Abstract
Mutational optimization of two long-range interactions first observed in Ac-WINGKWT-NH2 – a) bifurcated H-bonding involving the threonine amide HN and sidechain OH and the N-terminal acetyl carbonyl and b) an H-bond between the entgegen-HN of the C-terminal amide and the indole ring of Trp6 which stabilizes a face-to-edge indole/indole interaction between Trp1 and Trp6 – has afforded ≤10 residue systems that yield a remarkably stable fold in water. Optimization was achieved by designing a hydrophobic cluster that sequesters these H-bonds from solvent exposure. The structures and extent of amide H/D exchange protection for CH3CH2CO-WIpGXWTGPS (p = D-Pro, X = Leu or Ile) were determined. These two systems are greater than 94% folded at 298K (97.5% at 280K) with melting temperatures > 75 °C. The fold appears to display minimal fluxionality; a well converged NMR structure rationalizes all of the large structuring shifts observed and we suggest that these designed constructs can be viewed as microproteins.
Introduction
Few chemists need to be convinced that proteins are complex macromolecules with intriguing properties and the essential elements of biochemistry and, indeed, of life itself. But the complex nature of these structured chains of amino acids has an obvious drawback: proteins are typically more difficult to study and manipulate (whether in vivo, in vitro, or in silico) than smaller, simpler molecules. Studies of the protein folding phenomenon have increasingly focused on the design and analysis of small, cooperatively-folding systems that display particularly fast folding rates.1 The truncation and optimization of natural folds2 has been one source of peptides that can be designated as fast-folding miniproteins. In this category, the 21 residue N-terminal fragment of the villin headpiece2b is the smallest system to display a significant fold population. The mutational fold optimization of peptides displaying some tertiary structure has been the other source of miniproteins.3 Obtained in this way4, the Trp-cage fold, at an 18 – 20 residue length5 and displaying melting temperatures (Tm) as high as 63°C, meets all the criteria for a miniprotein: two state-folding with multiple elements of secondary structure, numerous tertiary contacts, and a hydrophobic core. Miniproteins have obvious advantages for MD folding simulations, as a confirmation of this; the Trp-cage quickly6 achieved protein folding paradigm status7. To our knowledge, the smallest system that has been designated8 as a miniprotein is chignolin (GYDPETGTWG), a 10-mer β hairpin with a particularly favorable 6-residue turn9 flanked and further stabilized by an aromatic/aromatic ring interaction 10,11. Chignolin was reported to have a fold population of 0.83 at 273K, with a Tm = 39 °C.9 As smaller and smaller structured polypeptides are designed, the question arises: what defines a protein? More stable hairpins, 8-mers with Tm = 65,14 12-mers with Tm = 7010c and 79,10b and 16-mers with Tm’s of 7010b and 85 °C15 have been reported. Does a stable β hairpin, without a context of multiple tertiary interactions as observed in proteins, constitute a miniprotein?
A peptide/protein boundary based on amino acid chain length no longer appears appropriate: long naturally occurring peptides lacking structure are known, there are stable folds reported for short peptides, and it is increasingly clear that there is residual structuring7b,16 in the denatured states of many proteins. The distinguishing characteristics of a prototypic protein appear to be a matter of folding mechanism and fold stability. While there are some instances of “downhill” folding17, 2-state folding remains the expectation for well-designed or -evolved proteins. This requires a deep, narrow energy well, with the energetically favorable folded state resting at the bottom. Ideally, the fold should resist entropic effects on warming, persisting above physiological temperatures (a high Tm). A deep energy well would only be possible with a uniquely stable conformation, and 2-state folding requires that this conformation contains and is determined by a network of strong, synergistic stabilizing interactions that can be found by a simple conformation search. These interactions should be dominated by longer range contacts. Probably as a consequence of this, most very small proteins have termini in close proximity; a result of the spatial realities associated with the establishment of stabilizing tertiary interactions.
These folding requirements lead to experimental diagnostics of protein-like behavior. A narrow and deep folding landscape implies a NOESY spectrum with no conflicting long-range cross-peaks and that all of the long-range through space contacts yield an NMR structure which predicts the vicinal coupling constants, short-range NOE intensities, and chemical shifts observed. Proteins, particularly if they contain aromatic residues, display well-dispersed NMR spectra with large structuring shifts (CSDs, chemical shift deviations). Cooperativity requires that all CSDs should melt out in concert.15,18 While CSDs have been used to measure changes in fold population associated with mutations and melting; they always present a dilemma with small folds: 100% folded values are difficult to access18b,19 since folding transitions are broad and pre-melting baselines are not available. This also applies to other folding measures such as fluorescence and CD exciton couplets. Cold denaturation has been documented15,18c,20 for both hairpins and helices; as one example, Cochran reports10b that a trpzip hairpin with a high Tm (79 °C) is less than 90% folded at 273K. As a result, it cannot be assumed that a low temperature plateau for spectroscopic parameters corresponds to 100% folded values. However, native proteins always have a significant fraction of backbone NH’s that are fully protected from H/D exchange in the folded state. Thus, H/D exchange protection factors can provide an indisputable measure of ΔGU for miniproteins21.
Here we present NMR (and other spectroscopic) data for a series of 7 – 10 residue peptides that appear to meet all of the criteria set forth above. The initial inspiration for the series was Ac-WNPATGKW-NH2, (ΔGU280 ≈ 0) a severally truncated trpzip peptide.10c The design strategy consisted of replacing the 6-residue turn with a 4-residue unit and then manipulating the termini of the sequence so as to introduce additional long-range interactions to stabilize the fold. Although these systems are β hairpins they display mutational effects and melting behavior that is fully protein-like. For example, even though hairpin stability is enhanced by favorable Coulombic interactions between the termini, 15,22 representatives of this new fold display the opposite effect: WINGKWTG is less than 15% folded while Ac-WINGKWTG-NH2 displays χF = 0.77 at 280K. The destabilizing effect could be traced to the deletion of the N-terminal acetyl which has dramatic effects on the spectroscopic parameters for sites at the C-terminus: deacetylation converts 2.71 ppm upfield structuring shifts at the Gly8 amide NH to statistical coil shift values. Amide NH H/D exchange data indicates that structured peptides produced from this study (for example, CH3CH2CO-WIpGXWTGPS, p = D-Pro, X = Ile or Leu) achieve 97.5+% folding in water at 280K. We argue that these can be viewed as “microproteins”.
Materials and Methods
All peptides were synthesized using fast FMOC chemistry on an ABI 433A peptide synthesizer. Cleavage from resin was accomplished by shaking in excess TFA with 2.5% water and 2.5% triisopropylsilane for 1.5 hrs. Acetylation, when required, was carried out prior to cleavage by reacting the resin-bound peptide with 3% acetic anhydride and 4.3% triethylamine, in DMF. Peptides were precipitated and washed three times in ether, and purified by reverse phase HPLC (C18 or C8 column) with a water (0.1% TFA)/acetonitrile (0.085% TFA) gradient. The identities of the peptides were verified by positive ion mass spectrometry on a Bruker Esquire ion trap spectrometer. Sequence and purity were further ensured upon analysis of 2D spectra and assignment of the NOESY connectivities.
CD Spectroscopy
Stock solutions were prepared by dissolving ~0.3 mg of peptide in 10 mM pH 6 potassium phosphate buffer with the actual peptide concentration determined based on the expectation UV absorption of Trp (ε = 5580 M−1 cm−1 per Trp) at 280 nm. Stock solutions were diluted with buffer (10 mM KxHxPO4, pH 6.4) and for one sample, a mix of buffer and 8.2 M guanidinium chloride stock solution (concentration verified by refractive index) to yield 30 μM solutions of peptide. Spectra were recorded on a Jasco J715 spectropolarimeter using 0.10 cm pathlength cells. CD data are reported in molar units (deg cm2 dmol−1), shown as degrees molar ellipticity throughout, rather than residue-molar ellipticity units (deg cm2 res-dmol−1) because of the dominance of a Trp/Trp exciton couplet rather than backbone amides to the net CD spectrum.10c The exciton couplet has a maximum at 227–229 nm, and the ellipticity at this wavelength was used (after subtraction of the random coil control value, see Supporting Information) to provide one estimate of fold population.
