Abstract
This study analyses the effect of host-specific pathogens on range restriction of their hosts across environmental gradients at population margins. Sterilizing diseases can limit host range by causing large reductions in population size in what would otherwise be the central area of a species range. Diseases showing frequency-dependent transmission can also pull back a population from its disease-free margin. A wide range of disease prevalence versus abundance patterns emerge which often differ from the classical expectation of increasing prevalence with increasing abundance. Surprisingly, very few empirical studies have investigated the dynamics of disease across environmental gradients or at range limits.
Keywords: parasitic castration, population regulation, spatial dynamics, sexually transmitted diseases, Microbotryum violaceum, Silene latifolia
1. Introduction
Although, the importance of marginal populations in ecology and evolution has been emphasized for many years (Mayr 1963; Antonovics 1976; Carter & Prince 1981; Watkinson 1985; Hoffmann & Blows 1994), only recently there has been an increase in the explicit and detailed study of such populations. Concern about climate change and invasive species has generated new interest in processes limiting species' distributions (Woodward 1987; Parmesan et al. 2005). It is now recognized that many ecological and evolutionary processes on a wide variety of spatial and temporal scales may be responsible for range limits (Lennon et al. 1997; Keitt et al. 2001; Ries et al. 2004; Holt et al. 2005).
Cases of disease-driven extinction or decline are well documented (De Castro & Bolker 2005). Most of these examples occur where one host species acts as a disease reservoir leading to the decline of a more susceptible host. The impact of host-specific diseases on range-limits has been less well studied. Classical disease models based on mass-action (or density-dependent) transmission predict that disease should fade out at low density, and thus host-specific diseases might not be expected to lead to extinction. Correspondingly, areas of low density around population or species margins should be relatively free of specialized pathogens and parasites. Even highly virulent pathogens are no exception because high host mortality decreases the duration of the infection and in extremis leads to a lower, rather than greater, impact on host numbers (Anderson & May 1981).
In this paper, I show that the ‘classical’ view that disease should be absent from marginal regions does not take into account the impact of pathogens that sterilize rather than kill their hosts and that have alternative transmission modes. Sterilizing pathogens have a severe effect on population size because the duration of the infection is unaffected (Anderson & May 1981), and extinction can occur in spatial models even with mass-action transmission (Boots & Sasaki 2001), especially where hosts are long-lived and effectively act as disease reservoirs. Sterilizing diseases have attracted much less attention than those that cause mortality, perhaps because they cause less alarm in humans. However, sterilization is widespread in natural populations, especially by animal parasites (parasitic castration: Kuris 1974; Baudoin 1975; Jaenike 1992; Lafferty & Kuris 1996), plant pathogens (Clay 1991; Wennstrom & Ericson 2003), as well as in sexually transmitted diseases (Lockhart et al. 1996).
In vector and sexually transmitted diseases, transmission is likely to be a function of frequency rather than density of infectious individuals, and this can lead directly to disease-driven extinction (Getz & Pickering 1983; Thrall et al. 1993; O'Keefe & Antonovics 2002). This transmission mode characterizes most sexually transmitted diseases (Lockhart et al. 1996), vector and pollinator transmitted diseases (Anderson 1981; Antonovics 2005), and is expected where host populations are sub-structured into social groups (Begon et al. 1999; Bjørnstad et al. 2002).
There are few studies of how environmental gradients interact with disease to determine dynamics at ecological boundaries. Hochberg & Ives (1999) showed that highly dispersed host-specific parasitoids could limit hosts to high resource areas, and Wilson et al. (1999) modelled a similar situation to explain the local restriction of a host-specific herbivore by its parasitoid. Travis et al. (2006) showed that the effect of positive or negative species interactions depended on whether environmental gradients affected mortality or reproduction.
This paper presents analytical and simulation results showing that host-specific pathogens have the potential to limit population ranges. Gradients in birth and death rates (probably at population margins) can also result in unexpected patterns of disease prevalence in relation to population size.
2. The model
The main study presents a stochastic simulation based on a lattice of interconnected patches. However, first a theoretical analysis of the relationship between prevalence and equilibrium population size is presented for single populations, representing each cell of the lattice.
