Abstract
In animal cells, cytokinesis is powered by a contractile ring of actin filaments (F-actin) and myosin-2. Formation of the contractile ring is dependent on the small GTPase, RhoA1,2, which is activated in a precise zone at the cell equator3. It has long been assumed that cytokinesis and other Rho-dependent processes are controlled in a sequential manner, whereby Rho activation via guanine nucleotide exchange factors (GEFs) initiates a particular event, and Rho inactivation via GTPase activating proteins (GAPs) terminates that event. MgcRacGAP is a conserved cytokinesis regulator thought to be required only at the end of cytokinesis4,5. Here we show that MgcRacGAP’s GAP activity is necessary early during cytokinesis for the formation and maintenance of the Rho activity zone. Disruption of GAP activity by point mutation results in poorly focused Rho activity zones, while complete removal of the GAP domain results in unfocused zones that display lateral instability and/or rapid side-to-side oscillations. We propose that the GAP domain of MgcRacGAP plays two unexpected roles throughout cytokinesis: first, it transiently anchors active Rho, and second it promotes local Rho inactivation resulting in the constant flux of Rho through the GTPase cycle.
Cytokinesis in animal cells relies on the centralspindlin complex, which is comprised of the kinesin MKLP-1 and the Rho GAP MgcRacGAP5,6. This complex is thought to activate Rho via Ect2, a Rho GEF that binds MgcRacGAP7,8. It is currently thought that the GAP activity of MgcRacGAP functions only at the end of cytokinesis to inactivate Rho and thereby disassemble the cytokinetic apparatus5,9. This model is consistent with standard ideas about Rho regulation in which GEFs and GAPs are thought to act sequentially. An alternative model proposes that GEFs and GAPs act simultaneously, so that Rho undergoes rapid flux through the GTPase cycle during cytokinesis and other processes that require rapid actomyosin dynamics3. In this “GTPase flux model”, the GAP activity of MgcRacGAP is envisioned as being employed throughout cytokinesis to help limit the spread of the Rho activity zone10.
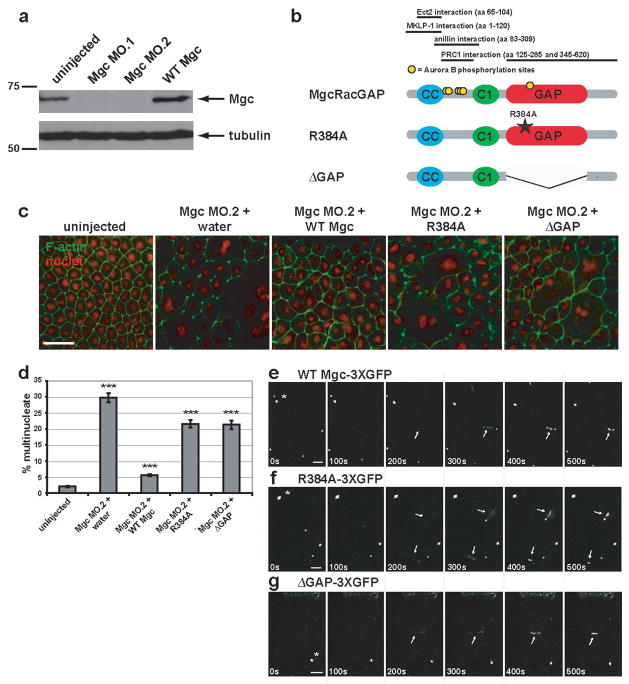
Previous studies addressed whether the MgcRacGAP’s GAP activity is required for cytokinesis but arrived at differing conclusions11–14; the role of the GAP activity in controlling the Rho zone was not monitored. To test the role of MgcRacGAP’s GAP activity, we employed Xenopus laevis embryos, a vertebrate system that permits imaging of Rho activity during cytokinesis3. The role of MgcRacGAP in this system was first tested by blocking expression of MgcRacGAP using one of two different antisense morpholinos; MO.1, which targets the beginning of the coding region or MO.2, which targets the 5′ UTR (Fig. S1a). Each morpholino resulted in more than 95% reduction in endogenous MgcRacGAP levels (Figs. 1a, S1d, S1e) and strongly inhibited cytokinesis as judged by quantifying multinuclearity (Fig. 1c, d and Fig. S1b, c), consistent with other systems4,5,8,13,14. In contrast, a 5 mispair control MO had virtually no effect (Fig. S1a–c).
Figure 1. MgcRacGAP’s GAP activity is required for cytokinesis in Xenopus embryos.
a, Western blot showing morpholino knock-down of MgcRacGAP protein levels by Mgc MO.1 and MO.2 (see Fig. S1a for morpholino target sites) as compared to uninjected or WT Mgc mRNA-injected Xenopus embryo lysates harvested 24 h post-injection. The top panel was immunoblotted with antibodies to Xenopus MgcRacGAP, and the bottom panel with antibodies to α-tubulin as a loading control. b, Domain diagram of MgcRacGAP and its GAP-DEAD mutants, R384A and ΔGAP. CC = coiled coil domain, C1 = putative phorbol ester/diacylglycerol binding domain, GAP = GTPase activating domain. Regions that interact with Ect2, MKLP-1, anillin, or PRC1 as well as sites phosphorylated by AuroraB are noted. c, Embryos were injected with MO.2 along with either water, WT Mgc mRNA, Mgc R384A mRNA, or Mgc ΔGAP mRNA. Embryos were fixed at 24 h post-injection and stained with propidium iodide to reveal the nuclei (red) and Alexa 488-phalloidin to visualize F-actin (green). Scale bar, 50 μm. d, The percent of multinucleate cells was quantified for gene replacement embryos from three independent experiments. Mean ± SE. Uninjected, n = 10 embryos; Mgc MO.2 + water, n = 10; Mgc MO.2 + WT Mgc, n = 11; Mgc MO.2 + R384A, n = 11, Mgc MO.2 + ΔGAP, n = 11. ***p < 0.005. e–g, Frames from time-lapse movies showing the localization patterns of WT Mgc-3XGFP (e), R384A-3XGFP (f), or ΔGAP-3XGFP (g). Asterisks indicate midbodies (from previous cell divisions), which are extremely bright. Arrows indicate the localization of the constructs at the closing cytokinetic apparati. The mutants are detectable at the equator at least as soon as WT MgcRacGAP (see 200s time point). Scale bars, 20 μm.
The fact that MO.2 targets the 5′ UTR allowed a gene replacement strategy in which this morpholino was microinjected along with MgcRacGAP mRNAs lacking the 5′ UTR of the endogenous mRNA. Three mRNAs were used: wild type MgcRacGAP (WT Mgc), MgcRacGAP with a point mutation that disrupts GAP activity (Mgc R384A)4, and MgcRacGAP lacking the GAP domain entirely (Mgc ΔGAP) (Fig. 1b). This approach permitted replacement of the endogenous MgcRacGAP with comparable levels of the three constructs (Fig. S1f–h). Phenotypic analysis revealed that while WT Mgc resulted in strong suppression of the multinuclearity phenotype, the GAP-DEAD mutants were significantly less effective (Fig. 1c, d). Further, as described below, time-lapse imaging revealed that many of the GAP-DEAD cells that do manage to divide do so abnormally. Together, these results indicate that MgcRacGAP’s GAP activity is indeed necessary for proper cytokinesis.
