SUMMARY
Attention has recently been drawn to Enterococcus faecium because of an increasing number of nosocomial infections caused by this species and its resistance to multiple antibacterial agents. However, relatively little is known about pathogenic determinants of this organism. We have previously identified a cell wall anchored collagen adhesin, Acm, produced by some isolates of E. faecium, and a secreted antigen, SagA, exhibiting broad spectrum binding to extracellular matrix proteins. Here, we analyzed the draft genome of strain TX0016 for potential MSCRAMMs (microbial surface component recognizing adhesive matrix molecules). Genome-based bioinformatics identified 22 predicted cell wall anchored E. faeciumsurface proteins (Fms) of which 15 (including Acm) have typical characteristics of MSCRAMMs including predicted folding into a modular architecture with multiple immunoglobulin-like domains. Functional characterization of one (Fms10, redesignated Scm for second collagen adhesin of E. faeciu m) revealed that recombinant Scm65 (A- and B-domains) and Scm36 (A-domain) bound efficiently to collagen type V in a concentration dependent manner, bound considerably less to collagen type I and fibrinogen, and differed from Acm in their binding specificities to collagen types IV and V. Results from far-UV circular dichroism of recombinant Scm36 and of Acm37 indicated that these proteins are rich in β-sheets, supporting our folding predictions. Whole-cell ELISA and FACS analyses unambiguously demonstrated surface expression of Scm in most E. faecium isolates. Strikingly, 11 of the 15 predicted MSCRAMMs clustered in four loci, each with a class C sortase gene; 9 of these showed similarity to Enterococcus faecalis Ebp pilus subunits and also contained motifs essential for pilus assembly. Antibodies against one of the predicted major pilus proteins, Fms9 (redesignated as EbpCfm), detected a “ladder” pattern of high-molecular weight protein bands in a Western blot analysis of cell surface extracts from E. faecium, suggesting that EbpCfm is polymerized into a pilus structure. Further analysis of the transcripts of the corresponding gene cluster indicated that fms1 (ebpAfm), fms5 (ebpBfm) and ebpCfm are co-transcribed, consistent with pilus-encoding gene clusters of other gram-positive bacteria. All 15 genes occurred frequently in 30 clinically-derived diverse E. faecium isolates tested. The common occurrence of MSCRAMM and pilus-encoding genes and the presence of a second collagen-binding protein may have important implications for our understanding of this emerging pathogen.
INTRODUCTION
Enterococcus faecium, a member of the normal commensal flora, has recently emerged as a prominent nosocomial pathogen causing serious infections, including infective endocarditis (Murray, 2000). Nosocomial infections due to E. faecium can be a life threatening challenge to physicians because of this organism’s multi-drug resistance (Murray, 2000; Rice, 2001). Furthermore, in addition to the selective advantage conferred by antibiotics, strains that emerge in the hospital setting more often carry putative virulence genes such as esp encoding enterococcal surface protein (Rice et al., 2003; Willems et al., 2001), hyl encoding a putative hyaluronidase (Rice et al., 2003) and a functional copy of the acm gene encoding a collagen adhesin (Nallapareddy et al., 2003; Nallapareddy et al., 2007b).
It is known that pathogenic bacteria have evolved a plethora of proteins to adhere to and invade host tissues and to resist host defenses (Pizarro-Cerda & Cossart, 2006). Among these is a family of surface proteins with well-established roles in host-pathogen adherence which have been termed MSCRAMMs (microbial surface component recognizing adhesive matrix molecules) (Patti et al., 1994). MSCRAMMs share several characteristics including i) an N-terminal signal peptide, ii) a non-repeated A-domain consisting of Ig-like fold(s), iii) a B-domain with a variable number of repeats among different strains, and iv) a C-terminal CWA (cell wall anchor) domain. Some of these MSCRAMMs were recently shown to be tethered to each other by a designated sortase to make up multimeric cell surface structures, named pili (Mora et al., 2005; Nallapareddy et al., 2006). Sortases, encoded by the srtA to srtD classes of genes (Dramsi et al., 2005), were originally described as membrane-bound transpeptidases that cleave the LPXTG-like motif in the CWA domain and covalently link them to the peptidoglycan (Marraffini et al., 2006). While Class A sortases appear to be ubiquitous and involved in cell surface anchoring of a large number of LPXTG containing proteins (Marraffini et al., 2006), the class C (subfamily 3) sortase enzymes, which have a more limited substrate specificity, have recently been shown to be involved in pilus biogenesis, in addition to their role in surface anchoring (Kemp et al., 2007; Scott & Zahner, 2006; Telford et al., 2006).
Studies that have characterized the binding interactions of staphylococcal and enterococcal MSCRAMMs identified that the ligand-binding A-domains consist of two to three subdomains (N1-N2/N3) each adopting an Ig-like fold (Liu et al., 2007; Nallapareddy et al., 2007a; Ponnuraj et al., 2003; Zong et al., 2005). Based on the crystal structures of prototype MSCRAMMs, two slightly different models have been proposed to explain their binding mechanisms to linear peptides of fibrinogen and to triple-helical collagen. In the “dock, lock and latch” binding model, a fibrinogen chain is inserted into a cleft between two Ig-folded subdomains and is then secured by a C-terminal N3 extension (latch) that is reoriented upon ligand binding and complements a β-sheet on the N2 subdomain (Ponnuraj et al., 2003). A variation of this two-subdomain binding model, “the collagen hug”, has been proposed for the collagen-binding MSCRAMMs (Liu et al., 2007; Zong et al., 2005).
Previous in silico analyses have identified a family of genes encoding MSCRAMM-like proteins in genomes of several gram-positive bacteria including our reports of the ebp (endocarditis and biofilm-associated pilus of Enterococcus faecalis) operon, Ace (adhesin of collagen from E. faecalis) and Acm (adhesin of collagen from E. faecium) (Bowden et al., 2005; Nallapareddy et al., 2000; Nallapareddy et al., 2003; Nallapareddy et al., 2006; Roche et al., 2003; Sillanpaa et al., 2004; Xu et al., 2004). Recently, Hendrickx et al. (2007) predicted 22 CWA proteins from E. faecium TX0016 (formerly, DO (Arduino et al., 1994)) genome, of which five were found to be enriched in isolates of the hospital-adapted clonal complex 17 (CC17). However, with the exception of the prototype MSCRAMM, Acm (Nallapareddy et al., 2003)), there has been no demonstration of other MSCRAMMs in E. faecium. Although our previous report identified an essential and secreted broad-spectrum adhesin, SagA, exhibiting binding to fibrinogen, collagen type I (CI), collagen type IV (CIV), fibronectin, and laminin, this protein lacks a CWA domain and other MSCRAMM characteristics (Teng et al., 2003).
In the present study, we identified 14 genes (in addition to acm) encoding predicted MSCRAMMs in the genome of E. faecium TX0016. Recombinant forms of one of these proteins, designated as Scm (for second collagen adhesin of E. faecium), were characterized to confirm the structural predictions and to identify its ligand. Cell surface expression of Scm by selected E. faecium strains was quantitated using FACS, and antibodies raised against a recombinant form of one of the major pilus proteins showed a high-molecular weight (HMW) ladder pattern characteristic of gram-positive pili. Co-transcription of one of the pilus-encoding gene clusters was demonstrated by Northern hybridization and RT-PCR. In addition, we determined the distribution of the MSCRAMM encoding genes among 30 diverse E. faecium clinical isolates.
METHODS
Strains, plasmids and cultivation of bacteria
Relevant characteristics of bacterial strains, growth conditions, and plasmids used in this study are summarized in Table 1 and in Supplementary Methods. All constructs were given TX numbers and plasmids from these constructs were assigned respective pTEX numbers (Table 1).
Table 1.
