Abstract
c-IAP1 (cellular inhibitor of apoptosis 1) has recently emerged as a negative regulator of the non-canonical NF-κB (nuclear factor κB) signalling cascade. Whereas synthetic IAP inhibitors have been shown to trigger the autoubiquitination and degradation of c-IAP1, less is known about the physiological mechanisms by which c-IAP1 stability is regulated. In the present paper, we describe two distinct cellular processes that lead to the targeted loss of c-IAP1. Recruitment of a TRAF2 (tumour necrosis factor receptor-associated factor 2)–c-IAP1 complex to the cytoplasmic domain of the Hodgkin's/anaplastic large-cell lymphoma-associated receptor, CD30, leads to the targeting and degradation of the TRAF2–c-IAP1 heterodimer through a mechanism requiring the RING (really interesting new gene) domain of TRAF2, but not c-IAP1. In contrast, the induced autoubiquitination of c-IAP1 by IAP antagonists causes the selective loss of c-IAP1, but not TRAF2, thereby releasing TRAF2. Thus c-IAP1 can be targeted for degradation by two distinct processes, revealing the critical importance of this molecule as a regulator of numerous intracellular signalling cascades.
Keywords: CD30, inhibitor of apoptosis (IAP), lymphoma, nuclear factor κB (NF-κB), second mitochondrial-derived activator of caspase (Smac)/direct inhibitor of apoptosis-binding protein with low pI (DIABLO), tumour necrosis factor receptor-associated factor (TRAF)
Abbreviations: ALCL, anaplastic large-cell lymphoma; CD30L, CD30 ligand; CHO, Chinese-hamster ovary; c-IAP, cellular inhibitor of apoptosis; DIABLO, direct inhibitor of apoptosis-binding protein with low pI; DN, dominant-negative; HA, haemagglutinin; HEK, human embryonic kidney; HL, Hodgkin's lymphoma; IAP, inhibitor of apoptosis; IBM, IAP-binding motif; LDS, lithium dodecyl sulfate; NF-κB, nuclear factor κB; NIK, NF-κB-inducing kinase; Ni-NTA, Ni2+-nitrilotriacetate; qRT-PCR, quantitative real-time PCR; RING, really interesting new gene; RIP1, receptor-interacting protein 1; siRNA, short interfering RNA; Smac, second mitochondrial-derived activator of caspase; TBS, Tris-buffered saline; TNF, tumour necrosis factor; TNFR, TNF receptor; TRAF, TNFR-associated factor; XIAP, X-linked IAP
INTRODUCTION
CD30, a member of the TNFR [TNF (tumour necrosis factor) receptor] superfamily, has been the subject of much investigation because of its greatly elevated expression in several types of leukaemia and lymphoma cells, including ALCL (anaplastic large-cell lymphoma) and HL (Hodgkin's lymphoma) [1,2]. Normally, CD30 expression is restricted to a small subset of activated B- and T-cells, and has been shown to induce downstream signalling events through TRAFs (TNFR-associated factors), similarly to other members of the TNFR family such as TNFR2 and CD40 [3]. In particular, the activated CD30 receptor leads to the induction of the NF-κB (nuclear factor κB) transcription factor cascade through a mechanism that is largely TRAF-dependent [4–7]. Induction of NF-κB-responsive genes is critical for immune responses, cell survival and proliferation, and the up-regulation of CD30 in leukaemia and lymphoma cells promote aberrant NF-κB activation driving lymphomagenesis [8,9]. Two major NF-κB pathways have been described, both of which are activated in response to CD30 stimulation [10]. The canonical pathway involves phosphorylation of IκBα (inhibitor of NF-κB α), and, in the alternative pathway, p100/NF-κB2 is proteolytically processed to p52 [11].
IAP (inhibitor of apoptosis) proteins are a broadly expressed group of intracellular factors that function in a wide range of cellular processes in addition to their initially described role in the suppression of programmed cell death [12]. A growing body of work is revealing alterations in both the expression and activity of numerous IAP proteins in neoplastic, lymphoproliferative and metabolic disorders, although given the diverse range of functions attributed to these proteins, the specific contributions of the IAP proteins to these disorders are still being elucidated [13–15].
IAP proteins contain one to three tandem repeats of an ∼70 residue BIR (baculoviral IAP repeat) domain [16]. In addition, several members of the IAP family, including the c-IAP (cellular IAP) proteins (c-IAP1 and c-IAP2), contain a C-terminal RING (really interesting new gene) domain that possesses E3 ubiquitin ligase activity [17,18]. Several IAP proteins have been implicated in the regulation of cell death through their ability to bind and enzymatically inhibit certain members of the caspase family, which are the primary effectors of the apoptotic programme. However, IAP proteins differ remarkably in their caspase-inhibiting ability, with XIAP (X-linked IAP) being the best described caspase inhibitor, whereas c-IAP1 and c-IAP2 can bind caspases but do not inhibit their enzymatic activity [19,20]. Originally identified through their association with TNFR2, c-IAP1 and c-IAP2 are recruited to TNFRs by TRAFs, primarily TRAF2 [21].
The c-IAP1 and c-IAP2 proteins were originally identified as TRAF-associated proteins [21], and c-IAP1 was identified in amplicons associated with squamous cell carcinoma and hepatocellular carcinoma, in which elevated expression correlates with resistance to radiotherapy [22,23]. c-IAP1 was also identified as a putative oncogene, functioning synergistically with c-Myc, in a murine hepatocellular carcinoma genetic screen [23], and, independently, c-IAP1 was also shown to target the c-Myc regulatory factor, MAD1 (mitotic arrest-deficient protein 1), for proteasome-mediated degradation [24]. Thus it is now evident that c-IAP1 can function in a range of cellular processes.
Previous reports have revealed several elegant mechanisms by which the activity of IAP proteins can be regulated. For instance, a number of mitochondrial proteins have been identified that are released into the cytosol during apoptosis, along with cytochrome c, which can bind to and antagonize IAP proteins [12,25]. These proteins share a conserved N-terminal domain known as the IBM (IAP-binding motif), and this motif has been shown to play a key role in the association with various IAPs. The best characterized mammalian IBM-containing protein is Smac (second mitochondrial-derived activator of caspase), known as DIABLO (direct IAP-binding protein with low pI) in mice. Although Smac was originally identified as an XIAP antagonist [26,27], it is also known to recognize other IAP proteins, including c-IAP1 [26–28]. Many of the binding properties of Smac can be recapitulated by a conserved tetrapeptide sequence, and small-molecule IAP antagonists that mimic the action of Smac have been designed to potentiate apoptosis in cancer cells [29–32]. Additionally, Smac can be selectively released from mitochondria in the absence of an apoptotic stimulus, and, indeed, in healthy cells, cytosolic Smac has been described, produced either as an alternatively spliced variant whose maturation has been shown to bypass mitochondria or through other less well-defined processes [33,34]. These findings suggest that the regulation of IAP proteins may be of central importance in the homoeostatic control of a number of physiological processes in addition to apoptosis.
