Abstract
Recurrent gene fusions between the androgen-regulated gene TMPRSS2 and the ETS transcription factor family members ERG, ETV1, and ETV4 have been identified as a critical event in prostate cancer development. In this study, we characterized the prevalence and diversity of these rearrangements in hormone-refractory metastatic prostate cancer. We employed a fluorescence in situ hybridization (FISH) split probe strategy to comprehensively evaluate TMPRSS2: ETS aberrations across 97 non-osseous metastatic sites of prostate cancer from 30 rapid autopsies of men who died of androgen independent disease. Tissue microarrays were constructed representing multiple metastatic sites from each patient, and split signal FISH probes for TMPRSS2, ERG, ETV1 and ETV4 were used to assess for TMPRSS2-ETS rearrangements. In patients exhibiting these aberrations, multiple sites from an individual case harbored the same gene fusion molecular sub-type suggesting clonal expansion of disease. The most common prostate cancer gene fusion, TMPRSS2-ERG, can be generated by the mechanism of intrachromosomal deletion (Edel) about 39–60% of the time in clinically localized disease (1, 2). Interestingly, we observed that all of the androgen independent metastatic prostate cancer sites harboring TMPRSS2-ERG were associated with Edel. These findings suggest that TMPRSS2-ERG with Edel is an aggressive, and in this study uniformly lethal, molecular sub-type of prostate cancer associated with androgen-independent disease.
Introduction
The majority of prostate cancer deaths are attributed to metastatic dissemination of the primary tumor. Androgen ablation is initially effective in the management of metastatic prostate cancer, however most patients develop an androgen independent state that is invariably fatal(3). To elucidate the biology of prostate cancer progression and metastasis, we have developed a “rapid” or “warm” autopsy program at the University of Michigan comprised of patients that died of androgen independent metastatic prostate cancer. Tumors are collected from metastatic sites and the prostate, when present, and thus constitute a valuable resource to study the natural history and evolution of prostate cancer (4, 5).
We recently identified the fusion of the 5’-untranslated region of TMPRSS2 (21q22.3) with the ETS family members, ERG (21q22.2), ETV1 (7q21.2), or ETV4 (17q21) in prostate cancer (6, 7) that has subsequently been confirmed by multiple groups(2, 8–10). More recently, we have identified several 5’ fusion partners, supporting the existence of additional molecular sub-types of ETS gene fusions(11). We also demonstrated that most prostate cancers harboring gene rearrangements can be identified using a 5’ and 3’ split probe FISH strategy with probes tightly flanking the TMPRSS2, ERG, ETV1 and ETV4 loci(1). Using this approach, 65% of clinically localized prostate cancers demonstrated TMPRSS2 rearrangement, with the majority (55%) being fused to ERG(1). Additionally, as TMPRSS2 and ERG are located approximately 3 Mb apart on chromosome 21, the rearrangement between them occurs either through a translocation between chromosome 21s, or by an intrachromosomal deletion(2, 9). Further, in our recent study of multifocal prostate cancers, the majority (70%) of cases demonstrated heterogeneous TMPRSS2 gene rearrangements between different tumor foci, thus supporting multifocal prostate cancer as a heterogeneous group of diseases(12). Similar observations were reported by Clark et al(13). We have also shown the oncogenic potential of ETS fusions through in vitro and in vivo expression of ETV1 (11) and ERG (unpublished data).
It is currently unknown to what extent lethal androgen independent metastatic prostate cancers exhibit gene rearrangements, or whether they harbor them uniformly at different metastatic sites. Hence we sought to characterize the frequency, mechanism, and significance of TMPRSS2: ETS aberrations in a rapid autopsy cohort of 30 American men who died of androgen independent metastatic prostate cancer.
Materials and Methods
Rapid Autopsy Tissue Procurement and Tissue Microarray Construction
The autopsies were conducted as previously described(4, 5) , they are referred to as “rapid’ or ‘warm’ because of the short average interval of 3 hours between death of patient and onset of autopsy. A tissue microarray (TMA) was constructed from 30 rapid autopsies representing all metastatic prostate cancer sites and tumor in the prostate (when present i.e., no prior radical prostatectomy). The rapid autopsy program is approved by University of Michigan Institutional Review Board and supported by SPORE (NCI grant CA 69568).