Bacterial strains and plasmids used in this study
| Strains/Plasmids | Relevant characteristics*; Origin; Yr of isolation | Reference or source |
|---|---|---|
| Strains | ||
| E. faecium | ||
| TX0016 (DO) | Endocarditis isolate; VanS; AmpR; Houston, Texas; 1992. (http://www.hgsc.bcm.tmc.edu/projects/microbial/microbial-detail.xsp?project_id=118) | (Arduino et al., 1994) |
| TX2535 | Endocarditis isolate; VanR; AmpR; Houston, Texas; 1995 | (Nallapareddy et al., 2003) |
| TX0068 | Endocarditis isolate, VanR, AmpR; Worcester, Massachusetts; 1994 | This study |
| TX0074 | Endocarditis isolate, VanR, AmpR; Valhalla, New York; 1995 | This study |
| E. coli | ||
| M15(pREP4) | E coli strain for expression of recombinant proteins | Qiagen |
| TX5432 | M15 (pTEX5432), Ampr, Kanr | This study |
| TX5628 | M15 (pTEX5628), Ampr, Kanr | This study |
| Plasmids | ||
| pQE30 | Expression vector | Qiagen |
| pTEX5432 | 1794 bp fragment from TX0016 scm (coding for complete A and B domains) cloned into pQE30 expression vector | This study |
| pTEX5628 | 921 bp fragment from TX2535 scm (coding for complete A domain) cloned into pQE30 expression vector† | This study |
| pTEX5630 | 1674 bp fragment from TX0016 ebpCfm (coding for mature EbpCfm without signal peptide or CWA domain) cloned into pQE30 expression vector | This study |
Ampr: ampicillin resistant; Vanr: vancomycin resistant.
The DNA sequence of scm cloned into pTEX5628 from strain TX2535 is 100 % identical with that of TX0016.
Escherichia coli and E. faecium cells were grown in Luria-Bertani (LB) broth/agar and Brain Heart Infusion broth/agar (Difco), respectively. For E. coli constructs, antibiotics were used at the following concentrations: 25 μg kanamycin ml-1 and 100 μg ampicillin ml-1.
Identification and structural analysis of CWA proteins
Bioinformatics methods used for genome-wide identification of CWA proteins are described in the Supplementary Methods. Domain architecture as well as fold-recognition analyses were carried out by comparing the protein sequences against domain databases as described in Supplementary Methods.
Construction of expression plasmids
Genomic DNA from E. faecium strains was isolated as described earlier (Wilson, 1994). DNA regions encoding amino acids 27 to 624 (A-domain and B-repeats) and 27 to 333 (A-domain) of Scm and aa 33 to 590 of EbpCfm (Fms9 was renamed as EbpCfm based on its 74% aa identity (84% similarity) with EbpC of E. faecalis over the entire protein) (mature protein without the signal peptide and the CWA domain) were amplified using primers listed in Supplementary Table S1 and cloned into the expression vector pQE30 (Qiagen) to obtain pTEX5432, pTEX5628 and pTEX5630 (Table 1). The corresponding expressed protein segments of Scm were designated as rScm65 and rScm36, respectively, based on their calculated molecular masses. Similarly, the cloned segment of EbpCfm was designated as rEbpCfm62. Constructs were confirmed by sequencing.
Purification of recombinant proteins
Recombinant proteins with N-terminal His6-tags were expressed and purified using nickel affinity chromatography followed by anion exchange chromatography (Nallapareddy et al., 2007a; Sillanpaa et al., 2004). Protein concentrations were determined by absorption spectroscopy at 280 nm using calculated molar absorption coefficient values. Molecular masses were determined with MALDI-TOF MS for rScm36 and rScm65.
Circular dichroism (CD) specta
Far-UV CD spectroscopy data were collected as described previously (Sillanpaa et al., 2004). Secondary structure compositions were quantitated with ContinLL, SELCON3 and CDSSTR (http://www.cryst.bbk.ac.uk/cdweb/html/home.html) (Lobley et al., 2002; Whitmore & Wallace, 2004).
ELISA-type solid phase ligand binding assays
Binding of the recombinant His-tag fusion proteins to components of the ECM (extracellular matrix) was tested using a previously described assay with minor modifications (Nallapareddy et al., 2000). In brief, 1 μg of each ECM protein (for source, see Supplementary Methods) was coated in 100 μl PBS in Immulon 2HB (Thermo Scientific) 96-well microplate wells. Wells were incubated with various concentrations of rScm and bound His-tag proteins were detected with anti-His6 mAb (GE Healthcare) followed by alkaline phosphatase-conjugated anti-mouse antibody (Bio-Rad). p-nitrophenyl phosphate (Sigma) was used for signal detection.
Production of polyclonal antibodies and purification of antigen-specific Igs
Polyclonal goat antibodies against rScm36 and rEbpCfm62 were raised at Bethyl Laboratories. Scm36- and EbpCfm62-specific Igs were eluted from CnBr-activated Sepharose 4B coupled with the corresponding antigen, according to the manufacturer’s protocol (GE Healthcare). 0.1 M glycine, pH 2.8, was used for elution of bound antibodies, which were immediately neutralized by 1 M Tris/HCl, pH 8.0, and dialyzed extensively against PBS. Antibody concentrations were determined using an estimated IgG molar absorption coefficient value of 210 000 M-1cm-1 and a molecular mass of 150 000 Da.
Whole-cell ELISA and FACS
Surface expression of Scm on E. faecium cells was detected by a whole-cell ELISA assay (Nallapareddy et al., 2003) using affinity purified rScm36-specific Igs. Control antiserum against formalin-killed TX0016-whole-cells (Rakita et al., 2000) was used as a positive control.
To quantitate surface expression of Scm by FACS analysis, bacteria grown in BHI 14 h from an inoculum of OD600nm = 0.01, were labeled with preimmune or affinity-purified anti-Scm-specific antibodies followed by donkey anti-goat IgG conjugated with F(ab’)2-fragment-specific R-phycoerythrin, as described earlier (Kemp et al., 2007). Cells were then fixed in 1 % paraformaldehyde in PBS and analyzed with a Coulter EPICSXL AB6064 flow cytometer (Beckman Coulter) and System II software.
Extraction of CWA proteins and Western blot analysis
CWA proteins were extracted from E. faecium strains grown for 8 h in BHI broth to late-exponential phase (starting inoculum, ∼0.01 OD600nm) with mutanolysin as described earlier (Nallapareddy et al., 2006). Equal amounts of concentrated mutanolysin extracts were separated using 4%-15% gradient SDS-PAGE gels (Bio-Rad) under reducing conditions and transferred to PVDF membranes according to the manufacturer’s protocol. Membranes were probed with affinity-purified anti-Scm36 and anti-EbpCfm62 antibodies (see above) followed by HRP-conjugated anti-goat IgG antibodies and, as a control, total IgG antibodies purified from preimmune goat sera were used. Signal was detected using SuperSignal West Pico Chemilumiscent detection reagents (Thermo Scientific).
Northern hybridization
Total RNA was isolated from TX0082 grown in BHI to mid-exponential phase using the RNAprotect Bacteria Reagent and RNeasy Mini Kit (Qiagen). Thirty micrograms of total RNA were separated using a formaldehyde-containing agarose gel under denaturing conditions and transferred to a Hybond-N+ membrane as described by the manufacturer (GE Healthcare). DNA probes obtained with primers listed in Supplementary Table S1 were radiolabeled by using the RadPrime DNA labeling system (Invitrogen). Hybridization was performed under high stringency conditions as detailed earlier for Southern blots (Murray et al., 1993).
RT-PCR
Total RNA (isolated as above for Northern hybridization) was treated twice with 20 U RQ1 DNase (Promega) for 30 minutes at 37°C. DNase was removed using the RNeasy Mini Kit and purification protocol (Qiagen). Total RNA was then reverse transcribed with specific primers (Supplementary Table S1) using the SuperScript One-Step RT-PCR with Platinum Taq kit (Invitrogen) according to the manufacturer’s instructions. DNA sequencing verified that the primer regions of TX0082 are 100 % identical to the corresponding sequences in TX0016. As an internal control, a 509-bp fragment of gyrA (encoding gyrase A) was amplified using the FmGyrF and FmGyrR primers (Supplementary Table S1). Reactions without RT were used as controls to verify lack of DNA contamination in the total RNA preparation.
