Abstract
Disease-mediated inbreeding depression is a potential cost of living in groups with kin, but its general magnitude in wild populations is unclear. We examined the relationships between inbreeding, survival and disease for 312 offspring, produced by 35 parental pairs, in a large, open population of cooperatively breeding American crows (Corvus brachyrhynchos). Genetic analyses of parentage, parental relatedness coefficients and pedigree information suggested that 23 per cent of parental dyads were first- or second-order kin. Heterozygosity–heterozygosity correlations suggested that a microsatellite-based index of individual heterozygosity predicted individual genome-wide heterozygosity in this population. After excluding birds that died traumatically, survival probability was lower for relatively inbred birds during the 2–50 months after banding: the hazard rate for the most inbred birds was 170 per cent higher than that for the least inbred birds across the range of inbreeding index values. Birds that died with disease symptoms had higher inbreeding indices than birds with other fates. Our results suggest that avoidance of close inbreeding and the absence of inbreeding depression in large, open populations should not be assumed in taxa with kin-based social systems, and that microsatellite-based indices of individual heterozygosity can be an appropriate tool for examining the inbreeding depression in populations where incest and close inbreeding occur.
Keywords: American crow, Corvus brachyrhynchos, disease, inbreeding, inbreeding depression, cooperative breeders
1. Introduction
Relatively high pathogen exposure and transmission rates are a potential cost of group living (Alexander 1974; Schmid-Hempel & Crozier 1999) that might be elevated in taxa with kin-based social groups (Spottiswoode 2008), if related individuals share similar susceptibility characteristics (Shykoff & Schmid-Hempel 1991; Hughes & Boomsma 2004) and/or if pathogens are locally adapted to common genotypes (Lively et al. 2004). Furthermore, in the absence of active inbreeding avoidance mechanisms, taxa that live in kin groups (particularly those with limited natal dispersal of both sexes) might have a higher probability of mating with kin than those that do not live in kin groups (Szulkin & Sheldon 2008), and offspring produced from these consanguineous matings might suffer even greater disease costs (Coltman et al. 1999). Inbred offspring have lower genome-wide heterozygosity than relatively outbred offspring and might therefore experience a disease-mediated reduction in fitness if (i) they are unable to recognize as wide a breadth of pathogens as more heterozygous individuals (overdominance) and/or (ii) pathogens are part of an environment that selects against individuals expressing deleterious recessive alleles (partial dominance; Coltman et al. 1999). Disease-mediated inbreeding depression might therefore represent a substantial cost to living and breeding with kin, potentially influencing the evolution of dispersal and incest avoidance (Charlesworth & Charlesworth 1987). Understanding the relationship between inbreeding and disease is also important for the preservation of small, declining populations, in which inbreeding is unavoidable (Hedrick & Kalinowski 2000; Keller & Waller 2002).
Because empirical data on inbreeding depression in wild populations is limited (Keller & Waller 2002), particularly in terms of disease (Spielman et al. 2004), the general magnitude and frequency of disease-mediated inbreeding depression are unclear. In laboratory settings, some studies suggest that inbreeding increases susceptibility to pathogens or parasites (Luong et al. 2007; Ilmonen et al. 2008), whereas others have found that the relationship between disease resistance and inbreeding varies with the nature of the immune challenge (Calleri et al. 2006). Among Gila topminnows (Poeciliopsis occidentalis), for example, the relationship between fluke infection and inbreeding varied with source population (Hedrick et al. 2001), and relatively inbred individuals from all populations had higher survival after experimental infection with a novel bacterium (Giese & Hedrick 2003). Relatively low disease costs might be expected among habitual inbreeders, particularly under benign environmental conditions (Armbruster & Reed 2005), if long-term inbreeding purges the population's genetic load of deleterious alleles (Barrett & Charlesworth 1991), although potential overdominance might limit the success of purging (Crnokrak & Barrett 2002), and purging does not appear to operate consistently in wild populations (Byers & Waller 1999).
Inbreeding depression measured in the captive populations or laboratory settings might underestimate costs in wild populations (Crnokrak & Roff 1999). Because adequate pedigree information is often unavailable, most recent studies of inbreeding depression in wild populations have relied on microsatellite heterozygosity to infer genome-wide heterozygosity and inbreeding (Hansson & Westerberg 2002; Keller & Waller 2002; Coltman & Slate 2003). Some of these studies have found a positive association between microsatellite-based estimates of individual homozygosity (and potential inbreeding) and ectoparasite burden (Whiteman et al. 2006), endoparasite burden (Coltman et al. 1999; MacDougall-Shackleton et al. 2005; Acevedo-Whitehouse et al. 2006; Rijks et al. 2008) and mortality during epidemics (Valsecchi et al. 2004; Ross-Gillespie et al. 2007). Likewise, Acevedo-Whitehouse et al. (2003) found an association between different diseases and marker-based estimates of individual heterozygosity in rehabilitated California sea lions (Zalophus californianus). Other studies, however, have found no association between marker-based heterozygosity and endoparasites (Cote et al. 2005), and negative results might be under-reported because of a publication bias towards significant correlations (Coltman & Slate 2003). It is difficult to assess the generality of disease-mediated inbreeding depression in natural populations with the available evidence.
The use of microsatellite markers to infer inbreeding coefficients could contribute to apparent variation in the relationship between disease and inbreeding among studies, because microsatellite heterozygosity is unlikely to predict the inbreeding coefficient in all systems (Hansson & Westerberg 2002; Balloux et al. 2004; Slate et al. 2004; DeWoody & DeWoody 2005). Microsatellite and genome-wide heterozygosity are expected to be most strongly correlated in very small populations with a high variance in inbreeding and a high proportion of incestuous matings, a scenario that is uncommon in nature (Balloux et al. 2004; Slate et al. 2004). If microsatellite and genome-wide heterozygosity are correlated in a given system, then heterozygosity estimated from one set of microsatellites should be positively correlated with heterozygosity from an independent set of microsatellites from the same individual (‘heterozygosity–heterozygosity correlations’ or HHCs; Balloux et al. 2004). Pedigree information and HHCs can be used together to examine (i) how well microsatellite and genome-wide heterozygosity are correlated in a given system, and (ii) whether heterozygosity–fitness correlations are likely to be explained (at least in part) by inbreeding.
