Abstract
Spatial structure has been identified as a major contributor to the maintenance of diversity. Here, we show that the impact of spatial structure on diversity is strongly affected by the ecological mechanisms maintaining diversity. In well-mixed, unstructured environments, microbial populations can diversify by production of metabolites during growth, providing additional resources for novel specialists. By contrast, spatially structured environments potentially limit such facilitation due to reduced metabolite diffusion. Using replicate microcosms containing the bacterium Escherichia coli, we predicted the loss of diversity during an environmental shift from a spatially unstructured environment to spatially structured conditions. Although spatial structure is frequently observed to be a major promoter of diversity, our results indicate that it can also have negative impacts on diversity.
Keywords: evolution of diversity, spatial structure, loss of diversity, facilitation, ecological interactions
1. Introduction
Niche theory posits that species diversity evolves and is maintained via trade-offs (Whittaker 1965), predominately mediated by ecological interactions such as competition, predation or mutualism (Doebeli & Dieckmann 2000; Schluter 2000). Spatial structure can promote diversity by localizing the impact of organisms on their environment (Amarasekare 2003). Locally depleted resources and limited diffusion of inhibitors generate a patchy environment, and the resulting multiple niches provide ecological opportunity for diversification (Chao & Levin 1981; Durrett & Levin 1994; Rainey & Travisano 1998; Czaran et al. 2002; Greig & Travisano 2004; Habets et al. 2006). However, spatial structure can also potentially limit niche generation by reducing resource availability; specifically, resources made available through facilitation (for example, as by-products of consumption). In such a case, primary consumers produce waste products that provide new resources. In a well-mixed environment, such resources are readily available to scavengers. By contrast, spatial structure localizes by-product resources to the immediate environment of primary consumers, potentially restricting resource availability to scavengers and thereby limiting ecological opportunity.
Laboratory populations of Escherichia coli and other bacteria rapidly adapt and diversify in spatially unstructured microcosms (Friesen et al. 2004), as predicted by niche theory, even in single-nutrient environments (Helling et al. 1987; Rozen & Lenski 2000). During growth on excess glucose or other carbohydrates, microbes release into the medium metabolites that serve as additional substrates for growth and can therefore give rise to the evolution of novel and coexisting types specializing on these metabolites, as has been repeatedly demonstrated (Rosenzweig et al. 1994; Turner et al. 1996; Doebeli 2002; Pfeiffer & Bonhoeffer 2004; Wolfe 2005). We therefore hypothesized that diversity maintained by such metabolite-mediated facilitation would decline after a shift to spatially structured conditions that restrict diffusion and, hence, the availability of metabolites.
2. Material and methods
(a) Evolution experiment
We evolved 12 populations of E. coli B in Davis minimal liquid medium supplemented with 410 μg ml−1 glucose (DM+Glu) for 1000 generations by daily diluting stationary-phase populations 100-fold into fresh medium. The populations were founded with two isogenic ancestral genotypes, which only differed in a selectively neutral marker (ara− and ara+; Levin et al. 1977; Lenski & Travisano 1994; Travisano & Lenski 1996) that can easily be distinguished on tetrazolium-arabinose (TA) indicator plates. We assessed colony size by plating a sample of each population at low density onto TA agar plates and incubating them for 48 hours, prior to measuring individual colony size (in pixels) using ImageJ (v. 1.31v, NIH). Diversity within a population was calculated as the corrected coefficient of variation (CV*) of colony size as CV*=(1+1/4n)×CV, where n is the sample size. The coefficient of variation, CV, is calculated as CV with the sample variance s and the sample mean (Sokal & Rohlf 1995). The corrected coefficient of variation corrects for small sample sizes compared to the coefficient of variation (Sokal & Rohlf 1995). We used CV* instead of the typical ‘binning’ measures of diversity (e.g. Shannon-Weaver or Simpson), as it provided an unbiased alternative to the difficulty of choosing appropriate bin sizes. We verified the genetic basis for colony size by plating single colonies, measuring their size on plates before and after 1 day of growth in liquid medium and subsequently calculating the correlation coefficient of mean colony size between the two sets of plates.