Colony hybridization
Preparation of colony lysate membranes and hybridization under high stringency conditions were performed as previously described (Coque et al., 1995; Singh et al., 1998). DNA probes were generated and radiolabeled as described above for Northern hybridization.
RESULTS AND DISCUSSION
Identification of putative CWA proteins with Ig-like folds
Our search of the nearly completed genome sequence of E. faecium endocarditis-derived strain TX0016 (GW, BEM, et al., unpublished data, http://www.hgsc.bcm.tmc.edu/) yielded a total of 22 ORFs encoding putative CWA proteins (designated Fms, for E. faeciumsurface proteins) with a tripartite pattern near the C-terminus (an LPXTG motif or variant, followed by a membrane-spanning hydrophobic region and a positively charged tail). This number is in the range of predicted CWA proteins of related gram-positive bacteria (Ponnuraj et al., 2003) which vary from 8 (Streptococcus mutans) to 41 (E. faecalis). While the number of CWA proteins identified here is the same as the number reported in a recent study (Hendrickx et al., 2007), which identified CWA proteins from a partial TX0016 genome sequence (available at NCBI), the published report did not include one of the ORFs here (Fms6) and classified a pseudogene (Fms15) as two ORFs (Supplementary Results).
Among these 22 putative CWA proteins, 18 contained a predicted N-terminal signal peptide sequence required for Sec-dependent secretion. Further analysis of the 5′-regions of the remaining four ORFs (Fms14, Fms15, Fms16, and Fms19) lacking a signal peptide revealed the presence of N-terminal signal peptides in the ORFs immediately upstream to Fms15, Fms16, and Fms19. Careful examination of the junction regions showed the presence of a premature stop codon due to a point mutation or frame-shift in fms15, fms16, and fms19 (Supplementary Results). Sequencing of fms16 and fms19 regions from additional isolates identified intact fms16 and fms19 genes in two of 10 E. faecium clinical isolates tested (Supplementary Results). However, we did not find an E. faecium strain with an intact fms15 gene (Supplementary Results).
Subsequently, using the fold recognition servers 3D-PSSM and PHYRE, we identified 15 of the predicted Fms proteins (Table 2) as containing one or more Ig-like folds enriched with β-sheets and only a small quantity of α-helices; similar secondary structure compositions have been found in the ligand binding A-regions of MSCRAMMs (Deivanayagam et al., 2002). One of these, Fms8, was previously identified in our laboratory as Acm (Nallapareddy et al., 2003). The percentage of E. faecium LPXTG-proteins containing Ig-like folds is greater than the percentages of other gram-positive bacteria (22 % - 45 %) (Ponnuraj et al., 2003). The Ig-like fold containing modules of the A-domain of a number of well characterized S. aureus MSCRAMMs (e.g., the fibronectin-, fibrinogen-, and elastin-binding protein, FnbpA; the fibrinogen-binding proteins, ClfA and ClfB; the collagen-binding protein, Cna) and the prototype MSCRAMM of E. faecium, Acm, have been shown to play a direct role in interactions with their respective ECM protein ligands (Nallapareddy et al., 2007a; Ponnuraj et al., 2003; Zong et al., 2005).
Table 2.
Summary of features of the predicted MSCRAMM proteins of E. faecium and the distribution of the corresponding genes in 30 E. faecium isolates associated with clinical infections
| Gene name | Accession no. | Protein length (aa) | Cell-wall anchoring motif | Best match to other CWA proteins (% similarity) | No. of isolates +ve (%) n = 30 |
|---|---|---|---|---|---|
| Genes Not Clustered | |||||
| acm* (fms8) | ZP_00604683 | 721 | LPKTS | Staphylococcus aureus Cna (62 %)* | 30 (100 %) |
| scm (fms10) | ZP_00604835 | 660 | LPKTG | Lactobacillus brevis hypothetical protein (54 %) | 30 (100 %) |
| fms15 † | ZP_00602631 | 1412 | LPKTG | Bacillus anthracis collagen binding protein, BA0871 (49 %) | 29 (97 %) |
| fms18 | ZP_00603098 | 1075 | FPQTG | Enterococcus faecalis EF1896 (91 %) | 24 (80 %) |
| Cluster 1 | |||||
| ebpAfm (fms1) | ZP_00602689 | 1129 | LPETG | E. faecalis EbpA (75 %) | 29 (97 %) |
| ebpBfm (fms5) | ZP_00602688 | 473 | LPKTN | E. faecalis EbpB (65 %) | 29 (97 %) |
| ebpCfm (fms9) | ZP_00602687 | 625 | LPSTG | E. faecalis EbpC (84 %) | 29 (97 %) |
| Cluster 2 | |||||
| fms11 | ZP_00604460 | 773 | LPSTG | Lactobacillus johnsonii hypothetical protein (46 %) | 29 (97 %) |
| fms19 ‡ | ZP_00604462 | 499 | FPKTS | E. faecalis EbpB (54 %) | 29 (97 %) |
| fms16 ‡ | ZP_00604464 | 470 | LPSTG | E. faecalis EbpC (46 %) | 29 (97 %) |
| Cluster 3 | |||||
| fms14 § | ZP_00603043 | 1313 | LPATG | E. faecalis EbpA (41 %) | 30 (100 %) |
| fms13 | ZP_00603041 | 565 | LPATG | E. faecalis EbpC (25 %) | 30 (100 %) |
| fms17 | ZP_00603042 | 437 | FPQTN | E. faecalis EbpB (47 %) | 30 (100 %) |
| Cluster 4 | |||||
| fms21 | ZP_00603529 | 658 | VPMTG | E. faecalis EbpC (41 %) | 28 (93 %) |
| fms20 | ZP_00603526 | 696 | VPKTG | E. faecalis EF2525 (41 %) | 22 (73 %) |
Identified in our previous studies (Nallapareddy et al., 2003).
In silico analysis predicts a one-nucleotide deletion in the B-repeat region.
These genes had a nucleotide substitution or deletion in E. faecium strain TX0016 vs. TX0068 and TX0074, leading to a pseudogene.
Fms14 lacks an identifiable signal peptide, but has the other characteristics of a CWA protein.
Structural organization of the E. faecium putative MSCRAMMs
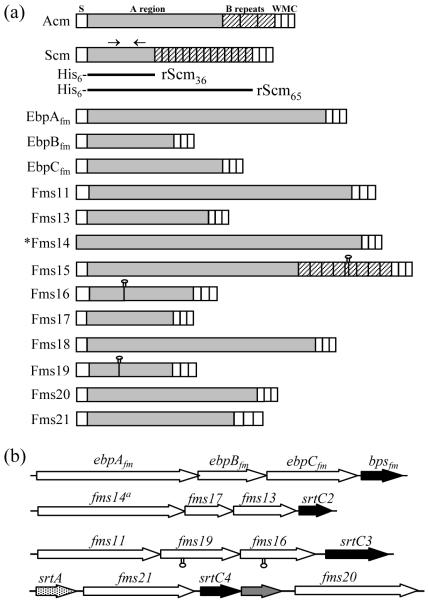
Because of our interest in surface adhesins, we next performed additional analyses on the subset of predicted Ig-like fold-containing proteins and identified that most possess a multi-domain architecture similar to the characterized MSCRAMMs of S. aureus and E. faecalis (Deivanayagam et al., 2002; Perkins et al., 2001; Sillanpaa et al., 2004) (Supplementary Results). A schematic of the predicted primary sequence organization of these proteins is depicted in Fig. 1. The N-terminal signal peptide sequences, found in all but Fms14, ranged from 24-56 aa in length. The majority had perfectly conserved canonical LPXTG motifs, while six had anchor motifs with a broader consensus (Table 2).
Fig. 1.
Domain organization of E. faecium CWA proteins with Ig-like folds. (a) S, signal peptide; W, cell-wall-spanning region; M, membrane anchor region; C, cytoplasmic tail with charged residues; Shaded A region, non-repetitive sequences; hatched B region, repetitive sequences; lollypop denotes presence of a stop codon due to a nucleotide substitution/deletion in Fms15, Fms16 and Fms19 of TX0016; solid black bars, His6-tag expressed regions of Scm (Fms10). Arrows above Scm show regions with predicted Ig-like folding. (b) Contiguously arranged ORFs encoding CWA proteins in E. faecium genome; srt, sortase. *Fms14 lacks an identifiable signal peptide, but has all the other characteristics of CWA MSCRAMMs.
