Abstract
PEPC [PEP(phosphoenolpyruvate) carboxylase] is a tightly controlled cytosolic enzyme situated at a major branchpoint in plant metabolism. Accumulating evidence indicates important functions for PEPC and PPCK (PEPC kinase) in plant acclimation to nutritional Pi deprivation. However, little is known about the genetic origin or phosphorylation status of native PEPCs from −Pi (Pi-deficient) plants. The transfer of Arabidopsis suspension cells or seedlings to −Pi growth media resulted in: (i) the marked transcriptional upregulation of genes encoding the PEPC isoenzyme AtPPC1 (Arabidopsis thaliana PEPC1), and PPCK isoenzymes AtPPCK1 and AtPPCK2; (ii) >2-fold increases in PEPC specific activity and in the amount of an immunoreactive 107-kDa PEPC polypeptide (p107); and (iii) In vivo p107 phosphorylation as revealed by immunoblotting of clarified extracts with phosphosite-specific antibodies to Ser-11 (which could be reversed following Pi resupply). Approx. 1.3 mg of PEPC was purified 660-fold from −Pi suspension cells to apparent homogeneity with a specific activity of 22.3 units · mg−1 of protein. Gel filtration, SDS/PAGE and immunoblotting demonstrated that purified PEPC exists as a 440-kDa homotetramer composed of identical p107 subunits. Sequencing of p107 tryptic and Asp-N peptides by tandem MS established that this PEPC is encoded by AtPPC1. Pi-affinity PAGE coupled with immunoblotting indicated stoichiometric phosphorylation of the p107 subunits of AtPPC1 at its conserved Ser-11 phosphorylation site. Phosphorylation activated AtPPC1 at pH 7.3 by lowering its Km(PEP) and its sensitivity to inhibition by L-malate and L-aspartate, while enhancing activation by glucose 6-phosphate. Our results indicate that the simultaneous induction and In vivo phosphorylation activation of AtPPC1 contribute to the metabolic adaptations of −Pi Arabidopsis.
Keywords: Arabidopsis, gene expression, Pi starvation, mass spectrometry, phosphoenolpyruvate carboxylase kinase (PPCK), protein phosphorylation
Abbreviations: Ab, antibody; anti-RcPEPC IgG, anti-(Ricinus communis PEPC) IgG; AtPPC1, Arabidopsis thaliana PEPC1; CAM, crassulacean acid metabolism; DTT, dithiothreitol; Glc-6-P, glucose 6-phosphate; MALDI, matrix-assisted laser-desorption ionization; MS medium, Murashige and Skoog medium; MS/MS, tandem MS; p107, 107-kDa PEPC polypeptide; oMALDI 2, orthogonal MALDI 2; PEP, phosphoenolpyruvate; PEPC, PEP carboxylase; +Pi, Pi-sufficient; −Pi, Pi-deficient; PP2A, protein phosphatase type-2A; PPCK, PEPC kinase; Q-TOF, quadrupole time-of-flight; QqTOF, quadrupole/quadrupole TOF; RT, reverse transcription
INTRODUCTION
PEPC [PEP (phosphoenolpyruvate) carboxylase] (EC 4.1.1.31) is a ubiquitous cytosolic enzyme in vascular plants that is also distributed widely in green algae and bacteria. It catalyses the irreversible β-carboxylation of PEP in the presence of HCO3− to yield oxaloacetate and Pi. PEPC plays a crucial role in C4 and CAM (crassulacean acid metabolism) photosynthesis, where it catalyses the initial fixation of atmospheric CO2. PEPC also fulfils several important non-photosynthetic functions, in particular the anaplerotic replenishment of tricarboxylic-acid-cycle intermediates consumed during biosynthesis and N2 assimilation. Owing to its location at a pivotal branchpoint in primary plant metabolism, PEPC is tightly controlled by a combination of fine metabolic controls, including allosteric effectors and reversible phosphorylation [1–3]. Allosteric inhibition by L-malate and activation by Glc-6-P (glucose 6-phosphate) are routinely observed, whereas phosphorylation at a conserved N-terminal serine residue of the 100–110-kDa PEPC subunit is catalysed by PPCK (PEPC kinase) [1–4]. Phosphorylation typically modulates PEPC sensitivity to allosteric effectors by relieving its inhibition by L-malate while simultaneously enhancing activation by hexose-phosphates. To date, all plant PPCKs that have been studied are novel approx. 31-kDa protein kinases that are controlled mainly at the level of synthesis and degradation. PPCK synthesis is mediated by endogenous circadian rhythms in leaves that undergo CAM, by light-sensing mechanisms in C4 leaves, or by the presence or absence of phloem-supplied sucrose in soya-bean root nodules or developing castor beans [3,5–7].
Phosphorus is an essential element for growth and metabolism because it plays a central role in nearly all metabolic processes. Plants preferentially absorb phosphorus from the soil in its fully oxidized anionic form, Pi (H2PO4−; orthophosphate). Despite its importance, Pi is one of the least available nutrients in many terrestrial and aquatic environments [8]. In soil, Pi is frequently complexed with Al3+, Ca2+ or Fe3+ cations and, therefore exists as insoluble mineral forms that render it unavailable for plant uptake. Agricultural Pi deficiency is alleviated by the massive application of Pi fertilizers, estimated to be approx. 40 million metric tons per year worldwide [8]. Therefore, studies of the remarkably adaptive mechanisms that contribute to the survival of −Pi (Pi-deficient) plants could facilitate the development of rational strategies and molecular tools for engineering Pi-efficient transgenic crops. Although these adaptations are not identical in all plants, particular aspects are conserved in a wide variety of plants from very different environments [8–11]. For example, enhanced levels of PEPC mRNA, protein and/or enzyme activity during Pi deprivation have been reported for diverse species, including Brassica nigra (black mustard), Brassica napus (rapeseed), Arabidopsis thaliana (thale cress), Cicer arietinum (chickpea), Triticum aestivum (wheat), Lupinus albus (white lupin), Nicotiana sylvestris (tobacco) and Lycopersicon esculentum (tomato) [12–20]. It has been suggested that PEPC provides a metabolic bypass (together with malate dehydrogenase and NAD-malic enzyme) to the ADP-limited cytosolic pyruvate kinase to facilitate continued pyruvate supply to the tricarboxylic acid cycle, while concurrently recycling the PEPC byproduct Pi for its reassimilation into the metabolism of the −Pi cells [4,9,12]. PEPC induction has also been correlated with the synthesis and consequent excretion of large amounts of malic acid and citric acid by roots during Pi stress [8,9,14–16]. This increases Pi availability to the roots by acidifying the rhizosphere to solubilize otherwise inaccessible sources of mineralized soil Pi [8].
