is also the precursor of butirosin B, which bears an (S)-4-amino-2-hydroxybutyrate substituent at the C-1 amine of the core aminocyclitol moiety. Conversion of ribostamycin to butirosin B requires the transfer of a N-protected (S)-4-amino-2-hydroxybutyrate from an ACP-bound intermediate by the acyltransferase BtrH followed by intramolecular cyclic deglutamylation (deprotection) of the product by the γ-glutamyl cyclotransferase BtrG [38,39]. These two reactions constitute the final steps in the biosynthesis of butirosin.
Biosynthesis of 2-epi-5-epi-valiolone-derived aminocyclitols
While earlier studies on the biosynthesis of 2-epi-5-epi-valiolone-derived aminocyclitols using isotope tracer experiments shed some light into the biosynthesis of this class of compounds [40], identification of their biosynthetic gene clusters has significantly improved our understanding of their modes of formation [6,41-45]. More recently, the biosynthetic gene clusters of salbostatin, a trehalase inhibitor from Streptomyces albus ATCC 21838 [46], and of cetoniacytone from the endosymbiotic Actinomyces sp. strain Lu 9419 have been reported [47]. A significant number of genes in the salbostatin cluster share high identity (51 – 72%) with genes from the acarbose cluster from Actinoplanes sp. SE50/110, suggesting that the two pathways are closely related [46]. On the other hand, the cetoniacytone cluster appears to be quite different from other known aminocyclitol clusters reported so far, except for the presence of the 2-epi-5-epi-valiolone synthase (cetA) and the 2-epi-5-epi-valiolone epimerase (cetB) genes. Consistent with the chemical structure of this unique aminocyclitol, the cluster contains numerous genes that code for proteins with unclear functions. Those include proteins from the cupin superfamily, which are mostly related to hydroxylation, epoxidation, decarboxylation, dehydration, and halogenation reactions. These genes are absent in other C7N-aminocyclitol clusters. However, BLAST search analysis using the newly isolated cet genes revealed an analogous suite of genes in the genome of the plant symbiotic nitrogen-fixing bacterium Frankia alni ACN14a, suggesting that this bacterium may be able to produce a secondary metabolite related to the cetoniacytones [47].
The key enzyme that unites all members of this family of aminocyclitols is the sugar phosphate cyclase, 2-epi-5-epi-valiolone synthase, which catalyzes the conversion of sedoheptulose 7-phosphate to 2-epi-5-epi-valiolone, the common precursor of all C7N-aminocyclitol natural products [6,41,44,46]. In the acarbose pathway, and presumably also in the salbostatin pathway, 2-epi-5-epi-valiolone is phosphorylated by the cyclitol kinase (AcbM) to 2-epi-5-epi-valiolone 7-phosphate [42]. Further downstream the pathway primarily involves phosphorylated intermediates. However, in the validamycin and cetoniacytone pathways, 2-epi-5-epi-valiolone is not phosphorylated, but instead epimerized to 5-epi-valiolone. This reaction is catalyzed by a class of metalloproteins that are highly similar to glyoxalases/bleomycin resistance proteins [47]. They appear to be a new type of α-hydroxyketo-epimerases that belong to the Vicinal Oxygen Chelate (VOC) superfamily. Structurally, they differ from the 2-epi-5-epi-valiolone 7-phosphate epimerase (AcbO), a homolog of sugar phosphate isomerases, previously identified in the acarbose pathway [43].
While the kinase (AcbM) from the acarbose pathway phosphorylates 2-epi-5-epi-valiolone, studies on the kinase (ValC) from the validamycin pathway indicated that the phosphorylation takes place later in the pathway [48]. ValC phosphorylates valienone and validone, but not 2-epi-5-epi-valiolone or 5-epi-valiolone. Interestingly, no similar kinase gene was found in the cetoniacytone cluster, suggesting that no phosphorylation step is necessary in its biosynthesis [47]. In contrast to validamycin formation, results of feeding studies indicate that valienone is not involved in cetoniacytone biosynthesis [6]. This suggests that the downstream modifications of 5-epi-valiolone in cetoniacytone biosynthesis are different from those in the validamycin pathway (Figure 4). However, no biochemical data are currently available to support the proposed pathway.
