Abstract
We have compared the efficacy of inhibition of the cytochrome bc1 complexes from yeast and bovine heart mitochondria and Paracoccus denitrificans by antimycin, ilicicolin H, and funiculosin, three inhibitors that act at the quinone reduction site at center N of the enzyme. Although the three inhibitors have some structural features in common, they differ significantly in their patterns of inhibition. Also, while the overall folding pattern of cytochrome b around center N is similar in the enzymes from the three species, amino acid sequence differences create sufficient structural differences so that there are striking differences in inhibitor binding to the three enzymes.
Antimycin is the most tightly bound of the three inhibitors, and binds stoichiometrically to the isolated enzymes from all three species under the cytochrome c reductase assay conditions. Ilicicolin H also binds stoichiometrically to the yeast enzyme, but binds approximately 2 orders of magnitude less tightly to the bovine enzyme and is essentially non-inhibitory to the Paracoccus enzyme. Funiculosin on the other hand inhibits the yeast and bovine enzymes similarly, with IC50 ∼10 nM, while the IC50 for the Paracoccus enzyme is more than 10 fold higher. Similar differences in inhibitor efficacy were noted in bc1 complexes from yeast mutants with single amino acid substitutions at the center N site, although the binding affinity of quinone and quinol substrates were not perturbed to a degree that impaired catalytic function in the variant enzymes.
These results reveal a high degree of specificity in the determinants of ligand binding at center N, accompanied by sufficient structural plasticity for substrate binding as to not compromise center N function. The results also demonstrate that, in principle, it should be possible to design novel inhibitors targeted toward center N of the bc1 complex with appropriate species selectivity to allow their use as drugs against pathogenic fungi and parasites.
1. Introduction
The cytochrome bc1 complex is a homodimeric, multisubunit enzyme embedded in the inner mitochondrial membrane of oxygen utilizing eukaryotic cells and the plasma membrane of many gram negative bacteria [1]. The catalytic core of the enzyme is formed by the cytochrome b subunit, the Rieske-iron sulfur protein and the cytochrome c1 subunit. In addition, the mitochondrial enzyme contains 6-8 supernumerary subunits lacking any cofactors, while the bacterial enzyme contains none or one extra subunit [2]. The protonmotive Q cycle describes the mechanism by which the enzyme couples the transfer of two electrons from quinol to two cytochrome c to the translocation of four protons across the membrane [3, 4]. The Q cycle involves two quinone binding sites in cytochrome b; the center P (“QP”) site catalyzes quinol oxidation, with one electron transferred to the Rieske protein en route to cytochromes c1 and c, while the second electron is transferred to the low potential b heme (bL1) and recycled inward through the membrane to the high potential heme (bH) to the opposite site of the membrane, where it reduces quinone to form a tightly bound semiquinone at the center N (QN) site.
Several inhibitory compounds are known that target specifically the QP or QN site [5]. These have been of use in the structural and functional studies of the individual catalytic sites. In addition, the QP inhibitor atovaquone [6] is of pharmaceutical relevance for treatment of some parasitic and fungal infections. Only a few center N specific inhibitors are known to date. These include antimycin, which is a natural product of various species of Streptomyces [7], ilicicolin H, isolated from the imperfect fungus Cylindrocladium ilicicola [8, 9], and funiculosin, produced by Penicillium funiculosum Thom [10, 11]. Structurally the inhibitors are clearly different, but they also share some similarities [9]. Upon binding to the QN site, they all displace semiquinone [12]. While ilicicolin H and antimycin both possess a phenol ring, funiculosin and ilicicolin H share a pyridone ring system. Based on a similar optical effect of the latter two inhibitors on the cytochrome bH spectrum, it was concluded that the ring system plays an important role in their binding [9]. On the other hand genetic studies in yeast indicate that the inhibitors have different modes of binding to the bc1 complex [13].
In this study we have compared the inhibitory action of the QN inhibitors, antimycin, funiculosin and ilicicolin H on the cytochrome bc1 complexes from three different species, Saccharomyces cerevisiae, Bos taurus, and Paracoccus denitrificans. We also examined the inhibitor sensitivity of yeast bc1 complexes with center N mutations obtained from ilicicolin H resistant [14] and respiratory deficient revertants [15, 16]. The results reveal striking differences in sensitivity of the enzymes from the different species, and comparing the cytochrome b sequences around the center N binding pocket allows some preliminary speculation regarding the structural basis of the differences in inhibitor efficacy between the species. The results also demonstrate the feasibility of designing new therapeutic drugs, targeted to the bc1 complex, with species selectivity based on structural differences in center N of pathogens and host enzymes.
2. Materials and Methods
2.1 Materials
Dodecyl maltoside was obtained from Anatrace. DEAE-Biogel and Tween-20 were obtained from Bio-Rad Laboratories. Antimycin, diisopropylfluorophosphate, horse heart cytochrome c, and decyl ubiquinone were purchased from Sigma Chemical Co. Dithionite was purchased from Fluka Biochemica. Decyl ubiquinol (DBH) was prepared as described before and quantified spectrophotometrically [17]. Funiculosin was a gift of Novartis, Basel, Switzerland. Ilicicolin H was obtained from the Merck sample repository. The QN inhibitors antimycin, ilicicolin and funiculosin were diluted in ethanol to a concentration of 50-200 μM. The extinction coefficients used to calculate the concentration of the stock solutions were, for antimycin, 4.8 mM-1 cm-1 at 320 nm, for ilicicolin H, 5.3 mM-1 cm-1 at 349 nm, and for funiculosin 5.5 mM-1 cm-1 at 293 nm [5, 9].
2.2 Purification of cytochrome bc1 complexes
The wild-type yeast strain with the KM91 background and the cytochrome b mutants of that strain, M221Q, M221E and W30C were obtained from Dr. Anne-Marie Colson (Universite Catholique de Louvain-La-Neuve, Belgium) and Dr. Gael Brasseur (CNRS Marseille, France) [16, 18]. The wild-type yeast strain with the W303 background and cytochrome b mutants of that strain, S20T, Q22E, Q22T and L198F, were described previously [14]. Bovine heart mitochondria were a gift from Dr. Chang-an Yu (Oklahoma State University).
