Abstract
We used parentage analysis to estimate seedling recruitment distances and genetic composition of seedling patches centred around reproductive trees of the animal-dispersed Neotropical canopy palm Iriartea deltoidea in two 0.5 ha plots within second-growth forest and one 0.5 ha plot in adjacent old-growth forest at La Selva Biological Field Station in north-eastern Costa Rica. Seedlings were significantly spatially aggregated in all plots, but this pattern was not due to dispersal limitation. More than 70 per cent of seedlings were dispersed at least 50 m from parent trees. Few seedlings were offspring of the closest reproductive trees. Seedling patches observed beneath reproductive trees originate from dozens of parental trees. Observed patterns of seedling distribution and spatial genetic structure are largely determined by the behaviour of vertebrate seed dispersers rather than by spatial proximity to parental trees.
Keywords: parentage, Iriartea deltoidea, seedling recruitment, dispersal limitation, second-growth forests, seedling shadow
1. Introduction
Forest regeneration requires both seed dispersal and seedling establishment, but our understanding of seedling establishment is much more advanced than our understanding of seed dispersal (Condit et al. 2000; Harms et al. 2000; Uriarte et al. 2004). Although they are distinct processes, seed dispersal and seedling establishment are closely linked. The Janzen–Connell model provides the major conceptual framework linking seed dispersal with seedling fate in tropical forests (Loiselle & Dirzo 2002), suggesting that seedling recruitment near conspecifics should be reduced as a result of density-dependent mortality. Thus, the probability of successful recruitment should increase with increasing distance from parent trees (Janzen 1970; Connell 1971; Schupp 1992).
Assessing the role of dispersal in the spatial patterns of seedlings remains a major challenge (Norden et al. 2007). Seedling aggregations beneath the crown of a reproductive tree are often interpreted as evidence for dispersal limitation (Franklin & Rey 2007). Extending this intuition, estimates of seed dispersal derived from inverse modelling (Ribbens et al. 1994; Clark et al. 1999), seed rain data or seedling spatial distributions are based on the assumption that dispersal is a declining function of distance from potential parent trees (Uriarte et al. 2005; Muller-Landau & Hardesty 2005). While this assumption may be valid for wind, gravity, or mechanically dispersed seeds, it may often be invalid for animal-dispersed seeds, which constitute more than 80 per cent of the woody flowering plants in tropical wet forests (Willson et al. 1989; Chazdon et al. 2003). An alternative view for animal-dispersed species is that seedling aggregations beneath fruiting trees reflect frugivore movement and behaviour. Recent studies using genetic markers have shown that dispersing animals often deposit seed below the canopy of conspecific trees (Jordano & Godoy 2002; Hardesty et al. 2006; Wang et al. 2007). Fruiting trees can also serve as foci of heterospecific seed deposition from a variety of vertebrate-dispersed species (Clark et al. 2004).
Here, we report results of a fine scale, spatially explicit investigation of genotyped, mapped seedlings of the canopy palm Iriartea deltoidea in relation to conspecific reproductive trees in old-growth and adjacent second-growth forest in the Atlantic lowlands of Costa Rica. We examined the genetic composition of seedling patches surrounding reproductive trees to test whether seedlings beneath reproductive trees are offspring of those trees. We also compared the observed patch size of seedlings in the field with the genetic patch size calculated from a previous analysis of spatial genetic structure. Unlike previous studies restricted to mature forests, we investigated seedling shadows in areas undergoing secondary forest regeneration, where relatively few animal-dispersed tree species have reached reproductive maturity (Vilchez et al. 2007). By focusing on established seedlings, our analysis allows us to assess the net effect of dispersal, germination and seedling establishment on the spatial distribution of offspring relative to the location of parents. Our analysis illustrates how the genetic composition of seedlings of a vertebrate-dispersed palm species in regenerating secondary forests is influenced by the behaviour of dispersal agents and by spatial patterns of genetic variation in the parental pool (Sezen et al. 2005).
2. Material and methods
(a) Study species
Iriartea deltoidea Ruiz and Pavon is a widespread Neotropical palm, found from Nicaragua in the north to Bolivia in the south. Its monoecious flowers are pollinated by native stingless bees (Trigona spp. and Melipona spp.) and introduced honeybees (Apis spp.) (Bawa et al. 1985; Henderson 1990; Moore 2001). Similar to most tropical woody forest species, I. deltoidea lacks a seed bank and depends on newly dispersed seed for establishment (Henderson 2002). Iriartea deltoidea produces fruits year-round and is a dependable food source for frugivorous animals (Henderson 2002). Seeds are dispersed by a variety of vertebrates including chestnut-mandibled toucans (Ramphastos swainsonii), keel-billed toucans (Ramphastos sulfuratus), tent-making bats (Artibeus spp.), white-faced monkeys (Cebus capucinus), spider monkeys (Ateles geoffroyi), peccaries (Tayassu spp.), tapirs (Tapirus spp.) and several species of rodents (Henderson 1990).