Genome-sequence annotation demonstrated that Arabidopsis contains four genes that encode PEPCs: AtPPC1, AtPPC2 and AtPPC3, which encode plant-type PEPCs, and AtPPC4, which encodes a distantly related bacterial-type PEPC [21]. The deduced AtPPC1–AtPPC3 polypeptides have a predicted size of 107–110 kDa and share considerable (85–91%) sequence identity, as well as the conserved N-terminal serine phosphorylation site that is characteristic of plant-type PEPCs. Although poor correlations exist for the majority of Pi-starvation-inducible genes identified in independent transcriptomic studies of −Pi Arabidopsis, several reports have consistently identified plant-type AtPPC genes as being Pi-starvation-inducible [14,22,23]. In addition, L-malate-inhibition studies of PEPC activity in desalted extracts from Arabidopsis suspension cells implied that PEPC becomes post-translationally activated by phosphorylation during Pi stress [23]. This was corroborated by the marked induction of PPCK polypeptides and/or transcripts by −Pi Arabidopsis [14,22,23]. The overall goal of the present study was to describe the genetic origin, biochemical and structural properties, and In vivo phosphorylation status of PEPC in −Pi Arabidopsis.
EXPERIMENTAL
Plant material
Heterotrophic A. thaliana (cv. Landsberg erecta) suspension cells were maintained in the dark at 25 °C in MS medium (Murashige and Skoog medium) (pH 5.7) containing 5 mM K2HPO4 (added from a sterile 1 M stock at the time of subculturing), as described previously [24,25]. Cells used in time-course studies were obtained by transferring 20 ml (0.04% packed-cell volume) of a 7-day-old suspension into 80 ml of fresh medium containing either 5 mM K2HPO4 [+Pi (Pi sufficient)] or no K2HPO4 (−Pi). Cells used for PEPC purification were obtained by scaling up the culture volume: 100 ml of a 7-day-old +Pi cell suspension was used to inoculate 400 ml of −Pi media in 3.2-l Fernbach flasks. Cells were harvested by filtration through Whatman 541 filter paper on a Büchner funnel, frozen in liquid N2 and stored at −80 °C. Arabidopsis (cv. Columbia) seedlings were cultivated axenically in 50 ml of half-strength MS liquid medium (pH 5.7) containing 1% (w/v) sucrose and 0.2 mM K2HPO4, as described previously [25]. At 7 days, the medium was replaced with fresh medium containing 3 mM (+Pi) or 20 μM (−Pi) K2HPO4. At 14 days, the medium was replaced with fresh medium, and at 21 days, roots and shoots of the +Pi and −Pi seedlings were harvested, quick-frozen in liquid N2, and stored at −80 °C.
RNA isolation and semi-quantitative RT (reverse transcription)–PCR
Total RNA was extracted and purified from Arabidopsis suspension cells and seedlings, as described previously [25]. RNA samples were assessed for purity (via their A260/A280 ratio) and integrity by resolving 1 μg of total RNA on a 1.2% (w/v) denaturing agarose gel. Normalization of RNA for RT was performed for each sample by density measurement of actin 2 bands from the above gel (scanned using ImageJ software from the National Institutes for Health, U.S.A.). Gene-specific primers were used to amplify Arabidopsis PEPCs (AtPPC1, AtPPC2 and AtPPC3) [21], PPCKs (AtPPCK1 and AtPPCK2) [5] and actin 2 [25]. RNA (5 μg) was reverse transcribed with Superscript III (Invitrogen) and non-competitive RT–PCR was performed as described in [26]. The amount of input cDNA necessary for non-saturating amplification for each primer pair was established by performing PCR using 0.17–50 ng of total RNA during first-strand cDNA synthesis.
Enzyme and protein assays and kinetic studies
PEPC activity was assayed at 25 °C by following NADH oxidation at 340 nm using a kinetics microplate reader (Molecular Devices) and the following optimized assay conditions: 50 mM Hepes/KOH (pH 8.0) containing 15% (v/v) glycerol, 2 mM PEP, 2 mM KHCO3, 5 mM MgCl2, 2 mM DTT (dithiothreitol), 0.15 mM NADH and 5 units·ml−1 of desalted porcine muscle malate dehydrogenase. One unit of activity is defined as the amount of PEPC resulting in the production of 1 μmol of oxaloacetate per min. All assays were linear with respect to time and concentration of enzyme assayed. Apparent Vmax, Km, and IC50 and Ka values (concentrations of inhibitors and activators producing 50% inhibition and activation, respectively) were calculated using the kinetics program as described by Brooks [27]. All kinetic parameters were the means of a minimum of three independent experiments and were reproducible within ±10% S.E.M. of the mean value. Stock solutions of PEP, amino acids and organic acids were made equimolar with MgCl2 and adjusted to pH 7.5. Protein concentrations were determined using a Coomassie Blue G-250 colorimetric method, as described previously [13].
Preparation of clarified extracts used in time-course studies
Quick-frozen tissues were ground to a powder in liquid N2 and homogenized [1:2 (w/v) for suspension cells and leaves; 1:3 (w/v) for roots] using a mortar and pestle and a small scoop of sand in ice-cold Buffer A, which contained 50 mM Hepes/KOH (pH 8.0), 5 mM EDTA, 1 mM EGTA, 50 mM NaF, 1 mM NaVO3−, 1 mM NaMoO4, 50 nM microcystin-LR, 0.1% (v/v) Triton X-100, 10% (v/v) glycerol, 2 mM DTT, 2 mM 2,2′-dipyridyl disulphide, 1 mM PMSF and 1% (w/v) poly(vinyl polypyrrolidone). Homogenates were centrifuged at 4 °C and 15000 g for 15 min, and the resulting clarified extracts were rapidly prepared for SDS/PAGE and immunoblotting, or assayed for total protein and PEPC activity.
Buffers used during PEPC purification
Buffers were degassed and adjusted to their respective pH values at 24 °C. All buffers contained protein-phosphatase inhibitors (5 mM EDTA, 5 mM NaPPi, 25 mM NaF, 1 mM NaMoO4 and 1 mM Na3VO4), whereas 50 nM microcystin-LR was added to all pooled column fractions (to inhibit further any co-purifying protein phosphatase activity). Buffer B contained 50 mM imidazole/HCl (pH 7.1), 1 mM DTT and 25% (saturation) (NH4)2SO4. Buffer C consisted of Buffer B lacking (NH4)2SO4 but containing 10% (v/v) ethylene glycol. Buffer D contained 100 mM Tris/HCl (pH 8.0), 1 mM DTT and 20% (v/v) glycerol. Buffer E contained 50 mM imidazole/HCl (pH 7.5), 5 mM KCl, 5 mM L-malate, 1 mM DTT and 15% (v/v) glycerol.
Purification of PEPC from −Pi Arabidopsis suspension cells
All chromatographic steps were carried out at room temperature (24 °C) using an ÄKTA FPLC system (GE Healthcare). Quick-frozen 7-day-old −Pi Arabidopsis suspension cells (240 g) were ground under liquid N2 using a mortar and pestle, homogenized in 300 ml of ice-cold Buffer A using Polytron, and centrifuged at 4 °C and 18 000 g for 20 min. The supernatant was brought to 30% (saturation) (NH4)2SO4, stirred for 20 min at 4 °C and centrifuged as above. The supernatant was adjusted to 60% (saturation) (NH4)2SO4, stirred and centrifuged as above. The 30–60% (saturation) (NH4)2SO4 pellets were resuspended in 100 ml of Buffer B containing 2.5 μl · ml−1 ProteCEASE-100 (G-Biosciences). After 15 min of centrifugation at 17500 g, the sample was loaded at 3 ml · min−1 on to a column (3.2×11.5 cm) of butyl-Sepharose 4 Fast Flow (GE Healthcare) pre-equilibrated with Buffer B. The column was washed until the A280 decreased to baseline, and PEPC was eluted with 500 ml of a linear gradient of decreasing concentrations of Buffer B (100–0%) and simultaneously increasing concentrations of Buffer C (0–100%).