In the present study, we examined the functional consequences reflected by the cellular interactions between c-IAP1, TRAF2 and Smac. We describe two apparently independent mechanisms by which c-IAP1 can be targeted for degradation. CD30-mediated cellular activation induces the degradation of a TRAF2–c-IAP1 complex, through a process that requires the RING domain of TRAF2. In contrast, Smac or a synthetic IAP antagonist triggers the selective degradation of c-IAP1, but not TRAF2, through the RING domain of c-IAP1. The consequence of c-IAP1 targeting by either of these processes is the activation of the non-canonical NF-κB pathway, consistent with previously proposed mechanisms for the role of c-IAP1 in this signalling cascade. Since NF-κB activation has frequently been associated with the transcriptional induction of gene signatures involved in cytoprotection, these findings indicate that the c-IAP proteins and IBM-containing proteins such as Smac can function in pro-apoptotic and anti-apoptotic signalling pathways.
EXPERIMENTAL
Materials
Reagents were obtained from the following sources: Ni-NTA (Ni2+-nitrilotriacetate)–agarose (Invitrogen), DMSO (Sigma–Aldrich), cell culture media (Mediatech), Ficoll-Paque PLUS (GE Healthcare) and QuikChange site-directed mutagenesis kit (Stratagene). AEG40730 was a gift from Aegera Therapeutics. Antibodies were obtained from the following sources: anti-TRAF2 (BD Pharmingen), anti-(COX IV) (cytochrome oxidase subunit IV) (Invitrogen), anti-Smac/DIABLO (Calbiochem), anti-GST (glutathione transferase) (Santa Cruz Biotechnology), horseradish-peroxidase-conjugated anti-HA (haemagglutinin), horseradish-peroxidase-conjugated anti-FLAG, anti-β-actin (Sigma–Aldrich), anti-p52/p100 (Upstate Biotechnology), and horseradish-peroxidase-conjugated anti-mouse, anti-rabbit and anti-rat (GE Healthcare). Anti-c-IAP1 was kindly provided by Dr John Silke (Department of Biochemistry, La Trobe University, Melbourne, Victoria, Australia) [35].
Plasmids
pEBB-HA c-IAP1 was subcloned from pEBG c-IAP1 [36]. Site-directed mutagenesis of c-IAP1 to generate c-IAP1 H588A, a mutant c-IAP1 deficient in E3 ubiquitin ligase activity, was performed using the QuikChange site-directed mutagenesis kit. pEBB-His6-Ub was a gift from Dr Ezra Burstein (University of Texas Southwestern Medical Centre, Dallas, TX, U.S.A.). Unless noted otherwise, plasmids used in the present study have been described previously [37–39].
Cell lines
HEK (human embryonic kidney)-293 cells were cultured in Dulbecco's modified Eagle's medium, Karpas 299 and L428 cells were cultured in RPMI 1640 medium, and CHO (Chinese-hamster ovary) cells were cultured in Ham's F12 nutrient medium. All media were supplemented with 10% fetal bovine serum and 2 mM L-glutamine and maintained at 37 °C in an atmosphere of 95% air and 5% CO2.
Transfections
HEK-293 cells were transfected with plasmids using a standard calcium phosphate transfection protocol. Both plasmids and siRNA (short interfering RNA) oligonucleotides were transfected into Karpas 299 cells using a Bio-Rad Gene Pulser II electroporator at 300 V and 950 μF.
CD30 stimulation and treatment with AEG40730
Stimulation of Karpas 299 cells with CD30L+ (CD30 ligand) CHO cells was performed as described previously [10]. Briefly, Karpas 299 cells were resuspended in a 1:1 mixture of RPMI 1640 and Ham's F12 nutrient media at a final concentration of 106 cells/ml. CHO cells (negative control) or CD30L+ CHO cells were seeded 24 h previously at a density of 0.8×106 cells/well, and 1 ml of Karpas 299 cells were layered on to CHO cells in six-well plates for 2 h. Karpas 299 cells were then removed from the CHO cells by gentle pipetting and collected by centrifugation at 200 g for 5 min. Cells were washed with 1 ml of PBS, and subsequently centrifuged at 200 g for 5 min, and resuspended in RIPA lysis buffer (PBS containing 1% Nonidet P40, 0.5% sodium deoxycholate and 0.1% SDS) supplemented with protease inhibitors. HEK-293 or Karpas 299 cells were treated with 0–25 nM AEG40730 for 24 h, or treated for 0–48 h with 25 nM AEG40730, as described in the Results section. DMSO was used as a control for all AEG40730 treatments.
Cell lysate preparation and immunoblot analysis
Cell lysates were prepared with RIPA lysis buffer, supplemented with protease inhibitors, for 30 min on ice to ensure complete lysis unless noted otherwise. Protein quantification was determined using a Bradford assay (Bio-Rad Laboratories). RIPA cell lysates of equal protein concentrations were prepared in LDS (lithium dodecyl sulfate) sample buffer, separated on denaturing NuPAGE 4–12% polyacrylamide gradient gels, and transferred on to 0.45 μM nitrocellulose membranes (Invitrogen). Membranes were blocked in 5% (w/v) dried non-fat skimmed milk powder in TBS (Tris-buffered saline) with 0.02–0.2% Tween 20 (Bio-Rad Laboratories), depending on the antibody requirements, followed by incubation with the indicated antibodies in 2.5% (w/v) dried non-fat skimmed milk powder for 1 h at room temperature (25 °C) or overnight at 4 °C. After washing with TBS containing 0.02–0.2% Tween 20, membranes were incubated with secondary antibodies for 1 h at room temperature. ECL® (enhanced chemiluminescence) (GE Healthcare) was used to visualize the blots.
Cellular fractionation preparation
Karpas 299 or L428 cells (106 cells/treatment) were prepared as described previously [40], with minor protocol modifications. Cells were harvested, washed with PBS, resuspended at 3×107 cells/ml in digitonin extraction buffer (PBS containing 250 mM sucrose, 70 mM KCl, 1 mM PMSF and 200 μg/ml digitonin) supplemented with additional protease inhibitors. Following incubation on ice for 5 min, samples were centrifuged at 1000 g for 5 min. RIPA lysis buffer was used to lyse sample pellets, as described above. Supernatants (cytoplasmic extracts) were collected and centrifuged again at 1000 g for 5 min to remove any residual debris. Extract protein concentrations were normalized using a Bradford assay.
RNA interference
Cells (107 cells/transfection) were transfected with 16 μg of siRNA oligonucleotides (Xeragon/Qiagen) by electroporation. Gene-specific targeting of Smac was performed using a previously described oligonucleotide corresponding to nucleotides 156–176 of the coding sequence of Smac [38]. As a negative control, a previously described oligonucleotide corresponding to nucleotides 322–342 of GFP (green fluorescent protein) was utilized [38]. At 24 h after electroporation, dead cells were removed by centrifugation at 400 g for 20 min on a Ficoll-Pique PLUS density cushion. At 48 h after transfection, CD30 was stimulated on the transfected cells for 2 h as described above. Following CD30 stimulation, total RNA was isolated from half of the cells and subjected to real-time reverse transcription–PCR. The remaining fraction of cells was used for whole-cell lysate preparation and Western blot analysis.
Real-time reverse transcription–PCR
Karpas 299 cells were transfected with plasmids or oligonucleotides, stimulated with CD30L for 2 h as described above and then washed with PBS. Total RNA was isolated using the RNeasy minikit (Qiagen) according to the manufacturer's instructions. Reverse transcription with random hexamer primers and MultiScribe™ reverse transcriptase (Applied Biosystems) was performed on 100 ng of total RNA. The indicated target assays were performed on 1 μl of cDNA using Taqman probes, according to the manufacturer's instructions, on an Applied Biosystems 7500 real-time PCR system. Each target assay was performed in triplicate and normalized to glyceraldehyde-3-phosphate dehydrogenase.