(a) Within population dynamics
For simplicity, I assume that: (i) the disease results in complete sterilization, and there is no added death rate of infected individuals, (ii) there is no recovery, and hence no immune class, and (iii) density impacts reproduction and/or establishment rather than mortality rate of adults.
The dynamics of a sterilizing disease can then be represented by the generalized difference equations
| (2.1) |
| (2.2) |
Throughout, b=birth rate, and d=death rate; X and Y symbolize numbers of healthy and diseased individuals, subscripted to time t, with total number N=X+Y and prevalence=Y/N; f(b,Nt) describes density-dependent birth/establishment, and is assumed to be of the form b/(1+kNt), where k represents the strength of density regulation. The disease-free equilibrium is
| (2.3) |
The term f(Yt,Nt) describes the rate at which healthy individuals become diseased (the ‘force of infection’) and is β(Yt/Nt) for frequency-dependent and βYt for density-dependent transmission; the two betas have different units (Antonovics et al. 1995). These equations give the following equilibria (Thrall et al. 1995).
(i) For frequency-dependent transmission
| (2.4) |
| (2.5) |
Using the above, equilibrium prevalence becomes
| (2.6) |
and equilibrium diseased population size
| (2.7) |
(ii) For density-dependent transmission
| (2.8) |
| (2.9) |
where, . Using the above, equilibrium prevalence becomes
| (2.10) |
And equilibrium diseased population size
| (2.11) |
(iii) Classical model
To compare the above outcomes with the ‘classical model’ (with disease-induced mortality and mass-action transmission) the difference equations used were
| (2.12) |
| (2.13) |
where α=disease-induced mortality, and k=density regulation acting on mortality. The analysis and equilibria of the differential equation equivalent of this model are in Anderson & May (1981; Model F).
(b) Spatially explicit simulations
The above difference equations described the dynamics within cells of a two-dimensional rectangular lattice, except that in the simulation the transmission was instantiated in exponential Nicholson–Bailey form, such that probability of infection=1−exp(f(YtNt)) to eliminate the possibility of extreme values generating more infectives than susceptibles. With these exponential transmission functions, explicit analytical solutions are not possible. Stochastic variation in transmission and death was imposed by sampling the expected proportions diseased or alive in each cell according to binomial expectations. Stochastic variation in the expected birth rate was imposed according to the Poisson distribution.
Migration among cells was deterministic and occurred at a rate m into the orthogonally adjacent four cells (m/4 immigration into each cell). Migration occurred as progeny for the host, and as spores for the pathogen. For simplicity, pathogen and host migration rates were assumed to be equal. Lattice sizes were varied between 10×50 and 50×50 cells (see §3), and boundaries were absorbing.
Environmental gradients in decreasing birth or increasing death rate were linear and started at 20 per cent of the distance from the left edge of the lattice (considered the ‘central’ region). They were scaled so the predicted disease-free population equilibria would be zero (i.e. marginal) on the right.
3. Results
(a) Deterministic equilibria
For frequency-dependent transmission, an increasing death rate (as might be expected at a range margin) results in a decrease in disease prevalence but the equilibrium population size does not change (equations (2.6) and (2.7)). However, a decreasing birth rate has no effect on equilibrium disease prevalence but causes a decrease in population numbers. At low birth rates (b<β), there is disease-based deterministic extinction.
For density-dependent transmission, an increasing death rate (equations (2.8)–(2.11)) has effects that are qualitatively similar to those for frequency-dependent transmission, disease prevalence declines but equilibrium density is unaffected. Decreasing birth rate, on the other hand, leads to both decreased population density and decreased prevalence, in line with classical expectations.
For the classical model, although an analysis of the possible equilibria for the difference equations was not carried out, numerical evaluation of equations (2.12) and (2.13) and examination of the differential equation equilibria (Anderson & May 1981) confirmed that increasing prevalence is always positively correlated with increasing diseased population size.