NMR Spectroscopy
NMR spectra were recorded on a 500 MHz Bruker NMR. Solvent suppression was accomplished in all cases by a WATERGATE23 pulse train. TOCSY spectra employed a 60 ms MLEV-17 spinlock24, and NOESY spectra had a 150 ms mixing time, except when intended for elucidation of an NMR structure. Aqueous samples were comprised of 0.8–1.6 mM peptide in 20–50 mM sodium phosphate buffer, pH 6-7, with 10% D2O and sodium 2,2-dimethyl-2-silapentane-5-sulfonate (DSS) as an internal standard. Glycol was added to some samples to increase the tumbling time of the smaller peptides and provide access to lower temperatures; these samples were identical to the aqueous samples except that they contained 20 vol-% d6 ethylene glycol.
All resonances displayed sharp signals: line widths consistent with the tumbling times of monomeric peptides. NMR fold population measures (measured at > 1.2 mM concentrations) are corroborated by CD fold population measures taken at 30μM, additional support for monomeric behavior of all peptides discussed.
H/D Exchange
In the case of Pr-WI(p/P)GLWTGPS, amide exchange protection was measured by recording, at 280K, serial NMR spectra (after dissolution in D2O, pD=3.84 and/or 4.74) of samples consisting of equimolar amounts of the microprotein with a D-Pro-Gly turn locus and the random coil control with an L-Pro-Gly ‘turn’ substitution. Processing involved recording integrals of relevant amide peaks normalized to a non-exchanging standard. Protection factors at each site were determined from the rate ratio kex(PG)/Kex(pG); with each kex resulting from an exponential fit to the peak intensity versus time plot. Due to 1-D overlap issues, the intensity measures for some peaks required deconvolution prior to further processing. For Pr-WIpGIWTGPS (pD=6.15), the solution did not contain the PG control; Molday factors25 were employed to convert the observed rates to protection factors.
NMR-Derived Structural Representations
NOESY spectra used for structure derivations were recorded at 500 MHz using a mixing time of 120ms; the sample contained 20% d6-glycol and the spectrum was recorded at 270K to increase the NOESY intensities through the viscosity effect on tumbling time. The structure ensemble generation procedure previously used14 for Ac-WINGKWTG-NH2 was applied to 144 NOE intensities observed for Pr-WIpGIWTGPS. The automated distance-constraint derivation, MD annealing protocols, and acceptance criteria were those described previously.10c,14 The file of distance constraints and the structure statistics appear in the Supporting Information.
Structure Validation
Indole/Indole ring exciton couplet calculation were based on the geometry derived from the NMR structural ensemble and used the method previously described by Woody12 using Bb transition moments from the literature26. Expectation ring current shifts were obtained using three different algorithms: that used in version 4.1 of the SHIFTS program of Case27 and the Johnson-Bovey and Heigh-Mallion methods incorporated in MolMol. All ring current calculations are the mean over the entire accepted NMR structure ensemble.
Results
Some of the data which eventually led to the design of the present series of microproteins appears in another research account14; these include some seminal spectroscopic observations that served to indicate that protein-like folding was increasingly observed with certain mutations. These results (summarized in Table 1) are given here as they relate to the microprotein design process. Our starting point (entry #1, Table I) is, like chignolin, a turn flanked, and stabilized by an EtF aryl/aryl interaction.11 In the case of W/W interactions, the EtF interaction produces an intense CD exciton couplet (with a ca. 228 nm maximum), and notable upfield shifts for the Hε3 and Hβ2 shifts of the ‘edge’ Trp. For typical WIZGKW loop hairpins (entry #2, provides an example), the N-terminal (S-2)10c Trp is the edge residue. When the 6-residue turn of peptide #1 was replaced by a 4-residue INGK turn (entry #3), the fold population decreased and the Trp Hε3 shifts indicated that there was some ambiguity regarding which Trp took on the edge position in the EtF W/W interaction. Reasoning that this ambiguity could be contributing to the decreased fold population, we looked for mutational effects that would restore a bias toward a unique W/W cluster conformation. Introduction of a Thr residue at the N-terminus (entry #5), decreased the hairpin fold population and placed the S-2 Trp as the dominant edge residue. In contrast, stabilizing the turn (by an N3 → D-Pro mutation, entry #4) or placing a Thr at the C-terminus (entry #6) increased the hairpin fold population and changed the Hε3 CSDs so as to indicate that the S+2 Trp (at the C-terminus of the turn) was the edge residue in the W/W interaction, a flipped ‘FtE’ interaction.11
Table 1.
Effects of Termini Changes on the NMR Structuring Diagnostics of –W-loop-W- Peptides.
| # | Sequence | Tm (°C)a | fFb | CSD (ppm) | ||
|---|---|---|---|---|---|---|
| 228 nm CD melt | <frCSD> | S-2 W Hε3c | S+2 W Hε3c | S+4 HNE | ||
| 1 | Ac-WNPATGKW-NH2 | ~0.45 (~0.48 CD)c | −0.873 | −0.179 | n. a. | |
| 2 | KAVW-INGK-WTVE | ~65 | ~0.83 | −1.922d | −0.280 | +0.55 |
| 3 | Ac-W-INGK-W-NH2 | ~5 | ≤0.45 (~.42 CD)c | −0.378 | −0.738 | n. a. |
| 4 | Ac-W-IpGK-W-NH2 | ≥ 45 | .80 ± 0.05 | −0.26 | −1.64 | n. a. |
| 5 | Ac-TW-INGK-W-NH2 | ≤0.30 (~.1 CD) | −0.570 | −0.384 | n. a. | |
| 6 | Ac-W-INGK-WT-NH2 | ~41 | 0.76 ± 0.04 | −0.380 | −1.680 | −2.7c |
| 7 | Ac-W-INGK-WTG-NH2 | ~38 | 0.77 ± 0.04 | −0.379 | −1.556 | −2.71 |
| 8 | W-INGK-WTG | 0.15 ± 0.10 | −0.052 | −0.108 | −1.11 | |
| 9 | W-INGK-WTG-NH2 | 0.09 ± 0.09 | −0.017 | −0.067 | −1.17 | |
| 10 | Ac-WTG-NH2 | −1.26 | ||||
| 11 | Ac-W-IpGK-WTG-NH2 | ≥ 65 | 0.94 ± 0.03 | −0.25 | −1.87 | −3.06 |
| 12 | Ac-W-IpnK-WTG-NH2 | ≥ 68 | < 0.97 > | −0.31 | −1.93 | −3.20 e |
These Tm estimates assume that 100% folded [θ]228 value is temperature independent and, with the exception of entries 2, 4 and 12, are derived from Figure S3 of reference 14. Variation in the 100%-folded value with sequence changesb,c, adds additional uncertainty to these estimates.
The reported <frCSD> values are the average (and standard error) of the 280K fF values derived from strand backbone HN and Hα sites displaying moderate to large CSDs (W1 Hα&HN, I2 Hα&HN, K5 Hα, W6 Hα&HN) assuming that Ac-WIPGKWTG-NH2 represents zero folding and that the best folded species (entry #12, Ac-WIpnKWTG-NH2) represents fF = 0.97. CD measures of fF were based on the magnitude of the 228 nm maximum of the W/W exciton couplet; the 100%-folded value calculated for entry #2 is, in Molar (not residue-Molar) terms, +446,000°14. The 100% [θ]228 value for entry #12 is +303,000°; an increase (to +418,000° at 278K) was observed for the optimized construct (Pr-WIpGIWTGPS) reported herein.
The largest [θ]228 value observed for prior-WNPATGKW- species is +434,000°,10c the CD folding estimate in entry #2 is based on this value. Other analogs with this W-loop-W motif have 100%-folded [θ]228 values as small as +380,000°. The maximum 100% value observed for W-(S/I)-(N/p)-GK-W species was +490,000°.10c Recent studies14, have documented both WIpGKW- and WINGKW-containing species in which the 100%-folded values are ≤ +300,000° (calibrated based on NMR measures of folding); it appears that magnitude of the exciton couplet can vary significantly.
CSDs for the shielded S±2 Trp Hε3 as large as −2.3 ppm have been observed for longer peptide hairpins incorporating the …W-turn-W … sequence (“turn”= NPATGK or INGK).10c,14 Variations in the preferred W/W interaction geometry can give rise to slightly different 100% folded values.14
The largest upfield shift observed for the Gly HN in prior WTG-NH2 terminated peptides was 3.42 ppm.14 In the present study, one construct shows a 3.91 ppm upfield shift under the same conditions (20 vol-% glycol at 270K.)