Toucans are the dominant seed dispersers in our study area. They swallow entire fruits (each weighing 6–11 g) and regurgitate intact seeds, with an average gut retention time of 30 min (Janzen 1983; Henderson 2002; Holbrook & Loiselle 2007). Toucans fly relatively long distances, even over pastures (Del Hoyo et al. 2002). Telemetry tracking data show that they frequently enter second-growth forest during foraging bouts (Graham 2001). In continuous forest, the core home range of R. sulfuratus is between 19 and 28 ha, but home ranges may be as large as 86 ha (Holbrook & Loiselle 2007).
(b) Field site and sampling
We carried out our study in the Caribbean lowlands of northeast Costa Rica, Sarapiquí County, Heredia Province, at La Selva Biological Station. In 2001, we established a 0.5 ha (50×100 m) seedling plot, nested within 20 ha of 24-year-old second-growth forest located near Lindero El Peje, the ‘near LEP plot’ (located approx. 200 m from the old-growth forest border). We mapped and collected genetic material from the leaf tissue of all 384 seedlings with a crown height of less than 1 m within the 0.5 ha plot. We estimate that seedlings were 1–4 years old (Sezen et al. 2007). During this period, we also collected genetic material from all trees of reproductive age, within the 20 ha of second-growth and 10 ha of the adjacent old-growth forest, covering a total area of 30 ha (196 trees total). In 2004, we sampled genetic material from a randomly selected subsample of seedlings in two additional 0.5 ha plots where all seedlings were mapped, one located in second-growth, the ‘far LEP plot’ (approx. 400 m from the old-growth forest border), and the other in old-growth (OG plot) forest (approx. 100 m from the old-growth forest border). The number of genotyped seedlings in the far LEP and the OG plots was 92 and 71, respectively (table 1). In 2004, we conducted a second census of adult trees and expanded the sample collection to include an additional 6 ha of old-growth forest at the LEP site (Sezen et al. 2007). We also sampled reproductive trees from a second site covering both forest types (4.5 ha in old-growth forest and 3.5 ha in second-growth forest) in a part of La Selva Biological Station called Lindero Occidental (LOC), approximately 2 km away. A total of 281 reproductive trees were used to resolve parent–offspring relationship, collected over an area of 43 ha (Sezen et al. 2007). Density of adult trees in the LEP site was highest at the old-growth boundary (119 trees, approx. 12 trees ha−1) and declined with increasing distance from the old-growth boundary (38 trees approx. 3.8 trees ha−1), whereas density in the adjacent old-growth forest was 8.5 trees ha−1.
Table 1.
Genetic composition of the seedling patches in the far LEP, near LEP and OG plots. (Patch numbers and locations are the same as given in figure legend 1.)
| far SG | near SG | OG | ||||
|---|---|---|---|---|---|---|
| patch 1 | patch 2 | patch 1 | patch 2 | patch 3 | 0.5 ha | |
| percentage from focal tree | 0 | 0 | 0 | 16.6 | 4.5 | 1.5 |
| percentage of full-sibling (self) | 18 (14) | 5 (5) | 40.9 (4.5) | 17.6 (0.09) | 21.2 (0) | 36 (20) |
| percentage of half-sibling (single parent) | 36 (7) | 40 (0) | 47.2 (5) | 61.7 (28.4) | 62.1 (31.8) | 45 (0) |
| percentage of single parent unique | 0 | 0 | 3.6 | 0.09 | 4.5 | 0 |
| percentage of parent pair unique | 11 | 14 | 3.6 | 0 | 0 | 3 |
| percentage of both parents outside | 36 | 40 | 4.5 | 19.6 | 12.1 | 16 |
| no. of unambiguous seedlings | 44 | 37 | 110 | 102 | 66 | 64 |
| no. of ambiguous | 3 | 8 | 26 | 27 | 53 | 7 |
| total no. of genotyped seedlings | 47 | 45 | 136 | 129 | 119 | 71 |
| no. of contributing parents | 23 | 24 | 62 | 38 | 32 | 27 |
| no. of mature trees | 1 | 1 | 1 | 1 | 1 | 8 |
| mean recruitment distance (m) | 445 | 425 | 246 | 254 | 151 | 214 |
| median recruitment distance (m) | 386 | 428 | 223 | 232 | 158 | 230 |
| mode recruitment distance (m) | 453 | 700 | 414 | 481 | 187 | 146 |
| minimum and maximum recruitment distance (m) | 233–929 | 229–796 | 28–553 | 1–486 | 1–495 | 1–543 |
| percentage of seeds travelled more than 50 m | 100 | 100 | 98 | 80 | 70 | 93 |
| seedling density (per 0.5 ha) | 157 | 384 | 137 | |||
(c) Parentage analysis and seedling recruitment distances
We used amplified fragment length polymorphisms (AFLPs) to generate DNA fingerprints. Of the 392 fragments initially isolated, we identified 141 fragments that could be reliably scored and that occurred in moderate frequency (Sezen et al. 2005). We used the parentage analysis program Famoz for reconstruction of parent–offspring relationships (Gerber et al. 2000). To avoid false assignment of sibling founder trees as parents, we excluded trees that first became reproductive during the 2004 field season from the parental pool of seedlings, as these individuals could not be parents of pre-existing seedlings (Sezen et al. 2007). Exclusion probabilities are 0.99954 for single parent and 1.0000 for parent pairs. Simulations show that cryptic gene flow accounts for at most 3 per cent (F=0.05) to 11 per cent (F=0.3) of the apparent gene flow.