(b) Spatially explicit simulations
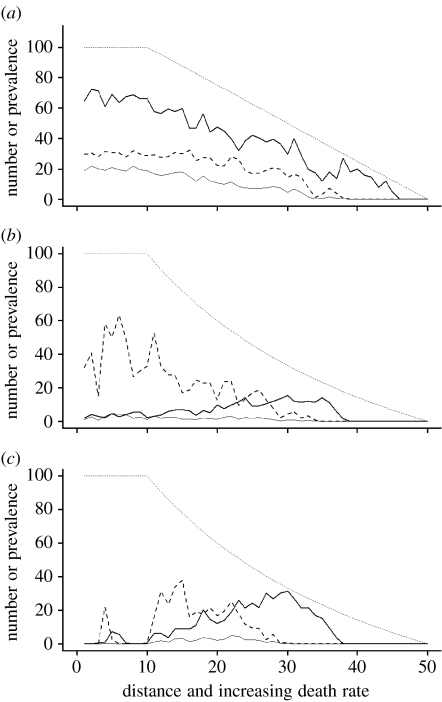
With classical within-cell dynamics (density-dependent transmission and disease induced mortality) host-specific pathogens had little effect on host distribution. With both a gradient of increasing death rate (figure 1a) and decreasing birth rate (not shown), there was a disease-free zone at the margin and a decreasing prevalence with decreasing host population size. Note that the figures show single, but representative, instantiations of the stochastic model.
Figure 1.
Population size and disease prevalence along a gradient of increasing death rate. Solid thick line, total population size; solid thin line, number diseased; dashed line, prevalence (scaled from 0 to 100% on y-axis); thin dotted line, disease-free deterministic population size. (a) Classical model with density-dependent transmission and disease induced mortality; α=0.25, b=2, β=0.3. (b) Sterilizing disease with density-dependent transmission; b=1, β=0.3. (c) Sterilizing disease with frequency-dependent transmission; b=1, β=3. Throughout: d (min)=0.5, m=0.2, K=100, lattice=10×50 cells.
With sterilizing diseases the patterns that emerged depended on the demographic gradient and on the transmission mode. When the gradient affected death rate and regardless of the transmission mode (figure 1b,c), the result was the inverse of the classical expectation; disease prevalence increased as host population density declined. This qualitative pattern was robust to varying the parameter values in the range permitting host-pathogen coexistence. In figure 1c, the parameters were chosen to reflect those observed in natural populations of anther-smut disease caused by Microbotryum violaceum on Silene latifolia (a sterilizing disease with frequency-dependent transmission; Antonovics et al. 1997), and so represent realistic magnitudes.
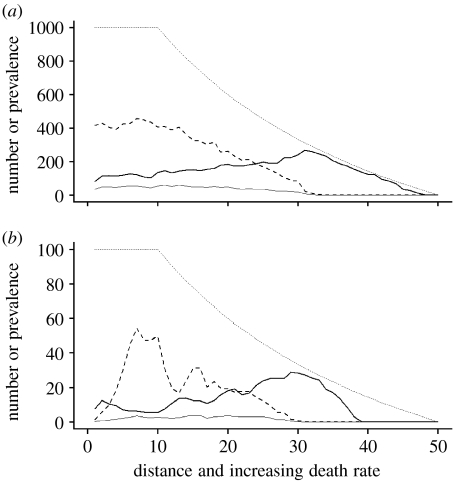
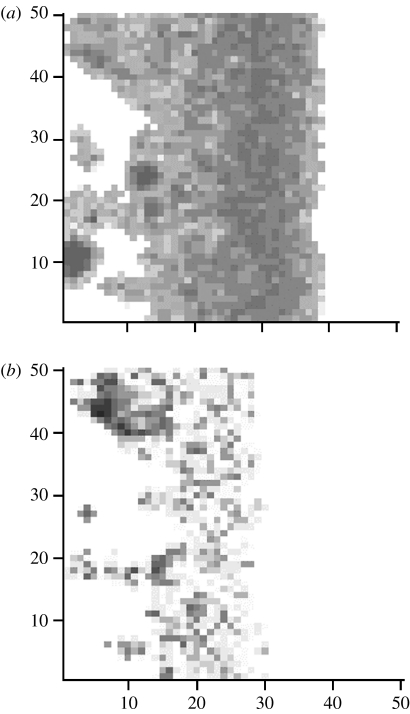
In the case of both transmission modes, lower population sizes occurred in central areas, i.e. those that had the highest host densities in the disease-free state. With frequency-dependent transmission (figure 1c), population range limits were frequently established in the central areas. This central region showed a high level of temporal and spatial heterogeneity in prevalence and population size, with chaotic probable fluctuations in numbers of diseased and healthy hosts. Thus over 20 independent simulations, the ‘central area’ showed mean occupancy of 18.7 per cent (s.d.=17.4) and a mean population size of 11.7 (s.d.=10.8), substantially less than the disease free state with 100 per cent occupancy and population size of 100. The presence of overall limits (with zero population sizes) was dependent on stochastic events and the dimensionality of the system (i.e. lattice width). When the disease-free population size within each cell was increased (figure 2a), there was a reduced extinction but still a severe decline in numbers in the central area. In wider lattices (figure 2b), there was increased transient persistence of local patches in the central region. This is illustrated in figure 3, where there are large areas where the host is absent, but also patches that can be largely healthy (bottom left-hand of the lattice) or heavily diseased (upper left).