Individually organized genes encoding putative MSCRAMMs
Fms10, Fms15, and Fms18, like Acm, are encoded by individual genes and are not clustered with other MSCRAMM-encoding genes. Among these, Fms18 has 94 % similarity (Table 2) to the E. faecalis MSCRAMM EF1896 which is predicted to be mostly Ig-folded and shows binding to a serum protein (J. S, S.R.N., B.E.M., and M. Hook, unpublished data). Scm (Fms10) of TX0016 has thirteen B-repeats of 19 aa in length and a partial repeat of 10 aa, while pseudogene fms15 predicts 8 B-repeats of 91 to 93 aa in length. The primary sequences of Scm and Fms15 each predict a single subdomain with an Ig-like fold in the A-domain, unlike the well-established MSCRAMMs which have two to three subdomains. Hence, we hypothesize that, if these proteins are adhesins, they may adopt a different binding mechanism, and we chose Scm for further characterization.
Characteristics of recombinant Scm protein segments
Total yields of the recombinant protein rScm65 (full mature protein consisting of the A-domain and B-repeats, aa 27-624) were low. Although we estimated that this protein was over 95 % pure immediately after eluting from the chromatographic columns as shown in Supplementary Fig. S1, it was susceptible to partial degradation even after short storage. rScm65 migrated as a larger band than expected from the calculated molecular weight of 64.5 kDa, likely due to the highly acidic nature (pI = 3.9) of the protein. To obtain a stable construct with higher yields, we next expressed rScm36 encompassing the complete A-domain (aa 27-333) (Supplementary Fig. S1). A MALDI-TOF mass spectrometry analysis determined the molecular mass of rScm36 as 35 711 Da, which is in good agreement with the calculated mass of 35 691 Da, and confirmed the absence of posttranslational modifications and homogeneity of the purified protein. As an additional confirmation of the identities of the purified protein segments and evidence of their intact N-termini, a monoclonal anti-His6 antibody detected the N-terminal His6- fusion in both purified protein segments in a Western blot assay (data not shown).
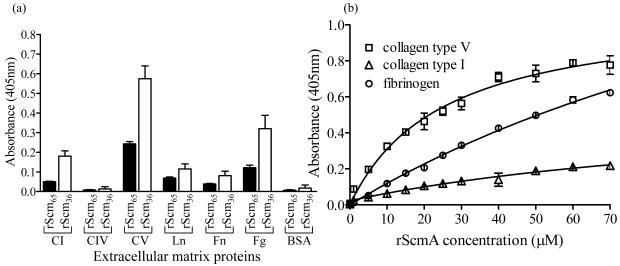
Recombinant Scm binds to collagen in a dose-dependent manner
Since the adhesive functions of previously studied MSCRAMM proteins have mostly been ascribed to binding to protein components of the host ECM, we examined the potential ECM-adhesive functions of Scm by testing its binding to a panel of individual ECM proteins in an ELISA-type solid-phase ligand binding assay. As seen in Fig. 2(a), both rScm65 (containing the full mature protein) and Scm36 (containing only the A-domain) exhibited significant binding to CV (35-fold and 82-fold, respectively, higher than the background level seen with immobilized BSA). Interestingly, binding of these rScm proteins to fibrinogen and CI (especially rScm36) was also observed compared to their binding to CIV, laminin, and fibronectin (Fig. 2a).
Fig. 2.
Binding of recombinant Scm proteins to ECM proteins in a solid phase ELISA-type ligand binding assay. (a) Binding of 20 μM rScm65 (aa 27-624) and rScm36 (aa 27-333) to a collection of immobilized ECM proteins. CI, type I collagen, Ln, laminin; Fn, fibronectin; Fg, fibrinogen; BSA, bovine serum albumin. Data points for rScm36 represent the means of A405nm values of 18 wells representing six independent experiments and including proteins from two different purifications, and for rScm65, 15 wells from five independent assays. (b) Binding of rScm36 to CI, CV and fibrinogen with increasing rScm36 concentrations. Data points represent the means of A405 nm values from three wells. The experiment was repeated up to six times using protein from two purifications. The reported KD value is the average ± standard error from the six assays. BSA values were subtracted from the respective collagen-binding values after which affinity calculations were performed with the one-ligand binding site model. The resulting curves are depicted in the figure.
Further analysis of the binding interactions of increasing concentrations of rScm36 with CI and CV as well as fibrinogen by dose response assays showed concentration dependent binding of rScm36 to CV which approached saturation with apparent half-maximal binding (KD) reached at 30.8 ± 1.8 μM (Fig. 2b). The apparent affinity of rScm36 for fibrinogen and CI was low and did not reach reliable saturation levels. In spite of partial degradation, Scm65 showed dose dependent binding with similar affinity for CV as Scm36 (data not shown). Although KD’s from steady-state assays are estimations, our affinity determination for the Scm A-domain is in the same range as previously shown for the minimal ligand binding domain of Ace to CI (48 ± 7 μM) (Rich et al., 1999), but lower than the A-domain of Acm to CI (3.8 μM) or CIV (12.8 μM) (Nallapareddy et al., 2003). Interestingly, Acm did not show appreciable binding to CV (data not shown) and bound efficiently to CI and, to a lesser degree to CIV, in agreement with our previous report (Nallapareddy et al., 2003). Taken together, our results suggest that CV is a binding ligand for Scm and this binding is mediated by the A-domain.
The three collagen types included in our assays have distinct structures and tissue distributions. CI is the most abundant form and the main component of collagen fibers that are widely distributed in human tissues, while CIV forms structurally different cross-linked networks and is found nearly exclusively in basement membranes. CV, although quantitatively less abundant, has a critical role in formation of the fibrillar collagen matrix and connecting interstitial collagen fibrils with membranous collagen networks (Nicholls et al., 1996; Wenstrup et al., 2004). Considering the diverse tissue localizations and structural differences of the various collagen types, possessing two collagen adhesins with different binding specificities to various collagen types could give E. faecium the ability to fine-tune its adherence phenotype to suit a given tissue. Since collagens, including CV, are also present in the intestinal submucosa (Liang et al., 2006), Scm could alternatively be involved in colonization and persistence in the intestinal tract, a major natural reservoir of both commensal and infection-associated strains of E. faecium in humans, or in facilitating translocation through damaged intestinal epithelium.
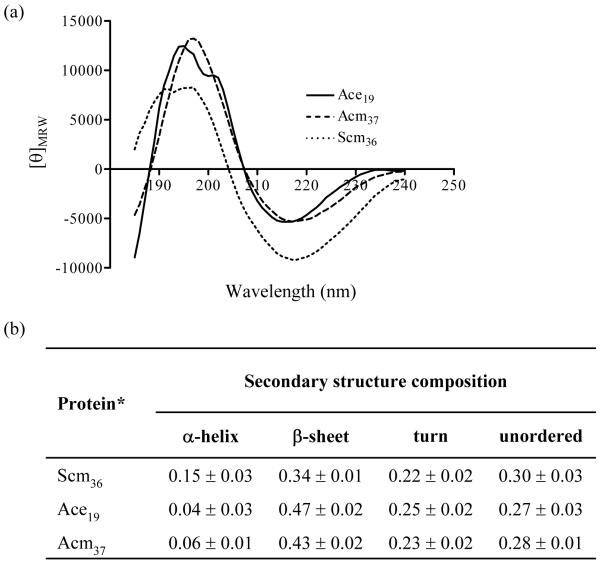
Structural analysis of recombinant Scm protein
To investigate the structural predictions made above, we analyzed the full A-domain of Scm with far-UV CD spectroscopy. The obtained spectra showed a maximum at 197 nm and another at 191 nm, and a minimum at 217 nm (Fig. 3a). A similar overall pattern was observed with the collagen-binding A-domain of the E. faecium prototype MSCRAMM Acm, which we have previously characterized (Nallapareddy et al., 2003; Nallapareddy et al., 2007a), and also with the minimal collagen binding region of Ace from E. faecalis, for which crystal structure studies have recently revealed that it folds into a similar DEv variant of the Ig-fold as previously found in the ligand binding A-domains of staphylococcal MSCRAMMs (Liu et al., 2007).