(b) Propagation in spatially structured environments
The microcosms were grown as lawns on 10 ml of DM supplemented with 410 μg ml−1 glucose and 14 mg ml−1 agar (DM+Glu+Agar) in small Petri dishes (62 mm diameter). Every day, the microcosms were propagated by 100-fold dilution of stationary-phase microcosms that were grown to a lawn. We extracted a plug of 13 mm diameter from the plate, dispersed the bacteria in 500 ml of 0.85 per cent saline and transferred 116 μl onto fresh medium. The plug included roughly one-twentieth of the area. Given the large size of the transferred population, this sampling procedure, in and of itself, did not lead to a loss of rare types and thus depress diversity. On the spatially structured selective medium (DM+Glu+Agar), different specialists were indistinguishable when plated at low density, as colony size did not vary at any time (24 or 48 hours after plating). On TA plates, colony size did vary. To assess diversity, we plated a sample of each microcosm at low density onto TA plates, measured the colony size after 48 hours of growth on the plate and calculated the corrected coefficient of variation (CV*) as a measure of colony size diversity.
(c) Frequency assay
Two colony morphotypes were isolated from each microcosm and competed through daily transfers into fresh medium for 5 days in liquid and for 3 days in solid medium at different initial frequencies (10 : 90 and 90 : 10). The method of propagation on a spatially structured environment was used as described above. In the spatially structured environment, the assays were stopped before one of the morphotype went extinct. At the beginning and end of competition, the frequencies of the two competitors were assessed and used to calculate the relative fitness per day (Lenski et al. 1991).
To study ecological relationships within cultures, we chose the most divergent colony morphs from each population, so that the morphs could be reliably distinguished in follow-up experiments. We subsequently grouped the morphotypes as ecotypes, depending on whether the colony variant persisted or vanished in the spatially structured environment.
(d) Cross-feeding analysis
To test whether cross-feeding was a possible mechanism by which diversity was maintained, we assessed the growth of different colony morphs in selective and depleted media. We generated depleted medium by inoculating 10 ml of the selective medium, DM+Glu, with 100 μl of stationary, conditioned cultures of each colony morph grown individually. After 7 and 24 hours of growth, we collected the populations and separated the bacteria from the medium by filter sterilization using a 0.22 μm PSE syringe filter. For the growth curve assays, we inoculated 200 μl of medium with 2 μl of conditioned, stationary-phase culture and grew the cultures in a 96-well plate for 24 hours in a plate reader. Every colony type was grown three times in three different media: in the selective medium DM+Glu; medium depleted by self; and medium depleted by conspecifics.
To estimate maximum growth rate, we log transformed the optical density data and determined maximum growth rate by fitting a linear slope over a sliding window and determining the maximum slope. Before testing whether the ecotypes grew differently in depleted media, we performed an analysis of variance (ANOVA) on the maximum growth rate in the ancestral medium, DM+Glu, with block and ecotype as factors. Ecotype was determined according to the performance of a colony morph during the experiment on spatially structured medium (persist or vanish). We observed no significant effect for block and subsequently performed a full factorial ANOVA with environment and ecotype as fixed factors. To test for a trade-off in fast growth on glucose and metabolite specialization, we performed a full factorial ANOVA on the time of maximal growth rate with ecotype and block as factors.
(e) Individual growth assays on spatially structured medium
To test for exploitative differences on spatially structured environments among different colony morphs, the different morphs were grown individually to stationary phase in liquid medium, before they were transferred to a spatially structured medium and grown for 24 hours under the experimental conditions. Before and after growth on the spatially structured medium, the population sizes were measured for both colony morph types and compared using a t-test with equal variance and eight degrees of freedom.
(f) Statistical analysis
To determine the effect of spatial structure on diversity, we performed a REML ANCOVA (restricted maximum likelihood analysis of covariance) using JMP (SAS Institute Inc., Cary, NC, USA) on the differences between CV* unstructured (control) and CV* structured (treatment) over 7 days, with a fixed intercept at zero. A full factorial analysis was carried out, with day (fixed), microcosm (random) and block (random) as factors. To test the effect of initial density, we performed an ANCOVA on the difference between CV* normal transfer density and CV* of 10-fold lower transfer density with day (fixed) and microcosm (random) as factors. As before, we set the intercept at zero. Since we tested a directional hypothesis, we converted the F-value of day into a one-tailed t-value.