Figure 2.
Population size and disease prevalence in a sterilizing disease with frequency-dependent transmission, along a gradient of increasing death rate. Parameters as in figure 1c, except for (a) K=1000, i.e. larger cell population numbers, (b) lattice=50×50 cells, i.e. wider area.
Figure 3.
Lattice view of simulation in figure 2b. Relative number of (a) healthy and (b) diseased individuals; black is high. The mortality gradient goes from low on the left to high on the right.
When the transmission was frequency-dependent, and the gradient was of decreasing birth rate, the population margin was pulled back substantially from its disease-free state (figure 4a). Over the gradient as a whole disease prevalence was independent of host population density. Again this new boundary showed chaotic fluctuations, with transient reinvasion by healthy individuals and disease. On the other hand, with density-dependent transmission there was a positive relationship between prevalence and density and no effect of disease on population limits (figure 4b).
Figure 4.
Population size and prevalence of disease along a gradient of decreasing birth rate. Lines as in figure 1. (a) Sterilizing disease with frequency-dependent transmission; β=0.3. (b) Sterilizing disease with density-dependent transmission; β=0.03. Throughout: b (max) =1, d=0.5, m=0.2, K=100, lattice=10×50 cells.
Throughout, qualitative outcomes were relatively insensitive to variation in the migration rate, as would be expected for gradual gradients in parameter values (cf. genetic differentiation along clines in evolutionary models; Endler 1977).
4. Discussion
This study shows that host-specific diseases can impact the range distribution of their hosts. These results are not surprising for frequency-dependent transmission, where the critical link between population density and transmission is lacking, and where pathogen-driven extinction is predicted even in deterministic non-spatial systems (Getz & Pickering 1983; Thrall et al. 1993; Lockhart et al. 1996). The ability of sterilizing diseases to generate range limits within regions where hosts would normally be most abundant was more surprising. However, this result is consistent with Boots & Sasaki (2001) who showed host extinction by sterilizing diseases using cellular automaton models and theory based on pair-approximation. The models in this study are somewhat different, being based on a lattice structure extended over an environmental gradient. They show that disease can restrict population ranges in what would otherwise (i.e. in a disease-free state) be considered marginal as well as central regions.
Stochastic effects played a large role in the extinction events observed. When disease reduces population size, demographic stochasticity is expected to accelerate local extinction (De Castro & Bolker 2005) and this can cascade into decreased patch occupancy (Antonovics et al. 1997; Antonovics 1999). However, extinction may only be partly caused by normal demographic stochasticity. Two other factors are likely to be important. First, finite host-pathogen systems show stochastically induced resonant cycles that can have surprisingly large amplitudes (McKane & Newman 2005; Alonso et al. 2007); these may substantially increase the chance of populations reaching critically low levels. Second, transient dynamics at the margins may lead to critically low numbers. In our study, populations at margins entered long-term chaotic-like fluctuations that appeared to be driven by host immigration into empty patches followed by epidemic-like reinvasion by the pathogen. This epidemic re-invasion led to large fluctuations leading to troughs in numbers that further increased the chance of extinction (Mollison 1991). The contribution of these different stochastic components in pathogen-driven extinction deserves further study. For populations threatened by disease, minimum viable population size estimates may be too high if based only on normal demographic stochasticity.