Figure 4.
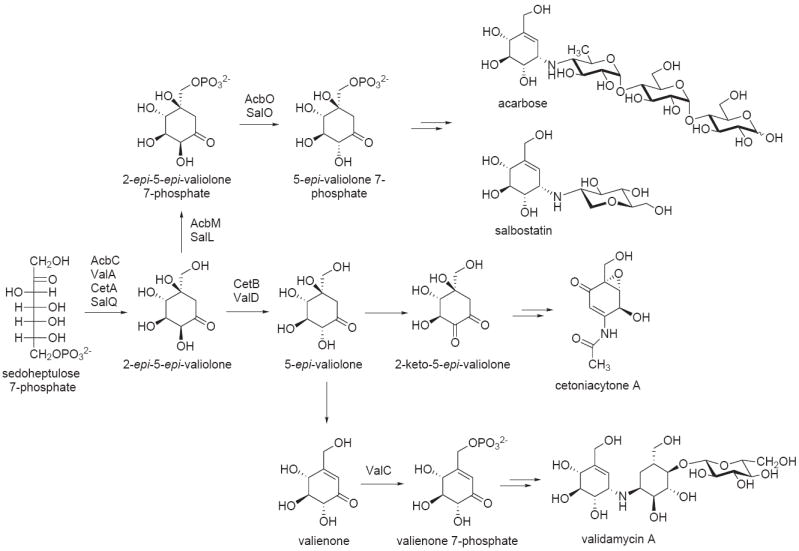
Biosynthetic pathways to various C7N-aminocyclitol-containing natural products. 2-epi-5-epi-Valiolone is the common intermediate involved in the biosynthesis of all C7N-aminocylitol natural products.
The genetic manipulation of the validamycin producer proved to be quite fruitful, enabling the construction of several key validamycin deletion mutants. Notably, the inactivation of the glycosyltransferase gene (valG) resulted in mutants that produce high levels of validoxylamine A, which can be used as a scaffold for the synthesis of validamycin analogs (Figure 5) [45,49]. The valG gene has been cloned and expressed in E. coli and the recombinant protein is capable of converting validoxylamine A to validamycin using UDP- or GDP-glucose, but not ADP- or dTDP-glucose, as sugar donors [45]. In addition, ValG also efficiently utilized UDP-galactose as sugar donor resulting in the production of an unnatural compound 4″-epi-validamycin A [49].
Figure 5.
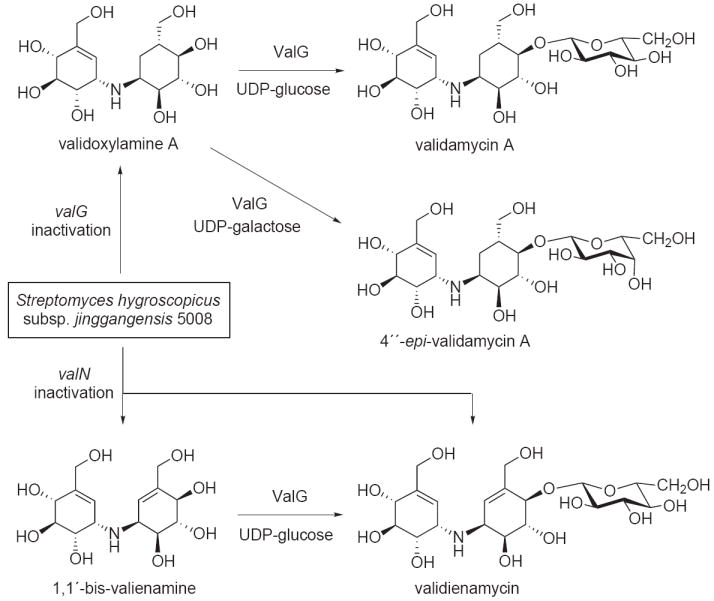
Inactivation of the validamycin glycosyltransferase valG and the putative cyclitol reductase valN genes resulted in mutant strains that produced new aminocyclitol products. Bioconversion of the resulting pseudodisaccharides using recombinant glycosyltransferase ValG gave their respective glycosidal products.