The C-terminal addition of a threonine (#6) also resulted in a new NMR structuring diagnostic. A large upfield CSDs for the entgegen amide proton was observed for the C-terminal CONH2 group. Stability was maintained on the C-terminal addition of glycine (though not for an Ala or Pro addition); the extreme upfield shift was now seen for the G8 amide proton (#7) and a well-resolved NMR structure was determined for the resulting 8-residue peptide.14 The NMR ensemble placed the Thr sidechain OH and the acetyl carbonyl in the geometry expected for an H-bonding interaction. Gly8 clearly plays an important role, a G8A mutation produced a peptide which aggregated rather than folded: we could obtain neither NMR nor CD data (at 30 μM) for the WTA-NH2 terminated species.
At this point we had 6–8 residue peptides displaying Tm’s as high as 45 °C. The importance of the N-terminal acetyl function for fold stabilization28 was established by examining the uncapped (#8) and deacetylated (#9) analogs. Both modifications reduced the values of the Gly8 HN shift to those observed for Ac-WTG-NH2. Turn optimization (entries #11 and #12) increased the diagnostic structuring shifts and increased the Tm to ≥65 °C. At the stage of the most stable construct of this series, Ac-WIpnKWTG-NH2, an additional feature, unprecedented for a peptide and even rare for proteins, appeared in the NMR spectrum: the Thr7 sidechain hydroxyl was observed at pH 7 in water at 280K, with 50 mM of exchange-catalyzing phosphate buffer29, disappearing due to exchange at higher temperatures. In further attempts to optimize this small protein-like folding motif, we looked for mutations which would: increase the S+2 Hε3 and G8HN CSDs (particularly at higher temperatures indicating an increase in Tm), extend the temperature range over which the T7-OH was observed, and retain the circa 1.5 ppm downfield structuring shift at Ile2 HN which reflects the cross-strand H-bond in the hairpin. The Tm’s observed in CD melts were also monitored. This process has yielded peptide sequences with hyperstable folds.
The C-terminus of Ac-WIpGKWTG-NH2 was amenable to mutation. The terminal NH2 was replaced by a Pro-Ser dipeptide unit, to mimic a YTGPS sequence known to adopt a local aryl-amide interaction in BPTI sequence fragments16a The G8 HN CSD increased as did the exchange protection at the threonine hydroxyl proton, which remains visible by 2-D NMR at 300K.
We had initially placed a lysine at position 5 for the sake of consistency with previously studied turn sequences, and to guarantee solubility; but hydrophobic burial and a polar, negatively charged C-terminus negated any potential benefit due to a positively charged residue at position 5. In the case of (Ac/Pr)-WI(N/p)GKWTGPS, replacement of K5 with Leu and Ile resulted in enhanced stability, due to hydrophobic packing and/or a propensity to adopt a local extended β-strand structure (see Supporting Information). The Ile appeared to be optimal, Ac-WINGIWTGPS (employing the “natural”, but le ss optimal, NG turn sequence) was seen to have CSDs indicative of a ≥90% fold population at 280K and, more tellingly, a visible T7 hydroxyl proton signal - a feature that had, previously, only been observed for species with D-Pro-Xaa units at the turn.
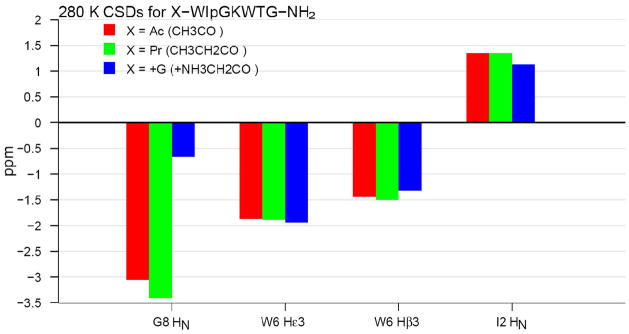
Mutations at the N-terminus were also examined. A selection of these appears in Figure 1. Significant fold improvement was noted upon replacing the acetyl group with a propanoyl. We attribute this to improved hydrophobic packing between the termini. It was clear that the propanoyl group was included in the structured motif: the βCH2 group of the propanoyl group appeared as two distinct signals in the NMR with a diastereotopic shift difference of 0.38 ppm; this implies significant restriction in the C1-C2 dihedral angle of the propanoyl group due to stereospecific hydrophobic interactions. To distinguish between hydrophobic and electronic effects, we also prepared a polar isostere with CH3CH2 replaced by +NH3CH2 (glycine), see Figure 1.
Figure 1.
Tracking the changes in chemical shift structuring diagnostics through N-terminal mutations.
The polar N-terminal glycines had little effect on the geometry (and presumably the extent of folding) at the FtE W/W interaction; although the diminished upfield shift at the cross-strand H-bonded I2HN may indicate a somewhat weaker H-bond. In contrast to these minor effects; the extreme upfield shift at G8HN completely disappeared: the observed CSD of − 0.66 ppm is in fact smaller than that observed for Ac-WTG-NH2 (see Table 1). Minor changes to one termini which affect CSDs at the opposite end of the polypeptide chain qualify as obvious bellwethers of protein-like structuring.30
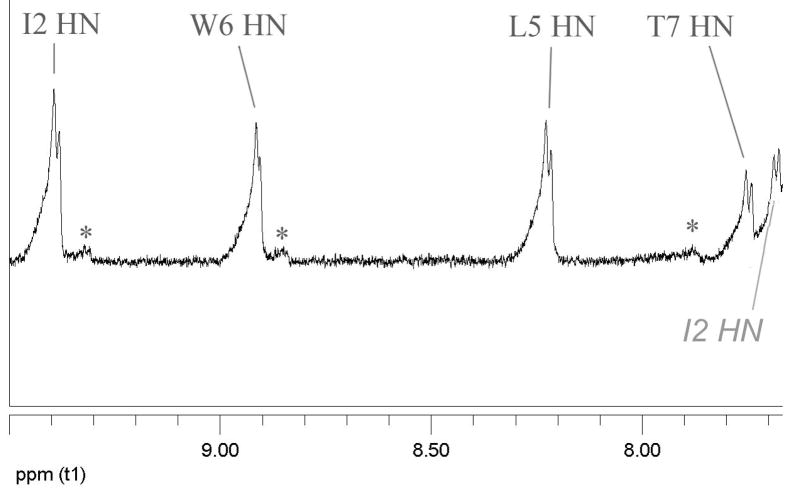
Combining the N-terminal propanoyl with the WTGPS C-terminus and a K5L mutation, yielded a hyperstable species (Pr-WIpGLWTGPS). Virtually no CSD melting was observed from 280 to 300K and the CD exciton couplet melt (228 nm) indicated a Tm in excess of 70°C. At this level of stability, fold population estimates based on CSDs are unreliable: there is no sound basis for determining whether a system is, for example, 94 or 98% folded. We turned to amide H/D exchange experiments21,25 at pD 3.84 and 4.74 for this purpose. Three backbone amides (I2, L5, G8) were highly protected from exchange (Figure 2), and displayed protection factors ≥45 at 280K (see Supporting Information) indicative of a fold population of circa 0.98. Validating the study, the W1, G4, and S10 amides exchanged at the same rate in the folded and reference peptides: protection factors 1.0, 2.6, and 0.96, respectively. Thr7 was modestly protected (17-fold). Under the more acidic conditions, W6 displayed a protection factor of 26 at pD 3.84, reflecting H-bonding to the Ile5 carbonyl rather than sequestration of Trp6 HN.
Figure 2.
Two time points in the D2O exchange (pD = 3.84) of a mixture of Pr-WIpGLWTGPS and its unfolded p3P mutant: A, 1.16 h; B, 15.6 h. Resonances of the unfolded control are labeled below the trace in panel A; labels above the trace are for the folded peptide; “x” denotes the location of resonances due to the cis-Gp amide species which is also folded. The Gly8 HN appearing at 4.57 ppm, not on the spectral segment shown here (see Supporting Information), is protected to comparable degree.