The extinction of populations in the centre of their disease-free range was also observed in simulations of host-parasitoid populations by Hochberg & Ives (1999), but for somewhat different reasons. Central populations went extinct in regions of high reproductive output as a result of unstable deterministic dynamics; extinction was only presumed when numbers dropped to a fractional individual. However, inspection of their figures also shows an inverse relationship between parasitoid prevalence and host abundance in these regions, and reflects the close conceptual and biological similarity between parasitoids and sterilizing diseases (Kuris 1974; Antonovics et al. 1995).
The other important outcome of this study is to seriously question the intuitive expectation of a positive relationship between host abundance and pathogen prevalence, derived from the classical model. Indeed, depending on the transmission mode and host demography, prevalence can stay constant or even increase as host abundance decreases. It is important to note that in the models, the transmission rate was always constant across the environmental gradient, and the only factors varying were the host birth and death rates.
In nature, there may be many other reasons why disease prevalence may increase in marginal populations. For example, transmission rate (e.g. vector abundance) may increases in peripheral regions, individuals may show increased disease susceptibility owing to physiological stress (Crawford 2008), or marginal populations may have a different age structure. Natural populations also do not show a uniformly decreasing abundance at the edges of their range (Gaston 2003), and different abundance structures may have different effects on pathogen-driven range limits. From a genetic perspective, marginal populations may also be more inbred or more uniform than central populations, thus predisposing them to higher disease levels. Co-evolutionary feedbacks between hosts and parasites can also impact on numerical dynamics (May & Anderson 1983; Antonovics 1994) and may drive populations to extinction (O'Keefe & Antonovics 2002). It is unknown under what conditions coevolutionary processes would stabilize or destabilize spatial dynamics at margins. For example, marginal populations may experience ‘virgin soil’ epidemics because if they transiently escape disease, they may lose immunological or genetic resistance and therefore be susceptible to epidemic re-invasion by pathogens. Indeed, host-shifts appear to be more common in marginal areas of host-ranges (Davies & Pedersen 2008).
It is surprising that there appear to have been only a handful of studies on disease incidence in marginal populations. However, the few precise distributional studies that have been done have shown contrasting patterns. Both decreased disease prevalence at margins (Brewer 1995; Alexander et al. 2007) as well increased prevalence have been observed (Antonovics et al. 2003; Briers 2003), but there is not enough information to match these contrasting patterns to the models presented here. Other than evidence of decreased species richness of parasites on islands (Gouy de Bellocq et al. 2002) or in invasive species (Wolfe 2002) there seems to be little direct empirical evidence for disease-free zones around the edges of a species range. The fact that different disease and host characteristics produce contrasting prevalence patterns argues for the potential value of gradients in understanding disease dynamics. For example, visual inspection of the simulations in this study reveals strong spatial sub-structuring towards the margin (cf. Antonovics et al. 2001, 2006), suggesting that cross-species covariances in abundance may change over the gradient and thus provide additional tools for interfacing models with field data.
In conclusion, this study has shown that sterilizing pathogens, especially those that are vector or sexually transmitted, can have a substantial impact on host distributions. There is a large need for empirically based studies on disease at population margins that go beyond simple documentation of abundance–prevalence relationships. Such relationships can provide useful signatures, but these signatures need careful interpretation in order to understand how disease impacts the distribution of a species.
Acknowledgments
I am grateful to Tim Newman for earlier models that inspired this study, to Linda Liu for help with programming and to Mike Boots, Alan McKane and Vijay Panjeti for stimulating discussions.
Footnotes
One contribution of 17 to a Special Issue ‘Geographic range limits of species’.