Furthermore, inactivation of the putative cyclitol reductase gene valN in S. hygroscopicus subsp. jinggangensis 5008 resulted in mutants that produced a symmetrical dimer of two unsaturated cyclitols, 1,1′-bis-valienamine, and its glucoside, validienamycin (Figure 5) [50]. The relationship between the two metabolites was confirmed by converting 1,1′-bis-valienamine to validienamycin with ValG, in the presence of UDP-glucose. The results suggest that valN is involved in the formation of the saturated moiety of validamycin A, and that the aminotransferase ValM, and other enzymes downstream the pathway, can recognize unsaturated intermediates as substrates. However, the lack of biochemical data impedes a definitive assignment of ValN function in the validamycin pathway.
Conclusion
The amazing architectural ability of Mother Nature to create diverse chemical entities exemplified in the biosynthesis of aminocyclitol-containing natural products remains an object of exploration. One of the intriguing features of aminocyclitol biosynthesis is the highly efficient assembly process that uses enzymes in a repetitive manner, which represents one of the most efficient biosynthetic machineries in microbial secondary metabolism. While more recent studies have provided additional insights into the biosynthesis of several aminocyclitols, the modes of formation of others remain somewhat elusive. Encouragingly, a relatively large quantity of genetic information for the aminocyclitol pathways is currently available and awaiting more detailed characterization and functional analysis. While the highly polar and stereochemically diverse intermediates involved in aminocyclitol biosyntheses may present significant challenges, this should not serve as a major impediment to assigning their critical biochemical roles.
Acknowledgments
The author thanks Drs. P. M. Flatt and T. M. Zabriskie for critical reading of this manuscript. This contribution was supported in part by a grant from the National Institutes of Health (R01 AI061528) and the Oregon State University College of Pharmacy General Research Funds.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Paper of particular interest, published within the period of review, have been highlighted as:
*of special interest
**of outstanding interest
- 1.Mahmud T. The C7N aminocyclitol family of natural products. Nat Prod Rep. 2003;20:137–166. doi: 10.1039/b205561a. [DOI] [PubMed] [Google Scholar]
- 2.Davies JE. Aminoglycosides: ancient and modern. J Antibiot (Tokyo) 2006;59:529–532. doi: 10.1038/ja.2006.73. [DOI] [PubMed] [Google Scholar]
- 3.Prasad R, Verma SK, Sahai S, Kumar S, Jain A. Efficacy and safety of kanamycin, ethionamide, PAS and cycloserine in multidrug-resistant pulmonary tuberculosis patients. Indian J Chest Dis Allied Sci. 2006;48:183–186. [PubMed] [Google Scholar]
- 4.Yamamura S, Kawada K, Takehira R, Nishizawa K, Katayama S, Hirano M, Momose Y. Prediction of aminoglycoside response against methicillin-resistant Staphylococcus aureus infection in burn patients by artificial neural network modeling. Biomed Pharmacother. 2008;62:53–58. doi: 10.1016/j.biopha.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Mahmud T, Flatt PM, Wu X. Biosynthesis of unusual aminocyclitol-containing natural products. J Nat Prod. 2007;70:1384–1391. doi: 10.1021/np070210q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *6.Wu X, Flatt PM, Schlorke O, Zeeck A, Dairi T, Mahmud T. A comparative analysis of the sugar phosphate cyclase superfamily involved in primary and secondary metabolism. Chembiochem. 2007;8:239–248. doi: 10.1002/cbic.200600446.. Describes a detailed comparative analysis of sugar phosphate cyclases and the identification of a new class of sugar phosphate cyclases that might regulate the biosynthesis of a novel set of secondary metabolites in fungi and cyanobacteria.