Fluorescence in situ hybridization (FISH)
We used previously documented 5’ and 3’ split probe strategy to detect TMPRSS2, ERG , ETV1 and ETV4 rearrangements (1) (Fig.1A). Interphase FISH was performed as previously described (1, 6, 7). Slides were examined using an ImagingZ1 microscope (Carl Zeiss; Meta Systems, Germany). FISH signals were scored manually (100x oil immersion) in morphologically intact nuclei by two pathologists (R.M. and R.B.S.), and a minimum of 50 cancer cells from each site were recorded. Cancer sites with very weak or no signals were recorded as insufficiently hybridized. Only the non-osseous metastatic sites were evaluated during this study, osseous metastatic sites were non-hybridizable due to dense bone structure and hence excluded.
Figure 1. FISH probe design and representative TMPRSS2-ETS chromosomal aberrations detected in androgen independent metastatic prostate cancer.
A, For all assays, the chromosomal location of the gene is indicated (boxes), with the direction of transcription indicated by the arrow. The 5’ and 3’ BACs are indicated in ovals, with the number identifying each respective BAC and the color indicating the probe color in the accompanying images.
B, Representative FISH images of the various TMPRSS2-ETS aberrations observed in this study. Green and red arrows show individual signals, and yellow arrows indicate colocalized signals in 4’6-diamidino-2-phenylindole (DAPI) stained nuclei. B1, Colocalized signals in metastatic prostate cancer cells in a case lacking TMPRSS2 rearrangement. B2, TMPRSS2 rearrangement positive case without Edel, as indicated by split 5’ and 3’ signals. B3, TMPRSS2 rearrangement positive case with Edel, as indicated by one pair of co-localized signals (yellow arrow) and the loss of one red labeled probe 3’ to TMPRSS2. B4, Co-localized signals (yellow arrows) in a representative case lacking ERG rearrangement. B5, An ERG rearrangement positive case with Edel showing loss of one red labeled probe 5’ to ERG. B6, ETV1 rearrangement positive case without deletion, as indicated by split 5’ and 3’ signals (green and red arrows). B7, ETV1 rearrangement positive case with deletion exhibiting loss of one red labeled probe 5’ to ETV1. B8, ETV1 amplification shown by multiple (7–9) copies of co-localized signals in case 30 B9, A case showing rearrangement of ETV4, as indicated by break apart probes. Numbered BACs are as follows: 1= RP11-35C4 (5’ to TMPRSS2) and 2= RP11-120C17 (3’ to 18 TMPRSS2), 3= RP11-95I21 (5’ to ERG) and 4= RP11-476D17(3’ to ERG), 5= RP11- 661L15 (5’ to ETV1) and 6= RP11-124L22(3’ to ETV1) and 7= RP11-100E5 (5’ to ETV4) and 8= RP11-436J4 (3’ to ETV4).
A cancer site was considered positive for a gene aberration when the tumor demonstrated a rearrangement for any of the split probes mentioned above (through split or deletion) (Fig.1B). A metastatic site was labeled as positive for fusion when TMPRSS2 rearrangement (through split or deletion) was concomitant with a rearrangement (through split or deletion) in any of its known 3’ partners, ERG, ETV1 or ETV4(1, 11).
Results and Discussion
This University of Michigan rapid autopsy cohort represents patients who died of androgen independent metastatic prostate cancer(5). Of this group, 28 of the 30 men were initially diagnosed with clinically localized prostate cancer, but later developed widely disseminated disease within 5–10 years. These patients exhibited progression to lethality despite the use of multiple therapeutic regimens (Table 1). In the context of our initial discovery of TMPRSS2: ETS fusions in prostate cancer (1, 6, 7), we sought to perform a detailed characterization of these gene aberrations in androgen independent metastatic prostate cancer.
Table1.