Ubiquitin precipitation
To detect His6–ubiquitin-conjugated proteins, cell lysates were prepared in 8 M urea lysis buffer (300 mM NaCl, 0.5% Nonidet P40, 50 mM NaPO4, 50 mM Tris/HCl, pH 8.0, and 1 mM PMSF) followed by sonication (25 pulses at output control 2.5, 75% duty cycle), normalized for protein content, and incubated with Ni-NTA–agarose beads for 2 h at 4 °C. Agarose beads were recovered by centrifugation at 500 g for 1 min and washed in 8 M urea lysis buffer, and precipitated proteins were eluted by adding LDS sample buffer (Invitrogen) and heating samples for 10 min at 95 °C. Recovered proteins were subsequently separated by electrophoresis, and examined by immunoblot analysis.
RESULTS
CD30 signalling activates the non-canonical NF-κB pathway and the degradation of both c-IAP1 and TRAF2
Ligand-mediated activation of many members of the TNFR superfamily, including CD30, initiates a series of intracellular signal transduction cascades [41]. The non-canonical NF-κB signalling pathway is activated by CD30, and is thought to utilize the signalling intermediate TRAF2, which binds directly to the cytoplasmic tail of CD30 [4–7]. In previous studies, we found that receptor activation rapidly induced the degradation of TRAF2 [10,37], and we proposed a model in which the depletion of TRAF2 serves both to ‘reset’ the signalling pathway and to facilitate cross-talk with other classes of receptors that also utilize TRAF2. Since c-IAP1 is a known TRAF2-binding protein that has been described recently as a regulator of NF-κB [32,35,42–44], we sought to determine whether c-IAP1 was also depleted following receptor activation, along with TRAF2, or whether the loss of TRAF2 displaced the c-IAP1 protein.
To compare the stability of c-IAP1 before and after receptor activation, we employed an experimental approach in which CD30+ ALCL cells are activated by CD30L, using an adherent cell line in which CD30L has been stably expressed [10]. ALCL cells were layered on to adherent CD30L-expressing cells or control cells, and c-IAP1 levels in the ALCL cells were subsequently examined between 0 and 5 h by immunoblotting. Interestingly, activation by CD30L resulted in a rapid loss of c-IAP1 (Figure 1A), concomitant with the loss of TRAF2, which we described previously using this system as a model to examine the induction of apoptosis, NF-κB activation and cell-cycle arrest in different cellular populations [10]. Importantly, CD30 activation also induced the non-canonical NF-κB pathway, as monitored by analysis of the processing of the p100 NF-κB subunit to its mature p52 form (Figure 1A, top panel). Under these conditions, a slight loss of p105 NF-κB was observed, presumably indicative of processing to p50, although the basal levels of p50 were found to be relatively high even in the absence of CD30 activation (Figure 1A, middle panel). In the light of models proposed recently in which c-IAP1 functions to suppress the levels and activities of factors involved in the non-canonical NF-κB pathway, notably NIK (NF-κB-inducing kinase) and RIP1 (receptor-interacting protein 1), through mechanisms thought to involve the E3 ubiquitin ligase activity of c-IAP1 [32,35,42–44], these findings suggest a mechanism by which CD30 signalling activates the non-canonical pathway by triggering c-IAP proteins for degradation.
Figure 1. CD30 activation induces the degradation of both c-IAP1 and TRAF2.
(A) Karpas 299 cells were layered on to CHO cells, or CD30L+ CHO cells, for the indicated periods of time. Cells were recovered, lysed in RIPA buffer and immunoblotted for p100/p52, p105/p50, c-IAP1 and TRAF2 as indicated. (B) HEK-293 cells were transfected with the indicated combinations of c-IAP1, wild-type TRAF2 and DN-TRAF2, together with a control plasmid or a constitutively active form of CD30. At 24 h after transfection, cell lysates were prepared, and immunoblot analysis was performed for HA–c-IAP1 and TRAF2. (C) HEK-293 cells were transfected with plasmids encoding TRAF2, HA–c-IAP1 or HA–c-IAP1 H588A, together with a control plasmid or a constitutively active form of CD30, as indicated. Cells were harvested and lysates were analysed by immunoblot as described in (B). Equivalent protein loading was verified by immunoblotting for β-actin.
Receptor-induced degradation of the TRAF2–c-IAP1 complex requires the E3 ubiquitin ligase activity of TRAF2, but not that of c-IAP1
Previous studies found that receptor-induced degradation of TRAF2 required an intact N-terminal RING domain [37], and that the TRAF2 RING functions as an E3 ubiquitin ligase [45]. To determine whether the RING domain of TRAF2 is required for the receptor-mediated loss of c-IAP1 protein, a previously established system was employed in which CD30 signalling is mimicked by a constitutively active CD28–CD30 chimaera [4]. HEK-293 cells were transiently transfected with plasmids encoding wild-type TRAF2 or a RING-deleted dominant-negative TRAF2 protein (DN-TRAF2 [46]), together with plasmids encoding c-IAP1 and activated CD30, as detailed in Figure 1(B). Co-expression of c-IAP1 with TRAF2 resulted in a stabilized form of c-IAP1 (Figure 1B, top panel, compare lanes 3 and 5) (R.A. Csomos and C.S. Duckett, unpublished work). Consistent with previous findings, DN-TRAF2 was not degraded when co-expressed with active CD30 (Figure 1B, middle panel, lanes 7 and 8). Interestingly, co-expression of activated CD30 with DN-TRAF2 consistently resulted in the detection of additional TRAF2 and c-IAP1 species with higher apparent molecular masses (Figure 1B, compare lanes 6 and 8), suggesting additional post-translational modification of these proteins. Importantly, under the same conditions and in the same lysates, c-IAP1 also did not degrade (Figure 1B, top panel, lanes 7 and 8), indicating that the RING domain of TRAF2 is required for the degradation of c-IAP1 by CD30.
Since c-IAP1 also functions as an E3 ubiquitin ligase, mediated by a C-terminal RING domain, we examined the involvement of the RING domain in the degradative process of c-IAP1, as mediated by receptor activation. The E3 ubiquitin ligase activity of c-IAP1 was rendered inert by changing a critical histidine residue at position 588 within the RING domain to alanine (H588A) [26,47]. HEK-293 cells were subsequently transfected with plasmids encoding either wild-type c-IAP1 or c-IAP1 H588A and TRAF2 in combination with a control plasmid or a constitutively signalling form of CD30 (Figure 1C). The E3 ubiquitin ligase-deficient c-IAP1 H588A was readily detected, and was not stabilized further by the inclusion of TRAF2 (Figure 1C, lane 3). Upon co-expression with the constitutively active CD30 and subsequent analysis of total detergent-soluble proteins by immunoblotting, cellular TRAF2 levels were observed to be markedly reduced relative to cells in which the control vector was included (Figure 1C, compare lanes 2–4 with lanes 6–8), consistent with previous reports [10,37]. Similarly, c-IAP1 levels were reduced in the presence of CD30 signalling (Figure 1B, compare lane 5 with lane 6, and Figure 1C, compare lane 3 with lane 7), supporting the data with endogenous c-IAP1 described in Figure 1(A). Interestingly, the c-IAP1 H588A RING mutant was also degraded (Figure 1C, compare lane 4 with lane 8), indicating that the E3 ubiquitin ligase activity of c-IAP1 is not required for the degradation of c-IAP1 or TRAF2, as induced by CD30 signalling. This finding that the E3 ubiquitin ligase activity of c-IAP1 is not required for degradation following CD30 activation contrasts with the requirement of the RING of TRAF2 for CD30-mediated degradation.
