Summary
Mis16 and Mis18 are subunits of a protein complex required for incorporation of the histone H3 variant CenH3 (Cnp1/CENP-A) into centromeric chromatin in Schizosaccharomyces pombe and mammals. How the Mis16-Mis18 complex performs this function is unknown. Here we report that the Mis16-Mis18 complex is required for centromere localization of Scm3Sp, a Cnp1-binding protein related to Saccharomyces cerevisiae Scm3. Scm3Sp is required for centromeric localization of Cnp1, whilst Scm3Sp localizes at centromeres independently of Cnp1. Like the Mis16-Mis18 complex but unlike Cnp1, Scm3Sp dissociates from centromeres during mitosis. Inactivation of Scm3Sp or Mis18 increases centromere localization of histones H3 and H2A/H2B, which are largely absent from centromeres in wild type cells. Whereas S. cerevisiae Scm3 is proposed to replace histone H2A/H2B in centromeric nucleosomes, the dynamic behavior of S. pombe Scm3 suggests that it acts as a Cnp1 assembly/maintenance factor that directly mediates the stable deposition of Cnp1 into centromeric chromatin.
Introduction
Centromeres are genomic loci that specify the sites for kinetochore assembly and microtubule attachment. They are essential for proper chromosome segregation during mitosis and meiosis. Centromeric DNA sequences vary greatly in size and composition among species, but they all share a specific chromatin structure in which canonical histone H3 is replaced by a histone variant known as CENP-A in mammals, Cse4 in Saccharomyces cerevisiae, and Cnp1 in Schizosaccharomyces pombe (Henikoff and Dalal, 2005; Morris and Moazed, 2007). Generically known as CenH3, these proteins contain a conserved histone fold domain (HFD) and an N-terminus that is variable in size and sequence (Smith, 2002). CenH3 provides an epigenetic mark for the centromere that is maintained during DNA replication and nuclear division. Deposition of CenH3 occurs during the S and G2 phases of the cell cycle in S. pombe (Takahashi et al., 2000; 2005), during anaphase in Drosophila embryos (Schuh et al., 2007) and during G1 in human cells (Jansen et al., 2007).
Studies of S. cerevisiae have been instrumental in characterizing centromere structure and function. Budding yeast has “point” centromeres consisting of ∼125 bp of DNA with a single Cse4-containing nucleosome. Centromere assembly requires binding of Ndc10 to a conserved DNA element (CDEIII) (Espelin et al., 1997; Espelin et al., 2003). Ndc10 does not associate with Cse4 but it does interact with Scm3 (Camahort et al., 2007; Mizuguchi et al., 2007; Stoler et al., 2007), a non-histone-like protein first identified as a high copy suppressor of a hypomorphic cse4 mutation (Chen et al., 2000). Scm3 binds to Cse4 and displaces H2A and H2B from H2A/H2B/Cse4/H4 octamers assembled in vitro (Mizuguchi et al., 2007). Coupled with the absence of H2A and H2B from the centromeric region, these observations suggest that Ndc10 establishes the centromeric nucleosome through its interaction with Scm3, a core subunit of an unusual hexameric nucleosome having two subunits each of Scm3, Cse4 and histone H4 (Mizuguchi et al., 2007).
The structure and biology of the point centromeres in budding yeast are quite different from the primary sequence-independent “regional” centromeres found in S. pombe and mammals. Fission yeast has ∼30-120 kb centromeric regions consisting of repetitive DNA elements that direct assembly of heterochromatin and subsequent transcriptional silencing. The centromeres consist of an inner central region (cnt and imr) that is comprised of the unique Cnp1-containing chromatin surrounded by highly repetitive sequences at the outer region (otr) (Takahashi et al., 1992). Cnp1 localization at centromeres requires a large number of proteins, none of which are related to Ndc10 (Hayashi et al., 2004). Central among the Cnp1 loading factors are Mis16 and Mis18, which appear to act as a complex that maintains the proper level of histone acetylation at the central region of the centromere, a condition that may be required for proper loading of Cnp1 (Hayashi et al., 2004; Fujita et al., 2007). Another critical factor is the Mis6 complex consisting of Mis6, Mis15, Mis17 and Sim4 (Hayashi et al., 2004; Pidoux et al., 2003). Both complexes localize to the central region of centromeres (Goshima et al., 1999; Hayashi et al., 2004; Pidoux et al., 2003; Saitoh et al., 1997). Although the recently described NASP-related Sim3 protein does not localize to centromeres, it interacts with Cnp1 and is required for its deposition at centromeres (Dunleavy et al., 2007). Apart from Sim3 there are no protein-protein interactions between any of these factors and Cnp1, hence the mechanism by which Cnp1 is deposited and maintained specifically at centromeres is unknown.
Many of the factors required for Cnp1 localization at centromeres in fission yeast are conserved in humans. The human homologs of Mis6 and Sim4 are CENP-I and CENP-H, respectively, both of which are constitutive centromere proteins (Saitoh et al., 1997; Sugata et al., 2000). The human homologs of Mis16 and Mis18 are RbAp46/RbAp48 and hMis18 (α and β isoforms), respectively (Hayashi et al., 2004; Fujita et al., 2007). These proteins are required for CENP-A localization at centromeres in human cells, and in Drosophila cells RbAp48 has been detected in a complex with CenH3 and histone H4 (Furuyama et al., 2006). RbAp46/48 is a component of the multi-subunit CAF-1 chromatin assembly complex (Verreault et al., 1996). Human RbAp46/48 and S. pombe Mis16 have a pan-chromosomal localization pattern although Mis16 is most highly concentrated at centromeres (Hayashi et al., 2004). This localization contrasts with Mis18 in S. pombe and humans, which is specifically found at centromeres. Significantly, Mis18 has a dynamic localization pattern in fission yeast and humans, delocalizing from centromeres in metaphase and reappearing there in telophase (Fujita et al., 2007). The kinetochore foci formed by S. pombe Mis16 have a similar cell cycle-regulated pattern of appearance (Hayashi et al., 2004). The delocalization of Mis16 and Mis18 from centromeres during mitosis is consistent with models in which the complex primes the centromere for Cnp1 deposition in the following cell cycle (Fujita et al., 2007), although Mis16-Mis18 may also be required for stable maintenance of Cnp1-containing nucleosomes at centromeres throughout interphase.