The within-population inbreeding coefficient (FIS) cannot be estimated from a single population using dominant markers (Holsinger et al. 2001). As a result, we carried out our analysis using Famoz at two inbreeding coefficients (0.05 and 0.3) that define a conservative range likely to include the true inbreeding coefficient for a woody, outcrossing, animal-pollinated species (Hamrick et al. 1992). We considered parentage to be unambiguous only when the same parents or parent pairs were assigned to offspring at both inbreeding coefficients. Error rates for mistyping and simulations were 0.1 per cent. Simulations with the assumption of large random mating population and tests of the gene flow parameters were carried out using 10 000 synthetic offspring and 1000 parents (Gerber et al. 2000). LOD score threshold values for single parent (Tp) and parent pair (Tc) decision cut-off points for all seedlings were 6.0 and 13.0, respectively. For saplings and seedlings Tp and Tc were 10.0 and 18.0, respectively.
Maternal and paternal parents cannot be distinguished based on AFLP markers. To provide a conservative estimate of distance from the maternal parent, we designated the nearer assigned parent as the maternal parent. The distance between the designated maternal parent and a seedling provides a lower bound on the seedling recruitment distance (Muller-Landau & Hardesty 2005). We conservatively use vertical distance to the nearest edge of the 36 ha LEP study area for those seedlings whose parents could not be assigned to genotyped trees within the entire 43 ha study area. As a crude estimate of seedling recruitment distance that might be calculated from an inverse model, we use the distance from a seedling to the nearest conspecific reproductive tree (Bustamante & Canals 1995).
(d) Spatial analysis
We used the spatial analysis program PASSAGE to visually explore and identify patches of all seedlings mapped in the three plots (Rosenberg 2001). In the OG plot, seedling patch structure was not spatially associated with locations of the trees; therefore, the whole plot was treated as a single seedling patch. Seedling recruitment distances, parental sources and genetic composition of patches were calculated from seedlings within the patches.
We used the spatial point pattern analysis program PPA to test for spatial randomness of all mapped seedlings in each plot based on Ripley's L(d) function (Ripley's K corrected for edge effects of the sampling plot; Chen & Getis 1998). Minimum and maximum 95% confidence interval envelopes were calculated from 250 permutations. Values above the envelopes indicate statistically significant spatial aggregation.
We calculated spatial autocorrelation among seedlings based on Moran's I using the geostatistical program package GS+ (Robertson 1998). We grouped individual seedling coordinates into abundances in 5000 (1×1 m) quadrats for each 0.5 ha plot. We analysed spatial distribution based on the Cartesian coordinates from the centre of each quadrat on a 50×100 m grid.
The genetic composition of seedling patches was assessed for all seedlings with unambiguous parentage located within the patches. We scored as full-siblings any seedlings having identical parent pairs identified within the 43 ha study area. We scored as half-siblings any seedlings sharing at least one parent sampled from the study area. Some individuals scored as half-siblings may actually be full-siblings sharing one parent outside the study area.
3. Results
(a) Spatial pattern
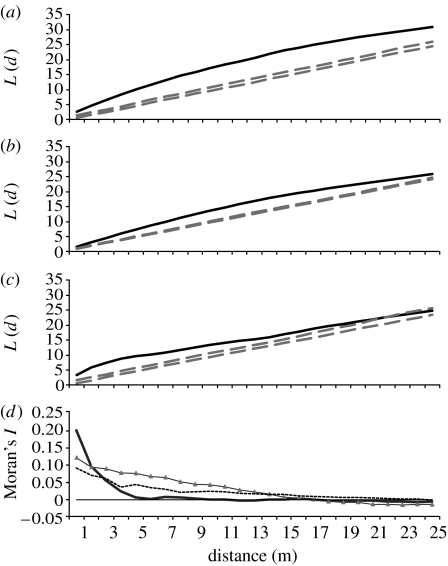
In both of the second-growth forest plots, seedlings demonstrated a highly aggregated spatial pattern with distinct patches around reproductive Iriartea trees (figures 1a, 2a and 3a). Only one tree in the far LEP plot was not associated with a seedling patch (figure 1a). By contrast, seedling patches in the OG plot were not associated with reproductive trees (figure 3a). Ripley's L shows significant spatial aggregation in all distance classes in all three plots (figure 4a–c). The observed seedling patch diameter in the near LEP plot measured by Moran's I (18 m) was quite similar to the genetic patch size (16 m) estimated using Tanimoto's D previously in this plot (figure 4d; Sezen et al. 2007). In the OG plot, Moran's I decayed steeply until 5 m and the genetic patch size was less than 18 m. In the far LEP plot, the observed spatial scale of patchiness was 24 m, whereas genetic structure was weakly expressed until 48 m.
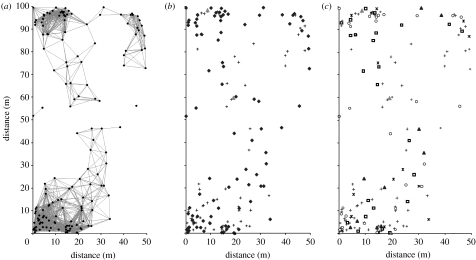
Figure 1.