Fig. 3.
Secondary structure analysis by far-UV circular dichroism spectroscopy. (a) Collected CD-spectra of Scm36, Ace19 and Acm37. Mean residue weight ellipticity is reported in deg cm2 dmol-1. (b) Summary of secondary structure compositions. *Scm36 contains the predicted A-domain of Scm (aa 27-333). Ace19 and Acm37 contain the corresponding minimal ligand binding regions of the collagen binding MSCRAMMs Ace (E. faecalis) and Acm (E. faecium), respectively.
Deconvolution of the collected data showed a secondary structure composition of 0.15 ± 0.03 of α-helix and 0.34 ± 0.01 of β-sheet for rScm36 (Fig. 3b). Although the helical content of this protein is slightly higher than in the two control proteins, these results generally resemble the overall secondary structure compositions of the ligand binding regions of Acm and Ace and are in good agreement with earlier CD analyses and crystal structure data of other MSCRAMM A-regions (Rich et al., 1999). While fold analyses and multiple alignments predicted a single Ig-folded subunit from aa 125 to 325 in the A-domain of Scm, our CD measurements indicate a high β-sheet composition for the entire Scm A-domain, thus suggesting that an Ig-folded or similar β-sheet-rich structure may extend over the whole A-domain (aa 27 to 333).
Cell surface expression of Scm
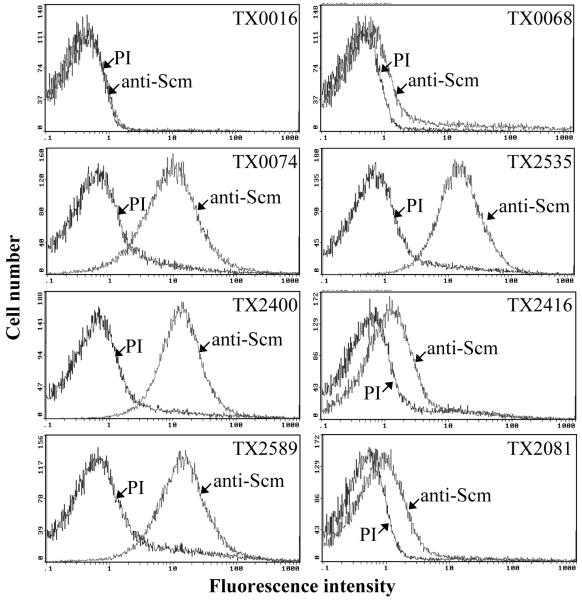
Using Scm-specific antibodies affinity purified (with rScm36) from goat anti-serum, we assessed the surface expression of Scm by 8 E. faecium clinical isolates (including the sequenced strain TX0016) using whole-cell ELISA. Two E. faecium community derived strains which lacked scm (identified below) were used as negative controls. Among 8 scm+ isolates, four (TX0074, TX2535, TX2400 and TX2589) were strongly positive, two (TX0068 and TX2081) were positive, one (TX2416) was weakly positive and one (TX0016) was negative in this assay (Supplementary Fig. S2), suggesting that Scm is produced on the cell surface of many E. faecium isolates during in vitro growth. Subsequent quantitation of Scm surface expression in these isolates by FACS (Fig. 4) showed variable levels of surface expression, with the percentage of cells expressing Scm ranging from 2.3 % to 96.9 %. In strains TX0074, TX2535, TX2400 and TX2589, Scm was detected on the surface of 90 to 97 % of cells and with relatively high fluorescence intensities, thus confirming our observations with whole-cell ELISA. Furthermore, these surface detection studies suggest that the majority of cells of scm+ isolates are actively producing Scm on the cell surface during in vitro growth. Consistent with our whole-cell ELISA and FACS results, a Western blot assay using the anti-Scm36 antibody identified a protein band corresponding to the expected size of mature Scm (∼ 63 kDa) in mutanolysin cell wall extracts from TX0074 and TX2535, but not from TX0016 (data not shown).
Fig. 4.
Quantitation of Scm surface expression by FACS analysis. Reactivity of both preimmune Igs (PI) and anti-Scm Igs for each strain was shown. Although some E. faecium isolates exhibit slight binding to goat PI, significant difference was observed between PI and anti-Scm Igs. Bacteria were analyzed using side scatter as the threshold for detection. Specific binding by anti-Scm antibodies is indicated as log fluorescence intensity on the X-axis. Each histogram represents 50 000 events of bacterium-sized particles.
Eleven of 15 putative MSCRAMM-encoding genes are clustered at four different genomic loci and nine are predicted to produce four distinct types of pili
Unlike Acm, Scm (Fms10), Fms15, and Fms18, the remaining 11 putative MSCRAMM genes were found clustered in four genomic loci. Nine of the clustered genes show significant similarity to pilus-associated proteins from other species (Table 2). Analysis of the nucleotide region of the locus spanning fms1, fms5 and ebpBfm (fms9) predicted the presence of four ORFs, all oriented in the same direction, including an srtC1 gene encoding a class C (subfamily 3) sortase (Fig. 1b). We subsequently renamed Fms1, Fms5, and SrtC1 as EbpAfm, EbpBfm, and Bpsfm, respectively, based on their high sequence identity with the E. faecalis Ebp cluster proteins (Nallapareddy et al., 2006) (Table 2). A similar arrangement of predicted pilus-associated proteins with an adjacent class C sortase was found in the fms14-17-13 and the fms11-19-16 clusters. Besides our prediction of ebpABCfm, fms14-17-13 and fms11-19-16 co-transcription, due to short or overlapping intergenic regions, we were unable to identify transcriptional terminator-like sequences in these gene clusters; thus, it is likely that these genes are in three operons (see also below). The genes encoding Fms20 and Fms21 were found to be located close to each other in the genome, separated by two ORFs. One of these ORFs encodes a predicted class C sortase (SrtC4). In addition to srtC4, this locus also contains one class A sortase-encoding gene (srtA) immediately upstream of fms21 (Fig. 1b). Another ORF located between SrtC4 and Fms20 shows 27 % similarity to the EbpB pilus protein and has a signal peptide sequence but no CWA motif. Of note, the E. faecium TX0016 genome also contains a sixth sortase gene (srtC5) predicting a class C sortase. However, none of the CWA protein genes were associated with the srtC5 locus.
Subsequent in silico predictions identified conserved pilin motifs and E-boxes of gram-positive pilus proteins in all four predicted major pilus protein homologues (EbpCfm, Fms13, Fms16, and Fms21) and all five of the predicted accessory protein homologues (EbpAfm, EbpBfm, Fms14, Fms17, and Fms19) associated with the four gene clusters described above (Supplementary Fig. S3). The lysine (K) residue of the pilin motif and the glutamic acid (E) of the E-box that were demonstrated to be essential in polymerization of Corynebacterium diphtheriae pilus (Ton-That & Schneewind, 2003; Ton-That et al., 2004) were found to be 100 % conserved. Two accessory proteins, namely EbpAfm and Fms14, have a von Willebrand factor type A-domain with a MIDAS-motif which is frequently found in pili-associated accessory proteins. Recently, crystal structure analyses of the minor pilin protein GBS52 of Streptococcus agalactiae and the major pilin Spy0128 of S. pneumoniae demonstrated that these proteins contain two Ig-like domains (Kang et al., 2007; Krishnan et al., 2007), consistent with our prediction of Ig-like folds in E. faecium pilus proteins. These features, together with the presence of an independent class C sortase (Class C sortases are used in the assembly of pili of related gram-positive bacteria) in each of the four clusters, indicate that E. faecium may harbor genes for multiple pilus-like structures. Recent studies on pili of C. diphtheriae, E. faecalis, group A and B streptococci, and pneumococci have demonstrated their role in bacterial adherence and biofilm formation, and their contribution to bacterial pathogenesis and modulation of the host immune system (Barocchi et al., 2006; Dramsi et al., 2006; Mandlik et al., 2007; Nallapareddy et al., 2006; Singh et al., 2007; Telford et al., 2006).