- 7.Llewellyn NM, Spencer JB. Biosynthesis of 2-deoxystreptamine-containing aminoglycoside antibiotics. Nat Prod Rep. 2006;23:864–874. doi: 10.1039/b604709m. [DOI] [PubMed] [Google Scholar]
- 8.Kudo F, Kasama Y, Hirayama T, Eguchi T. Cloning of the pactamycin biosynthetic gene cluster and characterization of a crucial glycosyltransferase prior to a unique cyclopentane ring formation. J Antibiot (Tokyo) 2007;60:492–503. doi: 10.1038/ja.2007.63. [DOI] [PubMed] [Google Scholar]
- 9.Sakuda S, Sugiyama Y, Zhou ZY, Takao H, Ikeda H, Kakinuma K, Yamada Y, Nagasawa H. Biosynthetic studies on the cyclopentane ring formation of allosamizoline, an aminocyclitol component of the chitinase inhibitor allosamidin. J Org Chem. 2001;66:3356–3361. doi: 10.1021/jo001629n. [DOI] [PubMed] [Google Scholar]
- 10.Sugiyama Y, Nagasawa H, Suzuki A, Sakuda S. Biosynthesis of the trehalase inhibitor trehazolin. J Antibiot (Tokyo) 2002;55:263–269. doi: 10.7164/antibiotics.55.263. [DOI] [PubMed] [Google Scholar]
- 11.Geiger JH, Jin X. The structure and mechanism of myo-inositol-1-phosphate synthase. Subcell Biochem. 2006;39:157–180. doi: 10.1007/0-387-27600-9_7. [DOI] [PubMed] [Google Scholar]
- 12.Knaggs AR. The biosynthesis of shikimate metabolites. Nat Prod Rep. 2003;20:119–136. doi: 10.1039/b100399m. [DOI] [PubMed] [Google Scholar]
- 13.Floss HG. Natural products derived from unusual variants of the shikimate pathway. Nat Prod Rep. 1997;14:433–452. doi: 10.1039/np9971400433. [DOI] [PubMed] [Google Scholar]
- 14.Ikeno S, Aoki D, Hamada M, Hori M, Tsuchiya KS. DNA sequencing and transcriptional analysis of the kasugamycin biosynthetic gene cluster from Streptomyces kasugaensis M338-M1. J Antibiot (Tokyo) 2006;59:18–28. doi: 10.1038/ja.2006.4. [DOI] [PubMed] [Google Scholar]
- 15.Flatt PM, Mahmud T. Biosynthesis of aminocyclitol-aminoglycoside antibiotics and related compounds. Nat Prod Rep. 2007;24:358–392. doi: 10.1039/b603816f. [DOI] [PubMed] [Google Scholar]
- 16.Thapa LP, Oh TJ, Liou K, Sohng JK. Biosynthesis of spectinomycin: heterologous production of spectinomycin and spectinamine in an aminoglycoside-deficient host, Streptomyces venezuelae YJ003. J Appl Microbiol. 2008;105:300–308. doi: 10.1111/j.1365-2672.2008.03788.x. [DOI] [PubMed] [Google Scholar]
- 17.Jo YY, Kim SH, Yang YY, Kang CM, Sohng JK, Suh JW. Functional analysis of spectinomycin biosynthesis genes from Streptomyces spectabilis ATCC 27741. J Microbiol Biotechnol. 2003;13:906–911. [Google Scholar]
- 18.Habib el SE, Scarsdale JN, Reynolds KA. Biosynthetic origin of hygromycin A. Antimicrob Agents Chemother. 2003;47:2065–2071. doi: 10.1128/AAC.47.7.2065-2071.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palaniappan N, Ayers S, Gupta S, Habib el S, Reynolds KA. Production of hygromycin A analogs in Streptomyces hygroscopicus NRRL 2388 through identification and manipulation of the biosynthetic gene cluster. Chem Biol. 2006;13:753–764. doi: 10.1016/j.chembiol.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 20.Dhote V, Gupta S, Reynolds KA. An O-phosphotransferase catalyzes phosphorylation of hygromycin A in the antibiotic-producing organism Streptomyces hygroscopicus. Antimicrob Agents Chemother. 2008;52:3580–3588. doi: 10.1128/AAC.00157-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *21.Truman AW, Huang F, Llewellyn NM, Spencer JB. Characterization of the enzyme BtrD from Bacillus circulans and revision of its functional assignment in the biosynthesis of butirosin. Angew Chem Int Ed Engl. 2007;46:1462–1464. doi: 10.1002/anie.200604194.. Description of the function of BtrD as a deacetylase and not a nucleotidyltransferase as reported previously.