Cytochrome bc1 complexes from all of the yeast strains and bovine heart mitochondrial membranes were purified as previously described [19]. Purified cytochrome bc1 complex from P. denitrificans was a gift from Dr. Bernd Ludwig (W. Goethe-Universität, Frankfurt am Main, Germany). The cytochrome c1 concentration in the bc1 complexes was determined from the difference spectrum of the ascorbate-reduced versus ferricyanide-oxidized enzymes, using an extinction coefficient of 17.5 mM-1 cm-1 at 554-539 nm. The cytochrome b concentration was determined from the difference spectrum of dithionite-reduced versus ferricyanide-oxidized enzymes. The extinction coefficients used were 50 mM-1 cm-1 at 562-578 nm for the yeast and bovine enzymes [20, 21], and 40 mM-1 cm-1 at 559-578 nm for the P. denitrificans bc1 complex.
2.3 Ubiquinol-cytochrome c reductase assay and inhibition by the center N inhibitors antimycin, ilicicolin and funiculosin
Ubiquinol-cytochrome c reductase activities of the purified cytochrome bc1 complexes from bovine heart, commercially available Red Star™ yeast, and P. denitrificans were measured at room temperature in assay buffer containing 50 mM potassium phosphate, pH 7.0, 250 mM sucrose, 1 mM sodium azide, 0.2 mM EDTA, 0.05 % dodecyl maltoside. For the purified cytochrome bc1 complexes from the other yeast strains, the assay buffer contained 50 mM potassium phosphate, pH 7.0, 1 mM sodium azide, 1 mM EDTA and 0.01 % Tween-20. The dodecyl maltoside and Tween-20 were included in the assay buffers because they improved the stability of the diluted enzyme during the prolonged incubation required for equilibration with the inhibitors. The enzymes were diluted first to ∼1 μM in 400 μL of the same assay buffer and incubated on ice for 30 minutes. For measuring the activities, an aliquot of the enzyme was diluted from the 1 μM sample to a final concentration of 2.5 nM in assay buffer containing 50 μM cytochrome c and 1 mM potassium cyanide and incubated for 1.5 min. The reaction was started by adding 50 μM DBH, and reduction of cytochrome c was monitored at 550-539 nm with the Aminco DW2a™ spectrophotometer in the dual wavelength mode. The extinction coefficient used to calculate the cytochrome c reduction was 21.5 mM-1 cm-1 at 550-539 nm. All measurements were done in duplicate.
For measuring the inhibitor titration curves, the activity was first measured without inhibitor and this was taken as 100 % activity. The concentration of inhibitor in the enzyme solution was increased incrementally by adding aliquots from the inhibitor stock solutions to the 1 μM enzyme sample. After addition, the enzyme-inhibitor solution was gently mixed and incubated for 2 min. on ice prior to measuring activity. The ethanol concentration in the solution did not exceed 5 %.
3. Results
3.1 Comparison of the QN sites from yeast, bovine and P. denitrificans bc1 complexes, and the location of yeast QN mutants
The QN site in the cytochrome bc1 complex from yeast [22] and bovine [23, 24] mitochondria is structurally highly conserved (Fig. 1). Comparing the sequences of residues2 16-38 and 195-230 that form the QN site shows that ∼50% of the amino acids are identical. No structural information is available for the complex from P. denitrificans, but the crystal structure of the complex from Rhodobacter capsulatus [25], which cytochrome b sequence is more than 80 % conserved with that of P. denitrificans, also shows a conserved structure of the QN site. However the sequence identity of the bacterial enzyme with those of the yeast and bovine enzymes is slightly lower, ∼35 and ∼45 %, respectively. Interestingly, the sequence identity of the residues that are present within 5 Å of the ubiquinone (in the yeast structure) is no higher than the overall identity in the center N pocket. It is striking that, even with this degree of sequence variation, the distance and orientation of the ubiquinone ring relative to the bH heme are essentially identical in the yeast and bovine enzymes (Fig. 1). Five of the most conserved residues near the ubiquinone are the axial histidine ligand to the heme iron, His 197, two residues implicated in quinone binding, His-202 and Asp-229, and Gly-33, Trp-30, and Lys-228 [26]. It is also striking that the conserved His-202 participates in a water-mediated hydrogen bond to the quinone ring in the yeast enzyme, but does not do so in the bovine enzyme where it is rotated closer to the quinone. We have studied here mutations of Trp-30, and four other residues, Ser-20, Gln-22, Leu-198, Met-221 that are less conserved, but are within 5 Å of the ubiquinone ring.
Fig. 1.
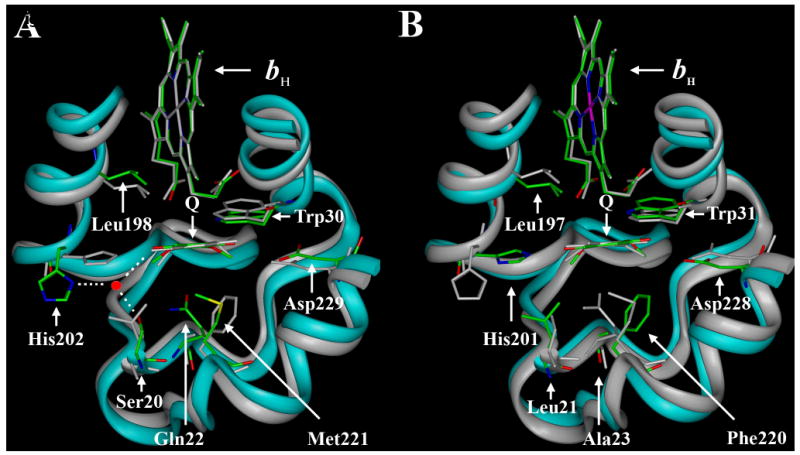
Comparison of the yeast and bovine cytochrome b protein structures in the region of the proteins surrounding the QN site. Both panels show the bH hemes, the ubiquinone ring and amino acid residues 16-38 and 194-230 of the yeast protein overlaid with residues 17-39 and 193-229 of the bovine protein. In panel A the carbon backbone of the yeast protein is colored cyan and the bH heme ring, ubiquinone ring, and selected amino acids are labeled and colored. Oxygen atoms are red, nitrogen atoms are blue, carbon atoms are green and sulfur is yellow. The carbon backbone and other features of the bovine protein are grey. Panel A also depicts the water-mediated hydrogen bond between His-202, Ser-20, and the ubiquinone ring that is present in the yeast, but not in the bovine, structure. In panel B the carbon backbone of the bovine protein is colored cyan and the heme ring, ubiquinone ring, and selected amino acids are labeled and colored according to the same scheme as in panel A, while the yeast protein is depicted in grey. The figures were created from the coordinates of the yeast (1EZV, Ref. 22) and bovine (1ppj, Ref. 24) enzymes.