3. Results
First, we generated diversity in well-mixed populations and confirmed that diversity was maintained by facilitation. From a single E. coli clone, we initiated 12 replicate microcosms and propagated them in shaken glucose-limited medium for 1000 generations. During the course of selection, the populations increased in fitness by 23 per cent relative to their common ancestor (mean fitness=1.23, 95% CI=0.029, n=12), which indicated adaptation to the selective environment. The fitness increase was greater during the first 500 generations than the second 500 generations (t11=3.56, p<0.01), a pattern frequently observed in selection experiments (Lenski & Travisano 1994). Similarly, phenotypic diversity evolved during the first 500 generations and remained stable for the remainder of the experiment (figure 1a). After 350 generations, we observed multiple colony morphotypes in all populations, when we sampled the microcosms onto solid, nutrient-rich indicator medium (Rosenzweig et al. 1994; Turner et al. 1996; Rainey & Travisano 1998; Friesen et al. 2004). Since all microcosms were seeded from a single ancestral clone, any diversity within microcosms arose through the appearance of de novo mutations during the selection experiment. We tested for a genetic basis of this phenotypic trait and observed a genetic basis for colony morphology, as the sizes of different colony variants isolated after 1000 generations were highly correlated before and after 24 hours of growth in liquid medium (r=0.92, n=27, p<0.0001).
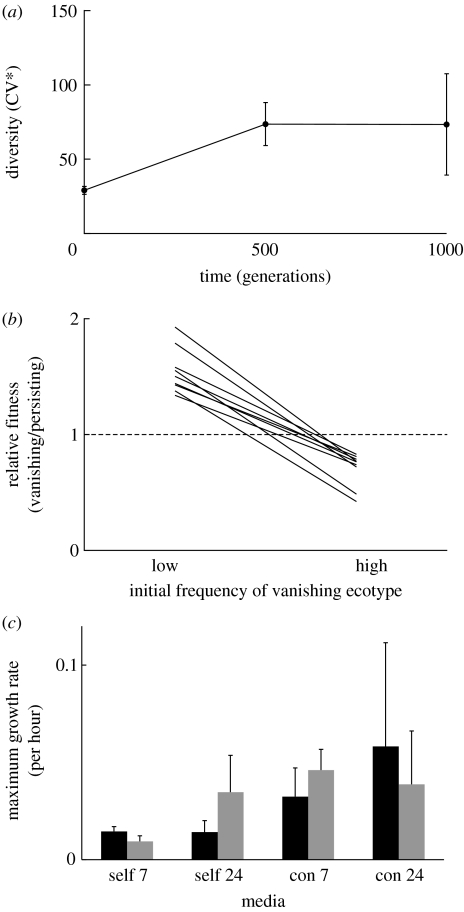
Figure 1.
Diversity evolved and persisted in the mass action liquid environment. (a) Colony size variation within microcosms evolved over the course of the selection experiment, as shown by the increase in average coefficient of variation for colony size (CV*) across microcosms (mean with 95% CI). Phenotypic diversity increased during the first 500 generations (F1,29=38.36, p<0.0001), without a significant change between generation 500 and 1000 (F1,59=0.0013, p=0.971). (b) Stabilizing frequency dependence between the persisting and the vanishing ecotype occurred in the well-mixed environment. Relative fitness of the vanishing ecotype was high at low initial frequency and low at high initial frequency (p<0.0001, paired t-test). Lines connecting the fitness values at low and high frequencies all cross the dashed line where both competitors have equal fitness. (c) Growth rate of the two ecotypes in DM+Glu and in media depleted for 7 or 24 hours by self or by the conspecific (averages with 95% CI). The maximal growth rate was measured over a 24-hour growth cycle for both the persisting (black bars) and the vanishing (grey bars) ecotype. In media depleted for 24 hours, the ecotypes did not differ in growth rate. In media depleted for 7 hours, we observed a significant effect of environment (media depleted by self or conspecific: F1,104=29.84, p<0.0001), with a significant interaction term between environment and ecotype (F1,104=6.96, p=0.0096).