Cytosolic Smac selectively induces autoubiquitination and proteasome-mediated degradation of c-IAP1, but not TRAF2
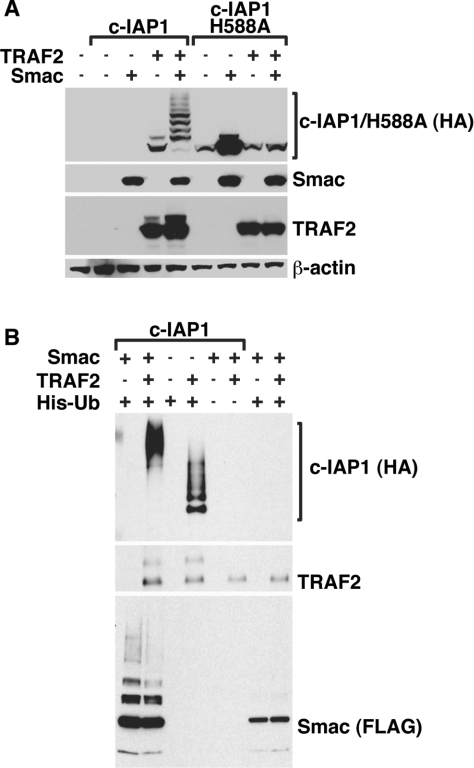
The c-IAP antagonist Smac has been shown to regulate the levels and activity of several IAP proteins following the release of Smac from mitochondria into the cytosol [48]. Interestingly, despite its mitochondrial targeting, cytosolic Smac was readily detectable in ALCL and HL cells (Supplementary Figure S1 at http://www.BiochemJ.org/bj/420/bj4200083add.htm). To determine whether the mechanism by which Smac targets c-IAP1 for degradation was related to CD30-induced degradation of c-IAP1, we used a mature cytosolic version of Smac that bypasses mitochondria [49]. Expression of mature Smac resulted in a decrease in unmodified c-IAP1 protein and a ladder-like appearance of higher c-IAP1 moieties (Figure 2A). In contrast, the expression of mature Smac had no significant impact on TRAF2 expression levels.
Figure 2. Cytosolic Smac triggers the autoubiquitination and degradation of c-IAP1, but not TRAF2.
(A) HEK-293 cells were transiently transfected with HA-tagged c-IAP1 or the c-IAP1 H588A mutant expression plasmid along with plasmids encoding TRAF2 and mature Smac. At 24 h after transfection, cell lysates were prepared, and immunoblot analysis was performed for HA–c-IAP1/H588A mutant, Smac and TRAF2. (B) HEK-293 cells were transfected with combinations of Smac–FLAG, HA–c-IAP1, TRAF2 and His6–ubiquitin (His-Ub). At 24 h after transfection, cell lysates were prepared in 8 M urea buffer and precipitated using Ni-NTA–agarose beads. The presence of HA–c-IAP1, TRAF2 and Smac–FLAG in the precipitate was determined by immunoblotting. Equivalent protein loading was verified in (A) and (B) by immunoblotting for β-actin.
Since c-IAP1 harbours E3 ubiquitin ligase activity, a likely possibility is that Smac might facilitate the autoubiquitination of c-IAP1. To test this possibility, HEK-293 cells were transfected with the c-IAP1 H588A mutant in the absence or presence of Smac. Unlike the pronounced ladder-like appearance of higher c-IAP1 moieties observed for wild-type c-IAP1 in the presence of Smac, expression of Smac had no effect on the levels of the E3 ubiquitin ligase-deficient c-IAP1 (Figure 2A). These findings suggest that the mechanism by which Smac regulates c-IAP1 expression is distinct from the mechanism by which c-IAP1 and TRAF2 are targeted following CD30 activation, since only c-IAP1 protein levels are altered in the presence of Smac, and an intact RING is necessary for Smac-mediated c-IAP1 degradation.
The laddering pattern of c-IAP1 that was observed in the presence of Smac is highly suggestive of polyubiquitination. To determine whether Smac could function to induce ubiquitination of c-IAP1, HEK-293 cells were transfected with a plasmid encoding His6–ubiquitin and combinations of c-IAP1, TRAF2 and Smac. Cellular lysates were prepared, and ubiquitinated proteins were precipitated with Ni-NTA–agarose beads. Smac expression induced the accumulation of high-molecular-mass ubiquitinated c-IAP1 (Figure 2B). Interestingly, the ubiquitination pattern of TRAF2 appeared to be unaffected by Smac (Figure 2B, middle panel). Since Smac could induce the polyubiquitination of c-IAP1, but not TRAF2, these data support a model in which Smac functions to specifically target c-IAP1 for autoubiquitination and degradation.
c-IAP1 is an inhibitor of the non-canonical NF-κB pathway, and Smac neutralizes this inhibition
Distinct targeting of c-IAP1 alone was observed in the presence of Smac, whereas CD30 activation resulted in the degradation of both c-IAP1 and TRAF2. Since the loss of c-IAP1 has been associated with the activation of the non-canonical NF-κB pathway, we evaluated the effect of Smac on non-canonical NF-κB activation by examining the processing of the p100 protein to its transcriptionally active p52 form. Expression of TRAF2 resulted in a significant increase in p100 processing to p52, confirming previous studies in which TRAF2 has been implicated in the non-canonical NF-κB pathway (Figure 3, lane 2) [10]. However, co-expression of c-IAP1 with TRAF2 inhibited the ability of TRAF2 to induce p100 processing (Figure 3, lane 3), consistent with reports that c-IAP1 functions to suppress the non-canonical pathway by targeting NIK for degradation [35,44]. Interestingly, expression of mature Smac alone induced p100 processing (Figure 3, compare lane 1 with lane 6), and Smac counteracted the inhibitory effects of c-IAP1 on p100 processing (Figure 3, lane 5). In the presence of Smac, the high-molecular-mass ladder of polyubiquitinated c-IAP1 was again observed, whereas TRAF2 appeared to be unaffected. Therefore Smac was found to activate the non-canonical NF-κB signalling pathway, and this activation occurred concomitantly with induced autoubiquitination of c-IAP1.
Figure 3. c-IAP1 is an inhibitor of the non-canonical NF-κB pathway, and Smac neutralizes c-IAP1-mediated NF-κB inhibition.
HEK-293 cells were transfected with mature Smac–FLAG, HA–c-IAP1 and TRAF2. At 24 h after transfection, cell lysates were prepared, and immunoblot analysis was performed for p100/p52, HA–c-IAP1, TRAF2 and Smac–FLAG. Equivalent protein loading was confirmed by immunoblotting for β-actin.