Pooled peak PEPC-activity fractions were concentrated to 3 ml using an Amicon YM-30 ultrafilter unit (Millipore), and brought to 5 ml with Buffer D and 5 μl · ml−1 ProteCEASE-100. After centrifugation at 17500 g for 15 min, the sample was brought to 20 ml with Buffer D and loaded at 2 ml · min−1 on to a column (1.6 cm×4.4 cm) of Fractogel EMD DEAE-650(S) (Merck) that had been pre-equilibrated with Buffer D. The column was washed with Buffer D until the A280 decreased to baseline, and PEPC was eluted by applying a linear 0–250 mM KCl gradient (160 ml in Buffer D). Fractions with PEPC activity were pooled and concentrated to 0.5 ml with an Amicon Ultra-15 100-kDa cut-off centrifugal filter device. The sample was adjusted to 1.2 ml with Buffer E, centrifuged (14000 g, 5 min) and applied at 0.3 ml · min−1 on to a Superdex-200 HR 16/60 gel-filtration column (GE Healthcare) that had been pre-equilibrated with Buffer E.
Peak activity fractions were immediately loaded at 0.75 ml · min−1 on to a Mono Q HR 5/5 column (GE Healthcare) pre-equilibrated with Buffer D. PEPC was eluted with a linear 0–300 mM KCl gradient (25 ml in Buffer D). Peak activity fractions were pooled, concentrated as described above to 0.7 ml, divided into 25-μl and 50-μl aliquots, frozen in liquid N2, and stored at −80 °C. The purified PEPC was stable for at least 6 months when stored frozen.
Determination of native molecular mass via Superdex-200 gel filtration
Native molecular-mass estimation for PEPC was performed during FPLC on the Superdex-200 column, as described above. Native molecular mass was estimated from a plot of Kav (partition coefficient) against log of molecular mass for the following protein standards: thyroglobulin (669 kDa), apoferritin (443 kDa), ferritin (440 kDa), catalase (232 kDa), aldolase (158 kDa), BSA (67 kDa) and ribonuclease (13.6 kDa).
In vitro dephosphorylation of PEPC
Clarified extracts were prepared in Buffer A lacking phosphatase inhibitors. Aliquots (39 μl containing 200 μg of protein) were incubated for 60 min at 30 °C with 8000 units · ml−1 of λ-phosphatase (New England BioLabs) with or without protein-phosphatase inhibitors (50 mM NaF, 10 mM NaVO3− and 50 mM EDTA) in a final volume of 50 μl according to the manufacturer's instructions. An aliquot of purified PEPC from the −Pi cells was dephosphorylated in 50 μl using 10 m-units · ml−1 of the catalytic subunit of bovine heart PP2A (protein phosphatase type-2A) as described previously [28]. PP2A was purified from 2 kg of freshly harvested bovine heart tissue following the protocol described by Tran et al. [29].
Electrophoresis and immunoblotting
SDS/PAGE and subunit molecular mass determination was performed using Mini-PROTEAN 3 gel electrophoresis (Bio-Rad) at 200 V for 50 min as described previously [13]. Pi-affinity SDS/PAGE was conducted at 75 V for 5 h, with the modification that the resolving gel (10% acrylamide) contained 75 μM Phos-tag™ acrylamide (www.phos-tag.com) and 150 μM MnCl2 [30]. Immunoblotting was performed by electroblotting proteins from gels on to PVDF membranes, followed by immunoreactive polypeptide visualization using an alkaline-phosphatase-conjugated secondary Ab (antibody) with chromogenic detection [13]. Mn2+ that was present following Pi-affinity PAGE was removed prior to electroblotting by incubating the gels for 10 min in transfer buffer containing 1 mM EDTA, and then for 10 min in transfer buffer lacking EDTA. Anti-RcPEPC [Ricinus communis (castor bean) PEPC] IgG was affinity-purified from corresponding rabbit immune serum raised against a homogeneous native Class 1 PEPC (RcPPC3) [26]. Anti-pSer-11 IgG immunoblots were probed with a polyclonal Ab raised against a synthetic phosphopeptide matching the conserved N-terminal Ser-11 phosphorylation domain of RcPPC3 (LEKLApSIDAQLR) [28]. The corresponding dephosphopeptide was used to block any non-specific antibodies raised against the non-phosphorylated sequence [28]. The relative amount of PEPC protein in clarified extracts from +Pi compared with −Pi cells and seedlings was estimated by quantification of the immunoreactive 107-kDa PEPC polypeptides (p107) on immunoblots by measuring A633 using an LKB Ultroscan XL laser densitometer and Gel Scan software (version 2.1) (Pharmacia LKB Biotech). Derived A633 values were linear with respect to the amount of the immunoblotted extract. All immunoblot results were replicated a minimum of three times, with representative results shown in the various Figures.
MS
Excised gel bands were destained, dried, reduced and alkylated as described in [31]. Subsequent digestion was performed at 37 °C overnight using 10 ng of sequencing-grade trypsin (Calbiochem) in 25 mM NH4HCO3 (pH 7.6) or 50 ng of Asp-N (Roche) in 10 mM Tris/HCl (pH 7.6). The resulting peptides were extracted and deposited on a MALDI (matrix-assisted laser-desorption ionization) target [31]. MALDI data were acquired using a QStar XL Q-TOF (quadrupole time-of-flight) mass spectrometer (Applied Biosystems/MDS Sciex) equipped with an oMALDI 2 (orthogonal MALDI 2) source and a nitrogen laser operating at 337 nm. Peptide sequencing of selected ions was carried out by MALDI–Q-TOF/QqTOF (quadrupole/quadrupole TOF)-MS/MS (tandem MS) measurements using argon as the collision gas. All peptide-fingerprinting masses and MS/MS ion searches were performed with the Mascot search engine (MatrixScience, http://www.matrixscience.com) using the NCBI (National Center for Biotechnology Information) non-redundant database (NCBInr, released 9 December 2008, which contains 7031513 protein sequences). These searches take into account one missed cleavage and the following modifications: carbamidomethylation; asparagine and glutamine deamidation to aspartic acid and glutamic acid; and N-terminal pyroglutamation and methionine oxidation. The mass tolerance between calculated and observed masses used for database searches was considered within the range of ±50 p.p.m. for MS peaks and ±0.1 Da for MS/MS fragment ions.