- 22.Popovic B, Tang X, Chirgadze DY, Huang F, Blundell TL, Spencer JB. Crystal structures of the PLP- and PMP-bound forms of BtrR, a dual functional aminotransferase involved in butirosin biosynthesis. Proteins. 2006;65:220–230. doi: 10.1002/prot.21076. [DOI] [PubMed] [Google Scholar]
- *23.Yokoyama K, Numakura M, Kudo F, Ohmori D, Eguchi T. Characterization and mechanistic study of a radical SAM dehydrogenase in the biosynthesis of butirosin. J Am Chem Soc. 2007;129:15147–15155. doi: 10.1021/ja072481t.. Describes the first example of oxidation of a hydroxyl group by a radical SAM enzyme. The mechanism is unique and is most likely absent in the biosynthetic pathways of other DOI-derived aminocyclitols.
- 24.Hirayama T, Kudo F, Huang Z, Eguchi T. Role of glutamate 243 in the active site of 2-deoxy-scyllo-inosose synthase from Bacillus circulans. Bioorg Med Chem. 2007;15:418–423. doi: 10.1016/j.bmc.2006.09.042. [DOI] [PubMed] [Google Scholar]
- *25.Huang F, Spiteller D, Koorbanally NA, Li Y, Llewellyn NM, Spencer JB. Elaboration of neosamine rings in the biosynthesis of neomycin and butirosin. Chembiochem. 2007;8:283–288. doi: 10.1002/cbic.200600371.. Describes the bifunctional roles of the oxidases and the aminotransferases in the neomycin and butirosin pathways.
- *26.Yokoyama K, Yamamoto Y, Kudo F, Eguchi T. Involvement of two distinct N-acetylglucosaminyltransferases and a dual-function deacetylase in neomycin biosynthesis. Chembiochem. 2008;9:865–869. doi: 10.1002/cbic.200700717.. Describes the distinct substrate specificity of the N-acetylglucosaminyltransferases and the bifunctional roles of the deacetylase from the neomycin pathway.
- 27.Thapa LP, Oh TJ, Lee HC, Liou K, Park JW, Yoon YJ, Sohng JK. Heterologous expression of the kanamycin biosynthetic gene cluster (pSKC2) in Streptomyces venezuelae YJ003. Appl Microbiol Biotechnol. 2007;76:1357–1364. doi: 10.1007/s00253-007-1096-4. [DOI] [PubMed] [Google Scholar]
- **28.Park JW, Hong JS, Parajuli N, Jung WS, Park SR, Lim SK, Sohng JK, Yoon YJ. Genetic dissection of the biosynthetic route to gentamicin A2 by heterologous expression of its minimal gene set. Proc Natl Acad Sci U S A. 2008;105:8399–8404. doi: 10.1073/pnas.0803164105.. Using heterologous expression of different combinations of putative 2-deoxystreptamine biosynthetic genes the authors discovered a subset of genes that is responsible for the biosynthesis of the core aminocyclitol of gentamicin. This in vivo study provided direct evidence for the biosynthetic pathway to gentamicin, allowing the unequivocal assignment of the biochemical role of each gene products.