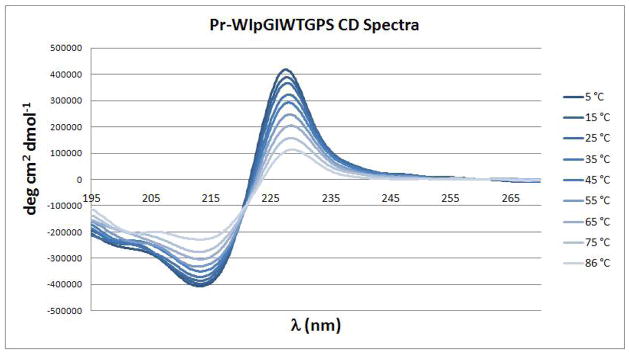
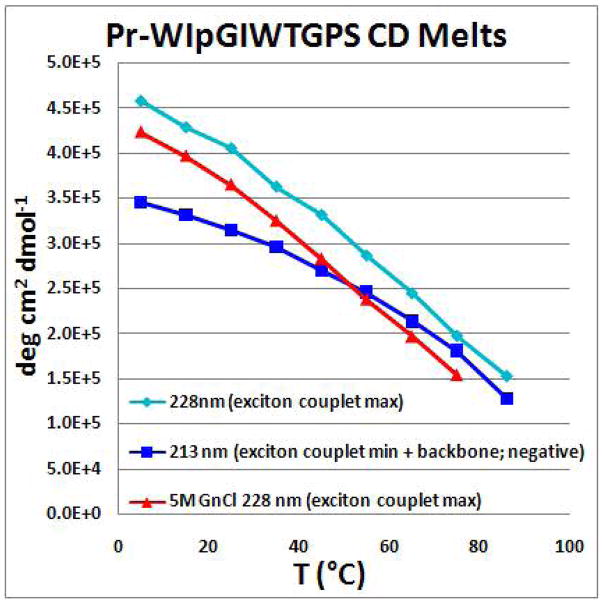
The species with an Ile at position 5 was also prepared. The HN exchange protection experiment at pD = 6.15 (Supporting Information) confirmed the presence of cross-strand hairpin H-bonds with protection factors above 50 (PF = 51 for I2HN and 91 for I5HN). CD and NMR shift melts also suggest some additional improvement in fold stability. The NMR melt comparison appears in the Supporting Information. The changes in the CD spectrum with temperature appear in Figure 3. Melting monitored at the two extrema of the exciton couplet gave somewhat different apparent Tm values (Figure 4), after factoring in the effects of the temperature dependence of [θ]228 for the folded state, a Tm of 80–85 °C represents our best estimate. Although we cannot ascertain whether the amplitude of the exciton couplet changes with Gdm Cl concentration, the CD melt observed in this denaturing medium suggests that this microprotein is very resistant to chemical denaturation, with a ≤ 11 °C drop in Tm at 5M Gdm Cl. Chignolin is also reported9 to retain its exciton couplet CD spectrum at high Gdm Cl concentrations.
Figure 3.
The CD melt of a Trp/Trp exciton couplet: spectra of Pr-WIpGIWTGPS were recorded in 10°C intervals.
Figure 4.
CD melting data for Pr-WIpGIWTGPS, adjusted (by subtracting the CD of Ac-WIPGKWTG-NH2 serving as an unfolded control) so that the zero degree line represents 0% folding, see Supporting Information. The actual melting temperature is expected to be in excess of 85°C, considering baseline effects. In the case of the plot of 213 nm data, a Tm > 80°C is required based on the second derivative method9c for defining Tm.
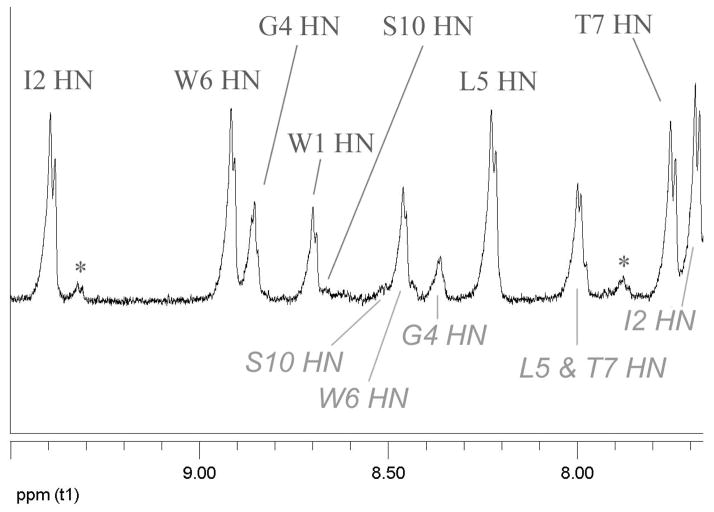
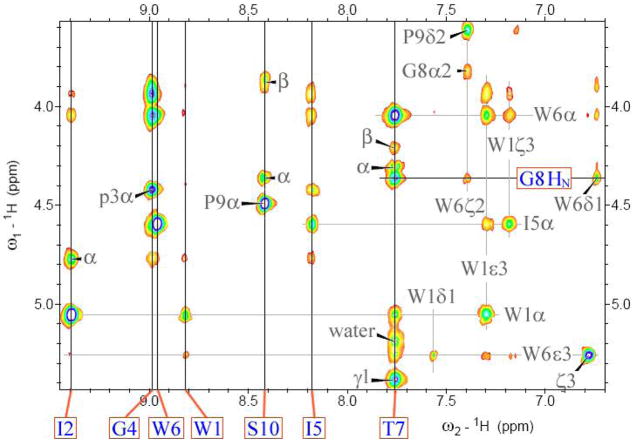
Additional data, including the generation of an NMR structure ensemble, was obtained for Pr-WIpGIWTGPS. The NOESY spectrum employed for this (see Figure 5) also serves to illustrate the remarkable chemical shifts and long- and medium-range connectivities observed for this motif. Of particular note are the extreme upfield locations of G8HN (4.36 ppm) and W6Hε3 (5.26 ppm). The chemical shift deviations, and melting behavior, observed during the final stages of fold optimization are compared to those observed in earlier –WTG-containing peptides in Figure 7 (vide infra) with the complete assignments for Pr-WIpG(I/L)WTGPS and Ac-WIpGKWTGPS appearing in the Supporting Information. There is a web of NOE connectivities between the two Trp residues which serves to define the geometry of the Trp/Trp interaction, the hairpin register is established by intense W1α/T7HN I5HN/I2HN and I5α/W1ζ3 cross-peaks, and the Me group of the propanoyl cap displays numerous NOEs to sites in residues near the C-terminus.
Figure 5.
The HN/Hα NOESY spectrum of Pr-WIpGIWTGPS at 270K. The HN resonance positions are labeled and indicated by solid lines; other resonance positions are shown by gray lines. Peaks due to the minor cis-GP isomer and water suppression artifacts have been deleted to clarify the assignment; the complete spectrum appears in the Supporting Information.
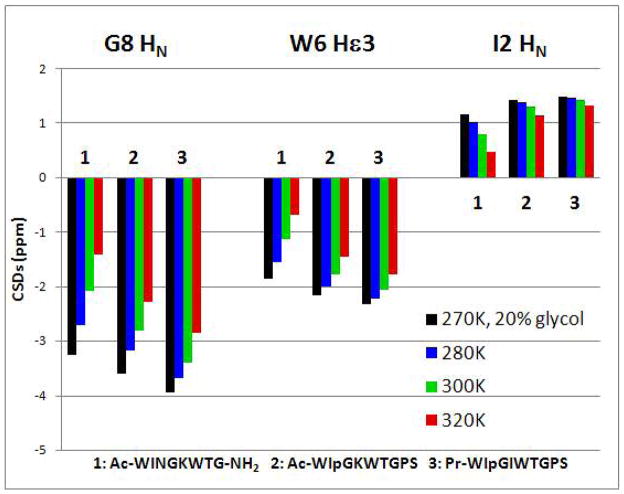
Figure 7.
Tracking three large structuring shifts and their melting behavior during fold optimization.
The NOESY spectrum was recorded at lower temperature in 20 vol-% glycol to increase the NOE growth rate by lowering the viscosity. An NMR structure ensemble calculated exclusively from NOE distance constraints, 148 inter-proton distances of which 48 are medium to long range i/i+n (n ≥ 4), appears in Figure 6.