RESULTS
Semi-quantitative RT–PCR analysis of AtPPC1–AtPPC3 and AtPPCK1–AtPPCK2 transcripts in +Pi compared with −Pi Arabidopsis suspension cells and seedlings
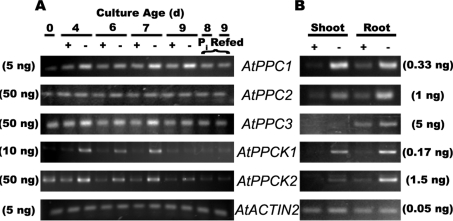
AtPPC1, AtPPC3, AtPPCK1 and AtPPCK2 transcripts were elevated in the −Pi cells, and this was reversed within 1 day of Pi resupply (Figure 1A). Similarly, AtPPC1, AtPPCK1 and AtPPCK2 transcripts were induced in shoots and roots of the −Pi seedlings (Figure 1B). By contrast, AtPPC2 transcripts were present at relatively low and constant levels in the +Pi and −Pi suspension cells, but appeared to be induced in the −Pi seedling tissues, whereas AtPPC3 transcripts were detected in the roots but not in shoots. These results are in general agreement with a variety of transcriptomic studies of AtPPC1–AtPPC3, and AtPPCK1 and AtPPCK2 in Arabidopsis seedlings or cell cultures [14,21–23,32]. For example: (i) quantitative real-time PCR demonstrated that AtPPC1 and AtPPC2 transcripts were abundant in Arabidopsis leaves, whereas those encoding AtPPC3 were negligible [32]; and (ii) AtPPCK1 and AtPPCK2 have been demonstrated to be amongst the most strongly induced genes in −Pi Arabidopsis [14,22,23].
Figure 1. Semi-quantitative RT–PCR analysis of AtPPC1–AtPPC3, and AtPPCK1 and AtPPCK2 gene expression in +Pi and −Pi Arabidopsis suspension cells (A) and seedlings (B).
Levels of mRNA were measured using primers specific for AtPPC1–AtPPC3, AtPPCK1 and AtPPCK2, and AtACTIN2. AtACTIN2 was used as a reference to ensure equal template loading. All PCR products were measured at cycle numbers determined to be non-saturating. Template concentrations needed to achieve non-saturating conditions for primer pairs as tested for −Pi cells or roots of −Pi seedlings are indicated in parentheses. Control RT–PCR reactions lacking reverse transcriptase did not show any bands. ‘Pi refed’ (A) denotes 7-day-old −Pi suspension cells that were supplied with 2.5 mM Pi and cultured for an additional 24 and 48 h. d, day.
Influence of Pi starvation on PEPC activity, amount and phosphorylation status in Arabidopsis cell cultures and seedlings
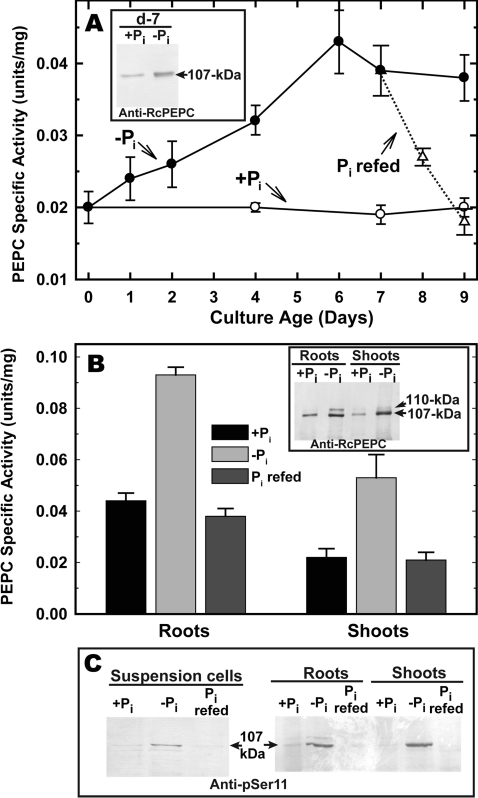
Pi deprivation resulted in a significant (>2-fold) up-regulation of PEPC specific activity in Arabidopsis suspension cells and seedlings (which was reversed following Pi resupply) (Figures 2A and 2B). Laser-densitometric quantification of immunoblots revealed a good correlation between extractable PEPC activity and the relative amount of p107 (Figures 2A and 2B, insets). Analogous results have been described for the PEPC of −Pi white-lupin proteoid roots or B. napus suspension cells relative to corresponding +Pi controls [13,16]. Immunoblotting with anti-pSer-11 [28] indicated that Pi starvation resulted in enhanced p107 phosphorylation In vivo, and that this was also reversed following Pi resupply (Figure 2C). Immunodetection of p107 with anti-pSer-11 was eliminated when immunoblots were probed in the presence of anti-pSer-11 containing 10 μg · ml−1 of the corresponding blocking phosphopeptide and/or following pre-incubation of clarified extracts with λ-phosphatase for 60 min (see Supplementary Figure S1 at http://www.BiochemJ.org/bj/420/bj4200057add.htm).
Figure 2. PEPC is reversibly up-regulated and phosphorylated by −Pi Arabidopsis.
All values represent the mean PEPC activities±S.E.M. of replicate determinations of clarified extracts from n=4 different cultures (A) or seedlings (B). At 0 days, 10-ml aliquots of suspension cells (A) cultured for 7 days in 5 mM Pi were subcultured into 40 ml of fresh MS medium containing 0 or 5 mM Pi (−Pi and +Pi respectively). Flasks of 7-day-old −Pi cultures were resupplied with 2.5 mM Pi (Pi refed) and cultured for an additional 2 days as shown. (A, inset) Immunological detection of PEPC from the 7-day-old (d-7) +Pi and −Pi cells. Clarified extracts were subjected to SDS/PAGE (5 μg of protein/lane) and blot-transferred on to a PVDF membrane. Blots were probed with affinity-purified anti-RcPEPC Ab [26] and immunoreactive polypeptides were detected using an alkaline-phosphatase-linked secondary Ab. Seedlings (B) were germinated and cultivated aseptically in liquid 0.5× MS medium containing 0.2 mM Pi for 7 days then transferred into fresh MS media containing 20 μM Pi (−Pi) or 3 mM Pi (+Pi) and cultivated for an additional 14 days as described in the Experimental section. At 21 days, replicate −Pi seedlings were resupplied with 3 mM Pi and cultivated for an additional 3 days (Pi refed). (B, inset) Immunological detection of PEPC from +Pi compared with −Pi roots and shoots (5 μg of protein/lane) using anti-RcPEPC Ab as described above. (C) Clarified extracts from the −Pi, +Pi and Pi-refed suspension cells or seedlings were subjected to SDS/PAGE and electroblotted on to a PVDF membrane. Immunoblots were probed with an anti-pSer-11 Ab raised against the conserved N-terminal Ser-11 phosphorylation domain of a plant-type RcPEPC isoenzyme, in the presence of 10 μg · ml−1 of the corresponding dephosphopeptide [28]. Each lane of (C) was loaded according to the PEPC activity measured in the corresponding clarified extracts, as determined using the optimal assay conditions outlined in the Experimental section (0.2 m-units/lane for suspension cells; 0.7 m-units/lane for roots and shoots).