Trp-30 forms a hydrogen bond with one of the propionate groups of the bH heme (Fig. 1). The substitution of tryptophan by a cysteine in the W30C mutant has no natural equivalent. Leu-198 is conserved in the yeast and bovine enzymes (Fig. 1, 2) and is located close to the bH heme and the quinone. The analogous residue in P. denitrificans is an isoleucine. The substitution of leucine by phenylalanine in the L198F mutant is native in Candida, but other hydrophobic residues are also observed. Interestingly the L198F mutation was reported to confer resistance towards the three center N inhibitors, antimycin, funiculosin and ilicicolin H in the yeast enzyme [14]. It is not known whether the Candida bc1 complex is intrinsically resistant to these center N inhibitors.
Fig. 2.
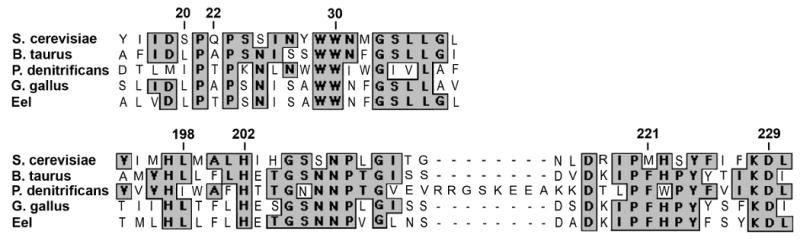
Sequence alignment of the cytochrome b proteins from bovine, yeast, P. denitrificans, chicken and eel in the regions forming the QN site. The sequences from the latter two species were included since the enzymes from those species have been reported to be resistant to funiculosin, as discussed in the text. The alignment was constructed using ClustalW and yeast numbering of residues 16-38 and 194-230. In the bovine enzyme the QN site is formed by residues 17-39 and 193- 229, and in the P. denitrificans enzyme by residues 31-53 and 208-252. The numbered arrows show the positions of the QN site yeast mutations that were analyzed, and the residues His-202 and Asp-229.
A hydrophobic or van der Waals interaction is observed between the residue at position 221 and the ubiquinone (Fig. 1). In the complex from bovine mitochondria, P. denitrificans and most other species the corresponding residue is a phenylalanine (Fig. 2) [26]. The occurrence of methionine in the yeast bc1 complex is an exception. The M221E and M221Q mutant enzymes studied here contain substitutions that have not been found in any cytochrome b sequences. The yeast strains with these mutations were previously obtained as revertants of a respiratory deficient mutant strain, M221K [27].
Ser-20 and Gln-22 from yeast are two of the least conserved residues, but the results of an ilicicolin H resistance screen showed their importance in center N function [14, 28]. In the QN site of yeast, these two residues are part of a hydrogen-bonding network that also includes His-202, one carbonyl group of the ubiquinone, and an active site water (Fig. 2A). In the bovine enzyme serine is substituted by leucine and glutamine by an alanine. A structural consequence of these substitutions appears to be rotation of the histidine ring towards the ubiquinone, a direct hydrogen bond between a ubiquinone carbonyl group and the imidazole nitrogen [24] or indirectly through a water molecule [29]. The Leu-21 (bovine numbering) may also stabilize this histidine conformation. In the P. denitrificans enzyme the residues at positions 20 and 22 are isoleucine and threonine, respectively. The crystal structure of the R. capsulatus enzyme does not contain a ubiquinone bound at the QN site, but the histidine has the same configuration as in the bovine structure. Interestingly, substitutions of Ser-20 by a leucine, or Gln-22 by a threonine rendered the yeast strains resistant to ilicicolin H. Because the S20L mutant bc1 complex was reported to be unstable [14], it has not been further characterized. However the S20T, Q22T and Q22E mutants have been investigated.
3.2 Inhibition of yeast, bovine and P. denitrificans bc1 complexes by center N inhibitors
Fig. 3 compares the inhibition of cytochrome c reductase activities of yeast, bovine and P. denitrificans bc1 complexes by antimycin, ilicicolin H and funiculosin. The titration curves in Fig. 3A show the inhibition of the three bc1 complexes by antimycin. With one equivalent of antimycin more than 90% of activity is inhibited in the bovine and yeast enzyme, as shown before [9]. The P. denitrificans enzyme shows some residual activity at higher concentration of antimycin, but the inhibitor still effectively inactivates the enzyme. We previously reported that ilicicolin H is a potent inhibitor for yeast bc1 complex, but not for the bovine enzyme [9]. Under the assay conditions used here, the residual catalytic activity is ∼10% with one equivalent of ilicicolin H per bc1 monomer. On the other hand, both the bovine and P. denitrificans enzymes are highly resistant towards this inhibitor (Fig. 3B), with IC50 values >100 nM for the bovine enzyme while the bacterial enzyme is barely inhibited even at the highest concentrations of inhibitor tested (data not shown). For the yeast enzyme, funiculosin is the least effective of the three inhibitors (Fig. 3C), and the titration curve for the bovine enzyme is essentially identical to that the yeast complex. The Paraccocus enzyme is only weakly inhibited by this inhibitor.
Fig. 3.
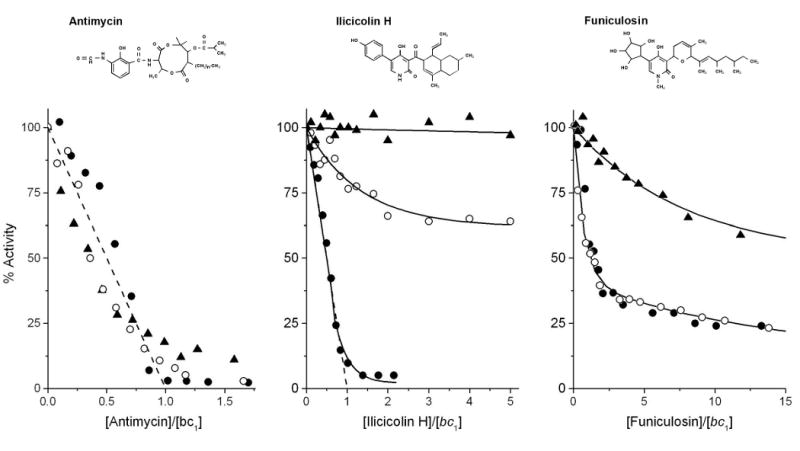
Inhibition of ubiquinol-cytochrome c reductase activity of yeast, P. denitrificans and bovine cytochrome bc1 complexes by the center N inhibitors antimycin (panel A), ilicicolin H (panel B) and funiculosin (panel C). Activities are expressed as percentage of the activity of the corresponding non-inhibited enzymes. The activities of the yeast bc1 complex are indicated with closed circles, those of the P. denitrificans bc1 complex with closed triangles, and those of the bovine bc1 complex with open circles. In the absence of inhibitor, the activities of the yeast, P. denitrificans and bovine enzymes were 125, 80 and 170 s-1, respectively. The titration curves were obtained as described in Materials and Methods. The dashed lines show the theoretical titration curves for the stoichiometric binding of one inhibitor (antimycin and iliciolin H) per bc1 complex. Structures of the inhibitors are shown above the titration curves.