- 29.Kim JY, Suh JW, Kang SH, Phan TH, Park SH, Kwon HJ. Gene inactivation study of gntE reveals its role in the first step of pseudotrisaccharide modifications in gentamicin biosynthesis. Biochem Biophys Res Commun. 2008;372:730–734. doi: 10.1016/j.bbrc.2008.05.133. [DOI] [PubMed] [Google Scholar]
- *30.Nango E, Kumasaka T, Hirayama T, Tanaka N, Eguchi T. Structure of 2-deoxy-scyllo-inosose synthase, a key enzyme in the biosynthesis of 2-deoxystreptamine-containing aminoglycoside antibiotics, in complex with a mechanism-based inhibitor and NAD+ Proteins. 2008;70:517–527. doi: 10.1002/prot.21526.. Describes the first crystal structures of 2-deoxy-scyllo-inosose synthase and a comparative study between the 2-deoxy-scyllo-inosose synthases and the dehydroquinate synthases.
- 31.Carpenter EP, Hawkins AR, Frost JW, Brown KA. Structure of dehydroquinate synthase reveals an active site capable of multistep catalysis. Nature. 1998;394:299–302. doi: 10.1038/28431. [DOI] [PubMed] [Google Scholar]
- 32.Yokoyama K, Kudo F, Kuwahara M, Inomata K, Tamegai H, Eguchi T, Kakinuma K. Stereochemical recognition of doubly functional aminotransferase in 2-deoxystreptamine biosynthesis. J Am Chem Soc. 2005;127:5869–5874. doi: 10.1021/ja0445948. [DOI] [PubMed] [Google Scholar]
- 33.Walker JB. Enzymatic synthesis of aminocyclitol moieties of aminoglycoside antibiotics from inositol by Streptomyces spp.: detection of glutamine-aminocyclitol aminotransferase and diaminocyclitol aminotransferase activities in a spectinomycin producer. J Bacteriol. 1995;177:818–822. doi: 10.1128/jb.177.3.818-822.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kudo F, Kawabe K, Kuriki H, Eguchi T, Kakinuma K. A new family of glucose-1-phosphate/glucosamine-1-phosphate nucleotidylyltransferase in the biosynthetic pathways for antibiotics. J Am Chem Soc. 2005;127:1711–1718. doi: 10.1021/ja044921b. [DOI] [PubMed] [Google Scholar]
- 35.Kudo F, Yamamoto Y, Yokoyama K, Eguchi T, Kakinuma K. Biosynthesis of 2-deoxystreptamine by three crucial enzymes in Streptomyces fradiae NBRC 12773. J Antibiot (Tokyo) 2005;58:766–774. doi: 10.1038/ja.2005.104. [DOI] [PubMed] [Google Scholar]
- 36.Yokoyama K, Ohmori D, Kudo F, Eguchi T. Mechanistic study on the reaction of a radical SAM dehydrogenase BtrN by electron paramagnetic resonance spectroscopy. Biochemistry. 2008;47:8950–8960. doi: 10.1021/bi800509x. [DOI] [PubMed] [Google Scholar]
- 37.Kudo F, Fujii T, Kinoshita S, Eguchi T. Unique O-ribosylation in the biosynthesis of butirosin. Bioorg Med Chem. 2007;15:4360–4368. doi: 10.1016/j.bmc.2007.04.040. [DOI] [PubMed] [Google Scholar]
- **38.Llewellyn NM, Li Y, Spencer JB. Biosynthesis of butirosin: transfer and deprotection of the unique amino acid side chain. Chem Biol. 2007;14:379–386. doi: 10.1016/j.chembiol.2007.02.005.. Describes a complete elucidation of ACP-mediated formation and transfer of (S)-4-amino-2-hydroxybutyrate side chain in butirosin biosynthesis. The pathway involves unusual protective biochemistry via γ-L-glutamylation of an ACP-bound substrate.