PEPC purification from −Pi Arabidopsis cells and its identification as AtPPC1
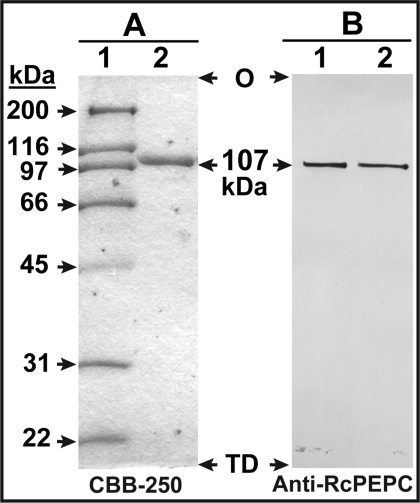
To assess the influence of Pi deprivation on Arabidopsis PEPC more thoroughly, approx. 1.3 mg of PEPC was purified 660-fold from 240 g of 7-day-old −Pi suspension cells, with an overall recovery of 23% (Table 1). A single peak of PEPC activity was resolved during all FPLC steps. The final specific activity of approx. 22 units·mg−1 compares favourably with values reported for homogeneous PEPCs from a range of plant sources [1,13,26]. When the final PEPC preparation was denatured and subjected to SDS/PAGE, a single Coomassie-Blue-staining polypeptide was obtained that strongly cross-reacted with the anti-RcPEPC and that co-migrated with the p107 subunit of purified RcPEPC (Figure 3). A native molecular mass of 430±15 kDa (mean±S.E.M., n=3) was estimated by gel-filtration FPLC on a calibrated Superdex-200 HR 16/60 column (see Supplementary Figure S2 at http://www.BiochemJ.org/bj/420/bj4200057add.htm). Thus, similar to most other plant PEPCs [1–3,13], the native enzyme from −Pi Arabidopsis suspension cells exists as a Class 1 PEPC homotetramer.
Table 1. Purification of PEPC from 240 g of –Pi Arabidopsis suspension cells.
| Step | Volume (ml) | Activity (units) | Protein (mg) | Specific activity (units·mg−1) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|---|
| Clarified extract | 375 | 120 | 6750 | 0.034 | 1.0 | 100 |
| (NH4)2SO4 fractionation | 126 | 74.3 | 3260 | 0.023 | 0.7 | 62 |
| butyl-Sepharose | 5.0* | 34.7 | 200 | 0.17 | 5.1 | 29 |
| DEAE-Fractogel | 1.2* | 46.3 | 17.0 | 2.7 | 80 | 39 |
| Superdex-200 FPLC | 5.5 | 40.0 | 2.8 | 14.5 | 450 | 33 |
| Mono Q FPLC | 0.7* | 28.0 | 1.3 | 22.3 | 660 | 23 |
*Concentrated pooled fractions.
Figure 3. SDS/PAGE and immunoblot analysis of purified PEPC from −Pi Arabidopsis suspension cells.
(A) SDS/PAGE: lane 1 contains various molecular mass standards (3 μg) whereas lane 2 contains 2 μg of the of the pooled peak fractions from the final purification step (Mono Q FPLC). Protein staining was performed using Coomassie Brilliant Blue R-250 (CBB-250). (B) Immunoblotting was performed using an anti-RcPEPC Ab [26]; lanes 1 and 2 contain 25 ng of the final preparation of −Pi Arabidopsis PEPC and homogeneous RcPEPC [26] respectively. O, origin; TD, tracking-dye front.
Discrimination between Arabidopsis AtPPC1–AtPPC3 via peptide mass fingerprinting is challenging, owing to the significant degree of sequence identity (>85%) between these closely related isoenzymes (Figure 4). Nevertheless, the use of MALDI–TOF-MS for mass fingerprinting of tryptic peptides derived from a PEPC immunoprecipitate of Arabidopsis rosette leaves led to the identification of all three plant-type PEPC isoenzymes [32]. Determination of the AtPPC isoenzyme that the final PEPC preparation of the present study corresponded to was also attempted by peptide mass fingerprinting using MALDI–QqTOF-MS of both tryptic and Asp-N digests (combined 78% sequence coverage) (see Supplementary Tables S1 and S2 at http://www.BiochemJ.org/bj/420/bj4200057add.htm), but proved inconclusive. Thus we employed MALDI–QqTOF-MS/MS to sequence numerous peptides derived from the p107 obtained following SDS/PAGE of the final PEPC preparation. The 21 tryptic peptides that were sequenced all matched AtPPC1 [TAIR (The Arabidopsis Information Resource; www.arabidopsis.org) accession number At1g53310; 45% sequence coverage] (Supplementary Table S1), with nine peptides unique to AtPPC1 (Figure 4). Similarly, all 12 Asp-N peptides that were sequenced matched AtPPC1 (41% sequence coverage) (Supplementary Table S2), with six peptides unique to AtPPC1 (Figure 4). In contrast, no peptides that were exclusive to any other polypeptide (including AtPPC2 or AtPPC3) were detected in tryptic or Asp-N digests of p107. These results are consistent with those demonstrating that Pi deprivation of Arabidopsis suspension cells or seedlings resulted in the marked (i) transcriptional induction of AtPPC1 (Figure 1) and (ii) accumulation of an anti-RcPEPC immunoreactive p107 (Figure 2). The collective results indicate that the simultaneous induction and In vivo phosphorylation of AtPPC1 form part of the metabolic adaptation of −Pi Arabidopsis.
Figure 4. Alignment of MALDI–QqTOF-MS/MS-sequenced tryptic and Asp-N peptides of purified PEPC from −Pi Arabidopsis suspension cells with selected regions of the deduced amino acid sequences of Arabidopsis plant-type PEPCs (AtPPC1–AtPPC3).
MALDI–QqTOF-MS/MS analysis of purified PEPC from the −Pi cells revealed 15 peptides unique to AtPPC1; nine were obtained following trypsin digestion (underlined), whereas six were obtained following Asp-N digestion (overlined). The Ser-11 phosphorylation site of AtPPC1 is marked in bold, the conserved N-terminal serine-phosphorylation motif typical of plant-type PEPCs [1–3] is outlined with a dotted box and identical amino-acid residues are indicated with an asterisk. The corresponding NCBI protein accession numbers are as follows: AtPPC1, NP_175738; AtPPC2, CAD58726; and AtPPC3, AAC24594.
Purified AtPPC1 from −Pi Arabidopsis cells is phosphorylated at Ser-11
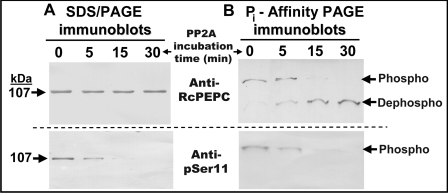
The use of anti-pSer-11 Ab for examining the phosphorylation status of p107 in purified AtPPC1 was complemented with: (i) an affinity-purified anti-RcPEPC Ab raised against a native PEPC (RcPPC3) from developing castor-oil seeds [28] that cross-reacts with phospho- and dephospho-forms of plant-type PEPCs; and (ii) Pi-affinity PAGE using Phos-Tag™ acrylamide (Figures 5A and 5B). The Phos-Tag™ ligand is a polyacrylamide-bound dinuclear Mn2+ complex that specifically captures phosphomonoester di-anions (-OPO32−), thereby reducing phosphoprotein mobility during SDS/PAGE relative to the corresponding dephosphoprotein [30]. Pre-incubation of purified AtPPC1 with PP2A for 30 min resulted in the time-dependent disappearance of immunoreactive p107 when the immunoblots were probed with anti-pSer-11, but not with anti-RcPEPC (Figures 5A and 5B). Pi-affinity PAGE indicated that nearly 100% of the p107 subunits of purified AtPPC1 migrated as a putatively monophosphorylated species that was converted into the faster-migrating dephospho-p107 during its pre-incubation with PP2A (Figure 5B). Plant PPCKs have been suggested to phosphorylate specifically the serine residue in the conserved PEPC N-terminal phosphorylation motif: (E/D)-(K/R)-X-Z-SIDAQLR, where X denotes M/H/L and Z denotes A/Q/S/H [2]. As the Ala-Gln residues in this motif have been substituted with Val-His in AtPPC1 (Figure 4), it has been implied that AtPPC1 may be incapable of undergoing regulatory phosphorylation [33]. However, our results demonstrate that AtPPC1 is an effective In vivo substrate for the endogenous PPCK activity of −Pi Arabidopsis.