Clinical Characteristics of the 30 consecutive subjects in our Rapid Autopsy cohort.
| Case # | Age at death (years) | Months to death after diagnosis | Gleason score at intial diagnosis | Treatment+ | Months Hormone Naïve | Months Hormone Refractory |
|---|---|---|---|---|---|---|
| 1 | 77 | 51.8 | 9 | hcr | 23 | 26 |
| 2 | 59 | 21.9 | Metastasis* | hcr | 12 | 21 |
| 3 | 53 | 20 | Metastasis* | hcr | 0 | 20 |
| 4 | 71 | 60.4 | 4 | hcr | 24 | 13 |
| 5 | 54 | 76.8 | 9 | hcr | 36 | 7 |
| 6 | 71 | 80.1 | 6 | hcrp | 30 | 2 |
| 7 | 65 | 65.6 | 9 | hcr | 31 | 34 |
| 8 | 68 | 84 | unknown | hcrp | 51 | 16 |
| 9 | 68 | 168.1 | unknown | hcrp | 144 | 21 |
| 10 | 78 | 84.2 | unknown | hcr | 24 | 12 |
| 11 | 67 | 39.8 | 8 | hcr | 12 | 5 |
| 12 | 79 | 176 | 7 | hc | 42 | 19 |
| 13 | 71 | 143.9 | unknown | hcr | 120 | 13 |
| 14 | 76 | 192.6 | 6 | hcr | 84 | 17 |
| 15 | 74 | 16 | 10 | hc | 4 | 3 |
| 16 | 70 | 37.7 | 7 | hcrp | 24 | 14 |
| 17 | 74 | 33.1 | 7 | hcr | 21 | 11 |
| 18 | 61 | 157 | 5 | hcrp | 60 | 14 |
| 19 | 84 | 121 | 7 | hcr | 69 | 28 |
| 20 | 66 | 92.5 | 9 | hcrp | 65 | 14 |
| 21 | 64 | 128.5 | 9 | hcrp | 25 | 30 |
| 22 | 64 | 30.5 | 8 | hcr | 22 | 9 |
| 23 | 72 | 96 | unknown | hcr | 27 | 45 |
| 24 | 76 | 55.7 | 7 | hcr | 13 | 9 |
| 25 | 66 | 45.8 | unknown | hr | 42 | 0 |
| 26 | 76 | 141.4 | unknown | hr | 60 | 35 |
| 27 | 74 | 94.3 | 7 | hrc | 80 | 15 |
| 28 | 86 | 168.6 | unknown | hcp | 119 | 61 |
| 29 | 77 | 112.8 | 8 | hcr | 102 | 8 |
| 30 | 71 | 44 | 9 | hcr | 13 | 16 |
Initial presentation as metastasis outside prostate
Treatment regimens: h- hormone ablation by bilateral orchiectomy and/or pharmacological blockage; c- chemotherapy; r- radiation; p- radical prostatectomy
Our FISH split probe strategy (Fig. 1A) revealed gene aberrations in 52% of the rapid autopsy cases. Overall, 48%, 37%, 11% and 4 % of the cases were positive for TMPRSS2, ERG, ETV1 and ETV4 split probe assays respectively (Fig. 1, 2). As observed in clinically localized disease(2, 8–10, 14), TMPRSS2: ERG fusion was the most common abnormality identified in hormone-refractory metastatic prostate cancer (37%). Additional fusions that were present included TMPRSS2:ETV1 (Case 26) and TMPRSS2: ETV4 (Case 8). In two cases, ETV1 was rearranged with an unknown partner (Case 4 and 23; the unknown partner in Case 23 was recently identified to be C15orf21 in an independent study(11)). Case 30 was found to be amplified with multiple copies of ETV1 (7–9) present in each nuclei. Fig.1B displays representative FISH images of the diversity of aberrations observed in this cohort.