Histone chaperones control the deposition of histones into chromatin to prevent non-specific aggregation and regulate their incorporation into DNA. Well-characterized examples include Asf1, CAF-1, Nap-1, and HIRA (Loyola and Almouzni, 2004). Some chaperones show selectivity for specific histone variants, a specificity that is often dependent on the timing of expression and incorporation of the specific histone isoform. For example, although mammalian H3.3 is constitutively expressed, it is incorporated into DNA at select loci in a replication-independent manner by the HIRA histone chaperone (Tagami et al., 2004). Such histone variants are deposited into specific genomic sites in order to regulate processes that include transcription and DNA repair (De Koning et al., 2007). The only known chaperone for CenH3 is RbAp48 (S. pombe Mis16), which mediates the assembly of H2A/H2B/CenH3/H4 complexes using purified components isolated from Drosophila (Furuyama et al., 2006) and is required for CenH3 centromeric localization in both S. pombe and humans (Hayashi et al., 2004).
Whilst progress has been made in identifying the proteins required for stable incorporation of Cnp1 into centromeric chromatin, still missing is a factor that both specifically localizes at centromeres and directly interacts with Cnp1. Here we describe a fission yeast ortholog of Scm3 as such a factor. Whereas budding yeast Scm3 is proposed to be a constitutive subunit of the centromeric nucleosome, our results together with data reported by Pidoux et al. (2008) show that fission yeast Scm3 associates dynamically with centromeres in a cell cycle-regulated manner similar to that of Mis16-Mis18 complex. Together, these studies support a model in which Mis16-Mis18 complex controls the localization of Scm3 at centromeres, which in turn mediates the stable deposition of Cnp1.
Results
Scm3 Interacts with Cnp1 in Fission Yeast
Since centromere structure and CenH3 localization mechanisms have diverged rapidly between S. cerevisiae and S. pombe, we decided to explore the function of an uncharacterized Scm3-related gene in fission yeast. SPAPB1A10.02 encodes a 336 amino acid protein that has an ∼50 amino acid region of sequence similarity that is shared with S. cerevisiae Scm3 and uncharacterized genes in other fungi (Figure 1A). The conserved N-terminal domains of these genes are followed by C-terminal regions that vary greatly in size and sequence (Aravind et al., 2007). We first investigated whether S. pombe Scm3 interacts with Cnp1 using a yeast two-hybrid (Y2H) assay (Figure 1B). Interactions in both bait and prey orientations were observed, while no interaction was detected between Scm3 and histone H4 (Figure S1). Truncation analysis revealed that the first 194 amino acids of Scm3 (including the homology domain) are required for the interaction with Cnp1 (Figure S2). The corresponding region of budding yeast Scm3 interacts with Cse4 (Mizuguchi et al., 2007). The Y2H analysis also detected a robust Scm3-Scm3 interaction (Figure 1B), indicating that Scm3 self-associates in vivo.
Figure 1. Fission Yeast Scm3 is a Conserved Fungal Protein that Physically Interacts and Co-localizes with Cnp1 at the Central Core of the Centromere.
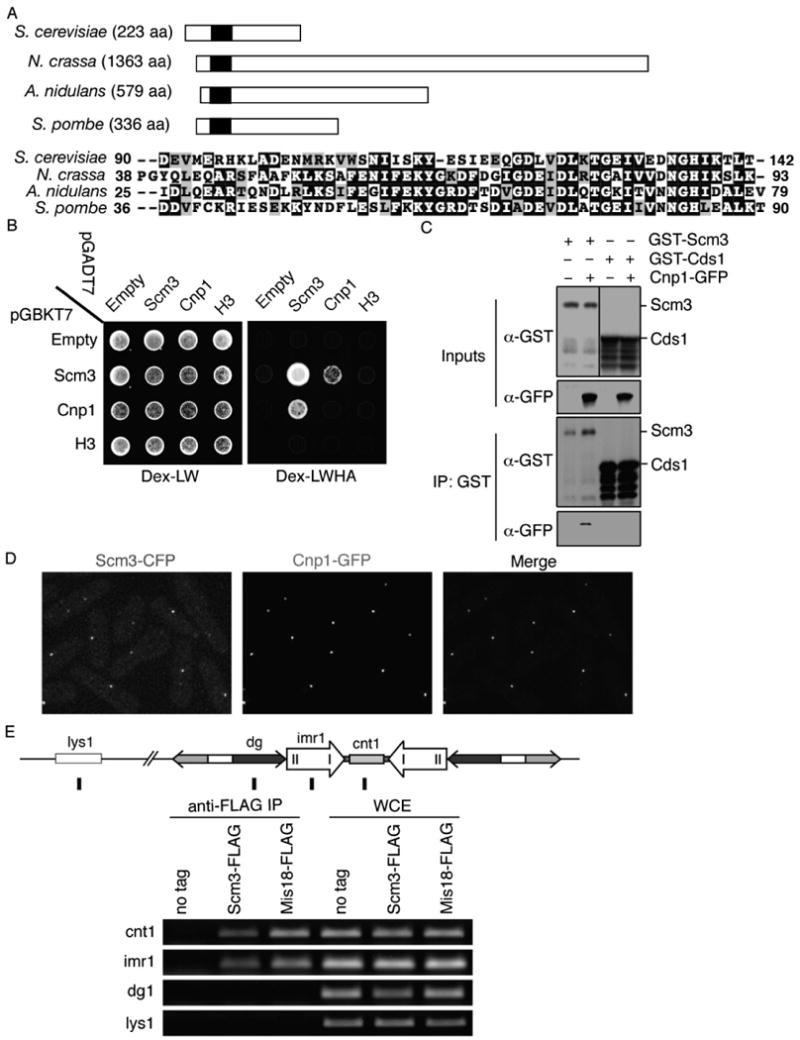
A. A schematic of Scm3 from selected fungal species showing the area of homology in the N-terminus consisting of approximately 50 amino acids. Alignment of this domain in S. pombe Scm3 and its homologs in A. nidulans, N. crassa, and S. cerevisiae is presented. Black background indicates identical residues, and light shading indicates similar residues.
B. Yeast two-hybrid interactions showing that Scm3 self-interacts and interacts with Cnp1.
C. Co-immunoprecipitation of GST-Scm3 with Cnp1-GFP. Extracts of cells expressing Cnp1-GFP and nmt1-GST-Scm3 or nmt1-GST-Cds11-190 (Boddy et al., 2000) were immunoprecipitated using glutathione sepharose. GFP-Cnp1 co-immunoprecipitated with GST-Scm3 but not with GST-Cds11-190.