A synthetic IAP antagonist selectively targets c-IAP1 for autoubiquitination
The data presented in Figures 2 and 3 indicate that Smac induces selective autoubiquitination of c-IAP1, while leaving TRAF2 unaffected, and that this process leads to the activation of the noncanonical NF-κB pathway by inducing the processing of the p100 NF-κB subunit. Since a number of synthetic cell-permeable IAP antagonists have been developed based on the Smac–IAP interface, we examined the possibility that an IAP antagonist, AEG40730 [32], would function similarly to mature Smac, by activating the autoubiquitination of c-IAP1, releasing TRAF2 and triggering p100 processing. Interestingly, AEG40730 was capable of degrading ectopically expressed c-IAP1 in a similar fashion to mature Smac, as increasing concentrations of AEG40730 resulted in decreasing levels of c-IAP1 (Figure 4A). Importantly, no differences in the levels of TRAF2 were observed.
Figure 4. The IAP antagonist AEG40730 induces the degradation of c-IAP1, but not TRAF2.
(A) HEK-293 cells were transfected with HA–c-IAP1 and TRAF2. At 24 h after transfection, cells were treated with 0, 1, 5, 10 or 25 nM AEG40730 or DMSO. At 24 h after treatment with AEG40730, cell lysates were prepared, and immunoblot analysis was performed for HA–c-IAP1 and TRAF2. Equivalent protein loading was confirmed by immunoblotting for β-actin. (B) HEK-293 cells were transfected with HA–c-IAP1 H588A. At 24 h after transfection, cells were treated with 0, 1, 5, 10 or 25 nM AEG40730 or DMSO. At 24 h after treatment with AEG40730, cell lysates were prepared and immunoblotted for HA–c-IAP1 H588A. As a control, c-IAP1 H588A was transfected with mature Smac–FLAG. Equivalent protein loading was verified by immunoblotting for β-actin.
Given the apparent similarities between the effects of mature Smac and the synthetic IAP antagonist, we sought to test whether AEG40730 could induce c-IAP1 autoubiquitination by analysis of the c-IAP1 E3 ubiquitin ligase mutant H588A in the presence of increasing doses of AEG40730. Similarly to the results observed for c-IAP1 H588A in the presence of mature Smac, the RING mutant of c-IAP1 was insensitive to AEG40730 treatment at any tested dosage (Figure 4B). These findings strongly support the notion that AEG40730 functions analogously to mature Smac in the ability to stimulate autoubiquitination of c-IAP1.
AEG40730 triggers processing of p100 to p52
Since the IAP antagonist AEG40730 appeared to trigger c-IAP1 autoubiquitination in a similar fashion to mature Smac, we next investigated the ability of the IAP antagonist to activate the non-canonical NF-κB pathway through c-IAP1 degradation. To determine whether AEG40730 was capable of activating the alternative NF-κB pathway, we examined p100 processing to p52. HEK-293 (Figure 5A) and Karpas 299 (Figure 5B) cells were treated for 24 h with increasing doses of AEG40730. As before, treatment with AEG40730 induced the loss of endogenous c-IAP1, whereas endogenous TRAF2 levels were unaffected (Figure 5). Importantly, treatment with AEG40730 also resulted in increased processing of p100 to p52, already evident by 6 h (Figure 5C). Although a slight reduction in TRAF2 levels was observed at the highest concentrations or longest time-points, c-IAP1 expression was essentially abrogated within minutes of AEG40730 addition, and at the lowest concentrations used. Therefore AEG40730 was capable of selectively promoting the degradation of c-IAP1, but not TRAF2, and resulted in the concomitant processing of p100, similar to the results observed with mature Smac.
Figure 5. Rapid degradation of c-IAP1, but not TRAF2, precedes activation of the non-canonical NF-κB pathway.
The IAP antagonist AEG40730 triggers the processing of p100 to its p52 form. (A) HEK-293 cells or (B) Karpas 299 cells were treated with DMSO or 0, 1, 5, 10 or 25 nM AEG40730 and incubated for 24 h. (C) Karpas 299 cells were treated for 0–48 h with 25 nM AEG40730 or DMSO. Following treatment with AEG40730, cell lysates were prepared, and immunoblot analysis was performed for c-IAP1, TRAF2 and p52/p100. Equivalent protein loading was confirmed by immunoblotting for β-actin. n.s., non-specific.
Both Smac and the IAP antagonist AEG40730 potentiate transcription of endogenous NF-κB target genes
The finding that both Smac and the synthetic IAP antagonist can induce the processing of p100 suggested that these agents might also modulate NF-κB-dependent transcription, as induced by a defined signal from a TNFR superfamily member such as CD30. To test this possibility, Karpas 299 cells were transfected with a plasmid encoding mature Smac or with an empty control plasmid, and subsequently stimulated with control CHO or CD30L+ CHO cells. Following stimulation, RNA was isolated and qRT-PCR (quantitative real-time PCR) analysis was performed on a panel of previously described NF-κB-responsive genes [10]. Interestingly, the induction of NF-κB-dependent genes by CD30 was augmented in cells expressing mature Smac (Figure 6A). Similarly, pre-treatment of Karpas 299 cells with AEG40730 before CD30 activation resulted in an augmentation of NF-κB-dependent transcription (Figure 6B), almost identical with that observed following ectopic expression of mature Smac. Therefore both Smac and the IAP antagonist AEG40730 were found to function indistinguishably in their ability to potentiate NF-κB-dependent transcription in response to CD30 activation.
Figure 6. Smac is a modulator of CD30-mediated signalling and activates transcription of endogenous NF-κB-dependent genes.
(A) Karpas 299 cells were electroporated with an expression plasmid encoding mature Smac–FLAG, or an empty control plasmid, as indicated. At 24 h after transfection, cells were layered on to CD30L+ CHO cells or control CHO cells for 2 h. Total RNA was extracted and reverse-transcribed, and cDNA was analysed by qRT-PCR using Taqman probes for the indicated NF-κB-dependent genes. (B) Karpas 299 cells were treated with 25 nM AEG40730 or DMSO. At 24 h after following treatment, cells were layered on to CD30L+ CHO cells or control CHO cells for 2 h. Total RNA was extracted and reverse-transcribed, and cDNA was analysed by qRT-PCR using Taqman probes for the indicated NF-κB-dependent genes. (C) Karpas 299 cells were electroporated with double-stranded siRNA oligonucleotides targeting Smac (siSmac) or an irrelevant sequence (siGFP), as indicated. At 48 h after transfection, cells were layered on to CD30L+ CHO cells or control CHO cells, as indicated, for 2 h. Total RNA was extracted and reverse-transcribed, and cDNA was analysed by qRT-PCR using Taqman probes for the indicated NF-κB-responsive genes. ICAM, intercellular adhesion molecule; IκBα, inhibitory κB α.
Although the findings shown in Figures 6(A) and 6(B) demonstrate a potentiation of NF-κB-dependent transcription in response to an IAP antagonist or overexpression of mature Smac, it remained unclear whether endogenous Smac could play a role in NF-κB activation. To address this issue, endogenous Smac expression was experimentally suppressed using an oligonucleotide-mediated siRNA approach, and the expression of defined NF-κB-target genes was subsequently analysed by qRT-PCR in response to CD30 activation. Interestingly, Karpas 299 cells with reduced Smac levels demonstrated a significant decrease in NF-κB-responsive genes following CD30 activation, compared with cells transfected with control oligonucleotides and stimulated in parallel through CD30L (Figure 6C). Altogether, these data indicate that Smac plays a role in the expression of NF-κB target genes, consistent with its role in controlling c-IAP1 levels.