Figure 5. In vitro dephosphorylation of purified AtPPC1 from −Pi suspension cells using the catalytic subunit of bovine PP2A.
Purified AtPPC1 was incubated with PP2A for the indicated times and subjected to (A) SDS/PAGE or (B) Pi-affinity PAGE [30], followed by immunoblot analysis with either anti-RcPEPC Ab [26] (25 ng of AtPPC1/lane) or the corresponding anti-pSer-11 Ab [28] (250 ng of AtPPC1/lane). ‘Phospho’ and ‘Dephospho’ denote the phosphorylated and dephosphorylated forms of p107 respectively.
It is notable that the aforementioned results were dependent upon the addition of a cocktail of protein-phosphatase inhibitors to all purification buffers and pooled column fractions generated during AtPPC1 isolation from the −Pi cells (see the Experimental section). Our initial purification trials employed a protein-phosphatase inhibitor cocktail in the extraction buffer and 20 mM NaF in column running buffers, but resulted in final AtPPC1 preparations displaying sub-stoichiometric p107 phosphorylation (as assessed by immunoblotting with anti-pSer-11, Pi-affinity PAGE, or kinetic studies with or without PP2A treatment) (results not shown). The combined results indicate that an endogenous protein-phosphatase activity capable of catalysing the In vitro dephosphorylation of AtPPC1 co-purified with AtPPC1, and that this activity was not fully inhibited by 20 mM NaF. This also provides a possible rationale for the absence of detectable phosphorylation in homogeneous PEPC preparations isolated from heterotrophic +Pi or −Pi B. napus suspension cells using a protocol that employed only 20 mM NaF in column running buffers to control unwanted phosphatase activity [13]. Our results support recommendations that effective In vitro suppression of endogenous protein-phosphatase activity necessitates the inclusion of a wide assortment of protein-phosphatase inhibitors (e.g. NaF, MoO4−, VO3−, PPi and microcystin-LR) in extraction and purification buffers [34].
Kinetic properties
Similar to other plant PEPCs [1,13,28,35], the purified AtPPC1 exhibited: (i) a broad pH/activity profile with a maximum between pH 8.0 and 9.0; PEPC activity at pH 7.0 was approx. 50% of that occurring at pH 8.5; (ii) an absolute dependence on a bivalent metal cation, with Mg2+ or Mn2+ satisfying this requirement; and (iii) hyperbolic PEP-saturation kinetics (results not shown). The AtPPC1 displayed a Km(PEP) value of 0.054±0.005 mM (mean±S.E.M., n=4) at pH 8.5. It has been amply demonstrated that phosphorylation of plant PEPCs reduces the sensitivity of the enzymes to L-malate when assayed at subsaturating PEP and suboptimal, but physiological pH values ranging from approx. pH 7.0 to 7.4 [1–3,28,35]. Phosphorylation can also relieve L-Asp inhibition of plant PEPCs, while simultaneously promoting binding of its substrate PEP and activator Glc-6-P at physiological pH (e.g. pH 7.3) [28,35]. Results presented in Table 2 demonstrate that In vivo phosphorylation of AtPPC1 during Pi stress also activates this enzyme at pH 7.3 by significantly lowering its Km(PEP) value and sensitivity to inhibition by L-malate and L-Asp, while increasing its activation by Glc-6-P.
Table 2. Kinetic properties of purified phosphorylated AtPPC1 compared with dephosphorylated AtPPC1.
Dephospho-AtPPC1 was obtained following In vitro dephosphorylation of purified AtPPC1 with bovine PP2A as described in the Experimental section and the legend to Figure 5. Ka and IC50 values were determined using subsaturating (0.25 mM) PEP. All values were determined at pH 7.3 and represent means±S.E.M. of n=4 separate determinations. All values are given in mM.
| Parameter | Phospho-AtPPC1 | Dephospho-AtPPC1 |
|---|---|---|
| Km(PEP) | 0.18±0.02 | 0.34±0.01 |
| Ka(Glc-6-P) | 0.028*±0.001 | ND*† |
| IC50(L-Asp) | 1.14±0.01 | 0.52±0.04 |
| IC50(L-malate)+0 mM Glc-6-P | 0.68±0.04 | 0.30±0.03 |
| IC50(L-malate)+0.2 mM Glc-6-P | 1.5±0.04 | 0.61±0.01 |
| IC50(L-malate)+2 mM Glc-6-P | 9.1±0.3 | 5.5±0.2 |
*Saturating Glc-6-P elicited 1.8- and 1.2-fold increases in the activity of phospho- and dephospho-AtPPC1 respectively.
†ND, not determined. Weak Glc-6-P activation prevented accurate determination of the Ka(Glc-6-P) value of dephospho-AtPPC1.
DISCUSSION
It has been suggested that PEPC plays three important roles in plant acclimation to Pi deficiency: (i) anaplerotic production of large quantities of organic acids for excretion via the roots into the rhizosphere to facilitate soil Pi solubilization; (ii) part of a glycolytic bypass to the ADP-limited cytosolic pyruvate kinase; and (iii) metabolic Pi recycling from PEP [4,8–20,23]. Despite substantial biochemical and transcriptomic evidence for the participation of PEPC in plant acclimation to Pi starvation, little research has been performed on native PEPCs to determine the specific isoenzymes(s) up-regulated during Pi stress, or the relationship between cellular Pi nutrition and the In vivo phosphorylation status of PEPC. The results of the present study are consistent with the hypothesis that the parallel induction and In vivo phosphorylation-activation of AtPPC1 at its conserved Ser-11 phosphorylation site plays an important role in the metabolic adaptations of −Pi Arabidopsis. However, the possible involvement of additional plant-type AtPPC isoenzymes in the PEP metabolism of −Pi Arabidopsis was indicated by (i) the semiquantitative RT–PCR results (Figure 1), and (ii) the PEPC immunoblots, which revealed the up-regulation and enhanced phosphorylation of a minor approx. 110-kDa immunoreactive PEPC polypeptide in planta, particularly in the −Pi roots (Figures 2B and 2C, and Supplementary Figure S1B). Additional studies will be required to assess the molecular and functional properties of this 110-kDa immunoreactive PEPC polypeptide and its relationship with AtPPC1.