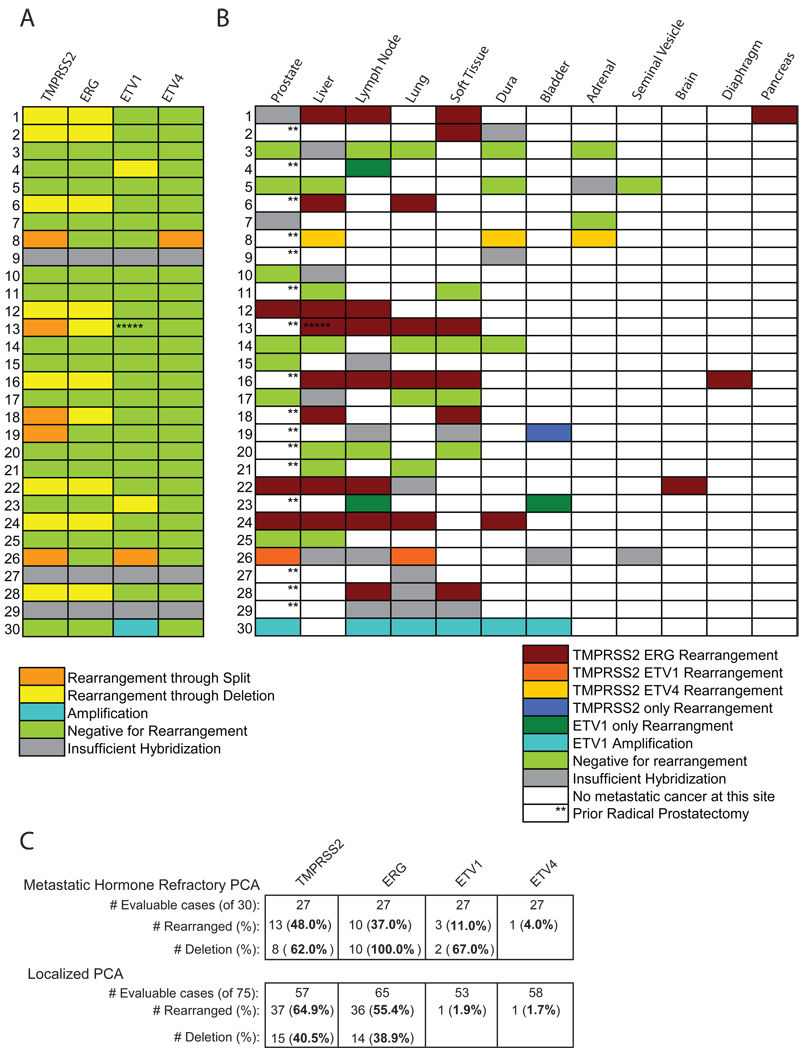
Figure 2. Diversity of TMPRSS2 and ETS rearrangements in androgen-independent metastatic prostate cancer.
A, Heat map representation of the TMPRSS2 and ETS aberrations in metastatic prostate cancers characterized in each subject. B, Heat map representation of TMPRSS2 and ETS rearrangement status of the metastatic sites and residual tumor in the prostate (when present) as evaluated in this cohort. **Prostate cancer tissues could not be procured at the time of autopsy because of previous radical prostatectomy and hence were not available for evaluation. No cancer focus was hybridizable for FISH probes in cases 9, 27 and 29.*****Case 13 which harbored a TMPRSS2:ERG rearrangement at metastatic foci within liver, lung, dura and soft tissue liver also showed a deletion of 5’ end of ETV1 in metastatic tumor within liver (presumably a non-specific secondary aberration). Color legend signifies respective aberrations. C. Table of results for rearrangements in TMPRSS2, ERG, ETV1 and ETV4 as detected by the assays shown in Figure 1. In this cohort, 27 of 30 cases were evaluable for at least one assay in various metastatic prostate cancer sites in this cohort, and the number of evaluable cases for each assay is indicated. The percentage (of evaluable cases for that assay) and number of cases with rearrangements for each assay is listed. For TMPRSS2, ERG, ETV1 and ETV4, the percentage (of rearrangement positive cases) and number of cases with assays consistent with intrachromosomal deletion between TMPRSS2 and ERG are given. The bottom panel contains a comparison of similar assays in a clinically localized prostate cancer cohort we previously evaluated (1). The number and percentage of cases with rearrangements for each assay is given, as well as the number and percentage of TMPRSS2 and ERG rearrangement positive cases with intrachromosomal deletion.