DISCUSSION
In the present study, we examined the cellular regulatory mechanisms for c-IAP1, which has been shown recently to function as an NF-κB inhibitory factor [32,42–44]. We found that c-IAP1 can be regulated by two distinct mechanisms (Figure 7). During CD30 signalling, c-IAP1 is targeted for degradation through its association with TRAF2 (Figure 1), consistent with a recent study examining the effects of signalling through another TRAF-binding TNFR superfamily member, FN14 [50]. On the other hand, cytosolic Smac selectively induces the autoubiquitination and proteasomal degradation of c-IAP1, whereas TRAF2 is unaffected by Smac (Figures 2 and 3). These constitute two endogenous cellular mechanisms by which c-IAP1 is controlled. Interestingly, a number of earlier reports describe the autoubiquitination and degradation of c-IAP proteins after treatment with IAP antagonists [35,44,48]. The present study indicates that endogenous Smac plays a similar role and places this factor in the regulation of NF-κB signalling through its effects on c-IAP1. In addition, the present study provides functional evidence in support of the original biochemical screen where c-IAP proteins were found to associate with TNFR2 [21].
Figure 7. Model of the two mechanisms by which the NF-κB regulatory factor c-IAP1 can be regulated.
Smac and IAP antagonists induce the selective autoubiquitination and degradation of c-IAP1. In the presence of mature Smac, c-IAP1, but not TRAF2, is degraded. Smac promotes processing of p100 to p52 and augments non-canonical NF-κB signalling. CD30 activation promotes the degradation of c-IAP1 and TRAF2. Degradation of c-IAP1 and TRAF2 does not require the E3 ubiquitin ligase activity of c-IAP1, but does requires the RING domain in TRAF2. CD30 signalling results in the induction of NF-κB through processing of p100 to the active p52 subunit.
The enhanced expression of CD30 in several forms of leukaemia and lymphoma suggest that CD30 signalling is probably involved in the pathogenesis of these tumours [8,9]. Previous studies have described CD30 as an activator of NF-κB signalling and showed that CD30 induces the degradation of TRAF2 [10,37]. However, the significance of TRAF2 depletion on NF-κB activation remained unclear. In the present study, we show that CD30 activation results in the degradation of not only TRAF2, but also c-IAP1 (Figure 1). Interestingly, the degradation of TRAF2 was shown previously to require the RING domain of TRAF2 [37], whereas the findings shown in Figure 1(B) indicate that the E3 ubiquitin ligase activity of the RING domain in c-IAP1 is not required for CD30-mediated degradation. Although the exact downstream targets of c-IAP1 remain unclear, c-IAP1 has been reported to function as an E3 ubiquitin ligase for NIK, RIP1 and NEMO (NF-κB essential modulator) and to function as an inhibitor of NF-κB activation [32,35,42–44,51]. In this context, receptor-mediated degradation of c-IAP1 is expected to facilitate NF-κB activation. These data also provide support for the rationale of targeting c-IAP proteins to promote cell death through the activation of an autocrine TNF-dependent pathway. Since several immunotherapeutic approaches are being tested for CD30+ malignancies, the targeting of IAP proteins in combination with CD30 might be especially effective.
The classical role described for IAP proteins was to protect against cell death. Although XIAP has been shown to bind to and inhibit caspases, the c-IAP proteins do not inhibit caspase activity. This suggests a non-apoptotic role for the c-IAP1–Smac interaction. Indeed, earlier reports have described a cytosolic localization of Smac in non-apoptotic cells. Deng et al. [33] reported the selective release of Smac from mitochondria in colon carcinoma cells following TNF treatment, and others reported the existence of an exclusively cytoplasmic variant of Smac, which is thought to be generated by alternative splicing of the Smac mRNA transcript [34]. After observing Smac in the cytosol of healthy ALCL and HL cells (Supplementary Figure S1), we examined the regulation of c-IAP1 by mature Smac or an IAP antagonist. We found that c-IAP1 underwent autoubiquitination in the presence of mature Smac or the IAP antagonist (Figures 2, 4 and 5), supporting previous studies [31]. Interestingly, Smac not only induced autoubiquitination of c-IAP1, but also activated the non-canonical NF-κB pathway (Figures 3, 5 and 6), as demonstrated by the ability of mature Smac to promote processing of p100 to p52 (Figures 3, 5 and 6).
A provocative earlier study revealed the ability of Smac to be recruited into the lymphotoxin-β receptor signalling complex [52]. Since we found that Smac caused increased p100 processing and existed in the cytosol of healthy cells, we examined whether Smac would affect CD30-mediated activation of NF-κB. Indeed, ectopic expression of Smac was found to potentiate transcription of endogenous NF-κB-responsive genes (Figures 6A and 6B). Suppression of Smac also resulted in a reduction of NF-κB induction (Figure 6C). Therefore, by targeting c-IAP proteins for degradation, Smac can function as a highly versatile molecule with the ability to regulate not only caspase-mediated apoptosis, but also caspase-independent signalling pathways including, but not restricted to, NF-κB activation. In this context, Smac might work differently to promote cell survival through NF-κB target genes. It will be of great importance to fully understand the intracellular cues that serve to trigger the abilities of Smac and potentially other IBM-containing proteins to function physiologically to regulate these processes.
ACKNOWLEDGEMENTS
We thank Dr Phil Barker (Department of Neurology and Neurosurgery, McGill University, Montreal, Quebec, Canada), Dr Ezra Burstein (University of Texas Southwestern Medical Centre, Dallas, TX, U.S.A.) and members of the Duckett laboratory for their insightful suggestions and reagents, Aegera Therapeutics for AEG40730 and Dr John Silke (La Trobe University, Melbourne, Victoria, Australia) for antibodies.
FUNDING
This work was supported in part by the University of Michigan Biological Scholars Program, Department of Defense IDEA Award [grant number W81XWH-04-1-0891xyr], National Institutes of Health [grant number GM067827] and the Sandler Foundation Award to C. S. D., Cancer Biology Pre-doctoral Training Award T32 [grant number CA09676] from the National Institutes of Health to R. A. C., Biomedical Research Council Post-Doctoral Award from the University of Michigan to C. W. W., Lung Immunopathology Training Grant T32 from the National Institutes of Health to S. G. and Department of Defense Award [grant number W81XWH-06-1-0429] to K. A. O.