D. Scm3 co-localizes with Cnp1. Live cells expressing both Scm3-CFP (red) and Cnp1-GFP (green) were photographed. Images were merged in the right panel.
E. ChIP analysis was performed using extracts expressing Scm3 tagged with the Flag epitope. The position of centromere probes cnt1, imr1, dg and euchromatic lys1 are indicated. Mis18-FLAG was used as a positive control.
The Scm3-Cnp1 Y2H interaction was confirmed by co-immunoprecipitation analysis. This experiment required over-expression of Scm3 because it was difficult to detect by immunoblotting when expressed at its endogenous level (Figure S3). GST-Scm3 and a negative control construct (GST-Cds1FHA) were produced in a strain in which Cnp1-GFP was expressed from the cnp1+ locus (Takahashi et al., 2000). Immunoblotting showed that only GST-Scm3 complexes contained Cnp1-GFP (Figure 1C). From these data we conclude that S. pombe Scm3 associates with Cnp1, indicating conserved functions for S. pombe and S. cerevisiae Scm3 proteins.
Scm3 Localizes to the Central Region of Centromeres
We next used microscopy and chromatin immunoprecipitation (ChIP) analysis to determine whether Scm3 localizes at centromeres. For the microscopy studies, Scm3-CFP (cyan fluorescent protein) was expressed from the scm3+ locus in cells that also expressed Cnp1-GFP. A single nuclear Scm3-CFP focus was observed in the majority of live cells (Figure 1D). This localization pattern is typical of clustered centromeres in fission yeast. The Scm3-CFP and Cnp1-GFP foci colocalized (Figure 1D). As described below, we also observed colocalization of Scm3 with Mis12 (Figure 6A), confirming that Scm3 localizes at centromeres.
Figure 6. Scm3 Dissociates from Centromeres During Mitosis.
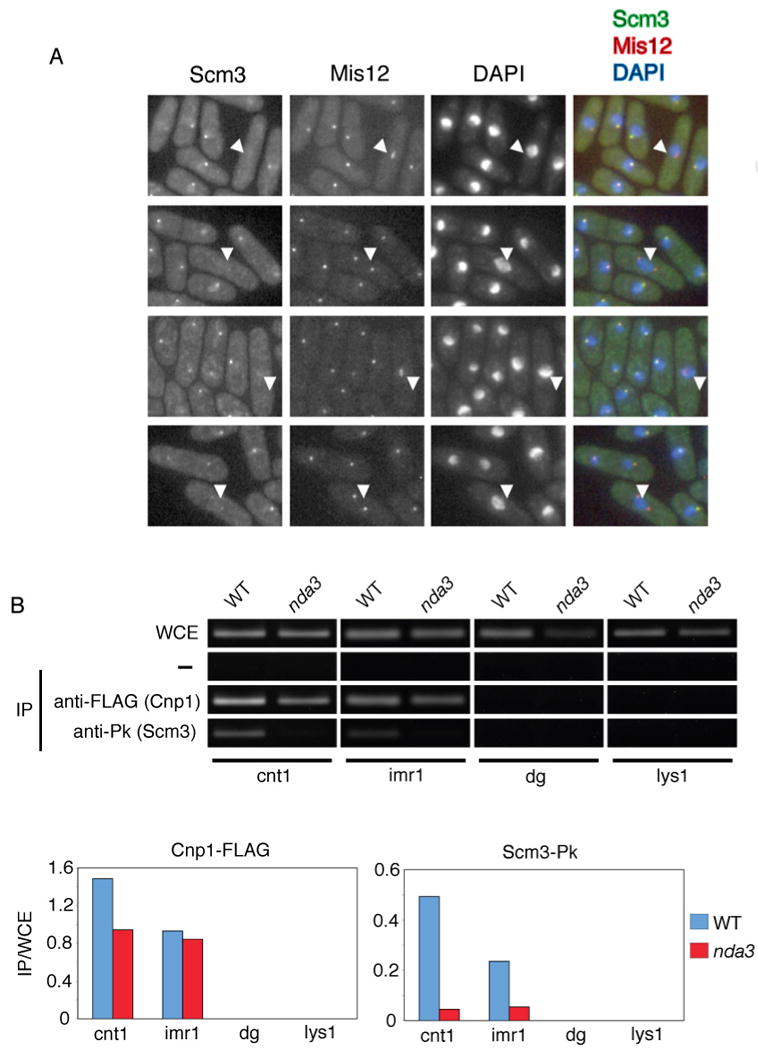
A. Scm3 is localized to centromeres during interphase, but not in mitosis. Colocalization of GFP-tagged Scm3 (green) and RFP-tagged Mis12 (red) was performed using cells grown at 20°C and fixed in methanol. The merged image is on the right. Representative images are displayed. White arrows indicate cells undergoing mitosis in which the nuclear dot corresponding to Scm3 has been lost but the dot for Mis12 persists.
B. Scm3 dissociates from centromeric chromatin during mitosis. ChIP assays were performed using wild type or nda3-KM311 cells expressing FLAG-tagged Cnp1 and Pk-tagged Scm3. Cells were grown at 20°C (restrictive temperature for the cold-sensitive nda3-KM311 mutation) for 8 h prior to harvesting. ChIP quantitation is provided below.
ChIP was performed using DNA probes corresponding to the different regions of the centromere to identify specific sites of Scm3 enrichment. Probes corresponding to the central region of centromere I (cnt1 and imr1) and the dg motifs of the outer regions were utilized (Takahashi et al., 1992; Figure 1E). Chromosomally integrated Scm3-FLAG was immunoprecipitated and DNA amplified using the indicated DNA probes, with Mis18-FLAG serving as a positive control (Fujita et al., 2007). The Scm3-FLAG and Mis18-FLAG immunoprecipitates were enriched at cnt1/imr1 but not at the dg motifs or the euchromatic lys1 probe (Figure 1E). This pattern mirrors that of other centromere-associated proteins including Cnp1, Mis6 and Mis16 (Hayashi et al., 2004; Saitoh et al., 1997). Taken together, these data demonstrate that Scm3 colocalizes with Cnp1, Mis12 and Mis18 at the central region of the centromere.
Scm3 is Required for Proper Chromosome Segregation
To address whether Scm3 is essential for viability in S. pombe, we generated heterozygous diploids in which one copy of scm3+ was replaced with the KanMX6 marker. Sporulation and dissection of tetrads showed that scm3Δ spores germinated and underwent 1 or 2 divisions before arresting growth (Figure 2A).