Maps of 0.5 ha far LEP seedling plot. (a) Visualization of the two seedling patches around two reproductive trees by 12 m connectivity matrix. Patches 1 and 2 are located in upper and lower halves of the plot. (b) Locations of genotyped seedlings and reproductive trees (pluses, seedlings; triangles, trees; diamonds, genotyped). (c) Genetic origins of seedlings according to the type of forest where a parent or parent pair is found within the entire 43 ha study area. No seedlings have originated from the reproductive trees in this plot. Black triangles, SG/SG (both parents in second-growth forest); asterisks, SG/OG (one parent in second-growth and the other in old-growth forest); plus squares, OG/OG (both parents in old-growth forest); circles, parent OUT (both parents outside the study area); pluses, seedlings; grey triangles, trees; grey diamonds, ambiguous.
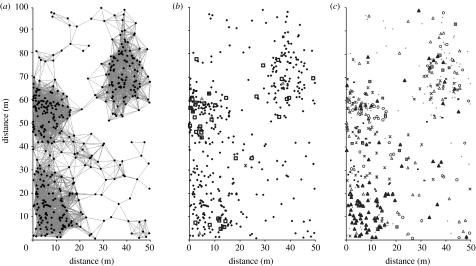
Figure 2.
Maps of 0.5 ha near LEP seedling plot. (a) Visualization of the three seedling patches around three reproductive trees by 9 m connectivity matrix. Locations of patches 1, 2 and 3 are lower left corner, middle and upper right corner, respectively. (b) Locations of locally produced seedlings by the reproductive trees at the centre of each patch. The only seedling from the tree I-85 is shown by a star (pluses, seedlings; triangles, trees; open squares, LEP35-05; plus squares, LEP32-09; asterisk, I-85). (c) Genetic origins of seedlings according to the type of forest where a parent or parent pair is found within the entire 43 ha study area. Black triangles, SG/SG; asterisks, OG/SG; plus squares, OG/OG; grey triangles, trees; circles, parent OUT; diamonds, ambiguous; open triangles, single SG; minuses, LOC mating; grey squares, single OG.
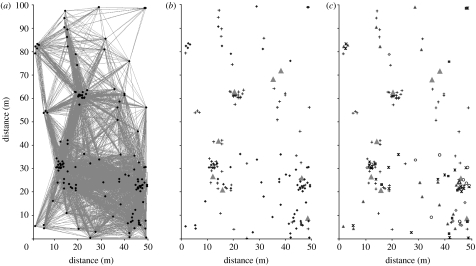
Figure 3.
Maps of 0.5 ha OG seedling plot. (a) Visualization of the seedling aggregations by 35 m connectivity matrix. (b) Locations of genotyped seedlings and reproductive trees (pluses, seedlings; triangles, trees; diamonds, genotyped). (c) Genetic origins of seedlings according to the type of forest where a parent or parent pair is found within the entire 43 ha study area. The only seedling from the tree I-39 is shown by a hollow triangle immediately below its crown. Black triangles, SG/SG; asterisks, SG/OG; plus squares, OG/OG; grey triangles, trees; circles, parent OUT; diamonds, ambiguous; open triangle, I-39; pluses, seedlings.
Figure 4.
(a–c) Edge corrected Ripley's K-test for spatial randomness of seedling distribution in three 0.5 ha plots ((a) Far LEP plot, (b) near LEP plot and (c) OG plot). Data values between the 95% confidence envelopes (dashed lines) indicate spatial randomness. (d) Moran's I spatial autocorrelation test for measuring spatial clumping in three 0.5 ha seedling plots (dashed line, far LEP plot; triangles, near LEP plot; solid line, OG plot).
(b) Seedling recruitment distances and sources
Although seedling patches in the two second-growth plots were centred on reproductive trees, these trees were the closest assigned parent of only a few seedlings within these patches (table 1). In the far LEP plot, none of the genotyped seedlings were offspring of the reproductive trees within the 0.5 ha sampling area (table 1). In the near LEP plot, the highest local tree contribution (16.6%) was in patch 2, and only 4.5 per cent of the seedlings in patch 3 were offspring of the focal reproductive tree. The reproductive tree (I-85) at the centre of patch 1 in the near LEP plot had only one (0.9%) contribution to the seedlings immediately below its crown (table 1). In the entire 0.5 ha OG plot, more than 98 per cent of the genotyped seedlings were assigned parents outside the plot (table 1).
Many seedlings recruited over long distances. Seedling recruitment distances were higher in the far LEP plot than in other plots. Minimum recruitment distances in both patches were more than 229 m. Moreover, 36 per cent of seedlings in one patch and 40 per cent in the other had parents outside the entire 43 ha study area (table 1). The nearest assigned parent was more than 50 m away from 70 to 98 per cent of the seedlings in the three patches in the near LEP plot and 93 per cent of the seedlings in the entire OG plot (table 1). Overall, the distribution of seedling recruitment distances derived from the parentage analysis differed greatly from the estimated distribution based on the assumption that the nearest reproductive conspecific tree is the maternal parent (Kolmogorov–Smirnov test, D=0.9291, p<2.2×10−16).