Online data
References
- 1.Stein H., Foss H. D., Dürkop H., Marafioti T., Delsol G., Pulford K., Pileri S., Falini B. CD30+ anaplastic large cell lymphoma: a review of its histopathologic, genetic, and clinical features. Blood. 2000;96:3681–3695. [PubMed] [Google Scholar]
- 2.Younes A., Carbone A. CD30/CD30 ligand and CD40/CD40 ligand in malignant lymphoid disorders. Int. J. Biol. Markers. 1999;14:135–143. doi: 10.1177/172460089901400303. [DOI] [PubMed] [Google Scholar]
- 3.Horie R., Watanabe T., Ito K., Morisita Y., Watanabe M., Ishida T., Higashihara M., Kadin M. Cytoplasmic aggregation of TRAF2 and TRAF5 proteins in the Hodgkin–Reed–Sternberg cells. Am. J. Pathol. 2002;160:1647–1654. doi: 10.1016/S0002-9440(10)61112-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duckett C. S., Gedrich R. W., Gilfillan M. C., Thompson C. B. Induction of nuclear factor κB by the CD30 receptor is mediated by TRAF1 and TRAF2. Mol. Cell. Biol. 1997;17:1535–1542. doi: 10.1128/mcb.17.3.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee S. Y., Lee S. Y., Kandala G., Liou M. L., Liou H. C., Choi Y. CD30/TNF receptor-associated factor interaction: NF-κB activation and binding specificity. Proc. Natl. Acad. Sci. U.S.A. 1996;93:9699–9703. doi: 10.1073/pnas.93.18.9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aizawa S., Nakano H., Ishida T., Horie R., Nagai M., Ito K, Yagita H., Okumura K., Inoue J., Watanabe T. Tumor necrosis factor receptor-associated factor (TRAF) 5 and TRAF2 are involved in CD30-mediated NF-κB activation. J. Biol. Chem. 1997;272:2042–2045. doi: 10.1074/jbc.272.4.2042. [DOI] [PubMed] [Google Scholar]
- 7.Boucher L. M., Marengere L. E. M., Lu Y., Thukral S., Mak T. W. Binding sites of cytoplasmic effectors TRAF1, 2, and 3 on CD30 and other members of the TNF receptor superfamily. Biochem. Biophys. Res. Commun. 1997;233:592–600. doi: 10.1006/bbrc.1997.6509. [DOI] [PubMed] [Google Scholar]
- 8.Horie R., Watanabe M., Ishida T., Koiwa T., Aizawa S., Itoh K., Higashihara M., Kadin M. E., Watanabe T. The NPM–ALK oncoprotein abrogates CD30 signaling and constitutive NF-κB activation in anaplastic large cell lymphoma. Cancer Cell. 2004;5:353–364. doi: 10.1016/s1535-6108(04)00084-4. [DOI] [PubMed] [Google Scholar]
- 9.Horie R., Watanabe T., Morishita Y., Ito K., Ishida T., Kanegae Y., Saito I., Higashihara M., Mori S., Kadin M. E., Watanabe T. Ligand-independent signaling by overexpressed CD30 drives NF-κB activation in Hodgkin–Reed–Sternberg cells. Oncogene. 2002;21:2493–2503. doi: 10.1038/sj.onc.1205337. [DOI] [PubMed] [Google Scholar]
- 10.Wright C. W., Rumble J. M., Duckett C. S. CD30 activates both the canonical and alternative NF-κB pathways in anaplastic large cell lymphoma cells. J. Biol. Chem. 2007;282:10252–10262. doi: 10.1074/jbc.M608817200. [DOI] [PubMed] [Google Scholar]
- 11.Hayden M. S., Ghosh S. Shared principles in NF-κB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 12.Srinivasula S. M., Ashwell J. D. IAPs: what's in a name? Mol. Cell. 2008;30:123–135. doi: 10.1016/j.molcel.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter A. M., LaCasse E. C., Korneluk R. G. The inhibitors of apoptosis (IAPs) as cancer targets. Apoptosis. 2007;12:1543–1568. doi: 10.1007/s10495-007-0087-3. [DOI] [PubMed] [Google Scholar]
- 14.Mufti A. R., Burstein E., Csomos R. A., Graf P. C., Wilkinson J. C., Dick R. D., Challa M., Son J. K., Bratton S. B., Su G. L., et al. XIAP Is a copper binding protein deregulated in Wilson's disease and other copper toxicosis disorders. Mol. Cell. 2006;21:775–785. doi: 10.1016/j.molcel.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 15.Rigaud S., Fondaneche M. C., Lambert N., Pasquier B., Mateo V., Soulas P., Galicier L., Le Deist F., Rieux-Laucat F., Revy P., et al. XIAP deficiency in humans causes an X-linked lymphoproliferative syndrome. Nature. 2006;444:110–114. doi: 10.1038/nature05257. [DOI] [PubMed] [Google Scholar]
- 16.Eckelman B. P., Salvesen G. S., Scott F. L. Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep. 2006;7:988–994. doi: 10.1038/sj.embor.7400795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaux D. L., Silke J. IAPs, RINGs and ubiquitylation. Nat. Rev. Mol. Cell Biol. 2005;6:287–297. doi: 10.1038/nrm1621. [DOI] [PubMed] [Google Scholar]
- 18.Yang Y., Fang S., Jensen J. P., Weissman A. M., Ashwell J. D. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]
- 19.Eckelman B. P., Salvesen G. S. The human anti-apoptotic proteins cIAP1 and cIAP2 bind but do not inhibit caspases. J. Biol. Chem. 2006;281:3254–3260. doi: 10.1074/jbc.M510863200. [DOI] [PubMed] [Google Scholar]
- 20.Deveraux Q. L., Roy N., Stennicke H. R., Van Arsdale T., Zhou Q., Srinivasula S. M., Alnemri E. S., Salvesen G. S., Reed J. C. IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases. EMBO J. 1998;17:2215–2223. doi: 10.1093/emboj/17.8.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothe M., Pan M.-G., Henzel W. J., Ayres T. M., Goeddel D. V. The TNFR2–TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell. 1995;83:1243–1252. doi: 10.1016/0092-8674(95)90149-3. [DOI] [PubMed] [Google Scholar]
- 22.Imoto I., Yang Z.-Q., Pimkhaokham A., Tsuda H., Shimada Y., Imamura M., Ohki M., Inazawa J. Identification of cIAP1 as a candidate target gene within an amplicon at 11q22 in esophageal squamous cell carcinomas. Cancer Res. 2001;61:6629–6634. [PubMed] [Google Scholar]
- 23.Zender L., Spector M. S., Xue W., Flemming P., Cordon-Cardo C., Silke J., Fan S. T., Luk J. M., Wigler M., Hannon G. J., et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu L., Zhu J., Hu X., Zhu H., Kim H. T., Labaer J., Goldberg A., Yuan J. c-IAP1 cooperates with Myc by acting as a ubiquitin ligase for Mad1. Mol. Cell. 2007;28:914–922. doi: 10.1016/j.molcel.2007.10.027. [DOI] [PubMed] [Google Scholar]
- 25.Vaux D. L., Silke J. Mammalian mitochondrial IAP binding proteins. Biochem. Biophys. Res. Commun. 2003;304:499–504. doi: 10.1016/s0006-291x(03)00622-3. [DOI] [PubMed] [Google Scholar]
- 26.Du C., Fang M., Li Y., Li L., Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 27.Verhagen A. M., Ekert P. G., Pakusch M., Silke J., Connolly L. M., Reid G. E., Moritz R. L., Simpson R. J., Vaux D. L. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell. 2000;102:43–53. doi: 10.1016/s0092-8674(00)00009-x. [DOI] [PubMed] [Google Scholar]
- 28.Hu S., Yang X. Cellular inhibitor of apoptosis 1 and 2 are ubiquitin ligases for the apoptosis inducer Smac/DIABLO. J. Biol. Chem. 2003;278:10055–10060. doi: 10.1074/jbc.M207197200. [DOI] [PubMed] [Google Scholar]