References
- Alexander H.M., Price S., Houser R., Finch D., Tourtellot M. Is there reduction in disease and predispersal seed predation at the border of a host plant's range? Field and herbarium studies of Carex blanda. J. Ecol. 2007;95:446–457. doi:10.1111/j.1365-2745.2007.01228.x [Google Scholar]
- Alonso D., McKane A.J., Pascual M. Stochastic amplification in epidemics. J. R. Soc. Interface. 2007;4:575–582. doi: 10.1098/rsif.2006.0192. doi:10.1098/rsif.2006.0192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R.M. Population dynamics of indirectly transmitted disease agents: the vector component. In: McKelvey J.J., Eldridge B.F., Maramorosch K., editors. Vectors of disease agents. Interactions with plants, animals, and man. Praeger; New York, NY: 1981. pp. 13–43. [Google Scholar]
- Anderson R.M., May R.M. The population dynamics of microparasites and their invertebrate hosts. Phil. Trans. R. Soc. B. 1981;291:451–524. doi:10.1098/rstb.1981.0005 [Google Scholar]
- Antonovics J. The nature of limits to natural selection. Ann. Miss. Bot. Garden. 1976;63:224–247. doi:10.2307/2395303 [Google Scholar]
- Antonovics J. The interplay of numerical and gene-frequency dynamics in host–pathogen systems. In: Real L., editor. Ecological genetics. Princeton University Press; Princeton, NJ: 1994. pp. 146–170. [Google Scholar]
- Antonovics J. Pathogens and plant population dynamics: the effects of resistance genes on numbers and distribution. In: Traynor P.M., Westwood H.H., editors. Ecological effects of pest resistance genes in managed ecosystems. Information Systems for Biotechnology; Blacksburg, VA: 1999. pp. 49–55. [Google Scholar]
- Antonovics J. Plant venereal diseases: insights from a messy metaphor. New Phytol. 2005;165:71–80. doi: 10.1111/j.1469-8137.2004.01215.x. doi:10.1111/j.1469-8137.2004.01215.x [DOI] [PubMed] [Google Scholar]
- Antonovics J., Iwasa Y., Hassell M.P. A generalized model of parasitoid, venereal, and vector-based transmission processes. Am. Nat. 1995;145:661–675. doi:10.1086/285761 [Google Scholar]
- Antonovics J., Thrall P.H., Jarosz A.M. Genetics and the spatial ecology of species interactions: the Silene–Ustilago system. In: Tilman D., Kareiva P., editors. Spatial ecology: the role of space in population dynamics and interspecific interactions. Princeton University Press; Princeton, NJ: 1997. pp. 158–180. [Google Scholar]
- Antonovics J., Newman T.J., Best B.J. Spatially explicit studies on the ecology and genetics of population margins. In: Silvertown J., Antonovics J., editors. Integrating ecology and evolution in a spatial context. Blackwell; Oxford, UK: 2001. pp. 91–116. [Google Scholar]
- Antonovics J., Hood M.E., Thrall P., Abrams J.Y., Duthie M. Herbarium studies on the distribution of anther-smut disease (Microbotryum violaceum) and Silene species in the eastern United States. Am. J. Bot. 2003;90:1522–1531. doi: 10.3732/ajb.90.10.1522. doi:10.3732/ajb.90.10.1522 [DOI] [PubMed] [Google Scholar]
- Antonovics J., McKane A.J., Newman T. Spatio-temporal dynamics at population margins. Am. Nat. 2006;167:16–27. doi: 10.1086/498539. doi:10.1086/498539 [DOI] [PubMed] [Google Scholar]
- Baudoin M. Host castration as a parasitic strategy. Evolution. 1975;29:335–352. doi: 10.1111/j.1558-5646.1975.tb00213.x. doi:10.2307/2407221 [DOI] [PubMed] [Google Scholar]