Figure 2. Fission Yeast Scm3 is Essential for Centromere Function.
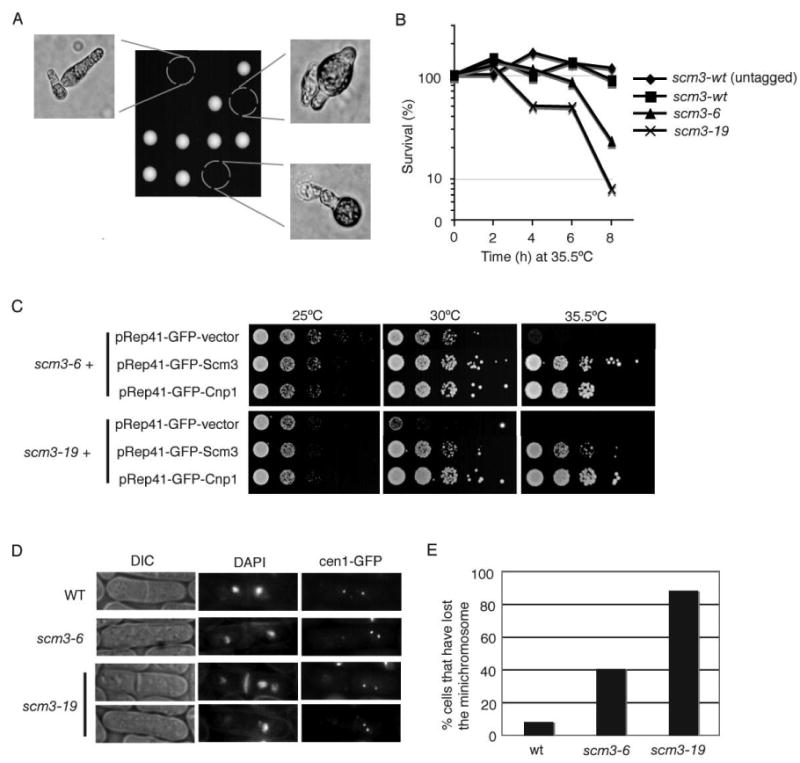
A. Scm3 is an essential protein. Tetrad analysis of an scm3+/scm3::kanMX6 diploid revealed that scm3::kanMX6 spores germinate but do not proliferate. All viable spores are Kan-.
B. Loss of viability of scm3-ts mutants at high temperature. The scm3-6 and scm3-19 mutants were grown at 25°C and then shifted to 35.5°C for the indicated periods of time. Cells were plated onto YES agar and colonies were counted following incubation at 25°C for 3 days. The untagged control strain is PR110.
C. The scm3-ts mutants are rescued by Cnp1 overexpression. The indicated strains expressing GFP-vector, Scm3, or Cnp1 under control of the nmt41 thiamine-repressible promoter were spotted in 10-fold serial dilutions onto selective medium containing thiamine. Leaky expression of Scm3 or Cnp1 from the nmt41 promoter in the presence of thiamine was sufficient to rescue scm3-6 and scm3-19. Rescue was also observed in the absence of thiamine (our unpublished data).
D. The scm3-ts mutants display unequal chromosome/centromere segregation. Missegregation of centromere I was observed in the scm3-6 and scm3-19 mutants grown for 6 hrs at 30°C. DAPI staining revealed binucleate cells with asymmetric nuclei.
E. Minichromosome 16 (Ch16) is unstable in scm3-ts mutants. The Ch16 loss assay (see Experimental Procedures) was performed following growth at 30°C for 16 h.
A PCR-based random mutagenesis screen was performed to identify scm3 temperature sensitive (ts) mutants. Two of these isolates were designated scm3-6 and scm3-19. DNA sequencing revealed an N100S mutation for scm3-6 and an N50S mutation for scm3-19. A liquid media growth assay showed that both mutants lost viability within 8 hrs after a temperature shift to 35.5°C (Figure 2B).
Genetic interactions between Scm3 and Cnp1 were explored by testing whether high-copy expression of Cnp1 rescues scm3-ts mutations. This analysis revealed a remarkably strong suppression of scm3-ts alleles by Cnp1 overexpression (Figure 2C). Since Cnp1-overexpression also rescues temperature sensitive mutants of mis6, 15, 16, 17 and 18 (Hayashi et al., 2004), these data suggest that Scm3 is required for proper Cnp1 function or localization.
Because Cnp1 inactivation or delocalization causes chromosome segregation defects (Hayashi et al., 2004; Takahashi et al., 2000), we examined chromosome segregation in the scm3-ts strains. This analysis was performed using a strain in which an array of Lac operator repeats are linked to centromere I. This centromere was visualized using the GFP-tagged Lac repressor. In centromere mutants, cytokinesis can occur in the absence of correct chromosome segregation (Hayashi et al., 2004; Pidoux et al., 2003; Takahashi et al., 2000). Following growth at 35.5°C for 4-6 hrs, chromosome segregation defects were observed in the scm3-ts mutants (Figure 2D; Figure S4). The scm3-ts cells often failed to segregate their chromosomes equally following mitosis, resulting in one daughter containing both copies of chromosome I (Figure 2D).
We also performed a genetic assay for chromosome loss (Prudden et al., 2003) and found that there was an enhanced loss rate of minichromosome 16 in the scm3-ts mutants following growth at the semi-permissive temperature of 30°C (Figure 2E). From these results we concluded that Scm3 is required for equal chromosome segregation.
Scm3 is Required for Centromere Localization of Cnp1
The Scm3-Cnp1 physical interaction and the rescue of scm3-ts alleles by Cnp1 overexpression suggested that Scm3 might play a role in regulating Cnp1 centromere localization. Cnp1-GFP localization was examined in scm3+ and scm3-ts strains following growth at either 20°C or 36°C. Cnp1-GFP forms nuclear foci at both the permissive and restrictive temperatures in scm3+ cells (Figure 3A). In contrast, Cnp1 foci formation was reduced in the scm3-6 and scm3-19 mutants at 20°C and abolished at 36°C (Figure 3A). A similar defect was observed in a mis18-ts mutant, as previously reported (Hayashi et al., 2004). The abundance of Cnp1-GFP was unaffected in the scm3-ts mutants (Figure 3B). Although Cnp1 localization is lost in the scm3-ts mutants, GFP-Scm3-6 and GFP-Scm3-19 retain the ability to form centromeric foci (Figure S5). These data show that Scm3 localization at centromeres does not depend on co-localization with Cnp1, a result that is confirmed by the ability of Scm3-GFP to form foci in a cnp1-1 mutant (Figure 5A).