Frequency-dependent selection is essential for the maintenance of diversity evolved through ecological specialization (Levene 1953; Rainey & Travisano 1998; Friesen et al. 2004). To test for frequency-dependent selection and niche specialization among microcosm conspecifics, we isolated single pairs of divergent colony morphotypes from nine microcosms at generation 1000 and competed them against one another. We excluded three microcosms from the analysis, due to difficulty in reliably differentiating colony variants. The experiments were conducted as reciprocal ‘invasion from rare’ assays, with one colony variant initially rare and the other common (Rainey & Travisano 1998). For diversity to be maintained by frequency-dependent selection, individuals of a given type must have a higher relative fitness when rare. This selective advantage should eventually subside as the type increases in frequency. Over 5 days of serial transfer, colony variants had high fitness when at low initial frequency and low fitness when at high initial frequency, relative to their conspecific competitors (paired t-test: t8=11.71, p<0.0001; figure 1b). This pattern indicates that diversity was maintained by stabilizing frequency dependence in the nine microcosms after 1000 generations of selection.
Diversity in single-resource environments can evolve through resource partitioning and facilitation, as has been shown in previous selection experiments with E. coli (Rosenzweig et al. 1994; Turner et al. 1996; Rozen & Lenski 2000; Friesen et al. 2004; Habets et al. 2006). During growth on glucose, E. coli secretes a variety of metabolites into the environment. These metabolites are taken up and metabolized when bacterial growth exhausts the glucose available in the medium. Glucose specialization results in a trade-off between rapid growth and metabolite use through the tricarboxylic acid (TCA) cycle (Helling et al. 1987; Rosenzweig et al. 1994; Turner et al. 1996; Rozen & Lenski 2000; Doebeli 2002; Friesen et al. 2004; Pfeiffer & Bonhoeffer 2004; Wolfe 2005). As a consequence, metabolites accumulate in the environment, providing resources for the evolution of metabolite specialists (Turner et al. 1996). To test for such cross-feeding, we measured the maximal growth rate of the different colony variants in media depleted by self and by the other colony morph (figure 1c). We collected media from two time points: during late exponential growth and during stationary phase, collecting media after 7 and 24 hours of incubation, respectively. First we tested whether the two ecotypes differed in their maximal growth rate in DM+Glu and determined that the two ecotypes did not differ in their maximal growth rate in the selective medium when grown in isolation (F1,106=0.05, p=0.82). This indicates that both types were able to grow on glucose-limited medium. Consistent with a trade-off in fast growth on glucose and specialization on metabolites, the two ecotypes differed in the timing of their maximal growth rate, with the persisting ecotypes reaching maximum growth before the vanishing ecotypes (ecotype: F1,102=10.93, p=0.0013; block: F2,102=5.4, p=0.006; ecotype×block: F2,102=0.34, p=0.714).
We also tested the growth in medium collected during stationary phase. We expected that the ecotypes would not differ in maximal growth rate, as the medium was equally depleted. As expected, we did not observe a significant effect of environment or ecotype on maximal growth rate (ecotype: F1,104=0.0013, p=0.97; environment: F1,104=2.46, p=0.119; ecotype×environment: F1,104=1.7, p=0.19). Lastly, we tested the growth in medium collected during late exponential growth phase. In this medium, we expected a difference in growth. We expected that the vanishing ecotype would be able to grow in medium depleted by the persisting ecotype, while the persisting ecotype would not grow in medium depleted by the vanishing ecotype. As expected, we observed a significant effect of environment (F1,104=29.84, p<0.0001), and more importantly a significant interaction between environment and ecotype (F1,104=6.96, p=0.0096), indicating that the different ecotypes grew differentially in media depleted by self or conspecific. Overall, we did not observe a significant difference among ecotypes (F1,104=0.144, p=0.70). These results suggest that cross-feeding is a very likely mechanism maintaining diversity in these microcosms.