In this study, we examine disease-mediated inbreeding costs in American crows (Corvus brachyrhynchos), a species that occupies a wide range of habitats and is capable of long-distance migration (Verbeek & Caffrey 2002). American crows in a wild, cooperatively breeding population in Ithaca, New York, exhibit natal philopatry and limited natal dispersal of both sexes, as well as incest and inbreeding (Townsend et al. 2009). We first assessed the appropriateness of our inbreeding index, estimated from a panel of 10 microsatellite markers, by comparing inbreeding indices with available pedigree information and parental relatedness coefficients, and through HHCs. We then explored the relationship between this inbreeding index and two indices of fitness: (i) survival within the duration of the study (2–50 months after banding, depending on the year in which an individual was sampled) and (ii) the probability of dying with disease symptoms.
2. Material and methods
(a) Field sampling
From 2004 to 2008, we collected blood via brachial venepuncture from 312 nestlings (table 1) belonging to 30 American crow family groups in a long-term study population in Ithaca, New York (described in McGowan 2001; Townsend et al. 2009). Offspring were marked individually with colour bands, aluminium bands and patagial tags on day 23–30 after hatching. We collected genetic samples from blood or passively moulted feathers from all members of the family groups of 283 out of these 312 nestlings (Townsend et al. 2009). We monitored marked focal offspring for survival and poxviral dermatitis lesions at least once per month from their initial marking until July 2008.
Table 1.
Number of offspring (n) marked in each year of the study and the maximum number of months that individuals from each cohort were monitored. (The number of months monitored represents a maximum because some individuals from each cohort died or disappeared before the endpoint of the study.)
| year | n | months monitored |
|---|---|---|
| 2004 | 35 | 50 |
| 2005 | 63 | 38 |
| 2006 | 73 | 26 |
| 2007 | 81 | 14 |
| 2008 | 60 | 2 |
(b) Genetic analyses
We genotyped 312 nestlings and associated family members at 10 microsatellite loci (Tarr & Fleischer 1998; Schoenle et al. 2006; Townsend et al. 2009). We assessed parentage of 283 nestlings with genotyped social parents using the maximum-likelihood method in the program Cervus v. 3.0 (Kalinowski et al. 2007), identifying probable genetic parents (within-pair and extra-pair) following criteria described in Townsend et al. (2009). For pairs identified as first-order kin by pedigree, we tested this degree of relatedness based on their genotypic information in the program KINGROUP (Rp=0, Rm=1; 100 000 simulations; α=0.05; Konovalov et al. 2004). We then assessed relatedness between the parental pair dyads for which we lacked pedigree information in KINGROUP, setting the selection criterion to identify pairs that were likely to be second-order kin (Rp=0, Rm=0.5). More markers (more than 17) would have been necessary to accurately assess deeper relationships (Goodnight & Queller 1999; Konovalov et al. 2004). We also used KINGROUP to estimate relatedness coefficients between all parental dyads. We estimated internal relatedness (IR), a microsatellite-based inbreeding index that accounts for background allele frequencies when estimating parental similarity from an offspring's microsatellite genotype (Amos et al. 2001), using IRMacroN4 (http://www.zoo.cam.ac.uk/zoostaff/amos/#ComputerPrograms). To examine the relationship between IR and parental relatedness, we regressed offspring IR values against the KINGROUP-generated relatedness coefficients of their respective genetic parents in a mixed model with parental pair as a random factor.
We used a combination of pedigree information and HHCs to examine how well microsatellite and genome-wide heterozygosity were correlated within individuals in this system. First, we divided offspring into three groups: offspring produced incestuously; offspring produced through second-order kin matings; and relatively outbred offspring. Following Balloux et al. (2004), we then generated HHCs for each of these three groups of offspring by (i) randomly splitting the 10 loci into two sets of five independent loci, (ii) calculating two IR values—one from each set of five loci—for each offspring, (iii) regressing the two IR values against one another for all offspring in each group and calculating the r2 value of the regressions, and (iv) repeating this procedure 50 times. We then used analysis of variance to compare the 50 r2 values generated for each of the three groups of offspring, predicting that mean r2 value would be highest for the offspring produced incestuously and close to zero for the relatively outbred offspring (Balloux et al. 2004).
(c) Fate determination
Dead crows were tested for West Nile virus (WNV) using reverse polymerase chain reaction (Clark et al. 2006). Birds that tested negative for WNV were necropsied with a complete external and internal examination. Dead crows discovered after November 2006 were subjected to gross examination and full necropsy, followed by the sampling of all major organs with fixation in 10 per cent neutral buffered formalin. Organs were sectioned using a tissue-cutting knife, embedded in paraffin, microtome sectioned at 4 or 5 μm and stained with haematoxylin and eosin using standard histological technique. Additional sections were also prepared for histochemical and immunohistochemical staining using the same protocol. All prepared sections were mounted with non-aqueous permanent mounting medium and analysed under light microscopy by two veterinary anatomic pathologists.