Figure 3. Centromere Localization of Cnp1 Requires Scm3.
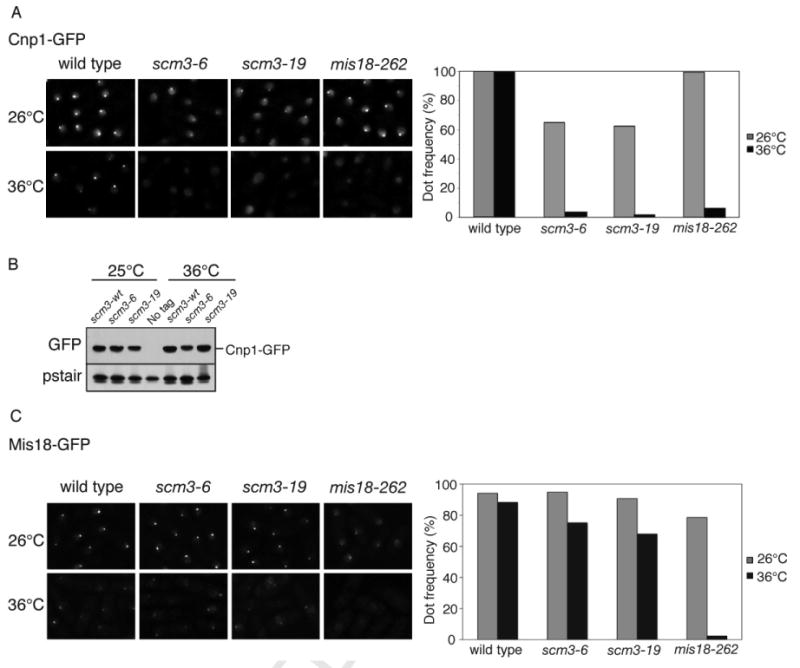
A. Cnp1 foci formation is abolished in the scm3-ts mutants at the restrictive temperature. Cnp1-GFP localization was determined in scm3-6, scm3-19 or mis18-262 strains grown at 26°C or shifted to 36°C for 8 h. Microscopy was performed following methanol fixation. Quantitation of cells with a Cnp1 dot is shown to the right.
B. Cnp1-GFP protein levels are not altered in the scm3-ts mutants at the restrictive temperature. Whole cell extracts were prepared following growth at 25°C or shift to 36°C for 8 h. Immunoblotting was performed with antibodies to GFP or Cdc2 (PSTAIR).
C. Mis18-GFP foci formation is unaffected by incubation of scm3-ts mutants at 36°C. Loss of Mis18-GFP foci in the mis18-262 cells is shown as a positive control.
Figure 5. Mis6, Mis16 and Mis18 are Required for Scm3 Localization at Centromeres.
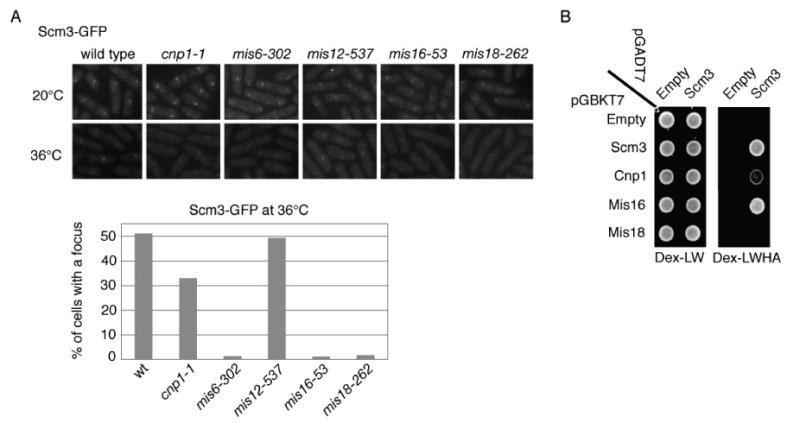
A. Localization of Scm3 at centromeres requires Mis6, Mis16, and Mis18. Strains were grown at 20°C and then shifted to 36°C for 6 h. Microscopy was performed following methanol fixation.
B. Yeast two-hybrid assay showing that Scm3 interacts with Mis16 and Cnp1 but not Mis18.
Mis16-Mis18 complex is required for the centromere localization of Cnp1, Mis6, Mis15, and Mis17. Mis16 localization is solely dependent on Mis18, and the reverse relationship is also true (Hayashi et al., 2004). In contrast to the dependence of Cnp1 localization on Scm3, Mis18-GFP foci persisted in the scm3-ts strains at the restrictive temperature (Figure 3C).
Genetic Interactions Involving Scm3, Cnp1 and Mis Proteins
Genetic analysis was performed to investigate the relationships between Scm3, Cnp1, and the Mis6, 12, 14, 15, 16 and 18 proteins. We first examined Scm3 overexpression effects on growth of mis-ts mutants. Plasmid expression using a pRep41-N-GFP-based plasmid was controlled by the presence (+Thi = repressed) or absence (-Thi = induced) of thiamine in the media. Scm3 overexpression was toxic in each of the mutant strain backgrounds compared to wild type, although to varying degrees (Figure 4A; Figure S6). For example, overexpression of Scm3 in the mis6-ts and mis15-ts mutants was toxic even at 26°C, whereas Scm3 overexpression did not significantly impair growth of the mis12-ts or mis14-ts mutants unless the temperature was raised to 33°C (Figure 4A; Figure S6). The strength of these interactions correlated with the functional classes of the Mis proteins. For example, Mis12 and Mis14 form a complex and localize at centromeres but are not required for centromeric localization of Cnp1 (Goshima et al., 1999; Goshima et al., 2003; Hayashi et al., 2004; Takahashi et al., 2000). A summary of these genetic interactions is presented in Figure 4A.
Figure 4. Synthetic Genetic Interactions Between scm3-ts and mis-ts Alleles.