(c) Genetic composition of seedling patches
Although seedling recruitment distances were very high, parentage data indicated that many seedlings were either half- or full-siblings (table 1). In both seedling patches in the far LEP plot, approximately half (45–54%) of genotyped seedlings were either full- or half-siblings. Based on earlier analyses, levels of seedling genetic diversity in this plot were similar to those in OG plot seedlings (Sezen et al. 2007). Although many seedlings in these patches were full- or half-siblings, the proportion of seedlings with unique parent pairs (seedlings with no overlapping parents with the others) was higher in the far LEP patches (11–14%) than elsewhere.
In the near LEP plot, 92–99 per cent of seedlings were either full- or half-siblings (table 1). Seedling patch 1 consisted of 110 seedlings, all with at least one parent unambiguously assigned. The patch is centred around reproductive tree I-85 (figure 2), yet 62 parents contributed offspring among which 41 per cent were full-siblings (of which 5% were the result of self-fertilization), 47 per cent were half-siblings and 7 per cent had at least one unique parent (table 1). Among those seedlings having unique parents, only 3.6 per cent had unique parent pairs and another 3.6 per cent had an unique single parent (seedlings for which only one parent could be assigned within the 43 ha study area and could be half-siblings by sharing a parent located outside the study area). Among the half-siblings, 5 per cent shared a single parent within the 43 ha study area and could be full-siblings if they share a parent outside the study area. Half-siblings had 17 parents in 18 paired combinations. Only 4.5 per cent of seedlings had parents outside the 43 ha study area, indicating the lowest rate of gene flow among all seedling patches.
The second seedling patch in the near LEP plot, surrounding the reproductive tree LEP35-05, consisted of 102 seedlings of which 18 per cent were full-siblings sharing 11 parents in paired combinations. Half-siblings formed 62 per cent, in which 28 per cent had only a single parent detected within the 43 ha study area (table 1). The tree at the centre of this patch was the parent of 17 per cent of seedlings immediately below its crown, the highest local contribution observed in this study (table 1; figure 2c). This patch also experienced the highest gene flow (20% of the total gene flow) where both parental contributions were detected.
In the third seedling patch in the near LEP, the reproductive focal tree LEP32-09 had genetic contribution to only 5 per cent of seedlings (table 1; figure 2c). Full-siblings formed 21 per cent of this patch and no self pollination was observed. Similar to the second seedling patch, a great majority (62%) of the seedlings were half-siblings and more than half of them (32%) had only a single parent within the study area (table 1). Twelve per cent of seedlings had both parents outside the entire 43 ha study area.
The 64 seedlings with unambiguous parentage in the OG plot did not show a distinct patch structure around any particular reproductive tree; therefore, we analysed the entire set of seedlings in the 0.5 ha plot as a single patch (figure 3). These seedlings represented 27 genetic parents. Only one of the eight reproductive trees found in the OG forest plot contributed to a single seedling within the plot (figure 3c). More than 98 per cent of the genotyped seedlings had their closest genetic parent outside the 0.5 ha plot (table 1). Yet, 81 per cent of the genotyped seedlings were either full-siblings (36%) or half-siblings (45%). Sixteen per cent of the seedlings had both parents outside the 43 ha study area (table 1).
4. Discussion
Our results demonstrate that proximity of seedlings to reproductive trees is a poor proxy for parentage in I. deltoidea. Apparent seedling shadows of individual reproductive trees of I. deltoidea in secondary forest plots are, in fact, clusters of recruited seedlings from many genetic sources. Reproductive trees spatially associated with seedling patches apparently serve as foci for seed deposition, but rarely contribute progeny to these patches. This is precisely the expected pattern for species with high rates of dispersal by highly mobile frugivores. Reproductive trees can serve as recruitment foci for conspecific (Russo & Augspurger 2004) and non-conspecific (Clark et al. 2004) seedlings, but few studies have investigated parentage within recruited seedling patches. Wang et al. (2007) found that 41 per cent of the seeds of Antrocaryon klaineanum found below fruiting trees in a protected old-growth rainforest of Cameroon were not offspring of the closest fruiting tree. Similarly, Hardesty et al. (2006) showed that 74 per cent of the genotyped Simarouba amara seedlings recruited beneath reproductive conspecific trees had a maternal parent that was not the nearest reproductive female tree.
We conducted the first, to our knowledge, detailed spatially explicit analysis of genetic composition of seedling patches in a secondary tropical forest (but see Cespedes et al. 2003). Seedling patches of I. deltoidea were genetic mixtures with contributions from 23 to 62 parents distributed over a large area within La Selva Biological Station (Sezen et al. 2007; table 1). Although seedling patches of I. deltoidea have multiple seed sources, 45–88 per cent of the seedlings within patches were either full- or half-siblings (table 1). These results imply that dispersers repeatedly carried multiple batches of seeds from the same source trees to the same deposition sites. Toucans are the primary dispersers with a core home range of 19–28 ha (Holbrook & Loiselle 2007). Given the density of reproductive trees in old-growth forests at our site (8.5 trees ha−1), between 160 and 240 reproductive trees might occur within the home range of a single toucan. With asynchronous fruiting throughout the year, I. deltoidea is the predominant fruiting tree of the young second-growth forest. Seeds of I. deltoidea were also commonly found beneath leaf tents of the tent-roosting bat Artibeus watsoni at La Selva and surrounding forest areas (F. P. L. Melo 2009, personal communication). Canopy trees and palms used by frugivores are more diverse and abundant in old-growth forest than in young secondary forest, thus increasing the number of focal trees for seed deposition (Vilchez et al. 2007). This higher abundance may explain the lack of spatial association between seedling patches and reproductive trees of I. deltoidea in the old-growth plot.