(d) IR and survival
To examine the relationship between IR and survival, we used Cox's proportional hazards regression and mark–recapture analyses. There was no evidence of non-proportionality in the data. Year and sex had non-significant effects and were removed from the final proportional hazards regression model. Capture-history matrices were constructed using resight data from 171 individuals for which we had the most consistent resight data from the 2005–2007 cohorts during the first 14 months after banding, divided into 10 time intervals (May, June, July, August–December, January, February, March, April, May and June–July). Multiple resights within intervals were treated as a single sighting. Survival (Φ) and recapture (p) parameters were estimated in the program MARK v. 5.1 (http://www.phidot.org/software/mark/index.html). Following Lebreton et al. (1992), model selection was made using the Akaike information criterion (AIC; Akaike 1973). First, we generated models to detect time (t), year (y) and sex (s) effects on Φ and p, starting with [Φ(s×t+y×t)p(t)] as the global model. We estimated a quasi-likelihood parameter by dividing the deviance estimate from the original data by the mean of the simulated deviances from a parametric goodness-of-fit test (1000 bootstrap samples), adjusting the overdispersion parameter to 1.15. We then constrained the best model with inbreeding index as an individual covariate. The model with the lowest quasi-AIC (QAIC) was accepted as the most parsimonious model for the data.
(e) IR and fate
We explored the relationship between inbreeding index and fate in a mixed model with family as a random factor and fate as a fixed factor. Inbreeding index was normally distributed. Year and sex had non-significant effects and were removed from the final fate model. To examine the influence of the most inbred birds on heterozygosity–fitness correlations, we then excluded from fate and survival analyses the 10 offspring (constituting 3% of the entire sample) that were known by pedigree to have been produced incestuously. Statistical analyses were conducted in JMP v. 7.0.
3. Results
(a) Genetic analyses
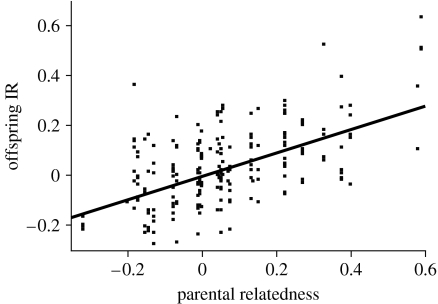
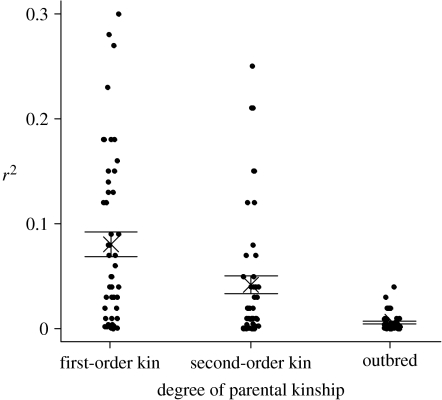
Thirty-five genetic parental pairs (including within-pair and extra-pair sires; Townsend et al. 2009) were identified for 230 offspring. Three of the 35 identified genetic parental pairs were first-order kin (mother–son extra-pair matings) by pedigree. KINGROUP identified these three genetic pairs as probable first-order kin, and identified five additional pairs as probable second-order kin, suggesting that eight out of the 35 identified genetic parental pairs (23%) were first- or second-order kin. First-order genetic pairs produced 10 out of these 230 offspring (4.3%), whereas probable second-order genetic pairs produced 33 out of the 230 offspring (14.3%). Mean relatedness coefficient between genetic parental pairs, estimated by KINGROUP, was 0.06 (range=−0.32–0.59). Individual IR of the 230 offspring with genotyped genetic parents was positively correlated with parental relatedness in a mixed model with parental pair as a random effect (0.54±0.05 s.e., F1,25.5=114.7, p<0.0001, R2=0.40; figure 1). There was significant variation in the strength of HHCs among offspring of different relatedness classes (F2,147=20.0, p<0.001, n=150): the strength of the correlation, as expected, decreased as parental relatedness decreased (Tukey's HSD, α=0.05; figure 2) and was close to zero for the relatively outbred birds.
Figure 1.
Relationship between offspring IR and parental relatedness coefficient. Ordinary least-squares regression illustrated (0.53±0.05 s.e., t228=11.7, p<0.001).
Figure 2.
HHCs for offspring produced by parents of different degrees of probable relatedness (e.g. first-order kin, second-order kin and relatively outbred parental pairs). r2 values in each group were obtained by randomly dividing the 10 loci into two groups of five loci, computing IR for both sets of loci for each offspring, regressing one against the other and resampling the data 50 times. Means and standard errors shown.
(b) Offspring fates
We placed the marked focal offspring into four fate categories: ‘alive’ (n=100) if they were seen within the last three months of the study; ‘trauma’ (n=67) if they died as a result of predation, car collisions, electrocution, shootings, and other violent deaths; ‘unknown’ (n=124) if they disappeared from the population, or if they were found dead but the cause of death was uncertain; and ‘diseased’ (n=21) if they had poxviral dermatitis lesions (n=14) or tested positive for WNV (n=3), bacterial infections (n=2), fungal pneumonia (n=1) or enteritis (n=1) when they died.