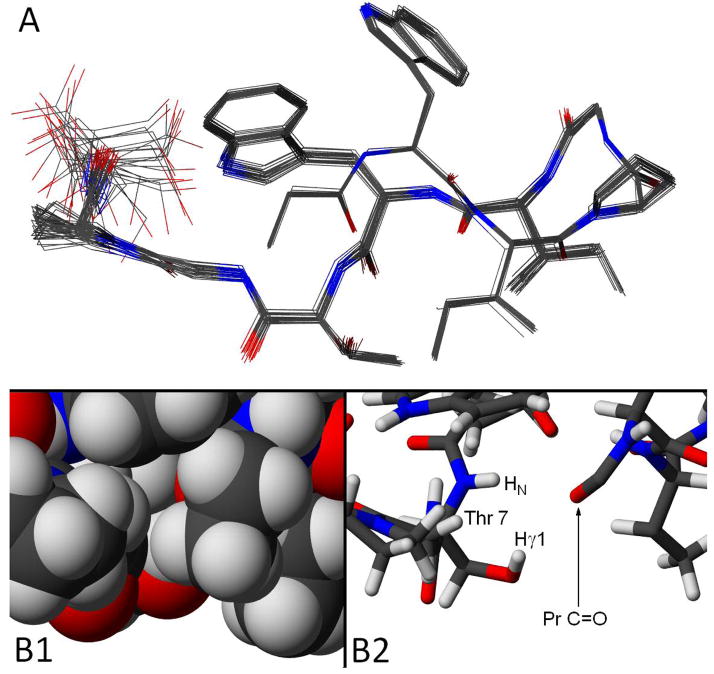
Figure 6.
Views of the NMR structure ensemble (backbone RMSD = 0.30 ± 0.13 Å) generated for Pr-WIpGIWTGPS. Panel A presents a view in which the twisted hairpin conformation is apparent; all heavy atoms are shown. This view also shows the I2/I5 alignment. Panel B shows two views of the Pr-C=O/Thr7 relationship in a representative structure. In the bonds-only view (right) the Me group of Thr, as well as the ethyl groups of Ile5 and the propanoyl cap, have been deleted. In the space-filling (CPK, left) view of the same region, these three alkyl groups (which effectively sequester the Thr7 OH proton from water) are included.
From Figure 6 it is apparent that the only remaining area of disorder in this microprotein is the sidechain of the C-terminal serine: the intra-ensemble heavy atom RMSD drops from 0.59 ± 0.20 to 0.17 ± 0.11 Å when Ser10 is excluded from the fit. The structure is another example of a hairpin stabilized by a turn-flanking Trp/Trp interaction. Prior studies14 suggest that FtE W/W interactions can occur with a variety of 2:2 and 2:4 WXXXXW turn geometries. In the present case, the turn region (φ/ψ) - D-Pro3 (+65 ± 1/+37.4 ± 0.5°), Gly4 (73 ± 1.6/12 ± 1°) - can be described as intermediate between type I′ and III′(two left-handed 310 helical sites). The orientation of the two indole rings is particularly well converged. The geometry in the ensemble was used to calculate the expectation exciton couplet using the method of Woody12 with indole transition moments taken from the literature26. The indole ring orientations from the NMR structure predict the sign of the observed CD couplet and its large magnitude. Chemical shifts were not employed in the generation of this structure ensemble; the indole ring orientations in the ensemble predict the following ring current shifts: the average calculated upfield ring current shifts, see Methods, (and observed CSDs; 270K) for G8HN, W6Hε3, W6Hβ3, G8Hα3, and the propanoyl Me were: 2.5 (vs 3.9), 2.9 by SHIFT 4.1 (vs 2.3), 2.2 (vs 1.6), 0.9 (vs 1.1), and 0.66 (vs 0.78 ppm). Since slight differences in the orientation of aryl rings translate to large changes in anisotropy effects, even for protein structures calculated ring current effects do not coincide perfectly with the observed shifts. In the case of G8HN, the greater shielding observed could also represent an additional contribution due to the absence (versus statistical coil circumstances) of inter-molecular H-bonding with the medium.
Discussion
The features of our microprotein constructs include: 1) a threonine with the hydroxyl H-bonding to an essential N-capping carbonyl, 2) cross-strand H-bonding indicating a 2:2 hairpin conformation, 3) a Gly-HN to indole ring H-bond, 4) a common single geometry for a FtE Trp1/Trp6 indole ring interaction, and 5) fold stability and rigidity improvements associated with the optimization of a hydrophobic core that include a turn residue as well as residues at both chain termini. These are discussed, in turn, below.
We first became aware of a stabilizing role for the threonine at position 7 when a T7→A mutation in the shorter “discovery” system (Ac-WINGKWT-NH2) resulted in significant destabilization (the CD melt indicated a 15°C drop in the Tm). Initially this seemed counterintuitive, because the near-zero ψ angle of residue 7 (as seen in the NMR structure ensemble of Ac-WINGKWTG-NH2)31 seemed better suited for alanine than β-branched threonine. The extreme upfield shift of one of the terminal amide hydrogens of Ac-WINGKWT-NH2 (CSD = −2.7 ppm) was greatly reduced in the A-NH2 terminated species (the larger of the two NH2 CSDs was −1.4 ppm). An explanation presented itself when a hydroxyl proton peak for the T7 sidechain appeared32 in the NMR spectra of more stable analogs, an indication of sequestration via H-bonding. The basis for this observation is clear in Figure 6, panel B. To probe this, we also examined mutations at Thr; the results appear in the Supporting Information. T7S, T7V, and T7Abu (α-aminobutyric acid) mutations were examined; all species retained a highly folded hairpin structure but Thr was, indeed, optimal at this position based on the G8HN CSD.
The Pr-C=O to Thr NH and OH H-bonding seen in Figure 6B is reminiscent of features observed35 for Ser and Thr sidechains in helices. In our hairpin constructs, the T7HN was less well protected than other backbone amide sites (≤17-fold versus 45–91-fold at the more protected hairpin cross-strand H-bonds); we attribute this to the bifurcated H-bond to the Pr C=O shared with the hydroxyl proton. The close proximity of the Thr-OH and HN is clearly evidenced by a very intense NOESY peak. Despite its visibility by NMR, the intrinsic rate of exchange of the hydroxyl proton in both the unfolded (and folded) states would be much greater than that of an amide, and thus one can envision a saturation transfer mechanism (H2O → Thr-OH followed by the NOE transfer to T7HN) as likely in the H2O medium and posit this as the source of the T7HN/H2O NOESY cross-peak in Figure 5.
The NMR ensemble revealed three tight cross-strand H-bonds in the β-hairpin with geometries like those observed in antiparallel β sheets in proteins. These provided our best estimates of ΔGU for our constructs: protection factors ≥ 45 were observed for both optimized constructs, implying ΔGU280 ≥ 8.9 kJ/mol. To our knowledge, this is the first observation of protein-like stability for a hairpin peptide.36 The I2 and I/L5 amide protons were protected to comparable extents, suggesting a 2:2 turn where the inner H-bond (I/L5-NH→O=C-I2) is largely present. This stands in contrast to many designed hairpin with IpGK and INGK turns in which the Lys NH appears upfield due to removal from solvent contact without compensation by a strong intra-molecular H-bond.18c The other intriguing H-bonding interaction is between the W6 indole ring and the G8 amide HN. This interaction does lead to exchange protection, but protection factor quantitation failed due to overlap with Hα signals. The best measure of this interaction is the upfield ring current shifts at Gly8. How this and the other large CSDs change during the fold optimization sequence is shown in Figure 7.
The CSDs at the G8HN and W6Hε3 sites are largely and completely, respectively, due to ring current effects while that at I2HN is due to H-bond formation and the alignment of the two β strands. In the case of Ac-WINGKWTG-NH2, a comparable loss of structuring shifts at all three sites occurs on warming. This represents melting, an increasing population of the statistical coil state at the higher temperatures. As the fold stability increases, melting is less evident, but temperature dependent loss of ring current shifts is still present due to increasing indole ring plane motions within the folded state (vide infra).