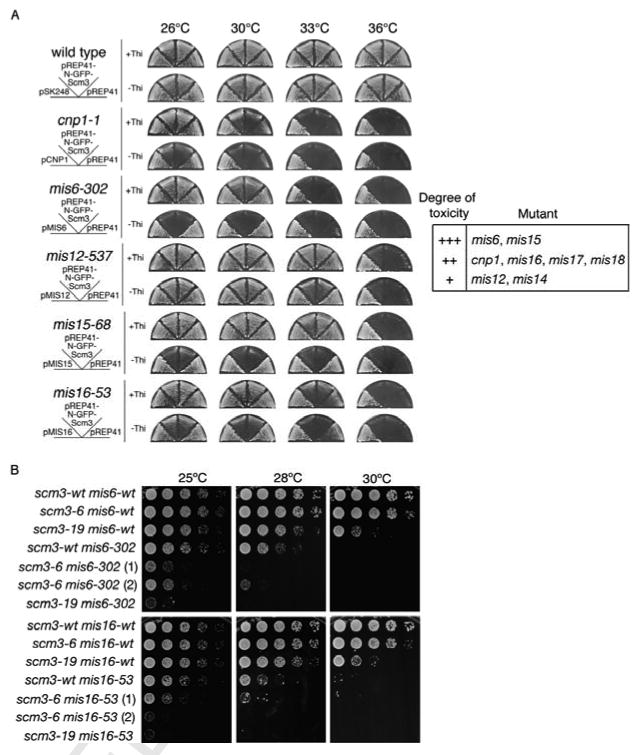
A. Overexpression of GFP-Scm3 is toxic in cnp1-ts and mis-ts strains. The indicated strains were transformed with pSK248 plasmids containing cnp1+ or mis+ genes, or pRep41-GFP plasmids containing scm3+ expressed from the nmt41 promoter. Cells were streaked onto minimal medium in the presence or absence of thiamine to repress (+Thi) or induce (-Thi) expression of GFP-Scm3. The table summarizes the toxicity of overexpression (+++ is most toxic).
B. Synthetic genetic interactions of scm3-ts with mis6-ts or mis16-ts mutations. Ten-fold serial dilutions of cells on YES agar were incubated at 25°C, 28°C or 30°C for 2-3 days. Two individual isolates for scm3-6 mis6-302 or scm3-6 mis16-53 double mutants are shown.
Double mutant strains were constructed to test for synthetic growth defects. Both scm3-6 and scm3-19 mutants show severe growth impairment when combined with mis6-302 or mis16-53 mutants (Figure 4B). In each case, a synthetic growth defect at 25°C was even more apparent at 28°C. Similar results were obtained upon crossing scm3-19 and mis18-262 (Figure S7), although this interaction was allele-specific because a similar effect was not seen with the scm3-6 mis18-262 double mutant. These genetic data support the idea that Scm3, Cnp1, and the Mis proteins share functional interactions at the centromere that are important for cell viability.
Scm3 Localization at Centromeres Requires Mis6, Mis16, and Mis18
The strong genetic interactions between scm3-ts and mis-ts mutants suggested that some of the Mis proteins might regulate Scm3 localization at centromeres. We therefore examined foci formation of Scm3-GFP expressed from the endogenous locus in mis6, mis12, mis16 and mis18-ts mutants incubated at 20°C or 36°C. All but Mis12 are required for Cnp1 localization at centromeres and all localize at centromeres themselves (Goshima et al., 1999; Hayashi et al., 2004; Saitoh et al., 1997; Takahashi et al., 2000). We observed that formation of Scm3-GFP foci was nearly abolished in the mis6, mis16 and mis18-ts mutants at 36°C (Figure 5A). In contrast, Scm3-GFP foci were unaffected by the mis12-ts mutation. Therefore, the same Mis proteins are required for Cnp1 and Scm3 localization at centromeres.
Scm3 Localizes at Centromeres Independently of Cnp1
The data described thus far are consistent with fission yeast Scm3 being either a component of centromeric nucleosomes as proposed for Scm3 in S. cerevisiae (Mizuguchi et al., 2007), or it being a non-nucleosome factor required for stable incorporation of Cnp1 into centromeric chromatin as shown for Mis16-Mis18 and the Mis6 complexes (Hayashi et al., 2004; Takahashi et al., 2000). The first possibility predicts that formation of Scm3 foci should require Cnp1, while the second model predicts that Scm3 foci should form independently of Cnp1. Scm3-GFP foci were readily detected in cnp1-1 cells incubated at 36°C, although the frequency was somewhat reduced compared to wild type (Figure 5A). These data indicate that Scm3 localization at centromeres is largely independent of Cnp1. Scm3 shares this property with Mis16 and Mis18 (Hayashi et al., 2004).
Scm3 Interacts with Mis16
Because Mis16 and Mis18 are absolutely required for Scm3 centromere localization, we tested whether they interact with Scm3 using Y2H analysis. Scm3 and Mis16 showed a robust Y2H interaction using either orientation of plasmids on high-stringency selective medium (Figure 5B, Figure S8A), and western blotting confirmed the expression of the bait plasmids (Figure S8B). The C-terminal 141 amino acids of Scm3 are required for interaction with Mis16 (Figure S2). Our attempts to co-immmunoprecipitate Scm3 and Mis16 from soluble cellular extracts were unsuccessful, possibly due to a dynamic interaction between these proteins. This may resemble the unstable interaction that appears to exist between the human Mis16 homologues, RbAp46 and RbAp48, and hMis18 (Fujita et al., 2007). We did not observe a positive Y2H interaction between Scm3 and Mis18 (Figure 5B), although we did find that Mis18 self-associates (Figure S8C).
Scm3 Delocalizes From Centromeres during Mitosis
The data described thus far establish that Mis6, Mis16 and Mis18 are required for centromere localization of Scm3, which itself is required for centromere localization of Cnp1. These observations suggest that Scm3 may act as an assembly or maintenance factor in the Mis16-Mis18 pathway for establishment of Cnp1 chromatin. To explore whether Scm3 shares other properties with Mis16 and Mis18, we examined whether Scm3 is dynamically localized at centromeres in cycling cells. Although Cnp1 and Mis12 display constitutive centromere localization throughout the cell cycle, Mis16 and Mis18 delocalize from centromeres from metaphase until telophase (Hayashi et al., 2004; Fujita et al., 2007).