EbpCfm is present as a HMW polymeric protein complex at the cell surface of E. faecium
To confirm the predicted location of EbpCfm on the surface of E. faecium as part of polymeric pili, we probed mutanolysin cell wall extracts of endocarditis-derived E. faecium strains TX0082 and TX0016 with affinity-purified anti-rEbpCfm62 antibodies. A ladder-like pattern of HMW bands with sizes greater than 200 kDa was detected from strain TX0082 (Fig. 5). In contrast, only a single band was seen with Acm in mutanolysin extracts of TX0082 (Nallapareddy et al., 2003). The HMW banding pattern is consistent with observations of a large number of pilus proteins from other gram-positive bacteria and suggests that EbpCfm is similarly assembled into multimeric pilus structures on the E. faecium cell surface. In contrast to TX0082, no signal was detected from the cell wall extract of TX0016, which is similar to our whole-cell ELISA results (data not shown) and indicates that the EbpCfm-containing pilus is not expressed in this strain under the growth conditions used.
Fig. 5.
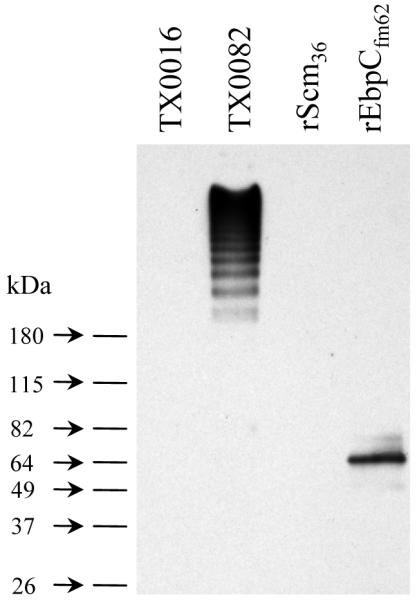
Western blot analysis of cell wall anchored proteins from E. faecium strains TX0082 and TX0016. The mutanolysin cell wall extracts (10 μg total protein/lane) were separated on 4-15% SDS-PAGE gels, transferred to PVDF membranes, and probed using affinity-purified anti-rEbpCfm62 antibodies. Purified rScm36 and rEbpCfm62 proteins (20 ng/lane) were included as controls.
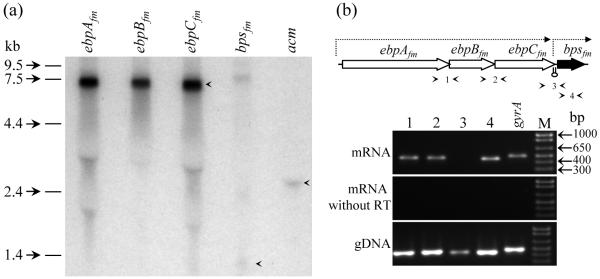
The ebpABCfm genes are transcribed as a single polycistronic mRNA
As stated above, bioinformatics analyses indicated that each of the four pilus-encoding gene clusters is likely to be organized as an operon. For the ebpAfm to bpsfm cluster, a 12-bp inverted repeat that could form a stem-loop structure (free energy of -22.55 kcal mol-1) located in the intergenic region between efbCfm and bpsfm, is likely to function as a ρ-independent transcriptional terminator, thus, predicting an efbAfm to efbCfm transcript and an independent bpsfm transcript. To validate this, we have carried out transcriptional analyses of the ebpAfm to bpsfm locus using Northern hybridization. The ebpAfm, ebpBfm, and ebpAfm probes all hybridized to a single RNA band of ∼ 7 kb (Fig. 6a) which corresponds to the expected size of a polycistronic ebpABCfm mRNA transcript, demonstrating that these three genes are co-transcribed. Probing with bpsfm detected a very low intensity band (∼ 1 kb), consistent with the expected size of a monocistronic bpsfm mRNA transcript, suggesting that this transcript may either be produced in low levels at this time point or has a relatively short half-life. RT-PCR using three independent sets of internal bpsfm primer pairs amplified fragments of the expected sizes (lane 4 of Fig. 6b) (data not shown for two additional primer pairs), thus confirming the expression of bpsfm. Another faint band of ∼8 kb was also observed with the bpsfm -probed Northern blots; while its identity is currently unclear, it might represent a related mRNA sequence such as one transcribed from the other predicted pilus operons.
Fig. 6.
Transcriptional analysis of the ebpAfm to bpsfm gene cluster. (a) Northern hybridization of total RNA (30 μg/lane) from TX0082 probed with intragenic fragments of ebpAfm, ebpBfm, ebpCfm, and bpsfm. The acm probe was used as a control. RNA bands with expected sizes are indicated with arrows. (b) RT-PCR analysis of the ebpAfm to bpsfm gene cluster. Location of each primer pair is shown with arrows in the schematic representation of the gene cluster. A lollypop between ebpCfm and bps depicts a predicted transcriptional terminator. Top gel, RT-PCR with DNase-treated total RNA (10 ng) as template; middle gel, control reaction of the same total RNA (100ng) preparation amplified without reverse transcriptase; bottom gel, control reaction amplified with genomic TX0082 DNA as template. An intragenic region of gyrase A was used as a control. M, molecular weight marker.
RT-PCR experiments of the ebpfm operon with primers designed to amplify intergenic regions of co-transcribed genes, confirmed the expected amplification between ebpAfm and ebpBfm as well as ebpBfm and ebpCfm, but not between ebpCfm and bpsfm (Fig. 6b). Taken together, our data unambiguously demonstrate that the E. faecium Ebpfm pilus-encoding gene cluster represents a three-gene operon, unlike the E. faecalis locus which produces a four-gene ebp-bps transcript in addition to an independent bps transcript (Nallapareddy et al., 2006).
Genes encoding MSCRAMM and pili proteins are commonly found in E. faecium isolates of clinical origin
We next examined 30 diverse (source/year/geographical location) human clinical isolates by colony hybridization using PCR generated DNA probes. E. faecium strain TX0016 and the acm gene served as controls for the hybridization studies. As shown in Table 2, the percentage of isolates showing hybridization to the individual gene probes varied from 73 % (fms20) to 100 %. The range in the number of probes hybridizing per isolate varied from 11 genes in one isolate (that lacked fms15, fms18, fms20, and fms21) to all 15 genes in 16 isolates (53 %). One isolate lacked the ebpABCfm (fms1-5-9) operon and another lacked the fms11-19-16 operon. The scm (fms10), fms14, fms17, and fms13 genes were present in all isolates tested, while fms15 was present in all but one. Among the endocarditis isolates included in this analysis, the multilocus sequence type 18 (ST18) isolate TX0068, belonging to the global epidemic hospital associated clonal complex CC17 (Leavis et al., 2003), had all 15 genes as did the sequenced ST18 isolate, TX0016. Another endocarditis isolate TX2535 (ST17) that also belongs to CC17, lacked fms18. A fourth endocarditis isolate, TX0074 (non-CC17, ST337), had 14 genes and lacked fms20. Thus, our results indicate broad but variable distribution of MSCRAMM-encoding genes in clinical isolates, consistent with a recent study of 22 E. faecium CWA protein-encoding genes in which four of the genes, fms11-19-16 (orf903-905-907) and fms18 (orf2430) were shown to be specifically enriched in CC17 isolates, while the remaining 9 genes were considered widespread in both CC17 and non-CC17 nosocomial isolates (Hendrickx et al., 2007). Of note, our preliminary data from screening for scm from several hundred of E. faecium isolates belonging to clinical and non-clinical groups indicate that scm is absent only rarely from clinical isolates but is frequently absent from stool isolates.