Since the GAP-DEAD mutations are outside the previously identified interaction domains of MgcRacGAP for MKLP-1, Ect2, anillin, or the N-terminal PRC1 binding site (Fig. 1b)6,8,15,16, it is unlikely that the deficits in cytokinesis result from failure of GAP-DEAD MgcRacGAP to bind these proteins and/or resultant defects in midzone organization. This point was tested via immunoprecipitation of MKLP-1 with MgcRacGAP, which showed that the two GAP-DEAD MgcRacGAP mutants immunoprecipitate MKLP-1 as well as WT Mgc (Fig. S2a). Further, localization analysis showed that WT Mgc and the GAP-DEAD mutants all localized to ingressing furrows during cytokinesis (Fig. 1e–g), indicating that failure of the GAP-DEAD mutants to rescue cytokinesis as well as WT Mgc does not result from failed subcellular targeting of MgcRacGAP. To further ensure that the effects of these mutations stemmed from their defects in the GAP domain, rather than in spindle midzone organization, spindles were characterized by immunofluorescence. We found no apparent differences in spindle shape, midzone density, or midzone organization in the mutants relative to either the WT replacement (Fig. S2b) or uninjected controls (not shown). As a more quantitative measure, we also compared the ratio of the pole-to-pole distance to total cell length in early anaphase spindles, since weakening or loss of the midzone results in greater separation of the poles17. No significant differences were found between spindles in the WT Mgc or GAP-DEAD replacement embryos (WT Mgc = 0.59 ± 0.07; R384A = 0.58 ± 0.05; ΔGAP = 0.56 ± 0.05; mean ± S.D. n = 20 spindles for each group). In addition, we monitored the distribution of several different proteins known to interact with MgcRacGAP and/or the central spindle: MKLP-1, Plk1, Aurora B, and anillin. We found that all of these players showed apparently normal early (anaphase) and late (telophase) localization to the central spindle in WT Mgc, R384A, and ΔGAP replacement embryos (Fig. S2c).
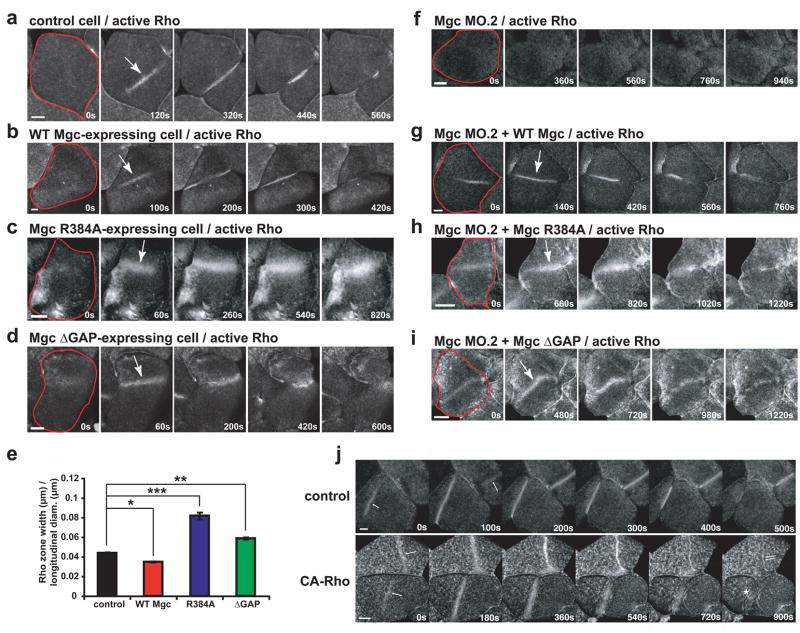
The above results indicated that MgcRacGAP’s GAP activity is required for cytokinesis. To understand the basis of this requirement, the effects of these mutations on the Rho activity zone were assessed using GFP-rGBD, which reports GTP-bound Rho3,18. Over-expression of WT Mgc led to Rho activity zones that were more focused (80% of the breadth of controls) and less intense than controls (Fig. 2a, b, e; Fig. S3a, b; and Movie S1). In contrast, expression of either Mgc R384A or Mgc ΔGAP resulted in Rho activity zones that were broader than those in control cells and often failed to ingress efficiently, leading to failed cytokinesis (Fig. 2c–e). Specifically, the Rho activity zones in cells expressing Mgc R384A were 1.9-fold broader and more intense than controls (Fig. 2c, e; Fig. S3a, b; and Movie S2). Similarly, Rho activity zones in cells expressing Mgc ΔGAP were also increased in breadth (1.3-fold broader than controls) and intensity (Fig. 2d–e; Fig. S3a, b; and Movie S2). Expression of either GAP-DEAD mutant also led to increased active Rho signal outside of the Rho zone (Fig. S3a, c). We also noted that the changes in Rho zone breadth increased relative to the longitudinal diameter of the Xenopus blastomeres, becoming particularly broad in GAP-DEAD expressing cells (Fig. S3d). In addition, the Mgc ΔGAP-expressing cells exhibited another phenotype: positional instability such that the zones moved laterally across the surface of the embryo. In extreme cases, this instability was manifest as rapid side-to-side oscillations (average oscillation velocity = 0.340 ± 0.004 μm/sec) (Fig. 2d and Movie S2).
Figure 2. Testing the GTPase flux model.
a–d, Embryos were injected with GFP-rGBD to monitor active Rho along with WT Mgc (b), Mgc R384A (c), or Mgc ΔGAP (d) and imaged during cytokinesis. Frames taken from time-lapse movies are shown. A single blastomere is outlined in red in the first frame, and the Rho activity zone is marked with an arrow in the second frame. Scale bars, 40 μm. e, Average Rho zone width / longitudinal diameter for multiple cells. Mean ± SE. Control, n = 12 embryos; WT Mgc, n = 9; R384A, n = 10; ΔGAP, n = 12. *p < 0.05; **p < 0.01; ***p < 0.005. f–i, Gene replacement experiments where embryos were injected with Mgc MO.2 to knock down endogenous MgcRacGAP along with mRNAs for WT Mgc (g), Mgc R384A (h), or Mgc ΔGAP (i). The embryos were also injected with GFP-rGBD to monitor active Rho. A single blastomere is outlined in red in the first frame, and the Rho activity zone is marked with an arrow in the second frame. Scale bars, 40 μm. j, Effects of constitutively-active Rho (CA-Rho) on Rho zone dynamics. Frames taken from time-lapse movies showing Rho activity dynamics during cytokinesis in control cells and cells expressing CA-Rho are shown. Cytokinetic Rho zones (arrows) are broader and brighter in cells expressing CA-Rho and are much slower to furrow. Moreover, the amount of Rho activity outside the equtorial regions is much higher in those cells expressing CA-Rho than the controls. Note that these phenotypes closely resemble those obtained with the R384A GAP-DEAD mutant. Scale bars, 40 μm.