(c) IR, survival and fate
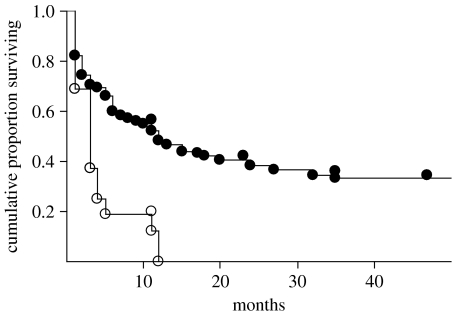
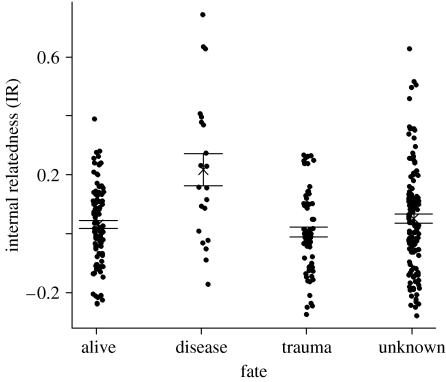
Mark–recapture estimates of survival during the first year after banding were lower for offspring with higher inbreeding indices, but only after individuals that died traumatically (deaths that were potentially independent of individual condition) were removed from the analyses: there was considerable support for a time-dependent model with inbreeding index as an additive effect (ΔQAIC=6.6; table 2). Survival analyses also suggested that survival probability in the first 2–50 months after banding was lower for relatively inbred birds (figure 3): the hazard rate for death or disappearance was 170 per cent (95% CI: 2–564%) higher for the most inbred birds across the range of IR values (model 1 in table 3), again after excluding birds that died traumatically. Inbreeding index had an even stronger effect on survival when we considered only the birds in the ‘alive’ and ‘diseased’ categories, the fate categories most likely to be influenced by individual condition (model 2 in table 3). IR varied with fate: in a mixed model with family as a random factor (F3,299.5=3.29, p=0.02), individuals that died with disease symptoms had significantly higher inbreeding indices than all other individuals (Tukey's HSD, α=0.05; figure 4). It is possible that death from WNV was independent of individual quality, because, in laboratory trials, all American crows died after WNV infection (Komar et al. 2003). Removing WNV-positive birds from the analysis did not change the association between inbreeding index and fate (mixed model with family as a random factor and fate as a fixed factor, F3,293.2=2.99, p=0.03, n=309). When the 10 offspring that were known, by pedigree, to have been produced incestuously were removed from the sample, inbreeding index had no effect on nestling survival (Χ2=2.0, p=0.16, n=235) and inbreeding index did not vary with fate (F3,290.9=2.3, p=0.08). Post hoc tests for local and/or direct effects (Hansson & Westerberg 2002), in which we reran the survival analysis and compared inbreeding index among the different fate categories with each locus sequentially removed (Hawley et al. 2005), yielded similar patterns, suggesting that these patterns were not driven by any single locus.
Table 2.
Candidate set of approximating models generated to fit American crow mark–recapture data. (np, number of parameters; Φ, survival; IR, internal relatedness; p, recapture; t, time; s, sex; y, year.)
| model | QAICc | ΔQAICc | QAICc weight | np | deviance |
|---|---|---|---|---|---|
| Φ(IR+t)p(t) | 1882.413 | 0 | 0.963 | 19 | 1843.921 |
| Φ(t)p(t) | 1889.014 | 6.601 | 0.035 | 18 | 1852.571 |
| Φ(t+s+y)p(t) | 1894.886 | 12.4739 | 0.002 | 22 | 1850.231 |
| Φ(t×y)p(t) | 1909.346 | 26.9338 | 0 | 45 | 1816.623 |
| Φ(t×s)p(t) | 1919.354 | 36.9411 | 0 | 36 | 1845.611 |
| Φ(t×s)+(t×y)p(t) | 1921.579 | 39.1662 | 0 | 54 | 1809.648 |
Figure 3.
Survival with inbreeding index. Kaplan-Meier plot showing that highly inbred birds (IR>0.35, n=18, open markers) had lower proportional survival than relatively outbred birds (IR<0.35, n=294, closed markers; log rank: Χ2=8.37, p=0.004). Although presented categorically here for the purpose of illustration, inbreeding index was treated as a continuous variable in all other analyses.
Table 3.
IR and risk of death or disappearance of American crows. (Model 1 includes crows that were alive, had died with disease symptoms or were of unknown fate by the end of the study, whereas model 2 includes just those that were alive or had died with disease symptoms by the end of the study.)
| model | na | d.f. | parameter estimate for IR | s.e. | Χ2 | p-value | risk ratio (RR) | 95% confidence limits for RR | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 245 | 1 | 1.0 | 0.47 | 4.0 | 0.047 | 2.70 | 1.02 | 6.64 |
| 2 | 121 | 1 | 4.8 | 1.0 | 19.1 | <0.001 | 126.7 | 16.46 | 851.96 |
Number of birds included in the analysis.
Figure 4.
IR of offspring in different fate classes. Means and standard errors shown.
4. Discussion
We have shown evidence for a substantial survival cost of inbreeding in this vast, contiguous and open crow population. Survival in the 2–50 months after banding was lower for relatively inbred birds: the hazard rate for the most inbred birds was 170 per cent higher than that for the least inbred birds across the range of inbreeding index values, after we excluded birds that died traumatically from the sample. Reduced survival for inbred birds appeared to be mediated, at least in part, by disease: birds that died with disease symptoms had higher inbreeding indices than those that lived for the duration of the study, died traumatically or whose cause of death or disappearance was ambiguous. This disease-mediated inbreeding depression represents the minimum cost of inbreeding in this population, because apparent costs tend to accumulate with life stages and fitness indices measured (Pusey & Wolf 1996). We did not account for potential inbreeding depression expressed early in development, whether inbred individuals were more likely to die embryonically, soon after hatching (Keller & Waller 2002) or later in life, nor whether inbred individuals had a lower probability of survival in later life stages, were less fecund (Spottiswoode & Moller 2004) or were less successful in acquiring mates (Pusey & Wolf 1996; Seddon et al. 2004; Hoffman et al. 2007).
Despite the high frequency of reported microsatellite-based heterozygosity–fitness correlations (Coltman & Slate 2003), a number of authors have suggested that microsatellite-based estimates of heterozygosity might generally be of limited use as indices of inbreeding (Balloux et al. 2004; Slate et al. 2004; DeWoody & DeWoody 2005). Our results suggest that they can be useful in populations where incestuous matings and matings between second-order kin occur. HHCs, as well as the positive correlation between parental relatedness coefficients and offspring IR, suggested that our microsatellite-based estimates of individual heterozygosity did indeed reflect inbreeding in this population. The relatively strong correlation between IR and genome-wide heterozygosity for the most inbred birds was not surprising, given that recent inbreeding events are expected to have a much larger effect on inbreeding coefficient than inbreeding events deeper in the pedigree (Balloux et al. 2004).