Our findings have implications for predicting future dynamics of forest regrowth and for modelling the spatial distribution and abundance of seedlings in secondary and mature forests. Dispersal models differentiate between dispersed and non-dispersed seeds (Russo et al. 2006). Our data suggest that the non-dispersed seed component of such models may be grossly overestimated, especially in tree species with vertebrate dispersers, as 70–100% of I. deltoidea seedlings in all patches were dispersed more than 50 m (table 1). In the animal-dispersed Neotropical tree S. amara (Simaroubaceae), inverse modelling predicted seed dispersal distances 10-fold lower than the true seedling recruitment distance (Hardesty et al. 2006).
Furthermore, our results show that successful dispersal away from parental trees does not necessarily enable escape from high densities of conspecific seedlings or adults, as predicted by the Janzen–Connell hypothesis. Our previous studies revealed that the founding adult cohort around the near LEP plot was largely composed of half- and full-sibling trees coming from only two reproductively dominant trees in the adjacent old-growth forest, and the spatial genetic structure demonstrated a marked decrease in genetic diversity (Sezen et al. 2005). Deposition of seeds at and/or around conspecific trees is a relatively unexplored component in distance-density predictions of the Janzen–Connell hypothesis. Dispersal, even under conspecific trees, may still offer some survival advantages. Seed handling by dispersers (gut treatment and removal of pulp before regurgitation) may enhance germination and reduce seed predation, thereby conveying a recruitment and survival advantage (Fragoso et al. 2003).
In conclusion, our results show that the spatial distribution of I. deltoidea seedlings in both old-growth and secondary forest areas is linked to their spatial genetic structure; yet this association is not due to dispersal limitation (table 1; figure 4). Owing to the activities of mobile frugivores, seedlings are distributed widely among habitat patches within the landscape, far from the location of parent trees, but still remain vulnerable to the effects of high local conspecific seedling density. Highly mobile frugivorous bird and bat species construct genetic mixtures of conspecific seedlings in patches beneath fruiting trees of I. deltoidea in secondary forests. These results do not conform to the expectation of minimum spatial aggregation or random spatial distributions of species with long-distance seed dispersal mechanisms (Seidler & Plotkin 2006). Finally, our results show that parentage analysis is necessary to distinguish dispersal limitation (failure to disperse seeds away from the parent tree) from frugivore-mediated seed deposition when seedlings are aggregated around conspecific trees (Jordano & Godoy 2002; Franklin & Rey 2007). Seed dispersal strongly affects local recruitment patterns in secondary forests, where contribution from non-local sources can be as high as 81 per cent (Uriarte et al. 2005). By focusing on established seedlings, we do not distinguish component processes such as dispersal, germination and seedling establishment, but we can assess their combined net effect on the spatial distribution of offspring relative to the location of parents. As the abundance and diversity of fruiting trees used by frugivores increase during secondary succession, we expect patches created by seed deposition to become less distinct, as we observed in the old-growth forest. Thus, the colonization of tree species used by frugivores plays a critical role in developing the spatial and genetic structure of seedling populations in regenerating forests.
Acknowledgments
Funding for this study was provided by the Andrew W. Mellon Foundation, the Garden Club of America, the Organization for Tropical Studies, and the Department of Ecology and Evolutionary Biology, University of Connecticut. We thank Rigoberto G. Vargas, Alvaro Redondo Brenes and Catherine Cardelús for their assistance with fieldwork and Chris Simon, John Cooley, Eldridge Adams, Bernard Goffinet and Elizabeth Jockusch for assistance in the laboratory and for maintenance of the ABI facility. Staff and administration at La Selva Biological Station provided much assistance and logistical support and we particularly thank Mahmood Sasa, Luis Diego Gomez and Robert Matlock.
References
- Bawa K.S., Bullock S.H., Perry D.R., Coville R.E., Grayum M.H. Reproductive biology of tropical lowland rain forest trees. II. Pollination systems. Am. J. Bot. 1985;72:346–356. doi:10.2307/2443527 [Google Scholar]
- Bustamante R.O., Canals L.M. Dispersal quality in plants: how to measure efficiency and effectiveness of a seed disperser. Oikos. 1995;73:133–136. doi:10.2307/3545736 [Google Scholar]
- Cespedes M., Gutierrez M.V., Holbrook N.M., Rocha O.J. Restoration of genetic diversity in the dry forest tree Swietenia macrophylla (Meliaceae) after pasture abandonment in Costa Rica. Mol. Ecol. 2003;12:3201–3212. doi: 10.1046/j.1365-294x.2003.01986.x. doi:10.1046/j.1365-294X.2003.01986.x [DOI] [PubMed] [Google Scholar]
- Chazdon R.L., Careaga S., Webb C., Vargas O. Community and phylogenetic structure of reproductive traits of woody species in wet tropical forests. Ecol. Monogr. 2003;733:331–348. doi:10.1890/02-4037 [Google Scholar]
- Chen, D. & Getis, A. 1998 Point pattern analysis (PPA) San Diego, USA: Department of Geography, San Diego State University.