In summary, this study identified genes for 14 new E. faecium CWA proteins with MSCRAMM-like features. We characterized the function of one of these proteins, Scm, and demonstrated that it is a second collagen binding protein for E. faecium with specificity to collagen type V. We further showed that the identified minimal ligand-binding region has a β-sheet-rich secondary structure composition characteristic of the A-domains of MSCRAMMs. Scm was shown to be expressed on the surface of many E. faecium clinical isolates during in vitro growth. Eleven of the 14 MSCRAMM-like protein encoding genes were clustered in four loci and 9 are predicted to encode the components needed to form multiple, distinct pilus-like structures. We characterized one of these clusters (ebpABCfm-bpsfm) and showed that the four genes of this cluster are expressed in TX0082 as two transcripts, ebpABCfm and bpsfm. Subsequent detection of a HMW ladder pattern with anti-EbpCfm in Western blots of cell surface extracts from TX0082 provides experimental evidence for the assembly of EbpCfm into polymeric structures, such as pili. Our hybridization results for all 14 genes indicate a broad distribution of MSCRAMM and pilus-encoding genes among clinical isolates; a future study will assess their distribution in natural E. faecium populations from a variety of sources. The ability of E. faecium to produce two surface-anchored collagen adhesins with different collagen type specificities and the common occurrence of the scm gene as well as the 13 other newly identified MSCRAMM- and pilus-encoding genes among clinical E. faecium isolates may have important implications for colonization and infection by this opportunistic pathogen.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported in part by NIH grant R01 AI067861 from the Division of Microbiology and Infectious Diseases, NIAID to Barbara E. Murray. Additional support for this work is funded by NIH grant R01 AI20624 from the NIAID to Magnus Hook.
Abbreviations
- CWA
cell wall anchor
- ECM
extracellular matrix
- MSCRAMMs
microbial surface component recognizing adhesive matrix molecules
- CI
collagen type I
- CIV
collagen type IV
- CV
collagen type V
- CD
circular dichroism
Footnotes
Publisher's Disclaimer: This is an author manuscript that has been accepted for publication in Microbiology, copyright Society for General Microbiology, but has not been copy-edited, formatted or proofed. Cite this article as appearing in Microbiology. This version of the manuscript may not be duplicated or reproduced, other than for personal use or within the rule of ‘Fair Use of Copyrighted Materials’ (section 17, Title 17, US Code), without permission from the copyright owner, Society for General Microbiology. The Society for General Microbiology disclaims any responsibility or liability for errors or omissions in this version of the manuscript or in any version derived from it by any other parties. The final copy-edited, published article, which is the version of record, can be found at http://mic.sgmjournals.org, and is freely available without a subscription.
REFERENCES
- Arduino RC, Murray BE, Rakita RM. Roles of antibodies and complement in phagocytic killing of enterococci. Infect Immun. 1994;62:987–993. doi: 10.1128/iai.62.3.987-993.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barocchi MA, Ries J, Zogaj X. A pneumococcal pilus influences virulence and host inflammatory responses. Proc Natl Acad Sci U S A. 2006;103:2857–2862. doi: 10.1073/pnas.0511017103. other authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden MG, Chen W, Singvall J, Xu Y, Peacock SJ, Valtulina V, Speziale P, Hook M. Identification and preliminary characterization of cell-wall-anchored proteins of Staphylococcus epidermidis. Microbiology. 2005;151:1453–1464. doi: 10.1099/mic.0.27534-0. [DOI] [PubMed] [Google Scholar]
- Coque TM, Patterson JE, Steckelberg JM, Murray BE. Incidence of hemolysin, gelatinase, and aggregation substance among enterococci isolated from patients with endocarditis and other infections and from feces of hospitalized and community-based persons. J Infect Dis. 1995;171:1223–1229. doi: 10.1093/infdis/171.5.1223. [DOI] [PubMed] [Google Scholar]
- Deivanayagam CC, Wann ER, Chen W, Carson M, Rajashankar KR, Hook M, Narayana SV. A novel variant of the immunoglobulin fold in surface adhesins of Staphylococcus aureus: crystal structure of the fibrinogen-binding MSCRAMM, clumping factor A. Embo J. 2002;21:6660–6672. doi: 10.1093/emboj/cdf619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dramsi S, Trieu-Cuot P, Bierne H. Sorting sortases: a nomenclature proposal for the various sortases of Gram-positive bacteria. Res Microbiol. 2005;156:289–297. doi: 10.1016/j.resmic.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Dramsi S, Caliot E, Bonne I, Guadagnini S, Prevost MC, Kojadinovic M, Lalioui L, Poyart C, Trieu-Cuot P. Assembly and role of pili in group B streptococci. Mol Microbiol. 2006;60:1401–1413. doi: 10.1111/j.1365-2958.2006.05190.x. [DOI] [PubMed] [Google Scholar]
- Hendrickx AP, van Wamel WJ, Posthuma G, Bonten MJ, Willems RJ. Five genes encoding surface-exposed LPXTG proteins are enriched in hospital-adapted Enterococcus faecium clonal complex 17 isolates. J Bacteriol. 2007;189:8321–8332. doi: 10.1128/JB.00664-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Coulibaly F, Clow F, Proft T, Baker EN. Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure. Science. 2007;318:1625–1628. doi: 10.1126/science.1145806. [DOI] [PubMed] [Google Scholar]
- Kemp KD, Singh KV, Nallapareddy SR, Murray BE. Relative contributions of Enterococcus faecalis OG1RF sortase-encoding genes, srtA and bps (srtC), to biofilm formation and a murine model of urinary tract infection. Infect Immun. 2007;75:5399–5404. doi: 10.1128/IAI.00663-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V, Gaspar AH, Ye N, Mandlik A, Ton-That H, Narayana SV. An IgG-like domain in the minor pilin GBS52 of Streptococcus agalactiae mediates lung epithelial cell adhesion. Structure. 2007;15:893–903. doi: 10.1016/j.str.2007.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavis HL, Willems RJ, Top J, Spalburg E, Mascini EM, Fluit AC, Hoepelman A, de Neeling AJ, Bonten MJ. Epidemic and nonepidemic multidrug-resistant Enterococcus faecium. Emerg Infect Dis. 2003;9:1108–1115. doi: 10.3201/eid0909.020383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang R, Woo SL, Takakura Y, Moon DK, Jia F, Abramowitch SD. Long-term effects of porcine small intestine submucosa on the healing of medial collateral ligament: a functional tissue engineering study. J Orthop Res. 2006;24:811–819. doi: 10.1002/jor.20080. [DOI] [PubMed] [Google Scholar]
- Liu Q, Ponnuraj K, Xu Y, Ganesh VK, Sillanpaa J, Murray BE, Narayana SV, Hook M. The Enterococcus faecalis MSCRAMM ACE binds its ligand by the Collagen Hug model. J Biol Chem. 2007;282:19629–19637. doi: 10.1074/jbc.M611137200. [DOI] [PubMed] [Google Scholar]
- Lobley A, Whitmore L, Wallace BA. DICHROWEB: an interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics. 2002;18:211–212. doi: 10.1093/bioinformatics/18.1.211. [DOI] [PubMed] [Google Scholar]
- Mandlik A, Swierczynski A, Das A, Ton-That H. Corynebacterium diphtheriae employs specific minor pilins to target human pharyngeal epithelial cells. Mol Microbiol. 2007;64:111–124. doi: 10.1111/j.1365-2958.2007.05630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Dedent AC, Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev. 2006;70:192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora M, Bensi G, Capo S, Falugi F, Zingaretti C, Manetti AG, Maggi T, Taddei AR, Grandi G, Telford JL. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc Natl Acad Sci U S A. 2005;102:15641–15646. doi: 10.1073/pnas.0507808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray BE, Singh KV, Ross RP, Heath JD, Dunny GM, Weinstock GM. Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. J Bacteriol. 1993;175:5216–5223. doi: 10.1128/jb.175.16.5216-5223.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray BE, Weinstock GM. Enterococci: new aspects of an old organism. Proc Assoc Am Physicians. 1999;111:328–334. doi: 10.1046/j.1525-1381.1999.99241.x. [DOI] [PubMed] [Google Scholar]