Kin matings in this population might occur incidentally from living in close proximity to sexually mature, opposite-sex kin. In order for limited natal dispersal of both sexes to have persisted in this population, we might expect that inbreeding costs are balanced by the benefits gained from living and/or breeding with kin (Alexander 1974). Various potential benefits have been proposed for living with kin, such as enhanced fitness of non-descendent kin (Emlen 1995; but see Caffrey 2000), nepotistic defence (Sherman 1981), enhanced survival (Ekman et al. 2000), lineage persistence (Marzluff & Balda 1990) and territorial inheritance (Woolfenden & Fitzpatrick 1978). Potential benefits of kin matings include kin selection (which can, in theory, outweigh surprisingly high inbreeding depression costs; Kokko & Ots 2006) and the maintenance of locally selected gene complexes, which could be disrupted through matings with individuals from other populations (‘outbreeding depression’; Shields 1982; Bateson 1983). In this American crow population, individuals exhibit habitat specificity, tending to breed in microhabitats (urban or rural) similar to their natal territory (McGowan 2001). Matings between birds adapted to the same microhabitat (such as kin) might promote offspring adaptation to a particular microhabitat.
In order to avoid ‘too much’ outbreeding, Bateson (1983) suggested that optimal mates might be those that are moderately related, particularly if any costs of mating with kin decline quickly with degree of parental relatedness. Disease-mediated costs of inbreeding might, indeed, have declined quickly with parental relatedness in this population: when we removed from our sample a small number of offspring that were known by pedigree to have been produced incestuously, significant patterns of survival and fate with IR disappeared. An alternative explanation for this result, however, is that we were only able to detect inbreeding depression in the most inbred offspring because our marker-based estimate of heterozygosity did not correlate well with genome-wide heterozygosity for relatively outbred offspring. Without additional molecular markers and pedigree information, we cannot determine whether relatively outbred birds did not suffer disease-mediated inbreeding costs or whether we were unable to detect these costs with our available marker set.
The results of this study are important for three reasons. First, although it often appears true that, when possible, incest and close inbreeding are avoided in cooperative breeders, particularly among birds (Koenig & Haydock 2004), our analyses suggest that close inbreeding is not uncommon in this open population of crows (see also Townsend et al. 2009). It is possible that incest and close inbreeding occur undetected in other taxa because of the expectation that it will not occur (Kokko & Ots 2006), or because limited marker sensitivity makes the detection of incest in taxa that live in kin groups challenging (McRae & Amos 1999). Second, the severity of disease-mediated inbreeding costs that we detected was surprising, given that close inbreeding, if strongly selected against, could presumably have been avoided in this large, open population. Third, we found evidence for a correlation between marker-based estimates of heterozygosity and actual inbreeding in this population. Simulations by Balloux et al. (2004) suggested that microsatellite heterozygosity would be most likely to reflect genome-wide heterozygosity in populations where there is a high proportion of consanguineous matings, such as might be found in very small or subdivided populations, or those with highly skewed mating systems. In our sample, even though 23 per cent of genetic pairs appeared to be first- or second-order kin, the proportion of offspring produced by these pairs was not high: only 4.3 per cent of offspring were produced by first-order kin dyads, and 14.3 per cent of offspring were produced by second-order kin dyads. Nevertheless, IR values estimated from our panel of 10 microsatellite markers appeared sufficient to reflect differences in genome-wide heterozygosity between these highly inbred offspring and relatively outbred offspring. In conclusion, microsatellite markers, when verified as an actual index of inbreeding by even very shallow pedigree information, can serve as a valuable tool for quantifying inbreeding and inbreeding depression in populations in which some level of incest and close inbreeding occurs. Care must be taken to evaluate the assumption of incest avoidance in each system, as well as the assumption that inbreeding costs will be low in large, open populations, particularly for taxa that live in kin groups.
Acknowledgments
All capture, handling, marking, observation and blood sampling of American crows was carried out under permit from the US Geological Survey Bird Banding Lab, NY State (no. 22263) and under protocols approved by the Binghamton University (nos 537-03 and 607-07) and Cornell University (no. 1988-0210) Institutional Animal Care and Use Committees.
We thank L. Shoenle, L. Stenzler, C. Makarewich and A. Talaba for contributions to the laboratory work; R. Heiss, J. Murray, J. Montagna, T. Wilson, Z. Adaila, G. Barr, J. McGowan and A. Tringali for their contributions in the field; A. Glaser for WNV assays; D. Robinson, J. Fitzpatrick and W. Koenig for advice; and J. Dickinson and the Dickinson lab for discussions and reading of the manuscript. Support for this work was provided by the National Science Foundation, the National Institute of Health, the Animal Behaviour Society, Cornell Sigma Xi Grant-in-Aid of Research, the Frank M. Chapman Memorial Fund, the Kieckhefer Adirondacks Fellowship, the Cooper Ornithological Society, the Wilson Ornithological Society, the Andrew W. Mellon Foundation, an Eloise Gerry Fellowship from Sigma Delta Epsilon/Graduate Women in Science and the American Association of University Women.