The propanoyl N-cap that provides the electron pair for the sequence remote H-bonds has a single geometry in our structure ensemble, a fully extended ‘ψ’ value (−159.8 ± 0.3°), provides additional (vs an acetyl group) exchange protection at the T7 sidechain hydroxyl, and also has profound effect (Figure 1) on the G8HN/W6 interaction (Figure 6, panel B). Based on NMR measures of β-hairpin strand alignment, deletion of the cap results in substantial fold destabilization, ΔΔGU = −6 ± 1 kJ/mol. Other structuring shifts associated with the fold are also affected. In the (Ac)-WINGKWTG-NH2 system, the Trp/Trp interaction is almost entirely abolished: the CD exciton couplet is reduced to near zero, with the W6 Hε3 CSD reduced from 1.556 to 0.067 ppm on deacetylation. The replacement of the propanoyl cap in Pr-WIpGIWTGPS by an isosteric but charged Gly unit has only a small effect of the hairpin fold population and the indole/indole interaction, but reduces the G8HN shielding to less than that observed in unfolded controls. The structure of Pr-WIpGIWTGPS suggests a rationale; we posit a flip of T7/G8 amide function so that Gly1 ammonium ion interacts with the T7 carbonyl. The Gly1 and Pr groups themselves appear to have comparable geometries in Pr-WIpGIWTGPS and GWIpGIWTGPS as both display a diastereotopic splitting of circa 0.38 ppm for the N-terminal -CH2-C=O group. The T7/G8 amide plane rotation would result in a greatly (but not completely) reduced ring current CSD, as well as additional solvent exposure for G8HN. Furthermore, shielding by the positively charged amine itself may reduce the CSD of G8HN, if the two remain close.
The large CSDs produced by the fixed geometries of the two indole rings vary slightly with temperature at a nearly-fixed fold population. For example, Pr-WIpGIWTGPS is expected to be ~98% folded at 280K in H2O, based on amide exchange data. At 270K in 20% d6 glycol, the diagnostic CSDs reflecting cross-strand H-bonding change very little, as would be expected for systems with fold populations varying between 97% and 99%. However, ring current CSDs increased an average of 5% (see Figure 7), suggesting that reduced librations of the indole rings at lower temperatures may result in greater shielding of nearby protons. An even greater temperature effect is seen for the CD exciton couplet, which is expected to be more sensitive to the precise orientation of the indole chromophores, and thus temperature-dependent changes in extent of “wiggling” of the two ring planes.
Due to the complex web of interactions across the termini of our microprotein constructs, it is difficult to quantitate the fold-stabilization associated with the indole/indole FtE interaction. FtE aryl/aryl interactions are common in proteins12, particularly for Trp/Trp and Trp/Tyr pairings. In the present case, the C-terminal WXGP sequence may be important since it serves to restrict the C-terminal Trp sidechain geometry. Our best estimate of the Trp/Trp interaction energy is from the turn Asn-HN CSD estimates of the fold destabilization associated with a W1T mutation in Ac-WINGKWTG-NH2. These imply a 5 kJ/mol fold stabilization (Supporting Information). The 2.3 kJ/mol stabilization seen in a mutational cycle in a WW domain2c likely represent the lower end of the Trp/Trp FtE interaction energy. Significant stabilization by a cross-strand Trp/Trp pair may require an additional interaction, with a turn residue, cationic sidechain moiety or H-bonding group that relies on the π base character of the indole ring. It should also be noted that theoretical studies13 suggest that FtE Trp/Trp interactions are not predominantly hydrophobic in character, and that specific aryl/aryl FtE interactions (including W/Y) have been observed38 in an organic solvent. The substantial π basicity of the indole ring of Trp, a key feature in both the FtE indole/indole interaction and the amide-HN to indole H-bond, has been the subject of a number of studies.39
Optimized sidechain packing interactions and the shielding of H-bonds from solvent exposure provide an explanation for the increase in stability observed in going from Ac-WIpGKWTG-NH2 to Pr-WIpGIWTGPS (the melting points differ by 15°C). Ac→Pr and K5→I/L mutations afforded improved ‘hydrophobic’ packing (with W6 and I2, respectively) as well as additional burial of the bifurcated Thr to Ac/Pr H-bonds. The addition of P9, in addition to reducing the χ1/χ2 flexibility at W6, may have provided some sequestration for the G8HN to indole H-bond. We are disinclined to describe these packing interactions as hydrophobic cluster formation since a significant positive ΔCpU is the hallmark of hydrophobically-driven folding in proteins. With our optimized constructs displaying Tm values of circa 80°C and yet having fold population of >97% at 7°C, the ΔCpU values must be less positive than +430 JK−1/mol, considerably smaller than values observed for many β-hairpin structures10c. We suspect that H-bond sequestration, associated with the burial of polar moieties (which would have a negative contribution to ΔCpU), is a more important factor for our constructs. This can be seen in the views of the H-bonding to the propanoyl-C=O appearing in panel B of Figure 6: the ethyl groups of the propanoyl group and isoleucine effectively block solvent access to this H-bonding network. We made no assumptions regarding H-bonding in deriving our NMR structure ensemble, rather it is supported by the wealth of NOE data. Since we have suggested fold-stabilizing roles for the methyl group of the propanoyl cap, a few of these NOEs warrant specific mention. The constraints for MD included ten distances involving the propanoyl Me. Of these the five shortest inter-residue distances (< 3.6 Å) are to: T7Hγ1(the OH) < G8Hα3 < W6Hζ3 < G8HN < T7HN. The threonine methyl also provides shielding from solvent, and this, combined with improved hydrophobic contact with I2, serves to rationalize the fold improvement for the T7 versus S7 analog. The propanoyl methyl group blocks solvent access to the G8HN→ indole H-bond. Furthermore, the structure suggests that W6Hε1 is partly buried; a PF of ~30 corroborates this. As more than just the cross-strand H-bonds are sequestered from solvent, the burial of polar moieties exceeds what would be expected of a β hairpin of this size.
Finally, what are the implications of a successful rational design of ≤10 residue constructs that have fold stabilities that are comparable to those of typical small protein domains? It lowers size limit for protein-like folding and suggests that quite small peptides could serve as scaffolds for the stereospecific display of pharmacophores in target-directed drug discovery. It illustrates the merits of monitoring chemical shifts during mutational fold optimization attempts, as maximizing CSDs corresponds to improving fold stability or rigidity. For small fast-folding systems, the CSD increases predominantly reflect fold population changes, but even for fully-folded system (or systems in slow exchange) increases in CSDs reflect fold optimization (in the latter case, a reduction in fluxionality.) With many small folds based on stabilizing hydrophobic interactions, positive, and relatively large, ΔCpU terms limit the maximum extent of folding that can be achieved in water at low temperatures. W/W interactions, which are not exclusively hydrophobic in character, and H-bonding optimization provided a means to avoid this limitation. The key to the optimization of folding in small constructs is likely to be the incorporation of multiple synergistic long-range interactions. In this regard, we are currently exploring the potential of Ac-W · · · WTGP termini as fold enhancing caps in other systems.
Supplementary Material
Acknowledgments
Dedicated to Professor E. J. Corey on the occasion of his 80th Birthday. Initial hairpin optimization studies were supported by a grant from the NSF (CHE0650318); the microprotein optimization work was accomplished under NIH support (GM059658). B. Kier acknowledges support as a predoctoral trainee of the Molecular Biophysics Training Program at U.W. (NIH grant 5 T32 GM008268). Purchase of the 600 MHz cryoprobe NMR used in the present study was facilitated by grants from the NSF (DBI 0439063) and NIH.
Footnotes
Supporting Information Available: Supporting information (5 figures, 3 tables, and textual comments) contains additional mutational studies, the distance constraints employed and structure statistics for the NMR ensemble generation, and full chemical shift assignments for 3 peptide sequences. The material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.Kubelka J, Hofrichter J, Eaton WA. Curr Opin Strct Biol. 2004;14:76–88. doi: 10.1016/j.sbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 2.(a) Struthers MD, Cheng RP, Imperiali B. Science. 1996;271:342–345. doi: 10.1126/science.271.5247.342. [DOI] [PubMed] [Google Scholar]; Dahiyat BI, Mayo SL. Science. 1997;278:82–7. doi: 10.1126/science.278.5335.82. [DOI] [PubMed] [Google Scholar]; Horng JC, Moroz V, Raleigh DP. J Mol Biol. 2003;326:1261–1270. doi: 10.1016/s0022-2836(03)00028-7. [DOI] [PubMed] [Google Scholar]; Anil B, Craig-Schapiro R, Raleigh DP. J Am Chem Soc. 2006;128:3144–3145. doi: 10.1021/ja057874b. [DOI] [PubMed] [Google Scholar]; Jager M, Zhang Y, Bieschke J, Nguyen H, Dendle M, Bowman ME, Noel JP, Gruebele M, Kelly JW. Proc Natl Acad Sci U S A. 2006;103:10648–10653. doi: 10.1073/pnas.0600511103. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Tang Y, Goger MJ, Raleigh DP. Biochemistry. 2006;45:6940–6946. doi: 10.1021/bi052484n. [DOI] [PubMed] [Google Scholar]; (c) Jager M, Dendle M, Fuller AA, Kelly JW. Protein Sci. 2007;16:2306–2313. doi: 10.1110/ps.072904107. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jager M, Nguyen H, Dendle M, Gruebele M, Kelly JW. Protein Sci. 2007;16:1495–1501. doi: 10.1110/ps.072775507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The term “miniprotein” has typically been applied to 20 – 45 residue polypeptides that form a stable fold; it has mostly been used for designed systems rather than naturally occurring ones. This term dates back to at least 1996 (Drug Design & Discovery), with a PNAS citation in 1998, and was in common usage prior to our application of it to the Trp-cage fold4. “Microprotein” appears to have two uses in the literature; an older one for specific urinary excretion products and, starting in 2004 (Current Opinions in Biotechnology), as a term for small natural cystine-knot proteins in the plant-cyclotide and conotoxin areas. Herein, following the usual view that “micro” is smaller then “mini”, we use “microprotein” to designated folds with protein-like stability that are much smaller than typical miniproteins.