Having obtained diverse populations, we then tested the impact of spatial structure on extant diversity within microcosms. Diversity maintained through facilitation should decrease if metabolite diffusion is limited, as that is the basis for facilitation in this system. We tested for the persistence of diversity during ecological disruption by comparing diversity of the 1000-generation microcosms maintained in well-mixed versus spatially structured environments for 7 days. Each of the nine evolved microcosms was used to initiate several new microcosms maintained on the same temporal daily transfer scheme as during the initial 1000-generation selection regime, but differing in spatial structure. The sole difference between the selective medium and the spatially structured medium was the addition of agar, an inert non-nutritive substance commonly used to solidify liquid media. The first set was maintained under the previous well-mixed shaken conditions in liquid medium, while the second and third sets were maintained on solid agar at the original or 10-fold lower densities at transfer, respectively. Lower initial density increases the distance between colonies and, hence, further reduces the potential for ecological interactions. Regardless of propagation density, densities on solid medium were high, and bacteria grew as a lawn, not as individual colonies. The imposition of spatial structure caused a 50 per cent reduction in diversity relative to the control in the liquid medium (day: F1,116=46.73, p<0.0001; microcosm: F8,116=11.46, p<0.0001; block: F1,116=8.52, p=0.004; figure 2a,b) when the microcosms were sampled on solid medium at low densities. As expected, greater reductions in ecological interactions among specialists owing to increased initial distance between colonies caused even greater losses in diversity (t54=1.87, p=0.039).
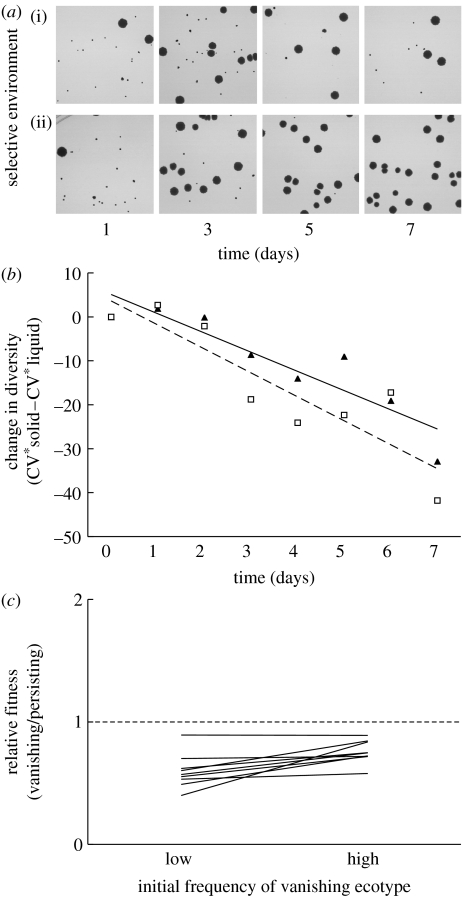
Figure 2.
Diversity declined in spatially structured environments. (a) Diversity in colony size of one population (ara- population 5) transferred (i) in liquid (DM+Glu) or (ii) in spatially structured medium (DM+Glu+Agar) plated on indicator plates (TA). By day 7, no small colonies were present in the population transferred in spatially structured medium, while small colonies persisted throughout the experiment in populations transferred in liquid medium. The presence of small colonies in the spatially structured populations by day 3 indicated that the total diversity was captured in the transfer regime used for this experiment. (b) Diversity declined after shifting from a well-mixed to a spatially structured environment both at normal transfer densities (triangles and solid line) and at 10-fold lower transfer densities (squares and dashed line). Change in diversity was calculated as the difference in diversity in solid and in liquid media (points are average of two measurements for nine populations at normal transfer density and the average of nine populations at 10-fold lower transfer density). Each line was determined by regression on independently collected data. (c) Stabilizing frequency dependence between the persisting and the vanishing ecotype did not occur in the spatially structured environment. Relative fitness of the vanishing ecotype was low, regardless of its initial frequency. None of the lines connecting the fitness at high and low initial frequency crossed the equal fitness line (dashed line).