- 39.Llewellyn NM, Spencer JB. Chemoenzymatic acylation of aminoglycoside antibiotics. Chem Commun (Camb) 2008:3786–3788. doi: 10.1039/b802248h. [DOI] [PubMed] [Google Scholar]
- 40.Mahmud T. Isotope tracer investigations of natural products biosynthesis: The discovery of novel metabolic pathways. J Labelled Comp Radiophar. 2007;50:1039–1051. [Google Scholar]
- 41.Stratmann A, Mahmud T, Lee S, Distler J, Floss HG, Piepersberg W. The AcbC protein from Actinoplanes species is a C7-cyclitol synthase related to 3-dehydroquinate synthases and is involved in the biosynthesis of the alpha-glucosidase inhibitor acarbose. J Biol Chem. 1999;274:10889–10896. doi: 10.1074/jbc.274.16.10889. [DOI] [PubMed] [Google Scholar]
- 42.Zhang CS, Stratmann A, Block O, Bruckner R, Podeschwa M, Altenbach HJ, Wehmeier UF, Piepersberg W. Biosynthesis of the C7-cyclitol moiety of acarbose in Actinoplanes species SE50/110. 7-O-phosphorylation of the initial cyclitol precursor leads to proposal of a new biosynthetic pathway. J Biol Chem. 2002;277:22853–22862. doi: 10.1074/jbc.M202375200. [DOI] [PubMed] [Google Scholar]
- 43.Zhang CS, Podeschwa M, Altenbach HJ, Piepersberg W, Wehmeier UF. The acarbose-biosynthetic enzyme AcbO from Actinoplanes sp. SE 50/110 is a 2-epi-5-epi-valiolone-7-phosphate 2-epimerase. FEBS Lett. 2003;540:47–52. doi: 10.1016/s0014-5793(03)00221-7. [DOI] [PubMed] [Google Scholar]
- 44.Yu Y, Bai L, Minagawa K, Jian X, Li L, Li J, Chen S, Cao E, Mahmud T, Floss HG, et al. Gene cluster responsible for validamycin biosynthesis in Streptomyces hygroscopicus subsp. jinggangensis 5008. Appl Environ Microbiol. 2005;71:5066–5076. doi: 10.1128/AEM.71.9.5066-5076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bai L, Li L, Xu H, Minagawa K, Yu Y, Zhang Y, Zhou X, Floss HG, Mahmud T, Deng Z. Functional analysis of the validamycin biosynthetic gene cluster and engineered production of validoxylamine A. Chem Biol. 2006;13:387–397. doi: 10.1016/j.chembiol.2006.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi WS, Wu X, Choeng YH, Mahmud T, Jeong BC, Lee SH, Chang YK, Kim CJ, Hong SK. Genetic organization of the putative salbostatin biosynthetic gene cluster including the 2-epi-5-epi-valiolone synthase gene in Streptomyces albus ATCC 21838. Appl Microbiol Biotechnol. 2008;80:637–645. doi: 10.1007/s00253-008-1591-2. [DOI] [PubMed] [Google Scholar]
- *47.Wu X, Flatt PM, Xu H, Mahmud T. Biosynthetic gene cluster of cetoniacytone A, an unusual aminocyclitol from the endosymbiotic Bacterium Actinomyces sp. Lu 9419. Chembiochem. 2009;10:304–314. doi: 10.1002/cbic.200800527.. Description of the first 2-epi-5-epi-valiolone epimerases that belong to the Vicinal Oxygen Chelate (VOC) superfamily.
- 48.Minagawa K, Zhang Y, Ito T, Bai L, Deng Z, Mahmud T. ValC, a new type of C7-cyclitol kinase involved in the biosynthesis of the antifungal agent validamycin A. Chembiochem. 2007;8:632–641. doi: 10.1002/cbic.200600528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu H, Minagawa K, Bai L, Deng Z, Mahmud T. Catalytic analysis of the validamycin glycosyltransferase (ValG) and enzymatic production of 4”-epi-validamycin A. J Nat Prod. 2008;71:1233–1236. doi: 10.1021/np800185k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu H, Yang J, Bai L, Deng Z, Mahmud T. Genetically engineered production of 1,1’-bis-valienamine and validienamycin in Streptomyces hygroscopicus and their conversion to valienamine. Appl Microbiol Biotechnol. 2009;81:895–902. doi: 10.1007/s00253-008-1711-z. [DOI] [PMC free article] [PubMed] [Google Scholar]