The potential effects of GAP-DEAD MgcRacGAP on Rac and Cdc42 activity were also examined. In control cells, elevated Rac and Cdc42 activity at cell-cell boundaries was observed, but we did not detect any cortical pattern of Rac or Cdc42 activity when Rho activity was rising at the equator (Fig. S4). Further, in contrast to the striking accumulation of active Rho caused by Mgc R384A, no obvious changes in Rac or Cdc42 activity were seen following expression of Mgc R384A (Fig. S4). These results suggest that in this system, MgcRacGAP is acting primarily on Rho rather than Rac or Cdc42 to control the assembly of the cytokinetic apparatus, although Rac and Cdc42 may participate in the cell-cell junction assembly that follows in the wake of the furrow19.
Rho activity was also examined using the gene-replacement approach. Rho activity zones were strongly reduced in cells where MgcRacGAP was knocked down with MgcRacGAP MO.2 (Fig. 2f), presumably due to the inability of the centralspindlin complex to localize Ect2 in the absence of MgcRacGAP7. Gene replacement with WT Mgc resulted in formation of focused Rho zones (Fig. 2g and Movie S3). In contrast, gene replacement with either of the GAP-DEAD mutants led to broader Rho activity zones with increased intensity (Fig. 2h, i; Fig. S3a, b; and Movie S4). Notably, gene replacement with Mgc ΔGAP led to Rho zones that were unstable and moved laterally (Fig. 2i and Movie S4), and gene replacement with either GAP-DEAD mutant resulted in increased Rho activity outside the Rho zone (Fig. S3a, c). Thus, the GAP activity of MgcRacGAP is needed to confine Rho activation to the equator.
The increase in Rho zone width and intensity observed with the GAP-dead mutants is consistent with the GTPase flux model where MgcRacGAP’s GAP activity is required throughout cytokinesis to keep the Rho activity zone focused. This model also predicts that constitutively-active Rho should mimic the GAP-DEAD MgcRacGAP phenotype. Indeed, constitutively-active Rho leads to excess Rho activity outside the furrow, broad Rho zones, and slower onset to furrowing, similar to the R384A mutant (Fig. 2j). In contrast, the sequential GEF/GAP model in which the GEF acts at the beginning of cytokinesis and the GAP acts at the end predicts that cytokinesis defects would not be observed until much later.
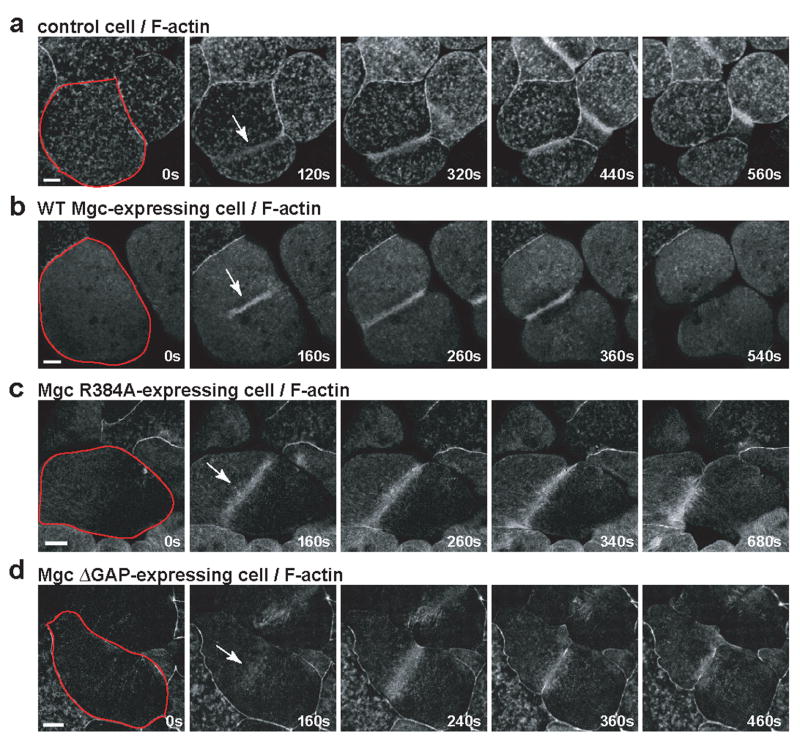
To determine the downstream consequences of having abnormal Rho zones, we imaged F-actin in live cells. Control cells exhibited transient F-actin patches throughout the cortex and concentrated F-actin signal at the furrow (Fig. 3a and Movie S5). Similar to control cells, cells expressing WT Mgc had focused accumulation of F-actin at the furrow (Fig. 3b and Movie S5). In contrast, Mgc R384A-expressing cells exhibited a broad accumulation of F-actin at the furrow (Fig. 3c and Movie S6). In cells expressing Mgc ΔGAP, F-actin accumulation was broader than in controls, and F-actin oscillated rapidly back and forth, as did the Rho activity zones (Fig. 3d and Movie S6). Thus, disruption of MgcRacGAP’s GAP activity leads to abnormal distribution and dynamics of downstream Rho targets. This failure to form an efficient contractile array likely leads to the observed cytokinesis failure.
Figure 3. Downstream consequences of abnormal GTPase flux.
a–d, Embryos were injected with GFP-UtrCH to monitor F-actin along with WT Mgc (b), Mgc R384A (c), or Mgc ΔGAP (d) and imaged during cytokinesis. Frames taken from time-lapse movies are shown. A single blastomere is outlined in red in the first frame, and the Rho activity zone is marked with an arrow in the second frame. Scale bars, 40 μm.