- Clark J.S., Silman M., Kern R., Macklin E., HilleRisLambers J. Seed dispersal near and far: patterns across temperate and tropical forests. Ecology. 1999;80:1475–1494. doi:10.2307/176541 [Google Scholar]
- Clark C.J., Pousen J.R., Connor E.F., Parker V.T. Fruiting trees as dispersal foci in a semi-deciduous tropical forest. Oecologia. 2004;139:66–75. doi: 10.1007/s00442-003-1483-1. doi:10.1007/s00442-003-1483-1 [DOI] [PubMed] [Google Scholar]
- Condit R., et al. Spatial patterns in the distribution of tropical tree species. Science. 2000;288:1414–1418. doi: 10.1126/science.288.5470.1414. doi:10.1126/science.288.5470.1414 [DOI] [PubMed] [Google Scholar]
- Connell J.H. On the role of natural enemies in preventing competitive exclusion in some marine animals and in rain forest trees. In: Den Boer P.J., Gradwell G.R., editors. Dynamics of populations. PUDOC; Wageningen, The Netherlands: 1971. pp. 298–312. [Google Scholar]
- Del Hoyo, J., Elliot, A. & Sargatal, J. (eds) 2002 Handbook of the birds of the world: jacamars to woodpeckers, vol. 7. Barcelona, Spain: Lynx Edicions.
- Fragoso J.M.V., Silvius K.M., Correa J.A. Long-distance seed dispersal by tapirs increases seed survival and aggregates tropical trees. Ecology. 2003;84:1998–2006. doi:10.1890/01-0621 [Google Scholar]
- Franklin J., Rey S.J. Spatial patterns of tropical forest trees in Western Polynesia suggest recruitment limitations during secondary succession. J. Trop. Ecol. 2007;23:1–12. doi:10.1017/S0266467406003774 [Google Scholar]
- Gerber S., Mariette S., Streiff R., Bodenes C., Kremer A. Comparison of microsatellites and amplified fragment length polymorphism markers for parentage analysis. Mol. Ecol. 2000;9:1037–1048. doi: 10.1046/j.1365-294x.2000.00961.x. doi:10.1046/j.1365-294x.2000.00961.x [DOI] [PubMed] [Google Scholar]
- Graham C. Habitat selection and activity budgets of keel-billed toucans at the landscape level. Condor. 2001;103:776–784. doi:10.1650/0010-5422(2001)103[0776:HSAABO]2.0.CO;2 [Google Scholar]
- Hamrick J.L., Godt M.J.W., Sherman-Broyles S.L. Factors influencing levels of genetic diversity in woody plant species. New Forests. 1992;6:95–124. doi:10.1007/BF00120641 [Google Scholar]
- Hardesty B.D., Hubbell S.P., Bermingham E. Genetic evidence of frequent long-distance recruitment in a vertebrate-dispersed tree. Ecol. Lett. 2006;9:516–525. doi: 10.1111/j.1461-0248.2006.00897.x. doi:10.1111/j.1461-0248.2006.00897.x [DOI] [PubMed] [Google Scholar]
- Harms K.E., Wright S.J., Calderon O., Hernandez A., Herre E.A. Pervasive density-dependent recruitment enhances seedling diversity in a tropical forest. Nature. 2000;404:493–495. doi: 10.1038/35006630. doi:10.1038/35006630 [DOI] [PubMed] [Google Scholar]
- Henderson A. Arecaceae. Part 1: introduction and the Iriarteinae. Flora Neotrop. Monogr. 1990;53:1–100. [Google Scholar]
- Henderson A. Botanical Garden Press; Bronx, NY: 2002. Evolution and ecology of palms. [Google Scholar]
- Holbrook K.M., Loiselle B.A. Using toucan-generated dispersal models to estimate seed dispersal in Amazonia Ecuador. In: Dennis A.J., Schupp E.W., Green R.J., Westcott D.A., editors. Seed dispersal: theory and its application in a changing world. CAB International; Wallingford, UK: 2007. pp. 300–321. [Google Scholar]
- Holsinger, K. E., Lewis, P. O. & Dey, D. K. 2001 A Bayesian approach to inferring population structure from dominant markers. Technical report, 01-03, Department of Statistics, University of Connecticut. [DOI] [PubMed]
- Janzen D.H. Herbivores and the number of tree species in tropical forests. Am. Nat. 1970;104:501–529. doi:10.1086/282687 [Google Scholar]
- Janzen D.H., editor. Costa Rican natural history. University of Chicago Press; Chicago, IL: 1983. [Google Scholar]
- Jordano P., Godoy J.A. Frugivore-generated seed shadows: a landscape view of demographic and genetic effects. In: Levey D.J., Silva W., Galetti M., editors. Frugivores and seed dispersal: ecological, evolutionary, and conservation issues. CAB International; Wallingford, UK: 2002. pp. 305–321. [Google Scholar]