This research supports recent studies demonstrating that suspension-cell cultures represent a valuable model for assessing the molecular and biochemical adaptations of Arabidopsis to suboptimal Pi nutrition [23–25]. In particular, a relatively large quantity of cells (and their surrounding media containing secreted proteins [24]) at a defined nutritional state can be obtained over a relatively short time period. Our results also corroborate L-malate inhibition studies of PEPC in Arabidopsis suspension-cell-culture extracts, and of a partially purified PEPC from proteoid lupin roots, suggesting that PEPC becomes phosphorylated In vivo during Pi starvation [17,23]. The striking reversible induction of AtPPCK1 and AtPPCK2 transcripts during Pi stress (Figure 1) [14,22,23] provides a logical rationale for the Pi-starvation-dependent AtPPC1 phosphorylation that we observed, and for the absence of detectable PEPC phosphorylation following Pi resupply to the −Pi cells or seedlings. Unlike many protein kinases that are allosterically controlled by second messengers such Ca2+ ions, control of plant PPCK activity, and hence the phosphorylation status of target PEPCs, appears to arise largely from PPCK synthesis/degradation [2,3,5]. Chen et al. [23] identified a transcription factor, BHLH32, that functions as a negative regulator of several Pi-starvation-inducible genes in Arabidopsis, including AtPPCK1 and AtPPCK2. Characterization of additional components of the signal-transduction pathways that link nutritional Pi status with the control of AtPPC1, and AtPPCK1 and AtPPCK2 transcription and translation, as well as AtPPC1, and AtPPCK1 and AtPPCK2 turnover, will be a fruitful area for future research.
Important insights into the Pi-starvation response of Arabidopsis and other vascular plants have been provided by the classification of genes displaying altered transcription during Pi deprivation [11,14,19,36–41]. However, the integration of transcriptomics with metabolomics, proteomics and native-enzyme biochemistry will be needed to achieve a thorough understanding of the intricate mechanisms by which plant metabolism acclimates to nutritional Pi deficiency. Future investigations of these pathways should provide further links between the molecular and biochemical control of plant metabolism. This knowledge is relevant to the applied efforts of agricultural biotechnologists to engineer transgenic crops exhibiting an improved resistance to abiotic stress, including nutritional Pi deprivation.
ACKNOWLEDGEMENTS
We are grateful to Professor Hugh Nimmo (Department of Systems Biology and Systems Medicine, University of Glasgow, Glasgow, U.K.) for helpful discussions and to Dr Srinath Rao (Department of Biology, Queen's University, Kingston, ON, Canada) for providing advice regarding RT–PCR.
FUNDING
This work was supported by the Natural Sciences and Engineering Research Council of Canada [grant numbers RG PIN/106349-2003 and RG PIN/106349-2008] and the Queen's Research Chairs program (to W. C. P.), and by a travel grant from the National Research Foundation of South Africa (to A. J. V.).
Online data
References
- 1.Chollet R., Vidal J., O'Leary M. H. Phosphoenolpyruvate carboxylase: a ubiquitous, highly regulated enzyme in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996;47:273–298. doi: 10.1146/annurev.arplant.47.1.273. [DOI] [PubMed] [Google Scholar]
- 2.Izui K., Matsumura H., Furumoto T., Kai Y. Phosphoenolpyruvate carboxylase: a new era of structural biology. Annu. Rev. Plant Biol. 2004;55:69–84. doi: 10.1146/annurev.arplant.55.031903.141619. [DOI] [PubMed] [Google Scholar]
- 3.Nimmo H. G. Control of the phosphorylation of phosphoenolpyruvate carboxylase in higher plants. Arch. Biochem. Biophys. 2003;414:189–196. doi: 10.1016/s0003-9861(03)00115-2. [DOI] [PubMed] [Google Scholar]
- 4.Plaxton W. C., Podestá F. E. The functional organization and control of plant respiration. Crit. Rev. Plant Sci. 2006;25:159–198. [Google Scholar]
- 5.Zhang X. Q., Chollet R. Phosphoenolpyruvate carboxylase protein kinase from soybean root nodules: partial purification, characterization, and up/down-regulation by photosynthate supply from the shoots. Arch. Biochem. Biophys. 1997;343:260–268. doi: 10.1006/abbi.1997.0190. [DOI] [PubMed] [Google Scholar]
- 6.Murmu J., Plaxton W. C. Phosphoenolpyruvate carboxylase protein kinase from developing castor oil seeds: partial purification, characterization, and reversible control by photosynthate supply. Planta. 2007;226:1299–1310. doi: 10.1007/s00425-007-0551-x. [DOI] [PubMed] [Google Scholar]
- 7.Fontaine V., Hartwell J., Jenkins G. I., Nimmo H. G. Arabidopsis thaliana contains two phosphoenolpyruvate carboxylase kinase genes with different expression patterns. Plant Cell Environ. 2002;25:115–122. [Google Scholar]
- 8.Vance C. P., Uhde-Stone C., Allan D. L. Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol. 2003;157:427–447. doi: 10.1046/j.1469-8137.2003.00695.x. [DOI] [PubMed] [Google Scholar]
- 9.Plaxton W. C. Biochemical adaptations of phosphate starved plants. In: Goodman R., editor. Encyclopedia of Plant and Crop Sciences. New York: Marcel Dekker; 2004. pp. 976–980. [Google Scholar]
- 10.Ticconi C. A., Abel S. Short on phosphate: plant surveillance and countermeasures. Trends Plant Sci. 2004;9:548–555. doi: 10.1016/j.tplants.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Raghothama K. G., Karthikeyan A. S. Phosphate acquisition. Plant Soil. 2005;274:37–49. [Google Scholar]
- 12.Duff S. M. G., Moorhead G. B., Lefebvre D. D., Plaxton W. C. Phosphate starvation inducible ‘bypasses’ of adenylate and phosphate dependent glycolytic enzymes in Brassica nigra suspension cells. Plant Physiol. 1989;90:1275–1278. doi: 10.1104/pp.90.4.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moraes T. F., Plaxton W. C. Purification and characterization of phosphoenolpyruvate carboxylase from Brassica napus (rapeseed) suspension cell cultures: implications for phosphoenolpyruvate carboxylase regulation during phosphate starvation, and the integration of glycolysis with nitrogen assimilation. Eur. J. Biochem. 2000;267:4465–4476. doi: 10.1046/j.1432-1327.2000.01495.x. [DOI] [PubMed] [Google Scholar]
- 14.Morcuende R., Bari R., Gibon Y., Zheng W., Pant B. D., Blasing O., Usadel B., Czechowski T., Udvardi M. K., Stitt M., Scheible W. R. Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ. 2007;30:85–112. doi: 10.1111/j.1365-3040.2006.01608.x. [DOI] [PubMed] [Google Scholar]
- 15.Neuman G., Romheld V. Root excretion of carboxylic acids and protons in phosphorus-deficient plants. Plant Soil. 1999;211:121–130. [Google Scholar]