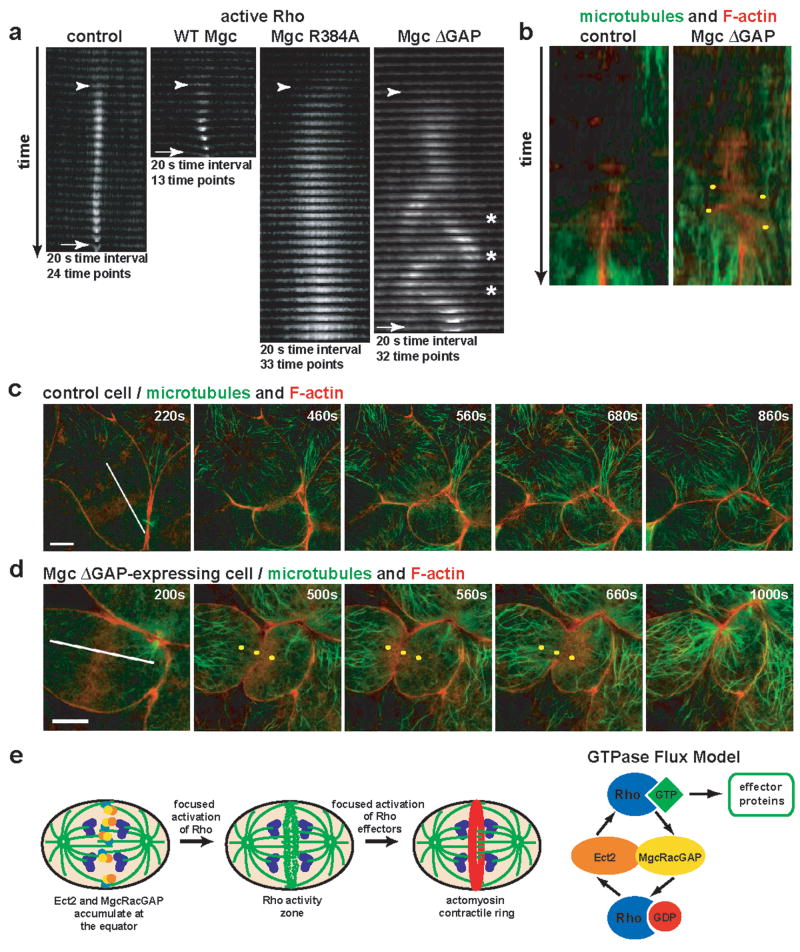
To better characterize the GAP-DEAD mutant phenotypes, Rho activity zones were analyzed in z-stack time-lapse montages (Fig. 4a), which revealed features not apparent from the en face views. In control cells, Rho zones formed and remained tightly focused as furrowing began, while in cells over-expressing WT Mgc the zone was even more tightly focused and furrowed faster. In contrast, the Rho zones in Mgc R384A-expressing cells were very broad, intense, and slow to furrow. In Mgc ΔGAP-expressing cells, the zone was likewise broader and slower to furrow than in controls. As the lateral oscillations began, the Rho zones moved in a wave-like fashion, leaving active Rho behind in its wake, such that at some time points, the zone appeared to split, although one of the zones quickly predominated (Fig. 4a). Two other striking features of the Mgc ΔGAP-expressing Rho zones were apparent from the z kymographs: first, the lateral distance traversed by the oscillating zones remained relatively constant, as if something defined boundaries between which the zone was confined. Second, the average maximum displacement of the oscillating zones (9.3% of the longitudinal diameter) was remarkably similar to the maximum breadth of the zones in R384A-expressing cells (8.2% of the longitudinal diameter, Fig. S5a).
Figure 4. Rho activity zones in MgcRacGAP ΔGAP-expressing cells oscillate.
a, z-stack cross-section kymographs of the region where the Rho activity zone is forming for control, WT Mgc-, Mgc R384A-, or Mgc ΔGAP-expressing cells. Appearance of the Rho activity zone is marked with an arrowhead, and the furrow is marked with an arrow. Asterisks indicate places where the Rho activity zone splits. b, Kymographs made along the lines shown in the first frame of c and d. Yellow dots indicate F-actin oscillations. c–d, Control (c) or Mgc ΔGAP-expressing cells (d) were co-injected with EMTB-3XGFP to monitor microtubules (green) and mChe-UtrCH to monitor F-actin (red) and imaged during cytokinesis. Frames from time-lapse movies are shown. Yellow dots in d are fiduciary marks to help visualize the F-actin oscillations. Scale bars, 20 μm. e, Model showing that equatorial accumulation of Ect2 and MgcRacGAP leads to focused activation of Rho, which in turn leads to focused activation of Rho effectors. Our results are consistent with the GTPase flux model, suggesting that Ect2 locally activates Rho, while MgcRacGAP counterbalances Ect2 by locally inactivating Rho, keeping Rho in a constant state of flux through the GTPase cycle and maintaining a focused Rho activity zone at the cell equator.
The GTPase flux model predicted broader and more intense Rho activity zones in GAP-DEAD mutants, but it did not predict zone instability and oscillation. The oscillations might represent Rho activity moving in association with moving elements of the contractile ring. However, the z kymographs indicated that oscillation started before actomyosin-powered ingression. Further, imaging of Rho and F-actin together showed that active Rho appeared before F-actin accumulation (Fig. S5b), while during oscillations, F-actin signal trailed the Rho signal as the zones moved laterally (Fig. S5c). A second possible explanation is that the oscillations reflect underlying oscillations of the spindle, which controls the Rho zone3. However, direct comparison of F-actin and microtubules showed that in ΔGAP-expressing cells, the microtubules of the mitotic spindle did not oscillate even when F-actin oscillations were apparent (Fig. 4b–d and Movie S7). Thus, spindle movement cannot be responsible for the oscillations.
It thus seems likely that the oscillations reflect a more direct role for MgcRacGAP in Rho activity zone control. We suggest that MgcRacGAP not only mediates Rho GTPase flux but also serves as a transient Rho-GTP anchor. In this view, MgcRacGAP would first bind to Rho-GTP generated by Ect2 and/or other GEFs20. Rho-GTP could then bind to a Rho effector, since the affinity of active Rho GTPases for their effectors is typically greater than for their GAPs21, or if no effectors are locally available, Rho-GTP would eventually be inactivated by the GAP activity of MgcRacGAP. This hypothesis is consistent with the available structural information and binding experiments, which indicate that the R384A substitution would suppress GAP activity without preventing interaction of MgcRacGAP with Rho-GTP22–26. Thus, the R384A mutant would still have the anchoring function but would not contribute to GTPase flux, explaining the progressive accumulation of active Rho. In contrast, the ΔGAP mutant, which completely lacks the GAP domain, would neither contribute to anchoring nor GTPase flux. Consequently, Rho could simply diffuse away from the equator following activation.
Oscillation is indicative of poorly damped feedback; thus, our results suggest that the anchoring function of MgcRacGAP normally protects Rho-GTP from unrestrained feedback. The mechanism of feedback remains to be determined, but our results show that it is spatially constrained in that both the R384A-induced Rho zone spreading and the ΔGAP-induced oscillations occupy remarkably similar maximal lateral dimensions. Because previous work has shown that limited suppression of microtubule polymerization results in broadening of cytokinetic Rho zones3, we suspect that microtubules spatially define the equatorial palette upon which Rho activation persists during cytokinesis. In addition, it was recently shown that positive feedback between the cortex and microtubules is essential for polarization during cytokinesis in monopolar cells27. The authors of that work suggest that this feedback may help focusing the furrow, a notion that is consistent with the results presented here.
While it had been assumed that the GEFs and GAPs regulating cytokinesis work in a sequential manner, our results demonstrate that the GAP activity of MgcRacGAP is necessary throughout cytokinesis in order to maintain a focused zone of Rho activity (Fig. 4e). This is consistent with the recent demonstration that Aurora B kinase, which phosphorylates MgcRacGAP at several sites (see Fig. 1b)9,16,28 and stimulates the Rho GAP activity of MgcRacGAP9, is itself activated in a gradient at the cell equator immediately after the onset of anaphase29. Thus, Rho zones do not simply represent sites of local Rho activation, but instead sites where Rho is being rapidly driven through the entire GTPase cycle. Given the similarities between cytokinesis and other GTPase-dependent processes such as wound healing, morphogenesis, and polarity establishment10, the current results also imply that GTPase flux may be a general feature of processes that depend on dynamic actomyosin arrays.