3.3 Inhibition of yeast cytochrome b mutants
The results in Fig. 4 show that none of the mutations effect the inhibition of the isolated enzymes by antimycin; more than 90 % of the catalytic activity is inhibited with a titer close to one inhibitor per bc1 complex monomer. Thus the IC50 is still at least an order of magnitude less than 2.5 × 10-9 M. The slight hysterisis in the inhibitor titration of the bc1 complex from wild-type yeast has been attributed by us to negative cooperativity between the two center N sites and inter-monomer electron transfer through the bL hemes [30-32]. This same behavior is seen with some of the cytochrome b mutant enzymes (Fig. 4). The lack of hysterisis in some of the mutant enzymes suggests that the negative cooperativity might have been weakened due to some of the mutations.
Fig. 4.
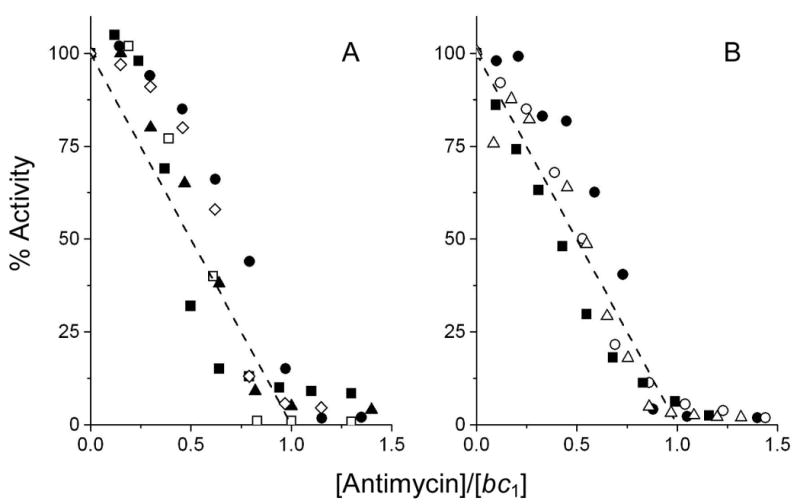
Inhibition of ubiquinol-cytochrome c reductase activity of yeast cytochrome b mutants by antimycin. Activities are expressed as percentage of the activity of the corresponding non-inhibited enzyme. In panel A the activities of W303 (wt) bc1 complex are indicated with closed circles, those of S20T with open diamonds, those of Q22T with closed triangles, those of Q22E with open squares and those of L198F with closed squares. In the absence of inhibitor, the activities of the W303 (wt), S20T, Q22T, Q22E and L198F enzymes were 290, 140, 250, 260, 260 s-1, respectively. In panel B, the activities of the KM91 (wt) bc1 complex are indicated with closed circles, those of M221Q with open circles, those of M221E with closed squares and those of W30C with open triangles. In the absence of inhibitor, the activities of the KM91, M221Q, M221E and W30C enzymes were 180, 150, 160 and 170 s-1, respectively. The inhibitor titration curves were obtained as described in Materials and Methods. The dashed lines show the theoretical titration curves for stoichiometric binding of one inhibitor (antimycin and ilicicolin H) per bc1 complex.
Ilicicolin H inhibits the cytochrome c reductase activity of the KM91 and W303 wild-type strains ∼90% with a titer of one inhibitor per bc1 monomer (Fig. 4), as is also observed with the bc1 complex from commercial yeast (Fig. 3B). The S20T, Q22E, Q22T and L198F mutations in the yeast bc1 complex that conferred resistance to ilicicolin H, indeed provide significant resistance towards this inhibitor (Fig. 5A). The Q22E mutant is completely resistant with up to 40 equivalents (100 nM) of inhibitor, whereas the other three mutations result in a gradual decrease in catalytic activity with increasing concentration of inhibitor. The M221E, M221Q and W30C mutants were not obtained as yeast strains with resistance to ilicicolin in vivo, but the purified enzymes from these mutants also do show resistance to inhibition by this inhibitor (Fig. 5B), although to a lesser extent than the other mutant enzymes.
Fig. 5.
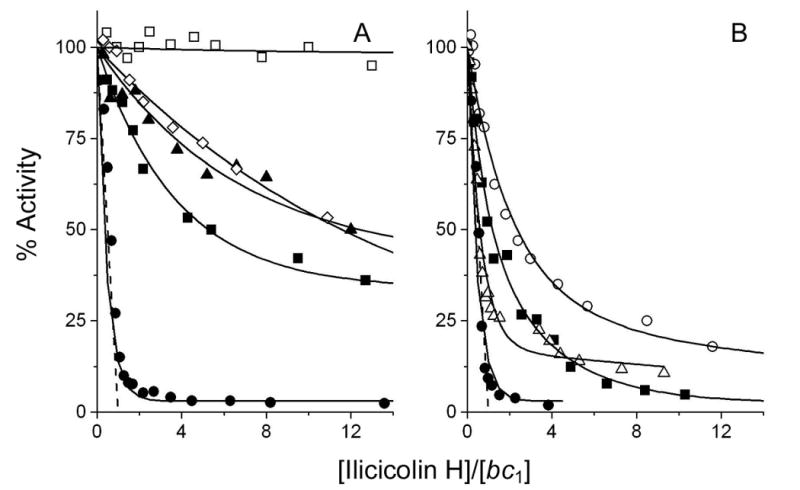
Inhibition of ubiquinol-cytochrome c reductase activity of yeast cytochrome b mutants by ilicicolin H. The symbols used for the mutated enzymes in panels A and B are the same as in Fig. 4. The dashed lines show the theoretical titration curves for the stoichiometric binding of one inhibitor per bc1 complex.