- 16.Johnson J. F., Vance C. P., Allan D. L. Phosphorus deficiency in Lupinus albus. Altered lateral root development and enhanced expression of phosphoenolpyruvate carboxylase. Plant Physiol. 1996;112:31–41. doi: 10.1104/pp.112.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uhde-Stone C., Gilbert G., Johnson J. M., Litjens R., Zinn K. E., Temple S. J., Vance C. P., Allan D. L. Acclimation of white lupin to phosphorus deficiency involves enhanced expression of genes related to organic acid metabolism. Plant Soil. 2003;248:99–116. [Google Scholar]
- 18.Penaloza E., Munoz G., Salvo-Garrido H., Silva H., Corcuera L. J. Phosphate deficiency regulates phosphoenolpyruvate carboxylase expression in proteoid root clusters of white lupin. J. Exp. Bot. 2005;56:145–153. doi: 10.1093/jxb/eri008. [DOI] [PubMed] [Google Scholar]
- 19.Toyota K., Koizumi N., Sato F. Transcriptional activation of phosphoenolpyruvate carboxylase by phosphorus deficiency in tobacco. J. Exp. Bot. 2003;54:961–969. doi: 10.1093/jxb/erg095. [DOI] [PubMed] [Google Scholar]
- 20.Pilbeam D. J., Cakmak I., Marschner H., Kirkby E. A. Effect of withdrawal of phosphorus on nitrate assimilation and PEP carboxylase activity in tomato. Plant Soil. 1993;154:111–117. [Google Scholar]
- 21.Sanchez R., Cejudo F. J. Identification and expression analysis of a gene encoding a bacterial-type phosphoenolpyruvate carboxylase from Arabidopsis and rice. Plant Physiol. 2003;132:949–957. doi: 10.1104/pp.102.019653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller R., Morant M., Jarmer H., Nilsson L., Nielsen T. H. Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol. 2007;143:156–171. doi: 10.1104/pp.106.090167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Z. H., Nimmo G. A., Jenkins G. I., Nimmo H. G. BHLH32 modulates several biochemical and morphological processes that respond to Pi starvation in Arabidopsis. Biochem. J. 2007;405:191–198. doi: 10.1042/BJ20070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tran H. T., Plaxton W. C. Proteomic analysis of alterations in the secretome of Arabidopsis thaliana suspension cells subjected to nutritional phosphate deficiency. Proteomics. 2008;8:4317–4326. doi: 10.1002/pmic.200800292. [DOI] [PubMed] [Google Scholar]
- 25.Veljanovski V., Vanderbeld B., Knowles V. L., Snedden W. A., Plaxton W. C. Biochemical and molecular characterization of AtPAP26, a vacuolar purple acid phosphatase up-regulated in phosphate-deprived Arabidopsis suspension cells and seedlings. Plant Physiol. 2006;142:1282–1293. doi: 10.1104/pp.106.087171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gennidakis S., Rao S., Greenham K., Uhrig R. G., O'Leary B., Snedden W. A., Lu C., Plaxton W. C. Bacterial- and plant-type phosphoenolpyruvate carboxylase polypeptides interact in the hetero-oligomeric Class-2 PEPC complex of developing castor oil seeds. Plant J. 2007;52:839–849. doi: 10.1111/j.1365-313X.2007.03274.x. [DOI] [PubMed] [Google Scholar]
- 27.Brooks S. A simple computer program for the analysis of enzyme kinetics. Biotechniques. 1992;13:906–911. [PubMed] [Google Scholar]
- 28.Tripodi K. E., Turner W. L., Gennidakis S., Plaxton W. C. In vivo regulatory phosphorylation of novel phosphoenolpyruvate carboxylase isoforms in endosperm of developing castor oil seeds. Plant Physiol. 2005;139:969–978. doi: 10.1104/pp.105.066647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tran H. T., Ferrar T. S., Ulke-Lemee A., Moorhead G. B. Purification of PP2Ac from bovine heart. Methods Mol. Biol. 2007;365:127–132. doi: 10.1385/1-59745-267-X:127. [DOI] [PubMed] [Google Scholar]
- 30.Kinoshita E., Kinoshita-Kikuta E., Takiyama K., Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell. Proteomics. 2006;5:749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Uhrig R. G., She Y. M., Leach C. A., Plaxton W. C. Regulatory monoubiquitination of phosphoenolpyruvate carboxylase in germinating castor oil seeds. J. Biol. Chem. 2008;283:29650–29657. doi: 10.1074/jbc.M806102200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gousset-Dupont A., Lebouteiller B., Monreal J., Echevarria C., Pierre J. N., Hodges M., Vidal J. Metabolite and post-translational control of phosphoenolpyruvate carboxylase from leaves and mesophyll cell protoplasts of Arabidopsis thaliana. Plant Sci. 2005;169:1096–1101. [Google Scholar]
- 33.Sánchez R., Flores A., Cejudo F. J. Arabidopsis phosphoenolpyruvate carboxylase genes encode immunologically unrelated polypeptides and are differentially expressed in response to drought and salt stress. Planta. 2006;223:901–909. doi: 10.1007/s00425-005-0144-5. [DOI] [PubMed] [Google Scholar]
- 34.Haystead T. A. G., Garrison J. C. Study of protein phosphorylation in intact cells. In: Hardie D.G., editor. Protein Phosphorylation: A Practical Approach. 2nd edn. Oxford: Oxford University Press; 2000. pp. 1–30. [Google Scholar]
- 35.Law R. D., Plaxton W. C. Regulatory phosphorylation of banana fruit phosphoenolpyruvate carboxylase by a copurifying phosphoenolpyruvate carboxylase-kinase. Eur. J. Biochem. 1997;247:642–651. doi: 10.1111/j.1432-1033.1997.00642.x. [DOI] [PubMed] [Google Scholar]
- 36.Hammond J. P., Broadley M. R., White P. J. Genetic responses to phosphorus deficiency. Ann. Bot. 2004;94:323–332. doi: 10.1093/aob/mch156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hernandez G., Ramirez M., Valdes-Lopez O., Tesfaye M., Graham M. A., Czechowski T., Schlereth A., Wandrey M., Erban A., Cheung F., et al. Phosphorus stress in common bean: root transcript and metabolic responses. Plant Physiol. 2007;144:752–767. doi: 10.1104/pp.107.096958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Misson J., Raghothama K. G., Jain A., Jouhet J., Block M. A., Bligny R., Ortet P., Creff A., Somerville S., Rolland N., et al. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc. Nat. Acad. Sci. U. S. A. 2005;102:11934–11939. doi: 10.1073/pnas.0505266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Penaloza E., Gutierrez A., Martinez J., Munoz G., Bravo L. A., Corcuera L. J. Differential gene expression in proteoid root clusters of white lupin (Lupinus albus) Physiol. Plant. 2002;116:28–36. doi: 10.1034/j.1399-3054.2002.1160104.x. [DOI] [PubMed] [Google Scholar]
- 40.Wasaki J., Shinano T., Onishi K., Yonetani R., Yazaki J., Fujii F., Shimbo K., Ishikawa M., Shimatani Z., Nagata Y., et al. Transcriptomic analysis indicates putative metabolic changes caused by manipulation of phosphorus availability in rice leaves. J. Exp. Bot. 2006;57:2049–2059. doi: 10.1093/jxb/erj158. [DOI] [PubMed] [Google Scholar]
- 41.Wu P., Ma L., Hou X., Wang M., Wu Y., Liu F., Deng X. W. Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol. 2003;132:1260–1271. doi: 10.1104/pp.103.021022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.