Methods
Constructs, morpholinos, and antibodies
GFP-rGBD (to observe active Rho), GFP-wGBD (to observe active Cdc42), GFP-pGBD (to observe active Rac), and CA-Rho constructs were described previously18. Xenopus MgcRacGAP constructs were generated using NCBI clone BC070771 (obtained from ATCC) as the template. The R384A point mutation was generated by PCR mutagenesis of the arginine 384 (CGA) codon to alanine (GCA). The Mgc ΔGAP deletion mutation was made by PCR using primers that flanked the splice site of the deleted region, amino acids 362–504. For untagged constructs, MgcRacGAP or the GAP-DEAD mutants were cloned into the pCS2+ vector using Bam HI and Eco RI restriction sites. For C-terminally tagged GFP or 3XGFP tagged constructs, MgcRacGAP or GAP-DEAD mutants were cloned into the pCS2+ C-GFP or C-3XGFP vector using Bam HI and Eco RI restriction sites. GST-MKLP-1 was made by PCR amplification of Xenopus MKLP-1 from NCBI clone BC84928 (obtained from ATCC) and cloning it into pGEX-4T-3 using the Sma I and Not I restriction sites; GST-MKLP-1 was then subcloned into pCS2+ using the Xho I and Xba I restriction sites. The GFP-UtrCH and mChe-UtrCH probes for F-actin were described previously30. EMTB-3XGFP, a probe for microtubules that was described previously, was a gift from Chloë Bulinski31; this probe was subcloned into pCS2+. mRNAs were transcribed in vitro from pCS2+-based vectors using the mMessage mMachine SP6 kit (Ambion). An antisense morpholino (Gene Tools) was made to target the last base of the 5′ UTR and the first 24 bases of the coding sequence (MO.1) with the sequence TAGGTTCATCAGGTTTGTCGCCATC; this morpholino targets both paralogs of Xenopus MgcRacGAP. A morpholino the same as MO.1 but with 5 mispairs (TAGcTTgATCAcGTTTcTCcCCATC) was made as a control. A second morpholino that targets entirely within the 5′ UTR (MO.2) was generated so that knockdown could be rescued by re-expression of mRNAs that have a different 5′ UTR. This morpholino is actually a mixture of two morpholinos, with the sequences GGCACTCTCAGTGACACGCAGCCG and TTCTAATGGCACGGGAAGCCGGAAC, to target the two Xenopus MgcRacGAP paralogs, which have differing 5′ UTRs. A Xenopus-specific polyclonal MgcRacGAP antibody was generated (Panigen) by immunizing rabbits with a synthetic peptide (generated by the University of Wisconsin Biotechnology Center) corresponding to amino acids 608–621 of Xenopus MgcRacGAP. The peptide was coupled to keyhole limpet hemacyanin, and antibodies were purified by peptide affinity chromatography and concentrated using a microcon column (Millipore). Other antibodies used include anti-GFP (clone JL-8, Clontech #632381), anti-GST (Amersham Biosciences #27-4577-01), anti-α-tubulin (DM1A, Sigma #T9026), anti-MKLP-1 (a gift from Todd Stukenberg, University of Virginia), anti-AuroraB (a gift from Johné Liu, University of Ottawa), anti-Plk1, and anti-anillin (both gifts from Aaron Straight, Stanford University).
Xenopus embryos and microinjections
Xenopus laevis females (obtained from NASCO) were induced to ovulate by injecting human chorionic gonadotropin (MP Biomedicals) into the dorsal lymph sac. Eggs were collected manually in a petri dish filled with a solution of 1X Marc’s Modified Ringers (MMR) (100 mM NaCl, 2 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 5 mM Hepes, pH 7.4). Fertilization was performed by removing buffer to 1/10 of the volume, eggs were exposed to testes isolated from Xenopus laevis males, and the petri dish was flooded with water to make a solution of 0.1X MMR. Fertilized eggs were dejellied with 2% cysteine in 1X MMR and washed extensively in 1X then 0.1X MMR. Fertilized eggs were stored in 0.1X MMR at 17°C. Embryos were generally injected at the 2-cell stage with 5 nl of mRNA (needle concentration = 0.5 mg/ml for GFP-rGBD, 0.25 mg/ml for GFP-UtrCH or mChe-UtrCH, 0.125 mg/ml for EMTB-3XGFP, 0.005–0.001 mg/ml for MgcRacGAP expression, 0.5 μg/ml for MgcRacGAP gene replacement); for double injections, the second injection was done at the 4-cell stage (5 nl) or the 8-cell stage (2.5 nl). For morpholino knock-down, both blastomeres at the two-cell stage were injected with 5 nl of 5 mM morpholino to ensure that MgcRacGAP would be knocked down throughout the embryo. Additional methods are available in Supplementary Information.
Immunofluorescence imaging of multinuclearity
Albino embryos were fixed overnight at room temperature 24 h post-injection in 3.7% formaldehyde, 0.25% glutaraldehyde, 0.2% Triton-X-100, and 66 nM Alexa 488-phalloidin (Molecular Probes) in the following buffer (80 mM K-PIPES, 5 mM EGTA, 1 mM MgCl2). Embryos were quenched 1 h at room temperature in PBS + 100 mM sodium borohydride and permeabilized 5 min in PBS + 0.5 % Triton-X-1000. Embryos were then subjected to RNAse treatment: first they were equilibrated in 2X SSC (0.3 M NaCl2, 0.03 M sodium citrate, pH 7.0), then they were incubated in 100 μg/ml DNAse free RNAse in 2X SSC for 20 min at 37°C, and they were washed three times in 2X SSC. Embryos were stained with 5 μM propidium iodide (Molecular Probes) in 2X SSC for 5 min and washed three times with 2X SSC. Embryos were stained with 66 nM Alexa 488-phalloidin in 2X SSC for 4 h on a nutator in the cold room and washed three times with 2X SSC.
Microscopy
Images were collected on either a Zeiss Axiovert 100 M confocal microscope with Bio-Rad 1024 Lasersharp Confocal software (Bio-Rad) equipped with 25X or 63X objectives or an Olympus Fluoview 1000 confocal microscope with FV10-ASW software equipped with 20X or 63X objectives. 4D imaging was performed as described3. Live imaging was generally carried out 3–8 hours post-fertilization (16 cell stage to mid blastula transition).