With funiculosin the titration curves of the wild-type enzymes with the two genetic backgrounds appear to show some difference; the W303 strain is more resistant towards this inhibitor than the KM91 strain, with half inhibition occurring at ∼8 vs. ∼2 equivalents of inhibitor per bc1 monomer, corresponding to IC50 values of 20 and 5 nM respectively. However, with the exception of L198F and W30C, the mutations appear to have no significant effect on the sensitivity towards funiculosin. The bc1 complex with the L198F cytochrome b mutation appears to be highly resistant, while the W30C mutation appears only to be more resistant at higher concentrations of funiculosin. Previous studies reported that both of these mutations confer resistance to this inhibitor [33, 34].
4. Discussion
In the current study we have compared the efficacy of three center N inhibitors, antimycin, ilicicolin H and funiculosin towards the purified cytochrome bc1 complexes from yeast, bovine heart, and P. denitrificans. We also tested the efficacy of the inhibitor against five variants of the yeast enzyme with mutations in cytochrome b. Comparison of the sequences and structures of the bc1 complexes from the three species showed some common features for the binding of quinone and the activity of center N. On the other hand, there are a number of residues near the quinone that are not conserved. Previous structural studies in the bovine enzyme [29] and genetic studies in yeast [13] showed that the binding sites for different QN inhibitors do not entirely overlap. We have probed the degree of overlap more extensively with the center N mutations Ser-20, Gln-22, Trp-30, Leu-198 and Met-221 in the yeast cytochrome b. Analyzing these mutant enzymes allows some preliminary conclusions to be made regarding the differences in efficacy of the center N inhibitors against enzymes from the three different species.
Antimycin has been known to bind with high affinity to the bovine enzyme with an indirectly measured Kd = 3.2 × 10-11 M [35]. In our experiments (Fig. 3A) we did not detect any difference in the efficacy of inhibiting the catalytic activity of the bc1 complexes from the three species by this inhibitor. Structural studies of the chicken [36] and bovine [24, 29] bc1 complexes with antimycin bound show that the inhibitor is similarly docked very close to the bH heme of cytochrome b in the two enzymes. Indeed, two of the most conserved residues (see Fig. 2 and ref. 26), Lys-228 and Asp-229, have been implicated in the strong binding of this inhibitor [29]. In the crystal structures of the chicken [36] and bovine [24, 29] bc1 complexes these two residues form hydrogen bonds with the inhibitor, directly or through structural waters. A hydrophobic interaction that is observed between ubiquinone and Met-221 (Phe in bovine and P. denitrificans, Figs. 1 & 2) may also contribute to the binding of this inhibitor, but this appears not to be essential (see below).
Likewise, none of the center N mutations in the yeast dramatically affect the sensitivity of the yeast enzyme for antimycin (Fig. 4). However, it should be noted that under the assay conditions employed in our experiments, an increase of as much as 50-fold from the Kd = 3.2 × 10-11 M estimated for the bovine enzyme would go undetected, since the concentration of enzyme is 2.5 ×10-9 M in the assay. This might explain why the L198F mutant, although resistant towards antimycin in vivo [14], was inhibited by a titer of ∼1 antimycin per bc1 monomer. The M221E and M221Q mutations also did not change the sensitivity to antimycin in our experiments, while a M221K mutation in the same parental strain blocked antimycin binding to the QN site [18].
We previously showed that the IC50 value for inhibition of the cytochrome c reductase activity of the bovine bc1 complex by ilicicolin H is ∼100-fold higher than for the yeast enzyme [9]. In the current experiments both the bovine and P. denitrificans enzymes appear to be highly resistant to this inhibitor (Fig. 3B). Thus the yeast enzyme may contain structural features important for the binding of this inhibitor that the other two are lacking, or the bovine and P. denitrificans enzymes may contain features that confer resistance to the binding of this inhibitor. Funiculosin is the weakest of the three inhibitors with all three species enzymes. It differs from ilicicolin H in that only the bc1 complex from P. denitrificans is less sensitive towards this inhibitor.
The L198F yeast cytochrome b mutation conferred cross-resistance towards all three inhibitors in vivo [14], but resistance was seen in vitro to only ilicicolin H and funiculosin, probably due to the high affinity of antimycin relative to enzyme concentration in the assay as discussed above. In particular, the effect of substitution of the leucine by a bulkier phenylalanine on the inhibition by funiculosin is dramatic. This suggests that this residue is an important binding determinant for this inhibitor. It is possible that an isoleucine at this position in the P. denitrificans enzyme may contribute to weakening of inhibition, but it is more likely that there are other determinants that are responsible for this difference. In this regard it is interesting to note that Degli Esposti and coworkers (26, 37) proposed that valine at the position equivalent to Val-194 in the yeast cytochrome b is one of the residues responsible for the intrinsic resistance of fish, rabbit and horse cytochrome bc1 complexes to funiculosin. However, the yeast and bovine enzymes, which have valine and alanine, respectively, at this position in their cytochrome b's, are equally sensitive to funiculosin (Figs. 2 & 3).
It should also be acknowledged that with all of the cytochrome b mutations the possibility remains that the effect of the mutational change is indirect, due to structural changes transmitted through the protein to a ligand-binding site some distance from the mutated residue. A precedent for such long-range effects might be found in mutational changes to Ala-126 in the yeast protein, which confer resistance to both center N and center P inhibitors (cf Ref. 37 and Table IV in Ref. 26). The distinction between direct local steric effects and long-range indirect effects can only be addressed when crystal structures of the enzymes with ilicicolin and funiculosin bound become available.
The W30C mutation slightly lowers the sensitivity to inhibition of the yeast bc1 complex by ilicicolin (Fig. 5), but has a more pronounced effect on funiculosin inhibition of the enzyme (Fig. 6). Trp-30 is itself not responsible for the difference in efficacy of funiculosin inhibition between the enzymes from P. denitrificans and the two other species, since it is conserved. However, the effects of the W30C mutation suggest that Trp-30 is part of a binding pocket for funiculosin, as was reported previously [34]. The W30C mutation was originally obtained as a second site mutation that corrects the respiratory-deficient phenotype of the S206L mutant [16]. Two other second site mutations in the same revertant screen were obtained, N208Y and N208K, and the double mutations also confer resistance towards funiculosin [38]. The three residues (Trp-30, Ser-206 and Asn-208) are within 5 Å from each other. The temperature-sensitivity of the three revertant strains suggests a structural change has occurred that makes these bc1 complexes more labile, and that may be near the funiculosin binding site and thus block the binding of funiculosin.