- 4.Neidigh JW, Fesinmeyer RM, Andersen NH. Nature Struc Biol. 2002;9:425–430. doi: 10.1038/nsb798. [DOI] [PubMed] [Google Scholar]
- 5.Barua B, Lin JC, Williams VD, Neidigh JW, Kummler P, Andersen NH. PEDS. 2008;21:171–185. doi: 10.1093/protein/gzm082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simmerling C, Strockbine B, Roitberg AE. J Am Chem Soc. 2002;124:11258–11259. doi: 10.1021/ja0273851. [DOI] [PubMed] [Google Scholar]; Snow CD, Zagrovic B, Pande VS. J Am Chem Soc. 2002;124:14548–14549. doi: 10.1021/ja028604l. [DOI] [PubMed] [Google Scholar]
- 7.(a) Searle MS, Ciani B. Curr Opin Struct Biol. 2004;14:458–464. doi: 10.1016/j.sbi.2004.06.001. [DOI] [PubMed] [Google Scholar]; (b) Mok KH, Kuhn LT, Goez M, Day IJ, Lin JC, Andersen NH, Hore PJ. Nature. 2007;447:106–109. doi: 10.1038/nature05728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Satoh D, Shimizu K, Nakamura S, Terada T. FEBS Lett. 2006;580:3422–3426. doi: 10.1016/j.febslet.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 9.Honda S, Yamasaki K, Sawada Y, Morii H. Structure. 2004;12:1507–1518. doi: 10.1016/j.str.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 10.(a) Favre M, Moehle K, Jiang L, Pfeiffer B, Robinson JA. J Am Chem Soc. 1999;121:2679–2685. [Google Scholar]; (b) Cochran AG, Skelton NJ, Starovasnik MA. Proc Natl Acad Sci U S A. 2001;98:5578–5583. doi: 10.1073/pnas.091100898. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Andersen NH, Olsen KA, Fesinmeyer RM, Tan X, Hudson FM, Eidenschink LA, Farazi SR. J Am Chem Soc. 2006;128:6101–6110. doi: 10.1021/ja054971w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aryl/aryl ring interactions are classified as parallel stacking or ‘T’ geometry interactions which are designated as edge-to-face (EtF) interactions. EtF aromatic/aromatic interactions have been recognized as contributors to the CD spectra of proteins since at least 199412. Although less common as specific cross-strand interactions in β sheets, this interaction has been seen in hairpin models9,10 and has become a strategy for hairpin stabilization.10c In the case of Trp/Trp interactions, quantum mechanical calculations13 indicate a strong preference for the EtF geometry and that the interactions is a multipole interaction rather than a classic hydrophobic interaction. For aryl/aryl interactions in hairpins, we make a further distinction based on the location, at the N-terminus versus C-terminus of the turn, of the aromatic ring that presents its edge to the face of the cross-strand aromatic. The interaction is “EtF” when the edge aromatic is located in the N-terminal strand and as “FtE” when the edge aromatic is in the C-terminal strand.
- 12.Grishina IA, Woody RW. Faraday Discuss. 1994;99:245–262. doi: 10.1039/fd9949900245. [DOI] [PubMed] [Google Scholar]
- 13.Guvench O, Brooks CL., III J Am Chem Soc. 2005;127:4668–4674. doi: 10.1021/ja043492e. [DOI] [PubMed] [Google Scholar]
- 14.Eidenschinck LA, Kier BL, Huggins K, Andersen NH. Proteins: Structure, Function & Genomics. 2008 in press. [Google Scholar]
- 15.Fesinmeyer RM, Hudson FM, Andersen NH. J Am Chem Soc. 2004;126:7238–7243. doi: 10.1021/ja0379520. [DOI] [PubMed] [Google Scholar]
- 16.(a) Kemmink J, Creighton TE. J Mol Biol. 1993;234:861–878. doi: 10.1006/jmbi.1993.1631. [DOI] [PubMed] [Google Scholar]; Kemmink J, van Mierlo CPM, Scheek RM, Creighton TE. J Mol Biol. 1993;230:312–322. doi: 10.1006/jmbi.1993.1144. [DOI] [PubMed] [Google Scholar]; (b) Noelting B, Golbik R, Soler-Gonzalez AS, Fersht AR. Biochemistry. 1997;36:9899–9905. doi: 10.1021/bi962879u. [DOI] [PubMed] [Google Scholar]; (c) Crowhurst KA, Tollinger M, Forman-Kay JD. J Mol Biol. 2002;322:163–178. doi: 10.1016/s0022-2836(02)00741-6. [DOI] [PubMed] [Google Scholar]; Robic S, Guzman-Casado M, Sanchez-Ruiz JM, Marqusee S. Proc Natl Acad Sci U S A. 2003;100:11345–11349. doi: 10.1073/pnas.1635051100. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) McCarney ER, Kohn JE, Plaxco KW. Crit Rev Biochem Mol Biol. 2005;40:181–189. doi: 10.1080/10409230591008143. [DOI] [PubMed] [Google Scholar]; (e) Li Y, Horng JC, Raleigh DP. Biochemistry. 2006;45:8499–8506. doi: 10.1021/bi052534o. [DOI] [PubMed] [Google Scholar]
- 17.(a) Eaton WA. Proc Natl Acad Sci U S A. 1999;96:5897–5899. doi: 10.1073/pnas.96.11.5897. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Garcia-Mira MM, Sadqi M, Fischer N, Sanchez-Ruiz JM, Munoz V. Science. 2002;298:2191–2195. doi: 10.1126/science.1077809. [DOI] [PubMed] [Google Scholar]
- 18.(a) Andersen NH, Barua B, Fesinmeyer RM, Hudson FM, Lin J, Euser A, White G. Chemical shifts, the ultimate test of polypeptide folding cooperativity. Peptides; Proc. 27th Eur. Peptide Symp.; Sorrento, Italy. 2002; Sorrento, Italy: Edizioni Ziino, Napoli, Italy; 2002. pp. 824–825. [Google Scholar]; (b) Santiveri CM, Santoro J, Rico M, Jiménez MA. J Am Chem Soc. 2002;124:14903–14909. doi: 10.1021/ja0278537. [DOI] [PubMed] [Google Scholar]; (c) Fesinmeyer RM, Hudson FM, White GWN, Olsen KA, Euser A, Andersen NH. J Biomol NMR. 2005;33:213–231. doi: 10.1007/s10858-005-3731-7. [DOI] [PubMed] [Google Scholar]; (d) Ferguson N, Sharpe TD, Schartau PJ, Sato S, Allen MD, Johnson CM, Rutherford TJ, Fersht AR. J Mol Biol. 2005;352:427–446. doi: 10.1016/j.jmb.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 19.Olsen KA, Fesinmeyer RM, Stewart JM, Andersen NH. Proc Natl Acad Sci U S A. 2005;102:15483–15487. doi: 10.1073/pnas.0504392102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersen NH, Dyer RB, Fesinmeyer RM, Gai F, Liu Z, Neidigh JW, Tong H. J Am Chem Soc. 1999;121:9879–9880. [Google Scholar]; Dyer RB, Maness SJ, Franzen S, Fesinmeyer RM, Olsen KA, Andersen NH. Biochemistry. 2005;44:10406–10415. doi: 10.1021/bi050698z. [DOI] [PubMed] [Google Scholar]
- 21.Lin JC, Barua B, Andersen NH. J Am Chem Soc. 2004;126:13679–13684. doi: 10.1021/ja047265o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.(a) Ramirez-Alvarado M, Blanco FJ, Serrano L. Prot Sci. 2001;10:1381–1392. doi: 10.1110/ps.52901. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Huyghues-Despointes BM, Qu X, Tsai J, Scholtz JM. Proteins. 2006;63:1005–1017. doi: 10.1002/prot.20916. [DOI] [PubMed] [Google Scholar]; (c) Wei Y, Huyghues-Despointes BM, Tsai J, Scholtz JM. Proteins. 2007;69:285–296. doi: 10.1002/prot.21494. [DOI] [PubMed] [Google Scholar]
- 23.Piotto M, Saudek V, Sklenar V. J Biomol NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 24.Bax A, Davis DG. J Magn Reson. 1985;65:355–360. [Google Scholar]
- 25.Bai Y, Milne JS, Mayne L, Englander SW. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.(a) Woody RW. European Biophysics J. 1994;23:253–262. doi: 10.1007/BF00213575. [DOI] [PubMed] [Google Scholar]; (b) Albinsson B, Norden B. J Phys Chem. 1992;96:6204–6212. [Google Scholar]
- 27.Chemical shift calculations were made using the SHIFTS program of David Case, version 4.1- Xu XP, Case DA. J Biomol NMR. 2001;21:321–333. doi: 10.1023/a:1013324104681.In the case of ring current shifts these calculations were supplemented by results obtained using the CalShift function, using both Johnson-Bovey and Heigh-Mallion models, within MolMol ( Koradi R, Billeter M, Wüthrich K. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4.