- Begon M., Hazel S.M., Baxby D., Bown K., Cavanagh R., Chantrey J., Jones T., Bennett M. Transmission dynamics of a zoonotic pathogen within and between wildlife host species. Proc. R. Soc. B. 1999;266:1939–1945. doi: 10.1098/rspb.1999.0870. doi:10.1098/rspb.1999.0870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørnstad O.N., Finkenstädt B., Grenfell B.T. Endemic and epidemic dynamics of measles: estimating epidemiological scaling with a time series SIR model. Ecol. Monogr. 2002;72:169–184. doi:10.1890/0012-9615(2002)072[0169:DOMEES]2.0.CO;2 [Google Scholar]
- Boots M., Sasaki A. Parasite-driven extinction in spatially explicit host–parasite systems. Am. Nat. 2001;159:706–713. doi: 10.1086/339996. doi:10.1086/339996 [DOI] [PubMed] [Google Scholar]
- Brewer L.G. Ecology of survival and recovery from blight in American chestnut trees (Castanea dentata (Marsh.) Borkh.) in Michigan. Bull. Torrey Bot. Club. 1995;122:40–57. doi:10.2307/2996402 [Google Scholar]
- Briers R.A. Range limits and parasite prevalence in freshwater snails. Proc. R. Soc. B. 2003;270(Suppl.):S178–S180. doi: 10.1098/rsbl.2003.0046. doi:10.1098/rsbl.2003.0046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter R.N., Prince S.D. Epidemic models used to explain biogeographical distribution limits. Nature. 1981;293:644–645. doi:10.1038/293644a0 [Google Scholar]
- Clay K. Parasitic castration of plants by fungi. Trends Ecol. Evol. 1991;6:162–166. doi: 10.1016/0169-5347(91)90058-6. doi:10.1016/0169-5347(91)90058-6 [DOI] [PubMed] [Google Scholar]
- Crawford R.M.M. Cambridge University Press; Cambridge, UK: 2008. Plants at the margin: ecological limits and climate change. [Google Scholar]
- Davies J.T., Pedersen A.B. Phylogeny and geography predict pathogen community similarity in wild primates and humans. Proc. R. Soc. B. 2008;275:1695–1701. doi: 10.1098/rspb.2008.0284. doi:10.1098/rspb.2008.0284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Castro F., Bolker B. Mechanisms of disease-induced extinction. Ecol. Lett. 2005;8:117–126. doi:10.1111/j.1461-0248.2004.00693.x [Google Scholar]
- Endler J. Princeton University Press; Princeton, NJ: 1977. Geographic variation, speciation, and clines. [PubMed] [Google Scholar]
- Gaston K.J. Oxford University Press; Oxford, UK: 2003. The structure and dynamics of geographic ranges. [Google Scholar]
- Getz W.M., Pickering J. Epidemic models: thresholds and population regulation. Am. Nat. 1983;121:892–898. doi:10.1086/284112 [Google Scholar]
- Gouy de Bellocq J., Morand S., Feliu C. Patterns of species richness of Western Palaearctic micro-mammals: island effects. Ecography. 2002;25:173–183. doi:10.1034/j.1600-0587.2002.250205.x [Google Scholar]
- Hochberg M.E., Ives A.R. Can natural enemies enforce geographical range limits? Ecography. 1999;22:268–276. doi:10.1111/j.1600-0587.1999.tb00502.x [Google Scholar]
- Hoffmann A.A., Blows M.W. Species borders: ecological and evolutionary perspectives. Trends Ecol. Evol. 1994;9:223–227. doi: 10.1016/0169-5347(94)90248-8. doi:10.1016/0169-5347(94)90248-8 [DOI] [PubMed] [Google Scholar]
- Holt R.D., Keitt T.H., Lewis M.A., Maurere B.A., Taper M.L. Theoretical models of species' borders: single species approaches. Oikos. 2005;108:18–27. doi:10.1111/j.0030-1299.2005.13147.x [Google Scholar]
- Jaenike J. Mycophagous Drosophila and their nematode parasites. Am. Nat. 1992;139:893–906. doi:10.1086/285365 [Google Scholar]
- Keitt T., Lewis M., Holt R. Allee effects, invasion pinning, and species' borders. Am. Nat. 2001;157:203–216. doi: 10.1086/318633. doi:10.1086/318633 [DOI] [PubMed] [Google Scholar]
- Kuris A.M. Trophic interactions: similarity of parasitic castrators to parasitoids. Quart. Rev. Biol. 1974;49:129–148. doi:10.1086/408018 [Google Scholar]
- Lafferty K.D., Kuris A.M. Biological control of marine pests. Ecology. 1996;77:1989–2000. doi:10.2307/2265695 [Google Scholar]