To verify that reduced ecological interactions were responsible for the loss of diversity, we performed two additional experiments. First, we assessed frequency dependence in the spatially structured environment using the same colony variant pairs used in the previous frequency assay in the well-mixed environment. Competition between conspecifics in the spatially structured environment was significantly different from the frequency-dependent selection observed in liquid medium (paired t-test on slopes: t8=−10.21, p<0.0001), with little frequency dependence in the spatially structured environment (figure 2c). Second, we assessed the ability of each of the different colony morphs to grow in the spatial environment in the absence of competition, and detected no difference between the different colony morphs to exploit the environment in the absence of competition (t8=1.31, p=0.23). These results strongly support the hypothesis that the decline of diversity in spatially structured environments is due to the interruption of ecological interactions.
4. Discussion
Spatial structure profoundly affects diversity. Spatial structure can promote diversity maintained by different ecological interactions including competition (Czaran et al. 2002), predator–prey interactions (Kerr et al. 2002) and even some forms of facilitation (Korona et al. 1994; Rainey & Travisano 1998). In these systems, imposition of spatial structure results in limited diffusion of resources, inhibitors, competitors or predators. Limited diffusion promotes increased diversity because of the generation of spatially distributed niches to which different specialists are adapted. Although it has frequently been observed to promote diversification, spatial structure is neither necessary nor beneficial for either the evolution or maintenance of diversity in our model system. Diversity in this study persists due to resource complexity as envisioned in Whittaker's niche model (Whittaker 1965), in which trade-offs in niche exploitation prevent a single most fit genotype (Lenski & Hattingh 1986). In this case, well-described trade-offs in central metabolic pathways generate a complex resource environment, as indicated by the differential growth in depleted media and the different time to maximal growth rate in the selective medium. Imposition of spatial structure alters resource complexity across the environment, shifting it to microenvironments that are dominated by a single genotype. Scavengers and specialists on by-products are selected against.
We used nutrient-rich indicator plates to distinguish genotypes within cultures, similar to the approach taken by Rainey & Travisano (1998). Assaying colony morphology on nutrient-rich agar surfaces was essential for our study, since different nutrient specialists formed colonies that were indistinguishable when sampled at low density onto the nutrient-limited medium used for the selection experiment. By plating the populations at low densities on nutrient-rich TA agar, we were able to distinguish different colony morphs and to estimate diversity without identifying post hoc discreet colony differences. Fortuitously, we were able to identify different ecotypes that differ by colony morphology. Even so, our estimate of ecotypes is likely to be an underestimate of phenotypic and genetic diversity, as we focused on only two divergent morphotypes per culture to carry out the competition experiments.
Ecotype colony morphology did vary depending upon the initial marker genotype: large colony morphs were lost in microcosms consisting of genotypes capable of using arabinose, while small colony morphs were lost in microcosms consisting solely of arabinose-minus genotypes (figure 2a). Even so, the arabinose-usage marker affects the morphotype lost, but not the loss of diversity. Arabinose was not provided at any time during the selection and was only present in the nutrient-rich medium used to distinguish morphotypes. The observed marker effect suggests that the niche-specific adaptation affects carbohydrate use generally, even on carbohydrates not present during selection. It also demonstrates that colony size, in the nutrient-rich environment used to distinguish morphs, was not a factor affecting loss of diversity during selection in the spatially structured environment.
Our experiments do not address the question of whether spatial structure is beneficial for the evolution of diversity, or the maintenance of diversity after evolution, on spatially structured environments. Theoretical models suggest that diversity based on cross-feeding can be maintained in a spatially structured environment under certain circumstances (Krone & Guan 2006). Habets et al. (2006) pursued this question experimentally by evolving populations in differently structured and unstructured media. They observed that spatial structure can promote the evolution of diversity, as long as the structure was maintained, even at transfer. Such conditions would certainly promote the evolution of facilitation and allow diversity to evolve. In our experiments, the populations were mixed at transfer. Presumably, diversity in a spatially structured environment could have decreased as a result of mixing the populations at transfer, hence interrupting established facilitation. Two lines of evidence support our claim that a spatially structured environment contributed more to the loss of diversity than mixing the populations at transfer. First, diversity was maintained in the liquid, constantly mixed environment. Second, when we transferred our diverse populations for 250 generations in the spatially structured environment, we again observed an initial decrease in diversity followed by an increase (J. Ho, G. Saxer and M. Travisano 2003, unpublished data), indicating that diversity could evolve despite the mixing of the populations at transfer. This diversity, however, was probably maintained by other mechanisms than facilitation.