A strain expressing Scm3-GFP and Mis12-RFP from endogenous loci was analyzed by fluorescence microscopy. As previously reported (Fujita et al., 2007; Goshima et al., 1999; Hayashi et al., 2004), Mis12 centromeric foci were present in each cell, regardless of cell cycle phase (Figure 6A). Scm3 formed nuclear foci that colocalized with Mis12 in interphase cells. However, like Mis16 and Mis18, Scm3 foci were not detectable in cells during metaphase or early to mid-anaphase (Figure 6A). To provide independent confirmation of these results, ChIP assays were performed on immunoprecipitates from nda3-KM311 cells arrested in metaphase by inactivation of β-tubulin (Hiraoka et al., 1984). Both Cnp1 and Scm3 are enriched at the central region of the centromere in the wild type strain (Figure 6B). However, the Scm3 signal is nearly abolished in the nda3-KM311 mutant at the restrictive temperature, whereas Cnp1 enrichment is largely unchanged. The centromere-association of Scm3 is therefore dynamic, a property shared with Mis16-Mis18 complex.
Scm3 and Mis18 Regulate Histone H3 and H2B Occupancy at Centromeres
Having established that Scm3 mediates the stable deposition of Cnp1 at centromeres, we lastly addressed the role of Scm3 in regulating the localization of canonical histones at centromeres. We were particularly interested in determining whether the absence of H2A/H2B from the point centromeres of budding yeast (Mizuguchi et al., 2007) is conserved for the regional centromeres of fission yeast. We first used ChIP to measure the centromere occupancy of Cnp1, H2B, H3, and both bulk and acetylated H4 in wild type, scm3-ts and mis18-ts mutants. As expected, the strong enrichment of Cnp1 at cnt1/imr1 was abolished in the scm3-ts and mis18-ts mutants. The loss of Cnp1 at cnt1/imr1 in the scm3-ts and mis18-ts mutants was accompanied by a large increase in the H3 signal in these regions. Total H4 signals at cnt1/imr1 in the scm3-ts strains were modestly decreased while acetylated histone H4 increased 2-3 fold (Figure 7A; Figure S9). Essentially identical effects were seen in the mis18-262 cells at 36°C. In wild type cells analyzed with H2B-specific antisera, H2B enrichment at cnt1/imr1 was reduced ∼10-fold relative to the outer repeat dg sequences and euchromatic lys1 (Figure 7A). In the scm3-ts and mis18-ts mutants the H2B signal at cnt1/imr1 increased ∼3-fold relative to wild type, whereas the H2B signal at the dg and lys1 sequences was essentially unchanged.
Figure 7. Scm3 and Mis18 Regulate Cnp1, H3 and H2B Occupancy at Centromeres.
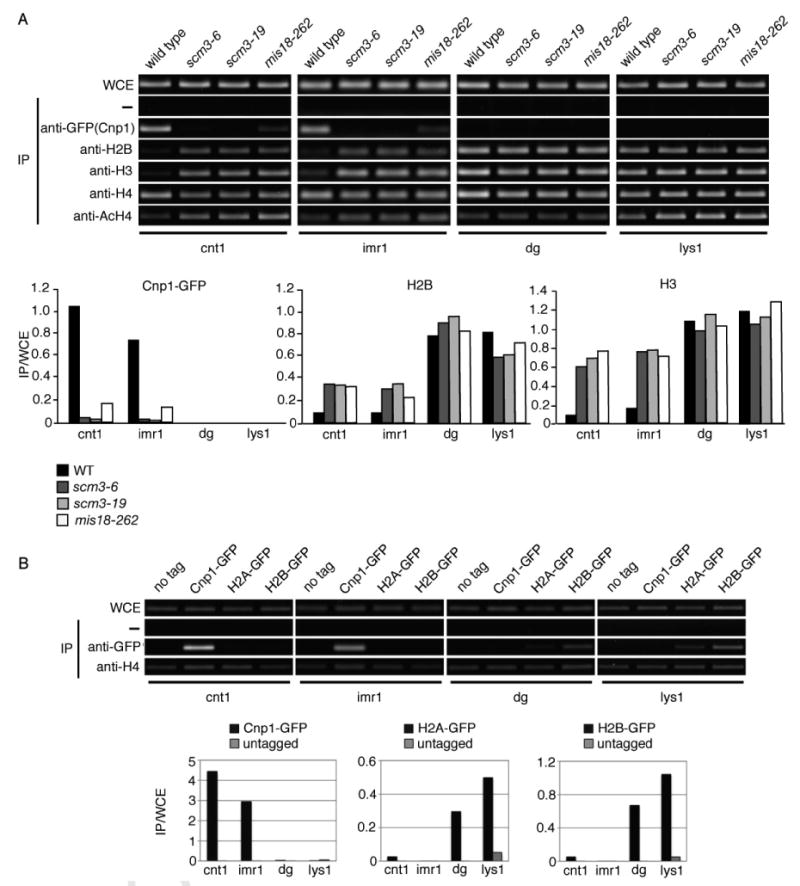
A. ChIP analysis of Cnp1 and canonical histone occupancy at the centromere in scm3-ts and mis18-262 cells. Cells were incubated at 36°C prior to harvesting. Compared to wild type, Cnp1-GFP enrichment at cnt1 and imr1 is reduced in the mutants while H2B and H3 enrichment is increased. Quantitation for Cnp1-GFP, H2B, and H3 is shown below; quantitation for H4 and acetylated-H4 is in Figure S9.
B. H2A and H2B are largely absent from the central core of the centromere. ChIP was performed using strains expressing Cnp1, H2A, or H2B tagged with GFP. The level of H4 at each of the centromere regions was determined as a control.
These data showed that both Mis18 and Scm3 are required for the enrichment of Cnp1 and the concomitant reduction of histones H2B and H3 at the central regions of centromeres, as well as for maintenance of a hypo-acetylated form of H4. To independently confirm that both H2A and H2B are underrepresented at cnt1/imr1, we performed ChIP with anti-GFP antisera using strains in which GFP-tagged Cnp1, H2A or H2B were expressed from their endogenous loci. Whereas Cnp1 was strongly enriched at cnt1/imr1 and essentially absent from the dg and lys1 regions, H2A and H2B displayed the opposite enrichment pattern (Figure 7B). These data indicate that H2A/H2B heterodimers are largely absent from the inner centromere regions that contain Cnp1-specific chromatin.