Fig. 6.
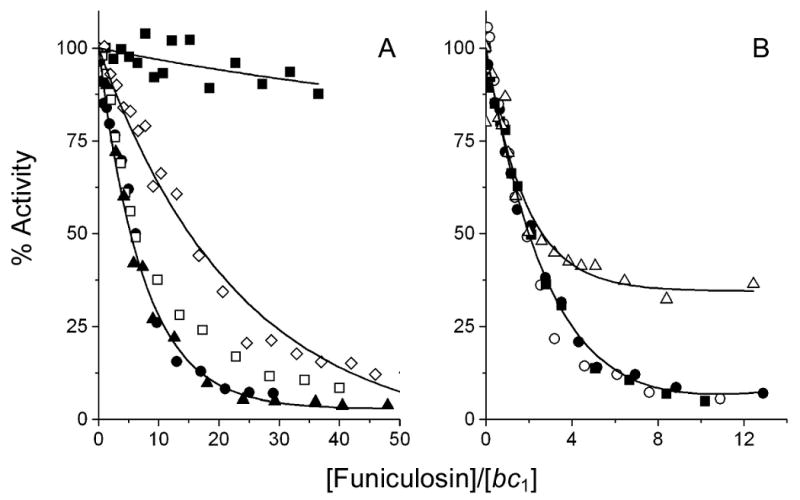
Inhibition of ubiquinol-cytochrome c reductase activity of yeast cytochrome b mutants by funiculosin. The symbols used for the mutated enzymes in panels A and B are the same as in Fig. 4.
The Ser-20, Gln-22 and Met-221 residues are within 5 Å of each other, on the opposite site of the quinone ring with respect to Trp-30 and Leu-198 (Fig. 1). As was discussed above, Met-221 in the yeast enzyme and the phenylalanine residue at the equivalent position in the bovine and P. denitrificans complexes (Figs. 1 & 2) stabilize the binding of the quinone at the QN site. The M221E and M221Q mutations render the yeast enzyme slightly resistant towards ilicicolin H (Fig. 5). One possible explanation is the importance of a hydrophobic interaction for the binding of ilicicolin H. However, the bovine and P. denitrificans enzymes are much less sensitive towards this inhibitor, even though a hydrophobic interaction may be present. The residues at positions 20 and 22 appear to be especially important for the binding of ilicicolin H, as discussed below. Thus, an alternative explanation for the results of the Met-221 variants is that these mutations have modified the conformation of Ser-20 and Gln-22 and thus weakened the binding of ilicicolin H.
Ser-20 and Gln-22 are two of the least conserved residues in the QN site of the three species. The results with the mutant yeast enzymes indicate that Ser-20 and Gln-22 play an important role in the different efficacies of ilicicolin H in inhibiting the bc1 complexes from the three species (Fig. 5). The purified bc1 complex from the S20T mutant and the enzyme in mitochondrial membranes from the S20L yeast strain [14] are both highly resistant towards ilicicolin H. Interestingly, leucine is the naturally occurring residue at the equivalent position in the bovine bc1 complex, while another hydrophobic residue, isoleucine, is present in the enzyme from P. denitrificans. Both the Q22T and Q22E mutations also render the yeast enzyme less sensitive to ilicicolin H. Threonine is again a naturally occurring residue in one of the species, P. denitrificans.
A straightforward explanation of these comparisons is that Ser-20 and Gln-22 are critical for the binding of the ilicicolin H to the yeast bc1 complex. In addition, the nature of the two residues at these positions appears to modulate the orientation of the imidazole ring of His-202. In the bovine and R. capsulatus (and possibly also P. denitrificans, see above) QN site the imidazole ring is rotated into the quinone-binding site. This configuration may also weaken the binding of ilicicolin. Replacement of ilicicolin H by antimycin, followed by monitoring the spectral changes in the ferro-cytochrome bH spectrum from the ilicicolin induced blue shift to the antimycin induced red-shift, indeed is much faster in the mutant enzymes than in the wild-type enzymes [Ref. 9 and unpublished results].
We previously reported the effect of these yeast mutations on the quinol oxidation reaction at the QN site [28]. Although there were significant differences in the kinetics of quinol oxidation at center N, these differences did not compromise the catalytic function of the enzymes. With the exception of the W30C mutation, the impairment in center N kinetics in these mutants was attributed to weakened affinity for the quinol substrate at center N. The altered affinity for the substrate correlates well with the resistance of the mutants towards ilicicolin H. This suggests that ilicicolin H occupies a similar binding site as the ubiquinol substrate in the yeast bc1 complex (Fig. 1) that involves the residues Ser-20, Gln-22, Leu-198 and Met-221. However ilicicolin is a much larger molecule than ubiquinol and structural perturbations by these mutations can have a more pronounced effect on the binding of the inhibitor. For example the L198F mutation has only a modest effect on the kinetics at center N, but increases the IC50 for ilicicolin H 12-fold. On the other hand the binding site for funiculosin appears to be slightly different, where the important residues Leu-198 and Trp-30C are near the bH heme. Ser-20, Gln-22 and Met-221 appear to play no direct role in the binding of this inhibitor, but are important for catalysis. The nature of the residues at positions 20 and 22 are important determinants for binding of ilicicolin H and can account for the different efficacies of inhibition by this inhibitor in the three species.
In conclusion, it should also be noted that the species differences noted here might have important consequences for drug design. Atovaquone, a hydroxynaphthoquinone inhibitor of the cytochrome bc1 complex, has been used therapeutically to treat Plasmodium falciparum malaria, Pneumocystis carinii pneumonia, and Toxoplasma gondii toxoplasmosis. This inhibitor kills these organisms by blocking ubiquinol oxidation at center P of the bc1 complex, although its therapeutic effectiveness has been compromised by spontaneously arising mutations that confer resistance to the drug [39]. In principle, it should also be possible to design drugs targeted to the quinone reduction site at center N of the enzyme, provided that the inhibitory compounds display appropriate species selectivity toward the pathogen versus host enzymes. This would allow the advantages of combination drug therapy directed toward a single essential enzyme target, thus minimizing the likelihood of side effects. The current results demonstrate that there are sufficient structural differences between fungal, bacterial, and mammalian center N sites to permit such drug design.