- 28.One a priori rationale for the loss of hairpin stability upon decetylation is that the acetyl carbonyl could be a cross-strand H-bond acceptor for T7HN. In the present instance, the peptide in entry 8 (Table 1) might have been expected to display increased stability due to the favorable Coulombic effects at the chain termini.15,22 The statistical observation of increased β-strand propensity after an N-terminal ammonium ion ( Pal D, Chakrabarti P. Biopolymers. 2000;53:467–475. doi: 10.1002/(SICI)1097-0282(200005)53:6<467::AID-BIP3>3.0.CO;2-9. is also contrary to the observation of increased β-hairpin formation upon acetylation (Table 1, entry 7 versus 9).
- 29.Liepinsh E, Otting G, Wüthrich K. J Biomol NMR. 1992;2:447–465. doi: 10.1007/BF02192808. [DOI] [PubMed] [Google Scholar]
- 30.Other N-terminal comparisons included TWINGKWTG and Ac-TWINGKWTG-NH2 vs Ac-WINGKWTG-NH2 (similar stability, but with less specificity in the W/W interactions, and a greatly reduced C-terminal Gly HN CSD) and (Ac-)PWIpGLWTGPS vs Ac-WINGKWTG-NH2. (The Ac-P version appeared to be a somewhat less stable hairpin, though the WTG interaction at the opposite terminus remained.) Curiously, the Ac-P amide bond was ~50% cis. The fact that proline is even remotely tolerated at this position is remarkable, considering that it is, in principle, at an H-bonding position in a β sheet. The N-terminal Pro species without the acetyl cap was a more stable hairpin based on the magnitude of the CSDs for protons that are directed inward toward the aligned strand (especially. I2HN). However, as was also the case for the Gly1 analog with a free N-terminal amine – it lacked the large upfield shift at the G8 HN.
- 31.The φ;/ψ values observed for T7 in the ensemble shown in Figure 6 are −132 ± 5° and +23 ± 7°, which still place the ψ value in a range that is less favorable for β-branched residues.
- 32.Hydroxyl protons of Ser and Thr sidechains in peptides and proteins dissolved in water can only be seen by NMR with special experiments for water suppression29 (see also - Jahnke W, Kessler H. J Biomol NMR. 1994;4:735–740. doi: 10.1007/BF00404281. or when well buried in the hydrophobic core of a proteins5,33 Based on the intrinsic exchange rates for OH groups, fold-populations in excess of ~0.90 are required for the observation of an H-bonded Thr OH by NMR under typical aqueous solvent conditions using standard water suppression techniques. The intrinsic OH exchange rate, and thus the required fold population required for observation of the OH signal, increases with temperature. We have not observed this resonance in peptides less than ~85–90% folded based on CSD-measure of folding.
- 33.A search of the BMRB assignment data for proteins (http://www.bmrb.wisc.edu/ref_info/statsel.htm#17) reveals that only 3.3 % of Thr residues include a shift assignment for Hγ1 (the OH); for Ser this statistic drops to 1.9%. An early and recent instance in which Ser and Thr-OH observations were particularly noted in protein studies appear here – Hammen PK, Scholtz JM, Anderson JW, Waygood EB, Klevit RE. Prot Sci. 1995;4:936–944. doi: 10.1002/pro.5560040513.Canadillas JMP, Tidow H, Freund SMV, Rutherford TJ, Ang HC, Fersht AR. Proc Natl Acad Sci U S A. 2006;103:2109–2114. doi: 10.1073/pnas.0510941103. To our knowledge, with the exception of a Trp-cage miniprotein5, hevein, a tetra-disulfide 43-mer,34 is the smallest protein to display sidechain OH signals in its NMR spectra under the conditions routinely used for protein resonance assignment.
- 34.(a) Andersen NH, Cao B, Rodriguez A, Arreguin B. Biochemistry. 1993;32:1407–1422. doi: 10.1021/bi00057a004. [DOI] [PubMed] [Google Scholar]; Cao B. PhD thesis. University of Washington; 1993. NMR structural studies of bound peptides and hevein, a small protein. [Google Scholar]; (b) Asensio JL, Cañada FJ, Bruix M, Rodriguez-Romero A, Jiménez-Barbero J. Eur J Biochem. 1995;230:621–633. doi: 10.1111/j.1432-1033.1995.tb20604.x. [DOI] [PubMed] [Google Scholar]
- 35.Walsh STR, Cheng RP, Wright WW, Alonso DOV, Daggett V, Vanderkooi JM, DeGrado WF. Prot Sci. 2003;12:520–531. doi: 10.1110/ps.0223003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chemical shift and CD melts, in most cases, cannot distinguish between 90 and 99% folded at low temperature due to ΔCp effects.10b,18b–c,20 To our knowledge, the only quantitative H/D exchange study of β-strand sites in an isolated hairpin is of the oxidized and reduced forms of the BPTI core hairpin37; the largest protection factors were on the order of 4-fold with at most a 50% increase associated with disulfide closure of the hairpin termini. Chignolin was reported9 to show a reversing loop proton (G7HN) with a protection factor above 20 at 277K.
- 37.Carulla N, Woodward C, Barany G. Biochemistry. 2000;39:7927–7937. doi: 10.1021/bi992927l. [DOI] [PubMed] [Google Scholar]
- 38.Mahalakshmi R, Raghothama S, Balaram P. J Am Chem Soc. 2006;128:1125–1138. doi: 10.1021/ja054040k.For proteins, a recent survey of aryl/Trp interactions ( Chakrabarti P, Bhattacharyya R. Prog in Biophys & Mol Biol. 2007;95:83–137. doi: 10.1016/j.pbiomolbio.2007.03.016.) confirmed the high propensity of Trp/Trp, Tyr/Tyr, and Tyr/Trp interaction to be EtF and indicates that Tyr is somewhat more likely to be the face residue in the latter interactions.
- 39.(a) Steiner T, Koellner G. J Mol Biol. 2001;305:535–557. doi: 10.1006/jmbi.2000.4301. [DOI] [PubMed] [Google Scholar]; (b) Hariharan M, Karunakaran SC, Ramaiah D. Org Lett. 2007;9:417–420. doi: 10.1021/ol062636m. [DOI] [PubMed] [Google Scholar]; (c) Yau WM, Wimley WC, Gawrisch K, White SH. Biochemistry. 1998;37:14713–14718. doi: 10.1021/bi980809c. [DOI] [PubMed] [Google Scholar]; (d) Hu J, Barbour LJ, Gokel GW. J Am Chem Soc. 2002;124:10940–10941. doi: 10.1021/ja020586k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.