References
- Acevedo-Whitehouse K., Gulland F., Greig D., Amos W. Disease susceptibility in California sea lions. Nature. 2003;422:35. doi: 10.1038/422035a. doi:10.1038/422035a [DOI] [PubMed] [Google Scholar]
- Acevedo-Whitehouse K., Spraker T.R., Lyons E., Melin S.R., Gulland F., Delong R.L., Amos W. Contrasting effects of heterozygosity on survival and hookworm resistance in California sea lion pups. Mol. Ecol. 2006;15:1973–1982. doi: 10.1111/j.1365-294X.2006.02903.x. doi:10.1111/j.1365-294X.2006.02903.x [DOI] [PubMed] [Google Scholar]
- Akaike H. Information theory as an extension of the maximum likelihood principle. In: Petrov B.N., Csaki F., editors. Second Int. Symp. on Information Theory. Akademiai Kiado; Budapest, Hungary: 1973. pp. 267–281. [Google Scholar]
- Alexander R.D. The evolution of social behavior. Annu. Rev. Ecol. Syst. 1974;5:325–383. doi:10.1146/annurev.es.05.110174.001545 [Google Scholar]
- Amos W., Worthington Wilmer J., Fullard K., Burg T.M., Croxall J.P., Bloch D., Coulson T. The influence of parental relatedness on reproductive success. Proc. R. Soc. Lond. B. 2001;268:2021–2027. doi: 10.1098/rspb.2001.1751. doi:10.1098/rspb.2001.1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster P., Reed D.H. Inbreeding depression in benign and stressful environments. Heredity. 2005;95:235–242. doi: 10.1038/sj.hdy.6800721. doi:10.1038/sj.hdy.6800721 [DOI] [PubMed] [Google Scholar]
- Balloux F., Amos W., Coulson T. Does heterozygosity estimate inbreeding in real populations? Mol. Ecol. 2004;13:3021–3031. doi: 10.1111/j.1365-294X.2004.02318.x. doi:10.1111/j.1365-294X.2004.02318.x [DOI] [PubMed] [Google Scholar]
- Barrett S.C.H., Charlesworth D. Effects of a change in the level of inbreeding on the genetic load. Nature. 1991;352:522–524. doi: 10.1038/352522a0. doi:10.1038/352522a0 [DOI] [PubMed] [Google Scholar]
- Bateson P. Optimal outbreeding. In: Bateson P., editor. Mate choice. Cambridge University Press; Cambridge, UK: 1983. pp. 257–277. [Google Scholar]
- Byers D.L., Waller D.M. Do plant populations purge their genetic load? Effects of population size and mating history on inbreeding depression. Annu. Rev. Ecol. Syst. 1999;30:479–513. doi:10.1146/annurev.ecolsys.30.1.479 [Google Scholar]
- Caffrey C. Correlates of reproductive success in cooperatively breeding western American crows: if helpers help, it's not by much. Condor. 2000;102:333–341. doi:10.1650/0010-5422(2000)102[0333:CORSIC]2.0.CO;2 [Google Scholar]
- Calleri D.V., Reid E.M., Rosengaus R.B., Vargo E.L., Traniello J.F.A. Inbreeding and disease resistance in a social insect: effects of heterozygosity on immunocompetence in the termite Zootermopsis angusticollis. Proc. R. Soc. B. 2006;273:2633–2640. doi: 10.1098/rspb.2006.3622. doi:10.1098/rspb.2006.3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B. Inbreeding depression and its evolutionary consequences. Annu. Rev. Ecol. Syst. 1987;18:237–268. doi:10.1146/annurev.es.18.110187.001321 [Google Scholar]
- Clark A.B., Robinson D.A., McGowan K.J. Effects of West Nile virus mortality on social structure of an American crow (Corvus brachyrhynchos) population in New York state. Ornithol. Monogr. 2006;60:65–78. doi:10.1642/0078-6594(2006)60[65:EOWNVM]2.0.CO;2 [Google Scholar]
- Coltman D.W., Slate J. Microsatellite measures of inbreeding: a meta-analysis. Evolution. 2003;57:971–983. doi: 10.1111/j.0014-3820.2003.tb00309.x. doi:10.1111/j.0014-3820.2003.tb00309.x [DOI] [PubMed] [Google Scholar]
- Coltman D.W., Pilkington J.G., Smith J.A., Pemberton J.M. Parasite-mediated selection against inbred Soay sheep in a free-living, island population. Evolution. 1999;53:1259–1267. doi: 10.1111/j.1558-5646.1999.tb04538.x. doi:10.2307/2640828 [DOI] [PubMed] [Google Scholar]
- Cote S.D., Stien A., Irvine R.J., Dallas J.F., Marshall F., Halvorsen O., Langvatn R., Albon S.D. Resistance to abomasal nematodes and individual genetic variability in reindeer. Mol. Ecol. 2005;14:4159–4168. doi: 10.1111/j.1365-294X.2005.02733.x. doi:10.1111/j.1365-294X.2005.02733.x [DOI] [PubMed] [Google Scholar]
- Crnokrak P., Barrett S.C.H. Purging the genetic load: a review of the experimental evidence. Evolution. 2002;56:2347–2358. doi: 10.1111/j.0014-3820.2002.tb00160.x. doi:10.1111/j.0014-3820-2002.tb00160.x [DOI] [PubMed] [Google Scholar]
- Crnokrak P., Roff D.A. Inbreeding depression in the wild. Heredity. 1999;83:260–270. doi: 10.1038/sj.hdy.6885530. doi:10.1038/sj.hdy.6885530 [DOI] [PubMed] [Google Scholar]
- DeWoody Y.D., DeWoody J.A. On the estimation of genome-wide heterozygosity using molecular markers. J. Hered. 2005;96:85–88. doi: 10.1093/jhered/esi017. doi:10.1093/jhered/esi017 [DOI] [PubMed] [Google Scholar]
- Ekman J., Bylin A., Tegelstrom H. Parental nepotism enhances survival of retained offspring in the Siberian jay. Behav. Ecol. 2000;11:416–420. doi:10.1093/beheco/11.4.416 [Google Scholar]