Acknowledgments
This research was supported by NIH Research Grant GM 20379.
Footnotes
Abbreviations used are bL heme, low potential b heme; bH heme, high potential b heme; DBH, decyl ubiquinol.
Yeast numbering of the cytochrome b amino acid sequence is used throughout, except when noted otherwise.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Trumpower BL. Cytochrome bc1 complexes of microorganisms. Microbiol Rev. 1990;54:101–129. doi: 10.1128/mr.54.2.101-129.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berry EA, Guergova-Kuras M, Huang LS, Crofts AR. Structure and function of cytochrome bc complexes. Annu Rev Biochem. 2000;69:1005–1075. doi: 10.1146/annurev.biochem.69.1.1005. [DOI] [PubMed] [Google Scholar]
- 3.Trumpower BL, Gennis RB. Energy transduction by cytochrome complexes in mitochondrial and bacterial respiration: the enzymology of coupling electron transfer reactions to transmembrane proton translocation. Annu Rev Biochem. 1994;63:675–716. doi: 10.1146/annurev.bi.63.070194.003331. [DOI] [PubMed] [Google Scholar]
- 4.Mitchell P. Possible molecular mechanisms of the protonmotive function of cytochrome systems. J Theor Biol. 1976;62:327–367. doi: 10.1016/0022-5193(76)90124-7. [DOI] [PubMed] [Google Scholar]
- 5.von Jagow G, Link TA. Use of specific inhibitors on the mitochondrial bc1 complex. Methods Enzymol. 1986;126:253–271. doi: 10.1016/s0076-6879(86)26026-7. [DOI] [PubMed] [Google Scholar]
- 6.Hudson AT, Dickins M, Ginger CD, Gutteridge WE, Holdich T, Hutchinson DB, Pudney M, Randall AW, Latter VS. 566C80: a potent broad spectrum anti-infective agent with activity against malaria and opportunistic infections in AIDS patients. Drugs Exp Clin Res. 1991;17:427–435. [PubMed] [Google Scholar]
- 7.Endo T, Yonehara H. Chemical studies on blastmycin. 3. Gas-liquid chromatography of antimycin A-blastmycin antibiotics. J Antibiot (Tokyo) 1970;23:91–95. doi: 10.7164/antibiotics.23.91. [DOI] [PubMed] [Google Scholar]
- 8.Hayakawa S, Minato H, Katagiri K. The ilicicolins, antibiotics from Cylindrocladium ilicicola. J Antibiot (Tokyo) 1971;24:653–654. doi: 10.7164/antibiotics.24.653. [DOI] [PubMed] [Google Scholar]
- 9.Gutierrez-Cirlos EB, Merbitz-Zahradnik T, Trumpower BL. Inhibition of the yeast cytochrome bc1 complex by ilicicolin H, a novel inhibitor that acts at the Qn site of the bc1 complex. J Biol Chem. 2004;279:8708–8714. doi: 10.1074/jbc.M311805200. [DOI] [PubMed] [Google Scholar]
- 10.Nelson BD, Walter P, Ernster L. Funiculosin: an antibiotic with antimycin-like inhibitory properties. Biochim Biophys Acta. 1977;460:157–162. doi: 10.1016/0005-2728(77)90162-1. [DOI] [PubMed] [Google Scholar]
- 11.Ando K, Suzuki S, Saeki T, Tamura G, Arima K. Funiculosin, a new antibiotic. I. Isolation, biological and chemical properties (studies on antiviral and antitumor antibiotics. 8) J Antibiot (Tokyo) 1969;22:189–194. doi: 10.7164/antibiotics.22.189. [DOI] [PubMed] [Google Scholar]
- 12.Covian RG, Zwicker K, Rotsaert FA, Trumpower BL. Asymmetric and redox-specific binding of quinone and quinol at center N of the dimeric yeast cytochrome bc1 complex: Consequences for semiquinone stabilization. J Biol Chem. 2007;281:3092–30932. doi: 10.1074/jbc.M700662200. [DOI] [PubMed] [Google Scholar]
- 13.Brasseur G, Saribas AS, Daldal F. A compilation of mutations located in the cytochrome b subunit of the bacterial and mitochondrial bc1 complex. Biochim Biophys Acta. 1996;1275:61–69. doi: 10.1016/0005-2728(96)00051-5. [DOI] [PubMed] [Google Scholar]
- 14.Ding MG, di Rago JP, Trumpower BL. Investigating the Qn site of the cytochrome bc1 complex in Saccharomyces cerevisiae with mutants resistant to ilicicolin H, a novel Qn site inhibitor. J Biol Chem. 2006;281:36036–36043. doi: 10.1074/jbc.M608026200. [DOI] [PubMed] [Google Scholar]
- 15.Coppee JY, Tokutake N, Marc D, di Rago JP, Miyoshi H, Colson AM. Analysis of revertants from respiratory deficient mutants within the center N of cytochrome b in Saccharomyces cerevisiae. FEBS Lett. 1994;339:1–6. doi: 10.1016/0014-5793(94)80373-0. [DOI] [PubMed] [Google Scholar]
- 16.Brasseur G, Coppee JY, Colson AM, Brivet-Chevillotte P. Structure-function relationships of the mitochondrial bc1 complex in temperature-sensitive mutants of the cytochrome b gene, impaired in the catalytic center N. J Biol Chem. 1995;270:29356–29364. doi: 10.1074/jbc.270.49.29356. [DOI] [PubMed] [Google Scholar]
- 17.Trumpower BL, Edwards CA. Purification of a reconstitutively active iron-sulfur protein (oxidation factor) from succinate. cytochrome c reductase complex of bovine heart mitochondria. J Biol Chem. 1979;254:8697–8706. [PubMed] [Google Scholar]
- 18.Brasseur G, Brivet-Chevillotte P. Characterization of mutations in the mitochondrial cytochrome b gene of Saccharomyces cerevisiae affecting the quinone reductase site (QN) Eur J Biochem. 1995;230:1118–1124. doi: 10.1111/j.1432-1033.1995.tb20663.x. [DOI] [PubMed] [Google Scholar]
- 19.Snyder CH, Trumpower BL. Ubiquinone at center N is responsible for triphasic reduction of cytochrome b in the cytochrome bc1 complex. J Biol Chem. 1999;274:31209–31216. doi: 10.1074/jbc.274.44.31209. [DOI] [PubMed] [Google Scholar]