To determine if the same gene rearrangement is shared across metastatic sites in these subjects, we examined 97 tumor foci from different organs as well as the prostate (when present) with the FISH split probe assays. Interestingly, we found that different metastatic cancer foci within an individual patient harbored the same gene aberration (Fig. 2B). Furthermore, the same gene rearrangement exhibited by the metastatic sites was present in tumor in the prostate (when available), as in Case 24 (Fig. 2B, 3A). Similarly, cases without TMPRSS2: ETS rearrangements were uniformly negative across all metastatic sites (Fig. 2B). The exception to this was Case 13, which harbored a TMPRSS2: ERG rearrangement at metastatic foci within liver, lung, dura and soft tissue. However, in this patient, metastatic tumor in liver also displayed a deletion of the 5’ end of ETV1, suggesting the possibility of a secondary rearrangement acquired during the evolution of metastatic prostate cancer at this site (Fig. 2A, 2B). While multi-focal prostate cancer may harbor different foci that may be TMPRSS2-ETS positive or negative(12, 13), our study suggests that a single clone evolves from clinically localized disease to seed the metastatic sites with cells harboring the same ETS gene rearrangement (Fig. 3B).
Figure 3. The clonal nature of TMPRSS2-ETS aberrations in the spread of metastatic prostate cancer.
A, Representative Case 24 of metastatic prostate cancer highlighting the clonal nature of cells harboring TMPRSS2-ETS gene fusions. In this case, the subject harbored TMPRSS-ERG with Edel in the prostate and metastatic sites in the liver, lung, dura, and lymph node (representative FISH images from each site are shown- using the multicolor FISH probe strategy a nucleus with an ERG deletion shows replacement of one juxtaposed red and green signal pair with a single green signal representing loss of the 5’ end (red probe)).
B, This schematic depicts that while multi-focal prostate cancer might harbor TMPRSS-ETS gene fusion positive nodes and negative nodes, only one “clone” or molecular subtype will progress and result in androgen-independent metastatic disease.
TMPRSS2-ERG gene rearrangement in metastatic disease is similar to what has been observed in clinically localized disease(1, 2, 8–10). In our University of Michigan cohort we observed TMPRSS2-ERG gene rearrangements in 54% of clinically localized prostate cancer(1) and 37% in metastatic disease. In clinically confined disease, 39% of TMPRSS2-ERG gene fusions are through intrachromosomal deletion (Edel) while 61% are not (and thus presumably caused by translocation of chromosome 21s). By contrast, 100% of androgen independent prostate cancers harboring the TMPRSS2-ERG gene fusion (10 out 10 subjects and 29 out of 29 metastatic sites) exhibit Edel (Fig. 2).
Taken together, this is the first study that comprehensively characterizes TMPRSS2: ETS aberrations in end-stage, androgen independent metastatic prostate cancer. Based on our split probe strategy(1), we found that 52% of the cases in this cohort harbored TMPRSS2-ETS rearrangements. This frequency (52%) is similar to the 65% frequency of gene aberrations reported by our group for clinically localized prostate cancer (1) and in agreement with other reports in U.S. prostatectomy cohorts(10, 15). This indicates that the high frequency of TMPRSS2: ETS aberrations in clinically localized prostate cancer is maintained during progression to an androgen independent metastatic state. Overall, TMPRSS2 was rearranged in 48% of the cases (Fig. 2). Gene fusions between 5’ TMPRSS2 and 3’ ERG were the most frequent form (37%) of TMPRSS2-ETS rearrangement in androgen independent prostate cancer, similar to the 40–55% previously reported in clinically localized prostate cancers (1, 2, 10, 16). The low frequency of TMPRSS2:ETV1 (3%) and TMPRSS2:ETV4 (3%) is also comparable to previous reports(1, 16).