Our results also show that positive ecological interactions can play a pivotal role in the evolution and maintenance of species diversity. Diversity in this single-resource environment could only occur through some form of niche partitioning (Stewart & Levin 1973). The potential for facilitation arises as an evolutionary response to competition for the primary nutrient and adaptation to rapid growth on glucose trade-offs with growth on metabolic products of glycolysis. The commonality of resource competition and trade-offs in resource use suggests that facilitation is likely to be a frequent outcome of niche partitioning and thereby a common mechanism for the origin and maintenance of diversity (Dieckmann & Doebeli 1999; Doebeli 2002).
Acknowledgments
We are grateful to R. Azevedo, C. Burch, B. Cole, O. Gilbert, E. Ostrowski, M. Quance, R. Quance, C. Rauter, J. Rudgers, J. Strassmann, D. Wiernasz, laboratory colleagues and numerous anonymous reviewers for their helpful comments. J. Ho and S. Durrani helped during the experiments and image analysis. T. Cooper kindly provided equipment and advice for the cross-feeding experiment. M.D. acknowledges the support of the Swiss National Science Foundation, the James S. McDonnell Foundation (USA) and NSERC (Canada). M.T. was supported by the US National Science Foundation.
References
- Amarasekare P. Competitive coexistence in spatially structured environments: a synthesis. Ecol. Lett. 2003;6:1109–1122. doi:10.1046/j.1461-0248.2003.00530.x [Google Scholar]
- Chao L., Levin B.R. Structured habitats and the evolution of anticompetitor toxins in bacteria. Proc. Natl Acad. Sci. USA. 1981;78:6324–6328. doi: 10.1073/pnas.78.10.6324. doi:10.1073/pnas.78.10.6324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaran T.L., Hoekstra R.F., Pagie L. Chemical warfare between microbes promotes biodiversity. Proc. Natl Acad. Sci. USA. 2002;99:786–790. doi: 10.1073/pnas.012399899. doi:10.1073/pnas.012399899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann U., Doebeli M. On the origin of species by sympatric speciation. Nature. 1999;400:354–357. doi: 10.1038/22521. doi:10.1038/22521 [DOI] [PubMed] [Google Scholar]
- Doebeli M. A model for the evolutionary dynamics of cross-feeding polymorphisms in microorganisms. Popul. Ecol. 2002;44:59–70. doi:10.1007/s101440200008 [Google Scholar]
- Doebeli M., Dieckmann U. Evolutionary branching and sympatric speciation caused by different types of ecological interactions. Am. Nat. 2000;156:S77–S101. doi: 10.1086/303417. doi:10.1086/303417 [DOI] [PubMed] [Google Scholar]
- Durrett R., Levin S. The importance of being discrete (and spatial) Theor. Popul. Biol. 1994;46:363–394. doi:10.1006/tpbi.1994.1032 [Google Scholar]
- Friesen M.L., Saxer G., Travisano M., Doebeli M. Experimental evidence for sympatric ecological diversification due to frequency-dependent competition in Escherichia coli. Evolution. 2004;58:245–260. doi:10.1111/j.0014-3820.2004.tb01642.x [PubMed] [Google Scholar]
- Greig D., Travisano M. The prisoner's dilemma and polymorphism in yeast SUC genes. Proc. R. Soc. Lond. B. 2004;271:S25–S26. doi: 10.1098/rsbl.2003.0083. doi:10.1098/rsbl.2003.0083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habets M.G.J.L., Rozen D.E., Hoekstra R.F., de Visser J.A.G.M. The effect of population structure on the adaptive radiation of microbial populations evolving in spatially structured environments. Ecol. Lett. 2006;9:1041–1048. doi: 10.1111/j.1461-0248.2006.00955.x. doi:10.1111/j.1461-0248.2006.00955.x [DOI] [PubMed] [Google Scholar]
- Helling R.B., Vargas C.N., Adams J. Evolution of Escherichia coli during growth in a constant environment. Genetics. 1987;116:349–358. doi: 10.1093/genetics/116.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr B., Riley M.A., Feldman M.W., Bohannan B.J.M. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature. 2002;418:171–174. doi: 10.1038/nature00823. doi:10.1038/nature00823 [DOI] [PubMed] [Google Scholar]