- Lennon J.J., Turner J.R.G., Connell D. A metapopulation model of species boundaries. Oikos. 1997;78:183–190. doi:10.2307/3545610 [Google Scholar]
- Lockhart A.B., Thrall P.H., Antonovics J. The distribution and characteristics of sexually transmitted diseases in animals: ecological and evolutionary implications. Biol. Rev. Camb. Phil. Soc. 1996;71:415–471. doi: 10.1111/j.1469-185x.1996.tb01281.x. doi:10.1111/j.1469-185X.1996.tb01281.x [DOI] [PubMed] [Google Scholar]
- May R.M., Anderson R.M. Epidemiology and genetics in the coevolution of parasites and hosts. Proc. R. Soc. B. 1983;219:281–313. doi: 10.1098/rspb.1983.0075. doi:10.1098/rspb.1983.1983.0075 [DOI] [PubMed] [Google Scholar]
- Mayr E. Belknap Press; Harvard, MA: 1963. Animal species and evolution. [Google Scholar]
- McKane A.J., Newman T.J. Predator-prey cycles from resonant amplification of demographic stochasticity. Phys. Rev. Lett. 2005;94:2 181 021–2 181 024. doi: 10.1103/PhysRevLett.94.218102. doi:10.1103/PhysRevLett.94.218102 [DOI] [PubMed] [Google Scholar]
- Mollison D. Dependence of epidemic and population velocities on basic parameters. Math. Biosci. 1991;107:255–287. doi: 10.1016/0025-5564(91)90009-8. doi:10.1016/0025-5564(91)90009-8 [DOI] [PubMed] [Google Scholar]
- O'Keefe K.J., Antonovics J. Playing by different rules: the evolution of virulence in sterilizing pathogens. Am. Nat. 2002;159:597–605. doi: 10.1086/339990. doi:10.1086/339990 [DOI] [PubMed] [Google Scholar]
- Parmesan C., Gaines S., Gonzalez L., Kaufman D.M., Kingsolver J., Peterson T., Sagarin R. Empirical perspectives on species borders: from traditional biogeography to global change. Oikos. 2005;108:58–75. doi:10.1111/j.0030-1299.2005.13150.x [Google Scholar]
- Ries L., Fletcher J., Battin J., Sisk T. Ecological responses to habitat edges: mechanisms, models, and variability explained. Annu. Rev. Ecol. Evol. Syst. 2004;35:491–522. doi:10.1146/annurev.ecolsys.35.112202.130148 [Google Scholar]
- Thrall P.H., Antonovics J., Hall D.W. Host and pathogen coexistence in vector-borne and venereal diseases characterized by frequency-dependent disease transmission. Am. Nat. 1993;142:543–552. doi:10.1086/285554 [Google Scholar]
- Thrall P.H., Biere A., Uyenoyama M.K. Frequency-dependent disease transmission and the dynamics of the Silene–Ustilago host–pathogen system. Am. Nat. 1995;145:43–62. doi:10.1086/285727 [Google Scholar]
- Travis J.M.J., Brooker R.W., Clark E.J., Dutham C. The distribution of positive and negative species interactions across environmental gradients on a dual-lattice model. J. Theor. Biol. 2006;241:896–902. doi: 10.1016/j.jtbi.2006.01.025. doi:10.1016/j.jtbi.2006.01.025 [DOI] [PubMed] [Google Scholar]
- Watkinson A.R. On the abundance of plants along an environmental gradient. J. Ecol. 1985;73:569–578. doi:10.2307/2260494 [Google Scholar]
- Wennstrom A., Ericson L. The concept of sexually transmitted diseases in plants: definition and applicability. Oikos. 2003;100:397–402. doi:10.1034/j.1600-0706.2003.12004.x [Google Scholar]
- Wilson W., Harrison S.P., Hastings A., McCann K. Stable pattern formation in tussock moth populations. J. Anim. Ecol. 1999;68:94–107. doi:10.1046/j.1365-2656.1999.00265.x [Google Scholar]
- Wolfe L.M. Why alien invaders succeed: support for the escape-from-enemy hypothesis. Am. Nat. 2002;160:705–711. doi: 10.1086/343872. doi:10.1086/343872 [DOI] [PubMed] [Google Scholar]
- Woodward F.I. Cambridge University Press; Cambridge, UK: 1987. Climate and plant distribution. [Google Scholar]