Western blots for morpholino knock-down
24 h post-injection, 20 embryos for each sample were lysed in 100 μl ice cold PHEME lysis buffer (60 mM K-PIPES, 25 mM Hepes, 10 mM EGTA, 2 mM MgCl2, pH 7.0 supplemented immediately before use with 1% Triton-X-100, 5 μg/ml leupeptin, 1 μg/ml aprotinin, 0.4 mM Pefabloc, 6 μg/ml chymostatin, 0.7 μg/ml pepstatin) by triteration through a P200 pipette tip in a 1.5 ml eppindorf tube. Samples were centrifuged at 14,000 rpm for 5 minutes at 4°C to stratify the yolk proteins. The cytoplasmic layer was transferred to an Ultrafree MC column (Millipore) and centrifuged at 5,000 rpm for 5 min a 4°C. The flow-through was mixed with boiling-hot 6X sample buffer and boiled for 10 min. Samples were loaded on 8% acrylamide gels, electrophoresed, and transferred to nitrocellulose membranes. Membranes were probed with anti-Xenopus MgcRacGAP antibodies (1:5000) or anti-α-tubulin antibodies (1:2500) overnight at 4°C, and appropriate HRP-conjugated antibodies (1:5000; Promega) were incubated with blots for 1.5 h at room temperature. Membranes were developed using an ECL detection kit (Pierce).
Image processing and quantification
Time-lapse images were processed using Volocity (Improvision) and converted to Quicktime movies; still images were processed in Photoshop; quantification was performed using Image J; and statistical analysis was completed in Microsoft Excel. The student’s t-test (2-tailed, 2 sample unequal variance) was used to compare data and to calculate p-values. The percentage of multinuclear cells was counted for multiple embryos of each class. Average Rho zone breadth for multiple cells was measured a three locations along the Rho zone for a given cell and averaged, while longitudinal diameter at the widest point perpendicular to the Rho zone was measured. Intensity measurements were made at three locations within the Rho zone and three locations outside the Rho zone and averaged.
Supplementary Material
Figure S1. Morpholino target sites and knockdown characterization and Western blots showing gene replacement. a, Domain diagram of MgcRacGAP showing the regions that the morpholinos MO.1, MO.2, and 5 mispair MO target. The region that the Xenopus-specific MgcRacGAP antibody recognizes is also noted. b, Xenopus embryos were injected with MO.1, Mgc 5 mispair MO, or water as a control. Embryos were fixed at 24 h post-injection and stained with propidium iodide to reveal the nuclei (red) and Alexa 488-phalloidin to visualize F-actin (green). Scale bar, 20 μm. c, The percent of multinucleate cells was quantified for at least two independent experiments. Mean ± SE, n ≥ 6 embryos, *p < 0.05, ***p < 0.005. d, Western blot of uninjected and WT Mgc mRNA-injected Xenopus oocyte lysates showing that the Xenopus MgcRacGAP antibody recognizes a specific band of ~69 kD. e, Western blot showing a time course for MgcRacGAP depeletion by the two morpholinos. MO.1 results in greater depletion of MgcRacGAP than MO.2 at the 6 h time point, although by later time points, the difference between the two is increasingly harder to detect, particularly when compared to the level of protein in uninjected cells. Corresponding Western blot of tubulin levels is shown as a loading control, f-h, Western blots showing morpholino knock-down of MgcRacGAP protein levels by MO.2 and gene replacement with WT Mgc (f) Mgc R384A (g), or Mgc Δ GAP (h) in Xenopus embryo lysates harvested 24 h post-injection.
Figure S2. Evidence that MgcRacGAP and its GAP-DEAD mutants interact with the kinesin MKLP and characterization of midzone microtubules and midzone proteins in gene replacement cells, a, Western blot showing that WT Mgc, Mgc R384A, and Mgc ΔGAP can all co-immunoprecipitate MKLP-1 from Xenopus oocyte lysates, suggesting that the centralspindlin complex in Xenopus functions similarly to that in other systems and that the GAP-DEAD mutations don’t compromise MgcRacGAP’s ability to interact with MKLP-1. b, Staining for microtubules in uninjected embryos or embryos where endogenous MgcRacGAP was knocked down with MO.2 and replaced with WT Mgc, Mgc R384A, or Mgc ΔGAP shows that midzone microtubules look similar in controls and GAP-DEAD cells. Scale bars, 20 μm. c, Staining for the cytokinesis regulators MKLP-1, anillin, Plk1, and AuroraB demonstates that their midzone localization early (anaphase) and late (telophase) in cytokinesis is the same in embryos where endogenous MgcRacGAP was replaced by WT Mgc or by GAP-DEAD MgcRacGAP. Scale bars, 20 μm.
Figure S3. Intensity quantification of Rho activity inside and outside of zones and plot of Rho activity zone width vs. blastomere longitudinal diameter, a, Cartoon showing the three stages of cytokinesis where intensity measurements were made. b, The mean intensity for Rho activity in zones was measured for control, WT Mgc-, R384A-, or ΔGAP-expressing cells and for R384A and ΔGAP gene replacement cells by averaging three locations along the Rho zone for each cell counted. c, The mean intensity of Rho activity outside of zones was measured by averaging three locations outside the Rho zone (approximately two Rho zone widths away from the zone) for each cell counted. Mean ± SE; control, n = 9 embryos, WT Mgc, n = 5, R384A, n= 5, ΔGAP, n = 8, Mgc MO.2 + R384A, n = 7, Mgc MO.2 + ΔGAP; *p < 0.05; **p < 0.01; ***p < 0.005. d, Plot of Rho activity zone width versus longitudinal diameter showing that Rho zone width increases with increasing blastomere size, becoming particularly broad in cells expressing the GAP-DEAD mutants.
Figure S4. Cytokinetic dynamics of active Rac and Cdc42. Frames from 4D movies show the localization of active Rac (a) and active Cdc42 (b) in control cells and cells expressing the GAP-DEAD mutant R384A. In all cases, active Rac and Cdc42 localize to cell-cell boundaries (indicated by asterisks at the 0s time point). Regions of ingressing furrows are indicated by arrows. Note that in contrast to the results with the probe for active Rho, virtually no active Rac or Cdc42 is visible along the top of the cell prior to or during furrow ingression, making the furrow hard to detect except when the sides of the cell are obviously pinching in. This reflects the fact that there is no significant surface signal from these reporters during cytokinetic apparatus assembly. Scale bars, 40 μm.
Figure S5. Additional characterization of the Mgc ΔGAP-expressing oscillations, a, The width of Rho activity zones in Mgc R384A-expressing cells versus cells size was compared with the maximum displacement of the Rho activity zone oscillation in the Mgc ΔGAP-expressing cells. In both cases, the width increases relative to longitudinal diameter, and the best-fit lines for the data are nearly identical, b and c, control (b) or Mgc ΔGAP-expressing embryos (c) were co-injected with GFP-rGBD to monitor active Rho (green) and mChe-UtrCH to monitor F-actin (red). The active Rho signal precedes the appearance of the F-actin signal, and in the case of Mgc ΔGAP-expressing cells, the F-actin signal trails just behind the active Rho signal as the zone moves laterally. Signal intensity line scans of active Rho and F-actin at two timepoints show that active Rho and F-actin are highly colocalized in control cells, but F-actin signal trails active Rho in ΔGAP-expressing cells (notice position of peaks and leading shoulders in active Rho line scans marked by asterisks). Scale bars, 40 μm.