- Emlen S.T. An evolutionary theory of the family. Proc. Natl Acad. Sci. USA. 1995;92:8092–8099. doi: 10.1073/pnas.92.18.8092. doi:10.1073/pnas.92.18.8092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese A.R., Hedrick P.W. Genetic variation and resistance to a bacterial infection in the endangered Gila topminnow. Anim. Conserv. 2003;6:369–377. doi:10.1017/S1367943003003445 [Google Scholar]
- Goodnight K.F., Queller D.C. Computer software for performing likelihood tests of pedigree relationship using genetic markers. Mol. Ecol. 1999;8:1231–1234. doi: 10.1046/j.1365-294x.1999.00664.x. doi:10.1046/j.1365-294x.1999.00664.x [DOI] [PubMed] [Google Scholar]
- Hansson B., Westerberg L. On the correlation between heterozygosity and fitness in natural populations. Mol. Ecol. 2002;11:2467–2474. doi: 10.1046/j.1365-294x.2002.01644.x. doi:10.1046/j.1365-294X.2002.01644.x [DOI] [PubMed] [Google Scholar]
- Hawley D.M., Sydenstricker K.V., Kollias G.V., Dhondt A.A. Genetic diversity predicts pathogen resistance and cell-mediated immunocompetence in house finches. Biol. Lett. 2005;1:326–329. doi: 10.1098/rsbl.2005.0303. doi:10.1098/rsbl.2005.0303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick P.W., Kalinowski S.T. Inbreeding depression in conservation biology. Annu. Rev. Ecol. Syst. 2000;31:139–162. doi:10.1146/annurev.ecolsys.31.1.139 [Google Scholar]
- Hedrick P.W., Kim T.J., Parker K.M. Parasite resistance and genetic variation in the endangered Gila topminnow. Anim. Conserv. 2001;4:103–109. doi:10.1017/S1367943001001135 [Google Scholar]
- Hoffman J.I., Forcada J., Trathan P.N., Amos W. Female fur seals show active choice for males that are heterozygous and unrelated. Nature. 2007;445:912–914. doi: 10.1038/nature05558. doi:10.1038/nature05558 [DOI] [PubMed] [Google Scholar]
- Hughes W.O.H., Boomsma J.J. Genetic diversity and disease resistance in leaf-cutting ant societies. Evolution. 2004;58:1251–1260. doi: 10.1554/03-546. doi:10.1111/j.0014-3820.2004.tb01704.x [DOI] [PubMed] [Google Scholar]
- Ilmonen P., Penn D.J., Damjanovich K., Clarke J., Lamborn D., Morrison L., Ghotbi L., Potts W.K. Experimental infection magnifies inbreeding depression in house mice. J. Evol. Biol. 2008;21:834–841. doi: 10.1111/j.1420-9101.2008.01510.x. doi:10.1111/j.1420-9101.2008.01510.x [DOI] [PubMed] [Google Scholar]
- Kalinowski S.T., Taper M.L., Marshall T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007;16:1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x. doi:10.1111/j.1365-294X.2007.03089.x [DOI] [PubMed] [Google Scholar]
- Keller L.F., Waller D.M. Inbreeding effects in wild populations. Trends Ecol. Evol. 2002;17:230–241. doi:10.1016/S0169-5347(02)02489-8 [Google Scholar]
- Koenig W., Haydock J. Incest avoidance. In: Koenig W., Dickinson J., editors. Ecology and evolution of cooperative breeding in birds. Cambridge University Press; Cambridge, UK: 2004. pp. 142–156. [Google Scholar]
- Kokko H., Ots I. When not to avoid inbreeding. Evolution. 2006;60:467–475. doi:10.1554/05-613.1 [PubMed] [Google Scholar]
- Komar N., Langevin S., Hinten S., Nemeth N., Edwards E., Hettler D., Davis B., Bowen R., Bunning M. Experimental infection of North American birds with the New York 1999 strain of West Nile virus. Emerg. Infect. Dis. 2003;9:311–322. doi: 10.3201/eid0903.020628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konovalov D.A., Manning C., Henshaw M.T. KINGROUP: a program for pedigree relationship reconstruction and kin group assignments using genetic markers. Mol. Ecol. Notes. 2004;4:779–782. doi:10.1111/j.1471-8286.2004.00796.x [Google Scholar]
- Lebreton J.D., Burnham K.P., Clobert J., Anderson D.R. Modeling survival and testing biological hypotheses using marked animals: a unified approach with case studies. Ecol. Monogr. 1992;62:67–118. doi:10.2307/2937171 [Google Scholar]
- Lively C.M., Dybdahl M.F., Jokela J., Osnas E.E., Delph L.F. Host sex and local adaptation by parasites in a snail–trematode interaction. Am. Nat. 2004;164:S6–S18. doi: 10.1086/424605. doi:10.1086/424605 [DOI] [PubMed] [Google Scholar]
- Luong L.T., Heath B.D., Polak M. Host inbreeding increases susceptibility to ectoparasitism. J. Evol. Biol. 2007;20:79–86. doi: 10.1111/j.1420-9101.2006.01226.x. doi:10.1111/j.1420-9101.2006.01226.x [DOI] [PubMed] [Google Scholar]
- MacDougall-Shackleton E.A., Derryberry E.P., Foufopoulos J., Dobson A.P., Hahn T.P. Parasite-mediated heterozygote advantage in an outbred songbird population. Biol. Lett. 2005;1:105–107. doi: 10.1098/rsbl.2004.0264. doi:10.1098/rsbl.2004.0264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff J.M., Balda R.P. Pinyon jays: making the best of a bad situation by helping. In: Stacey P., Koenig W., editors. Cooperative breeding in birds. Cambridge University Press; Cambridge, UK: 1990. pp. 197–238. [Google Scholar]