- Murray BE. Vancomycin-resistant enterococcal infections. N Engl J Med. 2000;342:710–721. doi: 10.1056/NEJM200003093421007. [DOI] [PubMed] [Google Scholar]
- Nallapareddy SR, Qin X, Weinstock GM, Hook M, Murray BE. Enterococcus faecalis adhesin, Ace, mediates attachment to extracellular matrix proteins collagen type IV and laminin as well as collagen type I. Infect Immun. 2000;68:5218–5224. doi: 10.1128/iai.68.9.5218-5224.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallapareddy SR, Weinstock GM, Murray BE. Clinical isolates of Enterococcus faecium exhibit strain-specific collagen binding mediated by Acm, a new member of the MSCRAMM family. Mol Microbiol. 2003;47:1733–1747. doi: 10.1046/j.1365-2958.2003.03417.x. [DOI] [PubMed] [Google Scholar]
- Nallapareddy SR, Singh KV, Sillanpaa J, Garsin DA, Hook M, Erlandsen SL, Murray BE. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J Clin Invest. 2006;116:2799–2807. doi: 10.1172/JCI29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallapareddy SR, Sillanpaa J, Ganesh VK, Hook M, Murray BE. Inhibition of Enterococcus faecium adherence to collagen by antibodies against high-affinity binding subdomains of Acm. Infect Immun. 2007a;75:3192–3196. doi: 10.1128/IAI.02016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallapareddy SR, Singh KV, Okhuysen PC, Murray BE. A Functional acm gene contributes to Enterococcus faecium infective endocarditis and correlates with the recent success of this emerging nosocomial pathogen; 47th Interscience Conference on Antimicrobial Agents and Chemotherapy (Abstract number B-1239b); Chicago, USA. 2007b. [Google Scholar]
- Nicholls AC, Oliver JE, McCarron S, Harrison JB, Greenspan DS, Pope FM. An exon skipping mutation of a type V collagen gene (COL5A1) in Ehlers-Danlos syndrome. J Med Genet. 1996;33:940–946. doi: 10.1136/jmg.33.11.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti JM, Allen BL, McGavin MJ, Hook M. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol. 1994;48:585–617. doi: 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- Perkins S, Walsh EJ, Deivanayagam CC, Narayana SV, Foster TJ, Hook M. Structural organization of the fibrinogen-binding region of the clumping factor B MSCRAMM of Staphylococcus aureus. J Biol Chem. 2001;276:44721–44728. doi: 10.1074/jbc.M106741200. [DOI] [PubMed] [Google Scholar]
- Pizarro-Cerda J, Cossart P. Bacterial adhesion and entry into host cells. Cell. 2006;124:715–727. doi: 10.1016/j.cell.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Ponnuraj K, Bowden MG, Davis S, Gurusiddappa S, Moore D, Choe D, Xu Y, Hook M, Narayana SV. A “dock, lock, and latch” structural model for a staphylococcal adhesin binding to fibrinogen. Cell. 2003;115:217–228. doi: 10.1016/s0092-8674(03)00809-2. [DOI] [PubMed] [Google Scholar]
- Rakita RM, Quan VC, Jacques-Palaz K, Singh KV, Arduino RC, Mee M, Murray BE. Specific antibody promotes opsonization and PMN-mediated killing of phagocytosis-resistant Enterococcus faecium. FEMS Immunol Med Microbiol. 2000;28:291–299. doi: 10.1111/j.1574-695X.2000.tb01489.x. [DOI] [PubMed] [Google Scholar]
- Rice LB. Emergence of vancomycin-resistant enterococci. Emerg Infect Dis. 2001;7:183–187. doi: 10.3201/eid0702.010205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice LB, Carias L, Rudin S, Vael C, Goossens H, Konstabel C, Klare I, Nallapareddy SR, Huang W, Murray BE. A potential virulence gene, hylEfm, predominates in Enterococcus faecium of clinical origin. J Infect Dis. 2003;187:508–512. doi: 10.1086/367711. [DOI] [PubMed] [Google Scholar]
- Rich RL, Kreikemeyer B, Owens RT, LaBrenz S, Narayana SV, Weinstock GM, Murray BE, Hook M. Ace is a collagen-binding MSCRAMM from Enterococcus faecalis. J Biol Chem. 1999;274:26939–26945. doi: 10.1074/jbc.274.38.26939. [DOI] [PubMed] [Google Scholar]
- Roche FM, Massey R, Peacock SJ, Day NP, Visai L, Speziale P, Lam A, Pallen M, Foster TJ. Characterization of novel LPXTG-containing proteins of Staphylococcus aureus identified from genome sequences. Microbiology. 2003;149:643–654. doi: 10.1099/mic.0.25996-0. [DOI] [PubMed] [Google Scholar]
- Scott JR, Zahner D. Pili with strong attachments: Gram-positive bacteria do it differently. Mol Microbiol. 2006;62:320–330. doi: 10.1111/j.1365-2958.2006.05279.x. [DOI] [PubMed] [Google Scholar]
- Sillanpaa J, Xu Y, Nallapareddy SR, Murray BE, Hook M. A family of putative MSCRAMMs from Enterococcus faecalis. Microbiology. 2004;150:2069–2078. doi: 10.1099/mic.0.27074-0. [DOI] [PubMed] [Google Scholar]
- Singh KV, Coque TM, Weinstock GM, Murray BE. In vivo testing of an Enterococcus faecalis efaA mutant and use of efaA homologs for species identification. FEMS Immunol Med Microbiol. 1998;21:323–331. doi: 10.1111/j.1574-695X.1998.tb01180.x. [DOI] [PubMed] [Google Scholar]
- Singh KV, Nallapareddy SR, Murray BE. Importance of the ebp (endocarditis- and biofilm-associated pilus) locus in the pathogenesis of Enterococcus faecalis ascending urinary tract infection. J Infect Dis. 2007;195:1671–1677. doi: 10.1086/517524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G. Pili in gram-positive pathogens. Nat Rev Microbiol. 2006;4:509–519. doi: 10.1038/nrmicro1443. [DOI] [PubMed] [Google Scholar]
- Teng F, Kawalec M, Weinstock GM, Hryniewicz W, Murray BE. An Enterococcus faecium secreted antigen, SagA, exhibits broad-spectrum binding to extracellular matrix proteins and appears essential for E. faecium growth. Infect Immun. 2003;71:5033–5041. doi: 10.1128/IAI.71.9.5033-5041.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton-That H, Schneewind O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol Microbiol. 2003;50:1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Marraffini LA, Schneewind O. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol Microbiol. 2004;53:251–261. doi: 10.1111/j.1365-2958.2004.04117.x. [DOI] [PubMed] [Google Scholar]
- Wenstrup RJ, Florer JB, Brunskill EW, Bell SM, Chervoneva I, Birk DE. Type V collagen controls the initiation of collagen fibril assembly. J Biol Chem. 2004;279:53331–53337. doi: 10.1074/jbc.M409622200. [DOI] [PubMed] [Google Scholar]
- Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems RJ, Homan W, Top J. Variant esp gene as a marker of a distinct genetic lineage of vancomycin-resistant Enterococcus faecium spreading in hospitals. Lancet. 2001;357:853–855. doi: 10.1016/S0140-6736(00)04205-7. other authors. [DOI] [PubMed] [Google Scholar]
- Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel FM, Brent R, Kingston RE, David DM, Scidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. Green Publishing Associates; Brooklyn, NY: 1994. pp. 2.4.1–2.4.2. [DOI] [PubMed] [Google Scholar]
- Xu Y, Liang X, Chen Y, Koehler TM, Hook M. Identification and biochemical characterization of two novel collagen binding MSCRAMMs of Bacillus anthracis. J Biol Chem. 2004;279:51760–51768. doi: 10.1074/jbc.M406417200. [DOI] [PubMed] [Google Scholar]
- Zong Y, Xu Y, Liang X, Keene DR, Hook A, Gurusiddappa S, Hook M, Narayana SV. A ‘Collagen Hug’ model for Staphylococcus aureus CNA binding to collagen. Embo J. 2005;24:4224–4236. doi: 10.1038/sj.emboj.7600888. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.