Acknowledgments
We thank George von Dassow and Julie Canman for helpful discussions; Chloë Bulinski for the EMTB-3XGFP construct; Todd Stukenberg, Johné Liu, and Aaron Straight for kindly giving us antibodies; John Wallingford and Esther Kieserman for advice on the midzone staining procedure; and members of the Bement Lab for advice and critical reading of the manuscript. A.L.M. is supported by postdoctoral fellowships from the American Cancer Society and the Helen Hay Whitney Foundation. W.M.B. is supported by an NIH grant.
Footnotes
Note: Supplementary information is available on the Nature Cell Biology website.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Kishi K, Sasaki T, Kuroda S, Itoh T, Takai Y. Regulation of cytoplasmic division of Xenopus embryo by rho p21 and its inhibitory GDP/GTP exchange protein (rho GDI) J Cell Biol. 1993;120:1187–95. doi: 10.1083/jcb.120.5.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drechsel DN, Hyman AA, Hall A, Glotzer M. A requirement for Rho and Cdc42 during cytokinesis in Xenopus embryos. Curr Biol. 1997;7:12–23. doi: 10.1016/s0960-9822(06)00023-6. [DOI] [PubMed] [Google Scholar]
- 3.Bement WM, Benink HA, von Dassow G. A microtubule-dependent zone of active RhoA during cleavage plane specification. J Cell Biol. 2005;170:91–101. doi: 10.1083/jcb.200501131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirose K, Kawashima T, Iwamoto I, Nosaka T, Kitamura T. MgcRacGAP is involved in cytokinesis through associating with mitotic spindle and midbody. J Biol Chem. 2001;276:5821–8. doi: 10.1074/jbc.M007252200. [DOI] [PubMed] [Google Scholar]
- 5.Jantsch-Plunger V, et al. CYK-4: A Rho family gtpase activating protein (GAP) required for central spindle formation and cytokinesis. J Cell Biol. 2000;149:1391–404. doi: 10.1083/jcb.149.7.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mishima M, Kaitna S, Glotzer M. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev Cell. 2002;2:41–54. doi: 10.1016/s1534-5807(01)00110-1. [DOI] [PubMed] [Google Scholar]
- 7.Yuce O, Piekny A, Glotzer M. An ECT2-centralspindlin complex regulates the localization and function of RhoA. J Cell Biol. 2005;170:571–82. doi: 10.1083/jcb.200501097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Somers WG, Saint R. A RhoGEF and Rho family GTPase-activating protein complex links the contractile ring to cortical microtubules at the onset of cytokinesis. Dev Cell. 2003;4:29–39. doi: 10.1016/s1534-5807(02)00402-1. [DOI] [PubMed] [Google Scholar]
- 9.Minoshima Y, et al. Phosphorylation by aurora B converts MgcRacGAP to a RhoGAP during cytokinesis. Dev Cell. 2003;4:549–60. doi: 10.1016/s1534-5807(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 10.Bement WM, Miller AL, von Dassow G. Rho GTPase activity zones and transient contractile arrays. Bioessays. 2006;28:983–93. doi: 10.1002/bies.20477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zavortink M, Contreras N, Addy T, Bejsovec A, Saint R. Tum/RacGAP50C provides a critical link between anaphase microtubules and the assembly of the contractile ring in Drosophila melanogaster. J Cell Sci. 2005;118:5381–92. doi: 10.1242/jcs.02652. [DOI] [PubMed] [Google Scholar]
- 12.Lee JS, Kamijo K, Ohara N, Kitamura T, Miki T. MgcRacGAP regulates cortical activity through RhoA during cytokinesis. Exp Cell Res. 2004;293:275–82. doi: 10.1016/j.yexcr.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 13.Yamada T, Hikida M, Kurosaki T. Regulation of cytokinesis by mgcRacGAP in B lymphocytes is independent of GAP activity. Exp Cell Res. 2006;312:3517–25. doi: 10.1016/j.yexcr.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 14.Goldstein AY, Jan YN, Luo L. Function and regulation of Tumbleweed (RacGAP50C) in neuroblast proliferation and neuronal morphogenesis. Proc Natl Acad Sci U S A. 2005;102:3834–9. doi: 10.1073/pnas.0500748102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gregory SL, et al. Cell division requires a direct link between microtubule-bound RacGAP and Anillin in the contractile ring. Curr Biol. 2008;18:25–9. doi: 10.1016/j.cub.2007.11.050. [DOI] [PubMed] [Google Scholar]
- 16.Ban R, Irino Y, Fukami K, Tanaka H. Human mitotic spindle-associated protein PRC1 inhibits MgcRacGAP activity toward Cdc42 during the metaphase. J Biol Chem. 2004;279:16394–402. doi: 10.1074/jbc.M313257200. [DOI] [PubMed] [Google Scholar]
- 17.Verbrugghe KJ, White JG. SPD-1 is required for the formation of the spindle midzone but is not essential for the completion of cytokinesis in C. elegans embryos. Curr Biol. 2004;14:1755–60. doi: 10.1016/j.cub.2004.09.055. [DOI] [PubMed] [Google Scholar]
- 18.Benink HA, Bement WM. Concentric zones of active RhoA and Cdc42 around single cell wounds. J Cell Biol. 2005;168:429–39. doi: 10.1083/jcb.200411109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danilchik MV, Brown EE. Membrane dynamics of cleavage furrow closure in Xenopus laevis. Dev Dyn. 2008;237:565–79. doi: 10.1002/dvdy.21442. [DOI] [PubMed] [Google Scholar]
- 20.Birkenfeld J, et al. GEF-H1 modulates localized RhoA activation during cytokinesis under the control of mitotic kinases. Dev Cell. 2007;12:699–712. doi: 10.1016/j.devcel.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang B, Chernoff J, Zheng Y. Interaction of Rac1 with GTPase-activating proteins and putative effectors. A comparison with Cdc42 and RhoA. J Biol Chem. 1998;273:8776–82. doi: 10.1074/jbc.273.15.8776. [DOI] [PubMed] [Google Scholar]
- 22.Cho YJ, et al. Abr and Bcr, two homologous Rac GTPase-activating proteins, control multiple cellular functions of murine macrophages. Mol Cell Biol. 2007;27:899–911. doi: 10.1128/MCB.00756-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu Y, Galan JE. A salmonella protein antagonizes Rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion. Nature. 1999;401:293–7. doi: 10.1038/45829. [DOI] [PubMed] [Google Scholar]
- 24.Leonard DA, Lin R, Cerione RA, Manor D. Biochemical studies of the mechanism of action of the Cdc42-GTPase-activating protein. J Biol Chem. 1998;273:16210–5. doi: 10.1074/jbc.273.26.16210. [DOI] [PubMed] [Google Scholar]
- 25.Rittinger K, et al. Crystal structure of a small G protein in complex with the GTPase-activating protein rhoGAP. Nature. 1997;388:693–7. doi: 10.1038/41805. [DOI] [PubMed] [Google Scholar]
- 26.Rittinger K, Walker PA, Eccleston JF, Smerdon SJ, Gamblin SJ. Structure at 1.65 A of RhoA and its GTPase-activating protein in complex with a transition-state analogue. Nature. 1997;389:758–62. doi: 10.1038/39651. [DOI] [PubMed] [Google Scholar]
- 27.Hu CK, Coughlin M, Field CM, Mitchison TJ. Cell polarization during monopolar cytokinesis. J Cell Biol. 2008;181:195–202. doi: 10.1083/jcb.200711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toure A, et al. Phosphoregulation of MgcRacGAP in mitosis involves Aurora B and Cdk1 protein kinases and the PP2A phosphatase. FEBS Lett. 2008;582:1182–8. doi: 10.1016/j.febslet.2007.12.036. [DOI] [PubMed] [Google Scholar]