- 20.Yu CA, Yu L, King TE. Preparation and properties of cardiac cytochrome c1. J Biol Chem. 1972;247:1012–1019. [PubMed] [Google Scholar]
- 21.Berden JA, Slater EC. The reaction of antimycin with a cytochrome b preparation active in reconstitution of the respiratory chain. Biochim Biophys Acta. 1970;216:237–249. doi: 10.1016/0005-2728(70)90215-x. [DOI] [PubMed] [Google Scholar]
- 22.Hunte C, Koepke J, Lange C, Rossmanith T, Michel H. Structure at 2.3 A resolution of the cytochrome bc1 complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment. Structure. 2000;8:669–684. doi: 10.1016/s0969-2126(00)00152-0. [DOI] [PubMed] [Google Scholar]
- 23.Iwata S, Lee JW, Okada K, Lee JK, Iwata M, Rasmussen B, Link TA, Ramaswamy S, Jap BK. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science. 1998;281:64–71. doi: 10.1126/science.281.5373.64. [DOI] [PubMed] [Google Scholar]
- 24.Huang LS, Cobessi D, Tung EY, Berry EA. Binding of the respiratory chain inhibitor antimycin to the mitochondrial bc1 complex: a new crystal structure reveals an altered intramolecular hydrogen-bonding pattern. J Mol Biol. 2005;351:573–597. doi: 10.1016/j.jmb.2005.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berry EA, Huang LS, Saechao LK, Pon NG, Valkova-Valchanova M, Daldal F. X-Ray Structure of Rhodobacter Capsulatus Cytochrome bc1: Comparison with its Mitochondrial and Chloroplast Counterparts. Photosynth Res. 2004;81:251–275. doi: 10.1023/B:PRES.0000036888.18223.0e. [DOI] [PubMed] [Google Scholar]
- 26.Esposti MD, De Vries S, Crimi M, Ghelli A, Patarnello T, Meyer A. Mitochondrial cytochrome b: evolution and structure of the protein. Biochim Biophys Acta. 1993;1143:243–271. doi: 10.1016/0005-2728(93)90197-n. [DOI] [PubMed] [Google Scholar]
- 27.Lemesle-Meunier D, Brivet-Chevillotte P, di Rago JP, Slonimski PP, Bruel C, Tron T, Forget N. Cytochrome b-deficient mutants of the ubiquinol-cytochrome c oxidoreductase in Saccharomyces cerevisiae. Consequence for the functional and structural characteristics of the complex. J Biol Chem. 1993;268:15626–15632. [PubMed] [Google Scholar]
- 28.Rotsaert FA, Covian R, Trumpower BL. Mutations in cytochrome b that affect kinetics of the electron transfer reactions at center N in the yeast cytochrome bc1 complex. Biochim Biophys Acta. doi: 10.1016/j.bbabio.2007.08.005. in press. [DOI] [PubMed] [Google Scholar]
- 29.Gao X, Wen X, Esser L, Quinn B, Yu L, Yu CA, Xia D. Structural basis for the quinone reduction in the bc1 complex: a comparative analysis of crystal structures of mitochondrial cytochrome bc1 with bound substrate and inhibitors at the Qi site. Biochemistry. 2003;42:9067–9080. doi: 10.1021/bi0341814. [DOI] [PubMed] [Google Scholar]
- 30.Covian R, Trumpower BL. Regulatory interactions between ubiquinol oxidation and ubiquinone reduction sites in the dimeric cytochrome bc1 complex. J Biol Chem. 2006;281:30925–30932. doi: 10.1074/jbc.M604694200. [DOI] [PubMed] [Google Scholar]
- 31.Covian R, Trumpower BL. Rapid electron transfer between monomers when the cytochrome bc1 complex dimer is reduced through center N. J Biol Chem. 2005;280:22732–22740. doi: 10.1074/jbc.M413592200. [DOI] [PubMed] [Google Scholar]
- 32.Covian R, Gutierrez-Cirlos EB, Trumpower BL. Anti-cooperative oxidation of ubiquinol by the yeast cytochrome bc1 complex. J Biol Chem. 2004;279:15040–15049. doi: 10.1074/jbc.M400193200. [DOI] [PubMed] [Google Scholar]
- 33.di Rago JP, Perea J, Colson AM. Isolation and RNA sequence analysis of cytochrome b mutants resistant to funiculosin, a center i inhibitor of the mitochondrial ubiquinol-cytochrome c reductase in Saccharomyces cerevisiae. FEBS Lett. 1990;263:93–98. doi: 10.1016/0014-5793(90)80713-s. [DOI] [PubMed] [Google Scholar]
- 34.Brasseur G. PhD thesis. University Marseille: 1986. Structure-function relationships in the bc1 complex of Saccharomyces cerevisiae: study of mutants affected in the QN center of quinone reduction and a nuclear mutant (ABC1) [Google Scholar]
- 35.Slater EC. The mechanism of action of the respiratory inhibitor, antimycin. Biochim Biophys Acta. 1973;301:129–154. doi: 10.1016/0304-4173(73)90002-5. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Z, Huang L, Shulmeister VM, Chi YI, Kim KK, Hung LW, Crofts AR, Berry EA, Kim SH. Electron transfer by domain movement in cytochrome bc1. Nature. 1998;392:677–684. doi: 10.1038/33612. [DOI] [PubMed] [Google Scholar]
- 37.Degli Esposti M, Ghelli A, Crdimi M, Baracca A, Solaini G, Tron T, Meyer A. Cytochrome b of fish mitochondria is strongly resistant to funiculosin, a powerful inhibitor of respiration. Arch Biochem Biophys. 1992;295:198–204. doi: 10.1016/0003-9861(92)90506-r. [DOI] [PubMed] [Google Scholar]
- 38.Brasseur G, Brivet-Chevillotte P. Specificities of the two center N inhibitors of mitochondial bc1 complex, antimycin and funiculosin: strong involvement of cytochrome b-asparagine-208 in funiculosin binding. FEBS Lett. 1994;354:23–29. doi: 10.1016/0014-5793(94)01077-3. [DOI] [PubMed] [Google Scholar]
- 39.Kessl JJ, Meshnick SR, Trumpower BL. Modeling the molecular basis of atovaquone resistance in parasites and pathogenic fungi. Trends in Parasitology. 2007 doi: 10.1016/j.pt.2007.08.004. in press. [DOI] [PubMed] [Google Scholar]