- Korona R., Nakatsu C.H., Forney L.J., Lenski R.E. Evidence for multiple adaptive peaks from populations of bacteria evolving in a structured habitat. Proc. Natl Acad. Sci. USA. 1994;91:9037–9041. doi: 10.1073/pnas.91.19.9037. doi:10.1073/pnas.91.19.9037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krone S.M., Guan Y. Spatial self-organization in a cyclic resource-species model. J. Theor. Biol. 2006;241:14–25. doi: 10.1016/j.jtbi.2005.11.005. doi:10.1016/j.jtbi.2005.11.005 [DOI] [PubMed] [Google Scholar]
- Lenski R.E., Hattingh S.E. Coexistence of two competitors on one resource and one inhibitor: a chemostat model based on bacteria and antibiotics. J. Theor. Biol. 1986;122:83–93. doi: 10.1016/s0022-5193(86)80226-0. doi:10.1016/S0022-5193(86)80226-0 [DOI] [PubMed] [Google Scholar]
- Lenski R.E., Travisano M. Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations. Proc. Natl Acad. Sci. USA. 1994;91:6808–6814. doi: 10.1073/pnas.91.15.6808. doi:10.1073/pnas.91.15.6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski R.E., Rose M.R., Simpson S.C., Tadler S.C. Long-term experimental evolution in Escherichia coli. I: adaptation and divergence during 2,000 generations. Am. Nat. 1991;138:1315–1341. doi:10.1086/285289 [Google Scholar]
- Levene H. Genetic equilibrium when more than one ecological niche is available. Am. Nat. 1953;87:331–333. doi:10.1086/281792 [Google Scholar]
- Levin B.R., Stewart F.M., Chao L. Resource-limited growth, competition, and predation: a model and experimental studies with bacteria and bacteriophage. Am. Nat. 1977;111:3–24. doi:10.1086/283134 [Google Scholar]
- Pfeiffer T., Bonhoeffer S. Evolution of cross-feeding in microbial populations. Am. Nat. 2004;163:E126–E135. doi: 10.1086/383593. doi:10.1086/383593 [DOI] [PubMed] [Google Scholar]
- Rainey P.B., Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. doi:10.1038/27900 [DOI] [PubMed] [Google Scholar]
- Rosenzweig R.F., Sharp R.R., Treves D.S., Adams J. Microbial evolution in a simple unstructured environment: genetic differentiation in Escherichia coli. Genetics. 1994;137:903–917. doi: 10.1093/genetics/137.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen D.E., Lenski R.E. Long-term experimental evolution in Escherichia coli. VIII: dynamics of a balanced polymorphism. Am. Nat. 2000;155:24–35. doi: 10.1086/303299. doi:10.1086/303299 [DOI] [PubMed] [Google Scholar]
- Schluter D. Oxford University Press; Oxford, UK: 2000. The ecology of adaptive radiation. [Google Scholar]
- Sokal R.R., Rohlf F.J. W. H. Freeman and Co; New York, NY: 1995. Biometry. [Google Scholar]
- Stewart F.M., Levin B.R. Partitioning of resources and outcome of interspecific competition: model and some general considerations. Am. Nat. 1973;107:171–198. doi:10.1086/282825 [Google Scholar]
- Travisano M., Lenski R.E. Long-term experimental evolution in Escherichia coli IV: targets of selection and the specificity of adaptation. Genetics. 1996;143:15–26. doi: 10.1093/genetics/143.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner P.E., Souza V., Lenski R.E. Tests of ecological mechanisms promoting the stable coexistence of two bacterial genotypes. Ecology. 1996;77:2119–2129. doi:10.2307/2265706 [Google Scholar]
- Whittaker R. Dominance and diversity in land plant communities. Science. 1965;147:250–260. doi: 10.1126/science.147.3655.250. [DOI] [PubMed] [Google Scholar]
- Wolfe A.J. The acetate switch. Microbiol. Mol. Biol. Rev. 2005;69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. doi:10.1128/MMBR.69.1.12-50.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]