- McGowan K.J. Demographic and behavioral comparisons of suburban and rural American crows. In: Marzluff J., Bowman R., Donelly D., editors. Avian ecology and conservation in an urbanizing world. Klumer Academic Press; Norwell, MA: 2001. pp. 365–381. [Google Scholar]
- McRae S.B., Amos W. Can incest within cooperative breeding groups be detected using DNA fingerprinting? Behav. Ecol. Sociobiol. 1999;47:104–107. doi:10.1007/s002650050655 [Google Scholar]
- Pusey A., Wolf M. Inbreeding avoidance in animals. Trends Ecol. Evol. 1996;11:201–206. doi: 10.1016/0169-5347(96)10028-8. doi:10.1016/0169-5347(96)10028-8 [DOI] [PubMed] [Google Scholar]
- Rijks J.M., Hoffman J.I., Kuiken T., Osterhaus A., Amos W. Heterozygosity and lungworm burden in harbour seals (Phoca vitulina) Heredity. 2008;100:587–593. doi: 10.1038/hdy.2008.18. doi:10.1038/hdy.2008.18 [DOI] [PubMed] [Google Scholar]
- Ross-Gillespie A., O'Riain M.J., Keller L.F. Viral epizootic reveals inbreeding depression in a habitually inbreeding mammal. Evolution. 2007;61:2268–2273. doi: 10.1111/j.1558-5646.2007.00177.x. doi:10.1111/j.1558-5646.2007.00177.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Hempel P., Crozier R.H. Polyandry versus polygyny versus parasites. Phil. Trans. R. Soc. Lond. B. 1999;354:507–515. doi:10.1098/rstb.1999.0401 [Google Scholar]
- Schoenle L.A., Townsend A.K., Lovette I.J. Isolation and characterization of microsatellite loci in a cooperatively breeding corvid, the American crow (Corvus brachyrhynchos) Mol. Ecol. Notes. 2006;7:46–48. doi:10.1111/j.1471-8286.2006.01520.x [Google Scholar]
- Seddon N., Amos W., Mulder R.A., Tobias J.A. Male heterozygosity predicts territory size, song structure and reproductive success in a cooperatively breeding bird. Proc. R. Soc. Lond. B. 2004;271:1823–1829. doi: 10.1098/rspb.2004.2805. doi:10.1098/rspb.2004.2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman P.W. Kinship, demography, and Belding's ground-squirrel nepotism. Behav. Ecol. Sociobiol. 1981;8:251–259. doi:10.1007/BF00299523 [Google Scholar]
- Shields W.M. State University of New York Press; Albany, NY: 1982. Philopatry, inbreeding, and the evolution of sex. [Google Scholar]
- Shykoff J.A., Schmid-Hempel P. Parasites and the advantage of genetic variability within social insect colonies. Proc. R. Soc. Lond. B. 1991;243:55–58. doi:10.1098/rspb.1991.0009 [Google Scholar]
- Slate J., David P., Dodds K.G., Veenvliet B.A., Glass B.C., Broad T.E., McEwan J.C. Understanding the relationship between the inbreeding coefficient and multilocus heterozygosity: theoretical expectations and empirical data. Heredity. 2004;93:255–265. doi: 10.1038/sj.hdy.6800485. doi:10.1038/sj.hdy.6800485 [DOI] [PubMed] [Google Scholar]
- Spielman D., Brook B.W., Briscoe D.A., Frankham R. Does inbreeding and loss of genetic diversity decrease disease resistance? Conserv. Genet. 2004;5:439–448. doi:10.1023/B:COGE.0000041030.76598.cd [Google Scholar]
- Spottiswoode C.N. Cooperative breeding and immunity: a comparative study of PHA response in African birds. Behav. Ecol. Sociobiol. 2008;62:963–974. doi:10.1007/s00265-007-0521-0 [Google Scholar]
- Spottiswoode C., Moller A.P. Genetic similarity and hatching success in birds. Proc. R. Soc. Lond. B. 2004;271:267–272. doi: 10.1098/rspb.2003.2605. doi:10.1098/rspb.2003.2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulkin M., Sheldon B.C. Dispersal as a means of inbreeding avoidance in a wild bird population. Proc. R. Soc. B. 2008;275:703–711. doi: 10.1098/rspb.2007.0989. doi:10.1098/rspb.2007.0989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarr C.L., Fleischer R.C. Primers for polymorphic GT microsatellites isolated from the Mariana crow, Corvus kubaryi. Mol. Ecol. 1998;7:253–255. doi:10.1046/j.1365-294x.1998.00385.x [PubMed] [Google Scholar]
- Townsend A.K., Clark A.B., McGowan K.J., Lovette I.J. Reproductive partitioning and the assumptions of reproductive skew models in the cooperatively breeding American crow. Anim. Behav. 2009;77:503–512. doi: 10.1016/j.anbehav.2008.10.030. doi:10.1016/j.anbehav.2008.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valsecchi E., Amos W., Raga J.A., Podesta M., Sherwin W. The effects of inbreeding on mortality during a morbillivirus outbreak in the Mediterranean striped dolphin (Stenella coeruleoalba) Anim. Conserv. 2004;7:139–146. doi:10.1017/S1367943004001325 [Google Scholar]
- Verbeek, N. A. M. & Caffrey, C. 2002 The American crow (Corvus brachyrhynchos). In Birds of North America, vol. 647 (eds A. Poole & F. Gill). Philadelphia, PA: Birds of North America, Inc.
- Whiteman N.K., Matson K.D., Bollmer J.L., Parker P.G. Disease ecology in the Galapagos Hawk (Buteo galapagoensis): host genetic diversity, parasite load and natural antibodies. Proc. R. Soc. B. 2006;273:797–804. doi: 10.1098/rspb.2005.3396. doi:10.1098/rspb.2005.3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfenden G.E., Fitzpatrick J.W. Inheritance of territory in group-breeding birds. Bioscience. 1978;28:104–108. doi:10.2307/1307423 [Google Scholar]