As TMPRSS2 and ERG are in close proximity (3 Mb) on chromosome 21, intrachromosomal deletion between these two genes is a common mechanism of TMPRSS2: ERG fusion in prostate cancer(2, 9, 17). In our previous study of clinically localized prostate cancers and TMPRSS2: ERG rearrangements in American men treated with radical prostatectomy, 39% were fused through deletion of 5’ end of ERG(1). Likewise, Perner et al demonstrated intronic deletions between ERG and TMPRSS2 resulting in TMPRSS2:ERG fusion in 60% of clinically localized cancers and 41% of hormone-naïve lymph node metastasis(2). Despite comparable frequencies of TMPRSS2:ERG rearrangement in end- stage androgen independent prostate cancer and clinically localized and hormone naïve prostate cancers, the mechanism of rearrangement between them is strikingly different. In comparison to clinically localized cancer and hormone naïve metastases, ERG rearrangement in androgen independent metastatic prostate cancers occurred exclusively through intrachromosomal deletion (100%). This suggests that prostate cancers with ERG fusion due to 5’ deletion harbor an aggressive molecular subtype that is susceptible to higher recurrence, evolves into an androgen independent state, and eventually progresses to metastasis. Other studies have documented the association of ERG fusions with a poor outcome(2, 17–19). Perner et al reported a significant association between clinically localized and hormone naïve locally advanced prostate cancers with TMPRSS2-ERG rearrangement through deletions and higher tumor stage and the presence of positive pelvic lymph nodes (2). Similarly, Attard et al demonstrated that patients with 5’ ERG deletion had a significantly worse cause-specific and overall survival (17). In this study, the presence of an identical ERG 5’ deletion in primary cancer within the prostate (when available) and all the metastatic sites from same individual strongly indicates that this molecular aberration occurred at the stage of localized disease before progression to a metastatic state (Case 24, Fig. 3A). Future studies will investigate the mechanism behind a preferential selection of ERG deleted (Edel) clinically localized prostate cancers to progress into an androgen independent phenotype.
This is the first study that demonstrates and characterizes a common gene rearrangement underlying localized prostate cancer and the metastatic foci in an individual patient (Fig. 2B, 3A). Our results showed that a unique clone of malignant cells in the prostate disseminated to seed different metastatic sites. Further, an aberrant clonal phenotype, acquired by clinically localized cancer, maintained its unique identity as the cancer metastized, as shown by the consistent presence of TMPRSS2: ETS throughout metastatic foci. Thus it is apparent that malignant cells from a single focus undergo clonal expansion and systemic dissemination to give rise to metastatic deposits in advanced prostate cancer. Such aberrations are presumably a primary initiating event, not secondarily accrued, since they are uniform across metastatic cancer foci. These findings support previous studies which suggested that untreated metastatic tumors contain the bulk of chromosomal alterations necessary for recurrence and eventual spread during anti-hormonal treatment(20).
Prostate cancer is usually a multifocal disease with considerable histologic, biologic, and molecular variations between different tumor foci(21). In our recent evaluation of multifocal prostate cancers, the majority of cases (70%) had heterogeneous TMPRSS2 gene rearrangements between different tumor foci suggesting independent clonal expansion (12). The histologic, immunophenotypic and molecular heterogeneity is persistent when prostate cancer progresses to an androgen independent phenotype(5). Despite the known heterogeneity of primary and advanced prostate cancers, we remarkably demonstrate identical TMPRSS2-ETS aberrations in all the metastatic samples and primary prostate cancer (when available) within a single individual patient. This strongly suggests that, although primary nodes of multifocal prostate cancers arise through separate clonal expansion, metastatic cancer arises through clonal expansion of malignant cells from a unique primary focus capable of dissemination. Our findings indicate that if a gene fusion positive cancer focus spreads from the prostate, the same clone is preferentially seeded across various organs to produce identical gene fusion characteristics. Similarly, all the metastatic foci are uniformly negative for these aberrations in TMPRSS2-ETS negative primary prostate cancers (Fig. 3A, 3B).