- 29.Arnt C. R., Chiorean M. V., Heldebrant M. P., Gores G. J., Kaufmann S. H. Synthetic Smac/DIABLO peptides enhance the effects of chemotherapeutic agents by binding XIAP and cIAP1 in situ. J. Biol. Chem. 2002;277:44236–44243. doi: 10.1074/jbc.M207578200. [DOI] [PubMed] [Google Scholar]
- 30.Schimmer A. D., Welsh K., Pinilla C., Wang Z., Krajewska M., Bonneau M. J., Pedersen I. M., Kitada S., Scott F. L., Bailly-Maitre B., et al. Small-molecule antagonists of apoptosis suppressor XIAP exhibit broad antitumor activity. Cancer Cell. 2004;5:25–35. doi: 10.1016/s1535-6108(03)00332-5. [DOI] [PubMed] [Google Scholar]
- 31.Wu H., Tschopp J., Lin S. C. Smac mimetics and TNFα: a dangerous liaison? Cell. 2007;131:655–658. doi: 10.1016/j.cell.2007.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertrand M. J., Milutinovic S., Dickson K. M., Ho W. C., Boudreault A., Durkin J., Gillard J. W., Jaquith J. B., Morris S. J., Barker P. A. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol. Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 33.Deng Y., Ren X., Yang L., Lin Y., Wu X. A JNK-dependent pathway is required for TNFα-induced apoptosis. Cell. 2003;115:61–70. doi: 10.1016/s0092-8674(03)00757-8. [DOI] [PubMed] [Google Scholar]
- 34.Roberts D. L., Merrison W., MacFarlane M., Cohen G. M. The inhibitor of apoptosis protein-binding domain of Smac is not essential for its proapoptotic activity. J. Cell Biol. 2001;153:221–228. doi: 10.1083/jcb.153.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vince J. E., Wong W. W., Khan N., Feltham R., Chau D., Ahmed A. U., Benetatos C. A., Chunduru S. K., Condon S. M., McKinlay M., et al. IAP antagonists target cIAP1 to induce TNFα-dependent apoptosis. Cell. 2007;131:682–693. doi: 10.1016/j.cell.2007.10.037. [DOI] [PubMed] [Google Scholar]
- 36.Duckett C. S., Li F., Wang Y., Tomaselli K. J., Thompson C. B., Armstrong R. C. Human IAP-like protein regulates programmed cell death downstream of Bcl-xL and cytochrome c. Mol. Cell. Biol. 1998;18:608–615. doi: 10.1128/mcb.18.1.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duckett C. S., Thompson C. B. CD30-dependent degradation of TRAF2: implications for negative regulation of TRAF signaling and the control of cell survival. Genes Dev. 1997;11:2810–2821. doi: 10.1101/gad.11.21.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilkinson J. C., Wilkinson A. S., Scott F. L., Csomos R. A., Salvesen G. S., Duckett C. S. Neutralization of Smac/Diablo by IAPs: a caspase-independent mechanism for apoptotic inhibition. J. Biol. Chem. 2004;279:51082–51090. doi: 10.1074/jbc.M408655200. [DOI] [PubMed] [Google Scholar]
- 39.Wilkinson J. C., Wilkinson A. S., Galban S., Csomos R. A., Duckett C. S. Apoptosis-inducing factor is a target for ubiquitination through interaction with XIAP. Mol. Cell. Biol. 2008;28:237–247. doi: 10.1128/MCB.01065-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilkinson J. C., Cepero E., Boise L. H., Duckett C. S. Upstream regulatory role for XIAP in receptor-mediated apoptosis. Mol. Cell. Biol. 2004;24:7003–7014. doi: 10.1128/MCB.24.16.7003-7014.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baud V., Karin M. Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol. 2001;11:372–377. doi: 10.1016/s0962-8924(01)02064-5. [DOI] [PubMed] [Google Scholar]
- 42.Gaither A., Porter D., Yao Y., Borawski J., Yang G., Donovan J., Sage D., Slisz J., Tran M., Straub C., et al. A Smac mimetic rescue screen reveals roles for inhibitor of apoptosis proteins in tumor necrosis factor-α signaling. Cancer Res. 2007;67:11493–11498. doi: 10.1158/0008-5472.CAN-07-5173. [DOI] [PubMed] [Google Scholar]
- 43.Petersen S. L., Wang L., Yalcin-Chin A., Li L., Peyton M., Minna J., Harran P., Wang X. Autocrine TNFα signaling renders human cancer cells susceptible to Smac-mimetic-induced apoptosis. Cancer Cell. 2007;12:445–456. doi: 10.1016/j.ccr.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Varfolomeev E., Blankenship J. W., Wayson S. M., Fedorova A. V., Kayagaki N., Garg P., Zobel K., Dynek J. N., Elliott L. O., Wallweber H. J., et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-κB activation, and TNFα-dependent apoptosis. Cell. 2007;131:669–681. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 45.Li X., Yang Y., Ashwell J. D. TNF-RII and c-IAP1 mediate ubiquitination and degradation of TRAF2. Nature. 2002;416:345–347. doi: 10.1038/416345a. [DOI] [PubMed] [Google Scholar]
- 46.Rothe M., Wong S. C., Henzel W. J., Goeddel D. V. A novel family of putative signal transducers associated with the cytoplasmic domain of the 75 kDa tumor necrosis factor receptor. Cell. 1994;78:681–692. doi: 10.1016/0092-8674(94)90532-0. [DOI] [PubMed] [Google Scholar]
- 47.Conze D. B., Albert L., Ferrick D. A., Goeddel D. V., Yeh W. C., Mak T., Ashwell J. D. Posttranscriptional downregulation of c-IAP2 by the ubiquitin protein ligase c-IAP1 In vivo. Mol. Cell. Biol. 2005;25:3348–3356. doi: 10.1128/MCB.25.8.3348-3356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Q. H., Du C. Smac/DIABLO selectively reduces the levels of c-IAP1 and c-IAP2 but not that of XIAP and livin in HeLa cells. J. Biol. Chem. 2004;279:16963–16970. doi: 10.1074/jbc.M401253200. [DOI] [PubMed] [Google Scholar]
- 49.Hunter A. M., Kottachchi D., Lewis J., Duckett C. S., Korneluk R. G., Liston P. A novel ubiquitin fusion system bypasses the mitochondria and generates biologically active Smac/DIABLO. J. Biol. Chem. 2003;278:7494–7499. doi: 10.1074/jbc.C200695200. [DOI] [PubMed] [Google Scholar]
- 50.Vince J. E., Chau D., Callus B., Wong W. W., Hawkins C. J., Schneider P., McKinlay M., Benetatos C. A., Condon S. M., Chunduru S. K., et al. TWEAK–FN14 signaling induces lysosomal degradation of a cIAP1–TRAF2 complex to sensitize tumor cells to TNFα. J. Cell Biol. 2008;182:171–184. doi: 10.1083/jcb.200801010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang E. D., Wang C. Y., Xiong Y., Guan K. L. A role for NF-κB essential modifier/IκB kinase-γ (NEMO/IKKγ) ubiquitination in the activation of the IκB kinase complex by tumor necrosis factor-α. J. Biol. Chem. 2003;278:37297–37305. doi: 10.1074/jbc.M303389200. [DOI] [PubMed] [Google Scholar]
- 52.Kuai J., Nickbarg E., Wooters J., Qiu Y., Wang J., Lin L. L. Endogenous association of TRAF2, TRAF3, cIAP1, and Smac with lymphotoxin β receptor reveals a novel mechanism of apoptosis. J. Biol. Chem. 2003;278:14363–14369. doi: 10.1074/jbc.M208672200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.