- 29.Fuller BG, et al. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 2008 doi: 10.1038/nature06923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burkel BM, von Dassow G, Bement WM. Versatile fluorescent probes for actin filaments based on the actin-binding domain of utrophin. Cell Motil Cytoskeleton. 2007;64:822–32. doi: 10.1002/cm.20226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faire K, et al. E-MAP-115 (ensconsin) associates dynamically with microtubules in vivo and is not a physiological modulator of microtubule dynamics. J Cell Sci. 1999;112 (Pt 23):4243–55. doi: 10.1242/jcs.112.23.4243. [DOI] [PubMed] [Google Scholar]
- 32.Kieserman EK, Glotzer M, Wallingford JB. Developmental regulation of central spindle assembly and cytokinesis during vertebrate embryogenesis. Curr Biol. 2008;18:116–23. doi: 10.1016/j.cub.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 33.Woolner S, O’Brien LL, Wiese C, Bement WM. Myosin-10 and actin filaments are essential for mitotic spindle function. J Cell Biol. 2008;182:77–88. doi: 10.1083/jcb.200804062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Morpholino target sites and knockdown characterization and Western blots showing gene replacement. a, Domain diagram of MgcRacGAP showing the regions that the morpholinos MO.1, MO.2, and 5 mispair MO target. The region that the Xenopus-specific MgcRacGAP antibody recognizes is also noted. b, Xenopus embryos were injected with MO.1, Mgc 5 mispair MO, or water as a control. Embryos were fixed at 24 h post-injection and stained with propidium iodide to reveal the nuclei (red) and Alexa 488-phalloidin to visualize F-actin (green). Scale bar, 20 μm. c, The percent of multinucleate cells was quantified for at least two independent experiments. Mean ± SE, n ≥ 6 embryos, *p < 0.05, ***p < 0.005. d, Western blot of uninjected and WT Mgc mRNA-injected Xenopus oocyte lysates showing that the Xenopus MgcRacGAP antibody recognizes a specific band of ~69 kD. e, Western blot showing a time course for MgcRacGAP depeletion by the two morpholinos. MO.1 results in greater depletion of MgcRacGAP than MO.2 at the 6 h time point, although by later time points, the difference between the two is increasingly harder to detect, particularly when compared to the level of protein in uninjected cells. Corresponding Western blot of tubulin levels is shown as a loading control, f-h, Western blots showing morpholino knock-down of MgcRacGAP protein levels by MO.2 and gene replacement with WT Mgc (f) Mgc R384A (g), or Mgc Δ GAP (h) in Xenopus embryo lysates harvested 24 h post-injection.
Figure S2. Evidence that MgcRacGAP and its GAP-DEAD mutants interact with the kinesin MKLP and characterization of midzone microtubules and midzone proteins in gene replacement cells, a, Western blot showing that WT Mgc, Mgc R384A, and Mgc ΔGAP can all co-immunoprecipitate MKLP-1 from Xenopus oocyte lysates, suggesting that the centralspindlin complex in Xenopus functions similarly to that in other systems and that the GAP-DEAD mutations don’t compromise MgcRacGAP’s ability to interact with MKLP-1. b, Staining for microtubules in uninjected embryos or embryos where endogenous MgcRacGAP was knocked down with MO.2 and replaced with WT Mgc, Mgc R384A, or Mgc ΔGAP shows that midzone microtubules look similar in controls and GAP-DEAD cells. Scale bars, 20 μm. c, Staining for the cytokinesis regulators MKLP-1, anillin, Plk1, and AuroraB demonstates that their midzone localization early (anaphase) and late (telophase) in cytokinesis is the same in embryos where endogenous MgcRacGAP was replaced by WT Mgc or by GAP-DEAD MgcRacGAP. Scale bars, 20 μm.
Figure S3. Intensity quantification of Rho activity inside and outside of zones and plot of Rho activity zone width vs. blastomere longitudinal diameter, a, Cartoon showing the three stages of cytokinesis where intensity measurements were made. b, The mean intensity for Rho activity in zones was measured for control, WT Mgc-, R384A-, or ΔGAP-expressing cells and for R384A and ΔGAP gene replacement cells by averaging three locations along the Rho zone for each cell counted. c, The mean intensity of Rho activity outside of zones was measured by averaging three locations outside the Rho zone (approximately two Rho zone widths away from the zone) for each cell counted. Mean ± SE; control, n = 9 embryos, WT Mgc, n = 5, R384A, n= 5, ΔGAP, n = 8, Mgc MO.2 + R384A, n = 7, Mgc MO.2 + ΔGAP; *p < 0.05; **p < 0.01; ***p < 0.005. d, Plot of Rho activity zone width versus longitudinal diameter showing that Rho zone width increases with increasing blastomere size, becoming particularly broad in cells expressing the GAP-DEAD mutants.
Figure S4. Cytokinetic dynamics of active Rac and Cdc42. Frames from 4D movies show the localization of active Rac (a) and active Cdc42 (b) in control cells and cells expressing the GAP-DEAD mutant R384A. In all cases, active Rac and Cdc42 localize to cell-cell boundaries (indicated by asterisks at the 0s time point). Regions of ingressing furrows are indicated by arrows. Note that in contrast to the results with the probe for active Rho, virtually no active Rac or Cdc42 is visible along the top of the cell prior to or during furrow ingression, making the furrow hard to detect except when the sides of the cell are obviously pinching in. This reflects the fact that there is no significant surface signal from these reporters during cytokinetic apparatus assembly. Scale bars, 40 μm.
Figure S5. Additional characterization of the Mgc ΔGAP-expressing oscillations, a, The width of Rho activity zones in Mgc R384A-expressing cells versus cells size was compared with the maximum displacement of the Rho activity zone oscillation in the Mgc ΔGAP-expressing cells. In both cases, the width increases relative to longitudinal diameter, and the best-fit lines for the data are nearly identical, b and c, control (b) or Mgc ΔGAP-expressing embryos (c) were co-injected with GFP-rGBD to monitor active Rho (green) and mChe-UtrCH to monitor F-actin (red). The active Rho signal precedes the appearance of the F-actin signal, and in the case of Mgc ΔGAP-expressing cells, the F-actin signal trails just behind the active Rho signal as the zone moves laterally. Signal intensity line scans of active Rho and F-actin at two timepoints show that active Rho and F-actin are highly colocalized in control cells, but F-actin signal trails active Rho in ΔGAP-expressing cells (notice position of peaks and leading shoulders in active Rho line scans marked by asterisks). Scale bars, 40 μm.