Further, to investigate the relationship between Androgen Receptor (AR) and TMPRSS2: ETS aberrations, we examined androgen receptor immunohistochemistry (IHC) staining status for multiple sites of these 30 rapid autopsy cases (as shown in supplementary information, Table S1). We did not observe any significant correlation between AR median staining and TMPRSS2:ETS or TMPRSS2:ERG rearrangements. Similar to AR IHC data, TMPRSS2:ETS and TMPRSS2:ERG aberrations also did not correlate with AR mRNA expression (supplementary information, Table S2). Hierarchical clustering of direct AR target genes yield similar patterns for cases with and without TMPRSS2:ETS rearrangements (supplementary information, Fig. S1A). This suggests that alternate mechanisms may be in play to maintain TMPRSS2:ETS expression in hormone-refractory prostate cancer. In a recent study, Hermans et al., identified cases of androgen-independent metastatic prostate cancer that harbored TMPRSS2:ERG fusions at the genomic level (as identified by aCGH), but did not express TMPRSS2:ERG fusion transcripts, leading them to suggest that ETS gene fusions are highly relevant in androgen-dependent metastatic cancer but are bypassed in androgen independent disease(22). Our results suggest that bypass of gene fusions may occur in some cases, however many androgen independent metastatic prostate cancers with TMPRSS2:ETS fusion as evidenced by aCGH or FISH strongly express fusion transcripts. For example, in our original report (6), both the index TMPRSS2:ERG (MET28; Case 28 in the present cohort) and TMPRSS2:ETV1 (MET26; Case 26 in the present cohort) cases were androgen independent cancers that strongly over-expressed ERG or ETV1, respectively
In summary, TMPRSS2: ETS rearrangements in end-stage androgen independent prostate cancers provide strong evidence that ERG fusion through deletion represents an aggressive molecular subtype of prostate cancer with a high susceptibility to evolve into androgen independent metastatic state. This study provides compelling evidence that metastatic prostate cancer arises through clonal expansion of a single focus of primary prostate cancer. Insight into the biology of these rearrangements will allow novel therapeutic regimens to be tailored to a susceptible molecular subtype of poor outcome cases.
Supplementary Material
Acknowledgments
We thank Jill Granger for editorial review of this manuscript and Michele LeBlanc, Alan Burgess, and Bo Han for technical support. Supported in part by Department of Defense (PC040517 to R.M., W81XWH-06-1-0224), the National Institutes of Health (Prostate SPORE P50CA69568, R01 CA102872), the Early Detection Research Network (UO1 CA111275-01), the Prostate Cancer Research Foundation and a sponsored research agreement from Gen-Probe, Inc. A.M.C. is supported by a Clinical Translational Research Award from the Burroughs Welcome Foundation. S.A.T. is supported by a Rackham Predoctoral Fellowship. S.A.T. is a Fellow of the Medical Scientist Training Program.
Footnotes
Disclosure: The University of Michigan has filed a patent on ETS gene rearrangements in prostate cancer, on which R.M., S.A.T., and A.M.C. are co-inventors, and the diagnostic field of use has been licensed to Gen-Probe Incorporated. Gen-Probe also has a sponsored research agreement with A.M.C., however Gen-Probe has not played a role in the in the design and conduct of the study, in the collection, analysis, or interpretation of the data, and in the preparation, review, or approval of the manuscript.
References
- 1.Mehra R, Tomlins SA, Shen R, et al. Comprehensive assessment of TMPRSS2 and ETS family gene aberrations in clinically localized prostate cancer. Mod Pathol. 2007;20:538–544. doi: 10.1038/modpathol.3800769. [DOI] [PubMed] [Google Scholar]
- 2.Perner S, Demichelis F, Beroukhim R, et al. TMPRSS2:ERG Fusion-Associated Deletions Provide Insight into the Heterogeneity of Prostate Cancer. Cancer Res. 2006;66:8337–8341. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 3.Taichman RS, Loberg RD, Mehra R, Pienta KJ. The evolving biology and treatment of prostate cancer. J Clin Invest. 2007;117:2351–2361. doi: 10.1172/JCI31791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rubin MA, Putzi M, Mucci N, et al. Rapid ("warm") autopsy study for procurement of metastatic prostate cancer. Clin Cancer Res. 2000;6:1038–1045. [PubMed] [Google Scholar]