- Loiselle B.A., Dirzo R. Plant–animal interactions and community structure. In: Chazdon R.L., Whitmore T.C., editors. Foundations of tropical biology: classic papers with commentaries. Chicago University Press; Chicago, IL: 2002. pp. 269–278. [Google Scholar]
- Moore D. Insects of palm flowers and fruits. In: Howard F.W., Giblin-Davis R., editors. Insects of palms. CABI Bioscience; Ascot, UK: 2001. pp. 233–266. [Google Scholar]
- Muller-Landau H.C., Hardesty B.D. Seed dispersal of woody plants in tropical forests: concepts, examples and future directions. In: Burslem D.F.R.P., Pinard M.A., Hartley S.E., editors. Biotic interactions in the tropics: their role in the maintenance of species diversity. Cambridge University Press; Cambridge, UK: 2005. pp. 267–309. [Google Scholar]
- Norden N., Chave J., Caubére A., Châtelet P., Ferroni N., Forget P.-M., Thébaud C. Is temporal variation of seedling communities determined by environment or by seed arrival? A test in a Neotropical forest. J. Ecol. 2007;95:507–516. doi:10.1111/j.1365-2745.2007.01221.x [Google Scholar]
- Ribbens E., Silander J.A., Pacala S.W. Seedling recruitment in forests: calibrating models to predict patterns of tree seedling dispersion. Ecology. 1994;75:1794–1806. doi:10.2307/1939638 [Google Scholar]
- Robertson G.P. Gamma Design Software; Plainwell, MI: 1998. GS+: geostatistics for the environmental sciences. [Google Scholar]
- Rosenberg, M. S. 2001 PASSAGE. Pattern analysis, spatial statistics, and geographic exegesis, v. 1.0. Tempe, AZ, USA: Department of Biology, Arizona State University.
- Russo S.E., Augspurger C.K. Aggregated seed dispersal by spider monkey limits recruitment to clumped patterns in Virola calophylla. Ecol. Lett. 2004;7:1058–1067. doi:10.1111/j.1461-0248.2004.00668.x [Google Scholar]
- Russo S.E., Portnoy S., Augspurger C.K. Incorporating animal behavior into seed dispersal models: implications for seed shadows. Ecology. 2006;87:3160–3174. doi: 10.1890/0012-9658(2006)87[3160:iabisd]2.0.co;2. doi:10.1890/0012-9658(2006)87[3160:IABISD]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Schupp E.W. The Janzen–Connell model for tropical tree diversity: population implications and the importance of spatial scale. Am. Nat. 1992;140:526–530. doi: 10.1086/285426. doi:10.1086/285426 [DOI] [PubMed] [Google Scholar]
- Seidler T.G., Plotkin J.B. Seed dispersal and spatial patterns in tropical trees. PLoS Biol. 2006;4:2132–2137. doi: 10.1371/journal.pbio.0040344. doi:10.1371/journal.pbio.0040344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezen U.U., Chazdon R.L., Holsinger K.E. Genetic consequences of tropical second-growth forest regeneration. Science. 2005;307:891. doi: 10.1126/science.1105034. doi:10.1126/science.1105034 [DOI] [PubMed] [Google Scholar]
- Sezen U.U., Chazdon R.L., Holsinger K.E. Multi-generational genetic analysis of tropical second-growth forest regeneration in an abundant canopy palm. Ecology. 2007;88:3065–3075. doi: 10.1890/06-1084.1. doi:10.1890/06-1084.1 [DOI] [PubMed] [Google Scholar]
- Uriarte M., Condit R., Canham C.D., Hubbell S.P. A spatially explicit model of sapling growth in a tropical forest: does the identity of neighbours matter? J. Ecol. 2004;92:348–360. doi:10.1111/j.0022-0477.2004.00867.x [Google Scholar]
- Uriarte M., Canham C.D., Thompson J., Zimmerman J.K., Brokaw N. Seedling recruitment in a hurricane-driven tropical forest: light limitation, density-dependence and the spatial distribution of parent trees. J. Ecol. 2005;93:291–304. doi:10.1111/j.0022-0477.2005.00984.x [Google Scholar]
- Vilchez, B., Chazdon, R. L. & Alvarado, W. 2007 Fenología reproductive de las especies del dosel superior en seis sitios de la region Huetar Norte de Costa Rica. Kuru: Revista Forestal Costa Rica 4(10): http://www.tec.cr/sitios/docencia/forestal/revista_kuru/anteriores/anterior10/pdf/articulo%202.pdf
- Wang B.C., Sork V.L., Leong M.T., Smith T.B. Hunting of mammals reduces seed removal and dispersal of the Afrotropical tree, Antrocaryon klaineanum (Anacardiaceae) Biotropica. 2007;39:340–347. doi:10.1111/j.1744-7429.2007.00275.x [Google Scholar]
- Willson M.F., Irvine A.K., Walsh N.G. Vertebrate dispersal syndromes in some Australian and New Zealand plant communities. Biotropica. 1989;21:133–147. doi:10.2307/2388704 [Google Scholar]