- 5.Shah RB, Mehra R, Chinnaiyan AM, et al. Androgen-independent prostate cancer is a heterogeneous group of diseases: lessons from a rapid autopsy program. Cancer Res. 2004;64:9209–9216. doi: 10.1158/0008-5472.CAN-04-2442. [DOI] [PubMed] [Google Scholar]
- 6.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 7.Tomlins SA, Mehra R, Rhodes DR, et al. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66:3396–3400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- 8.Clark J, Merson S, Jhavar S, et al. Diversity of TMPRSS2-ERG fusion transcripts in the human prostate. Oncogene. 2007;26:2667–2673. doi: 10.1038/sj.onc.1210070. [DOI] [PubMed] [Google Scholar]
- 9.Iljin K, Wolf M, Edgren H, et al. TMPRSS2 fusions with oncogenic ETS factors in prostate cancer involve unbalanced genomic rearrangements and are associated with HDAC1 and epigenetic reprogramming. Cancer Res. 2006;66:10242–10246. doi: 10.1158/0008-5472.CAN-06-1986. [DOI] [PubMed] [Google Scholar]
- 10.Wang J, Cai Y, Ren C, Ittmann M. Expression of Variant TMPRSS2/ERG Fusion Messenger RNAs Is Associated with Aggressive Prostate Cancer. Cancer Res. 2006;66:8347–8351. doi: 10.1158/0008-5472.CAN-06-1966. [DOI] [PubMed] [Google Scholar]
- 11.Tomlins SA, Laxman B, Dhanasekaran SM, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 12.Mehra R, Han B, Tomlins SA, et al. Heterogeneity of TMPRSS2 gene rearrangements in multifocal prostate adenocarcinoma: molecular evidence for an independent group of diseases. Cancer Res. 2007;67:7991–7995. doi: 10.1158/0008-5472.CAN-07-2043. [DOI] [PubMed] [Google Scholar]
- 13.Clark J, Attard G, Jhavar S, et al. Complex patterns of ETS gene alteration arise during cancer development in the human prostate. Oncogene. 2007 doi: 10.1038/sj.onc.1210843. [DOI] [PubMed] [Google Scholar]
- 14.Rajput AB, Miller MA, De Luca A, et al. Frequency of the TMPRSS2:ERG gene fusion is increased in moderate to poorly differentiated prostate cancers. J Clin Pathol. 2007;60:1238–1243. doi: 10.1136/jcp.2006.043810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lapointe J, Kim YH, Miller MA, et al. A variant TMPRSS2 isoform and ERG fusion product in prostate cancer with implications for molecular diagnosis. Mod Pathol. 2007;20:467–473. doi: 10.1038/modpathol.3800759. [DOI] [PubMed] [Google Scholar]
- 16.Soller MJ, Isaksson M, Elfving P, Soller W, Lundgren R, Panagopoulos I. Confirmation of the high frequency of the TMPRSS2/ERG fusion gene in prostate cancer. Genes Chromosomes Cancer. 2006;45:717–719. doi: 10.1002/gcc.20329. [DOI] [PubMed] [Google Scholar]
- 17.Attard G, Clark J, Ambroisine L, et al. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2007 doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Demichelis F, Fall K, Perner S, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007 doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 19.Nami RK, Sugar L, Wang Z, et al. Expression of TMPRSS2:ERG gene fusion in prostate cancer cells is an important prognostic factor for cancer progression. Cancer Biol Ther. 2007;6:40–45. doi: 10.4161/cbt.6.1.3489. [DOI] [PubMed] [Google Scholar]
- 20.Cher ML, Bova GS, Moore DH, et al. Genetic alterations in untreated metastases and androgen-independent prostate cancer detected by comparative genomic hybridization and allelotyping. Cancer Res. 1996;56:3091–3102. [PubMed] [Google Scholar]
- 21.Arora R, Koch MO, Eble JN, Ulbright TM, Li L, Cheng L. Heterogeneity of Gleason grade in multifocal adenocarcinoma of the prostate. Cancer. 2004;100:2362–2366. doi: 10.1002/cncr.20243. [DOI] [PubMed] [Google Scholar]
- 22.Hermans KG, van Marion R, van Dekken H, Jenster G, van Weerden WM, Trapman J. TMPRSS2:ERG fusion by translocation or interstitial deletion is highly relevant in androgen-dependent prostate cancer, but is bypassed in latestage androgen receptor-negative prostate cancer. Cancer Res. 2006;66:10658–10663. doi: 10.1158/0008-5472.CAN-06-1871. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.