Discussion
The experiments described in this paper demonstrate that S. pombe Scm3 is required for establishment of Cnp1 chromatin at the centromere. In this regard Scm3 joins the Mis16-Mis18 complex, the Mis6 complex and the NASP-related protein Sim3. However, amongst these proteins Scm3 has a unique set of properties. It both associates with Cnp1 and specifically localizes at centromeres. Importantly, Scm3 localization at centromeres requires Mis6, Mis16 and Mis18, whilst Mis18 localization at centromeres does not require Scm3. None of these proteins require Cnp1 for their localization at centromeres; indeed, the mutant Scm3 proteins encoded by the scm3-ts alleles remain in foci even when the Cnp1 foci dissolve. If the mechanism of Cnp1 incorporation into centromeric DNA is viewed as a stepwise pathway, then these data indicate that Scm3 acts downstream of Mis16-Mis18 and Mis6 complexes but upstream of Cnp1. With these attributes Scm3 is well positioned to be the missing link in the pathway that establishes Cnp1 centromeric chromatin. The most straightforward interpretation of these findings is that Mis16-Mis18 and Mis6 complexes direct the recruitment of Scm3 to centromeres, and Scm3 in turn mediates the stable deposition of Cnp1 into centromeric chromatin. Scm3 may act as an assembly or maintenance factor for Cnp1 that is escorted to centromeres in a complex with Sim3. This model is fully consistent with data presented by Pidoux et al. (2008), in which they independently characterize fission yeast Scm3 as being required for assembly of Cnp1-centromeric chromatin in cooperation with Mis16-Mis18.
A critical observation in support of this model is that Scm3 dissociates from centromeres in early mitosis and then returns in late mitosis. This dynamic localization behavior parallels that of Mis16 and Mis18. Since Mis16 and Mis18 are required for centromere localization of Scm3, it is tempting to propose that the dynamic pattern of Scm3 localization during the cell cycle is directly regulated by Mis16-Mis18. This proposal gains support from our Y2H studies showing an interaction between Scm3 and Mis16. The cell cycle regulated pattern of localization of Scm3, Mis16 and Mis18 is unlike that of other centromere proteins in fission yeast. Most importantly it is unlike that of Cnp1, which maintains a centromere localization pattern throughout the cell cycle even when Scm3, Mis16 and Mis18 disassemble from centromeres during mitosis. These distinct localization properties are maintained even during a long-term arrest in mitosis as carried out with the nda3-KM311 mutant, in which we observed nearly complete loss of the Scm3 signal at cnt1/imr1 with very little change in the Cnp1 signal.
The dynamic localization behavior of Scm3 most closely resemble those of Mis16 and Mis18, and therefore we propose that the most parsimonious interpretation of our data is that fission yeast Scm3 functions as a factor required for stable incorporation of Cnp1 into centromeric nucleosomes, as opposed to being a constitutive subunit of these nucleosomes. An alternative model is that Scm3 functions as a core subunit of Cnp1 nucleosomes specifically during interphase. According to this model, Scm3 would have to be stripped from centromeric nucleosomes at the onset of mitosis. Alternatively, there may be replacement of centromeric nucleosomes during this brief window of the cell cycle. In the absence of evidence for such a rapid reorganization or replacement of centromeric nucleosomes at the onset of mitosis, we favor the idea that fission yeast Scm3 is not a core subunit of centromeric nucleosomes. Consistent with this possibility, our immunoblot data showing that there is large excess of Cnp1 relative to Scm3 in total cell extracts (Figure S3) is not easily reconciled with a model in which Scm3 is a stoichiometric subunit of Cnp1-containing nucleosomes. This result correlates well with the detection of much brighter Cnp1-GFP foci relative to Scm3-GFP foci in cells that express these proteins from genomic loci (Figure S10), although we cannot know with certainty whether the intensity of these signals accurately reflects the relative abundance of these proteins at centromeres.
Taken together, our data in combination with results presented by Pidoux et al. (2008) are inconsistent with a role for Scm3 as a core component of centromeric nucleosomes in fission yeast, unlike the model proposed for budding yeast Scm3 (Mizuguchi et al., 2007). The hypothesized different functions of Scm3 proteins in these two distantly related organisms may be consistent with the very weak sequence relationship between the Scm3 orthologs. The extreme divergence in sequence and size amongst Scm3-related proteins in fungi is unexpected if these proteins are generally core subunits of specialized nucleosomes. Histones are among the most highly conserved of eukaryotic proteins, and the histone-fold domain of CenH3 orthologs is also well conserved in eukaryotes despite the large diversity of centromere sequences.
The conventional nucleosome is octameric in nature and is composed of a tetramer of histones H3 and H4 that associate with two H2A/H2B dimers. The CenH3-H4 tetramer is structurally more compact and rigid than a tetramer of H3 and H4, and this may confer a specialized nucleosome structure at the centromere in mammalian cells (Black et al., 2004). However, recent evidence suggests that nucleosome composition at the centromere may vary in a species-specific manner. As mentioned above, Mizuguchi et al. (2007) proposed an alternate nucleosome consisting of a hexamer of two copies each of Cse4, H4, and Scm3 at the point centromere of S. cerevisiae, whereas Henikoff and colleagues (Dalal et al., 2007) performed in vivo crosslinking experiments to detect heterotetrameric nucleosomes composed of CenH3, H4, H2A, and H2B in Drosophila. Inner centromeric chromatin in fission yeast has strong signals for Cnp1 and histone H4 but only very weak signals for histones H2A, H2B and H3 in our ChIP analysis (Figure 7). Strong Cnp1 signals correlate with weak H2A, H2B and H3 signals. Indeed, inactivation of Scm3 or Mis18 causes a reversal of these patterns — the Cnp1 signal is strongly diminished while the H3 and H2B signals increase (Figure 7A). This remarkable correspondence between the lack of H2A/H2B and H3 occupancy at the inner centromere regions, and their antithetical relationship to Cnp1, indicates that histones H2A/H2B are not core subunits of inner centromeric nucleosomes in fission yeast. We cannot conclude that all centromeric nucleosomes lack H2A/H2B and H3 completely, but certainly they are substantially under-represented by ChIP analyses. An alternative possibility that we cannot formally exclude is that H2A, H2B and H3 have a weak or more dynamic association with nucleosomes in the central region of the centromere that prevents them from being efficiently detected by ChIP.
The precise role of Scm3 in mediating the stable deposition of Cnp1 into centromeric chromatin remains to be determined. Scm3 may function as a loading factor for Cnp1, similar to the loading function of Asf1 for histones H3 and H4 (De Koning et al., 2007). Scm3 and Mis16-Mis18 may together coordinate correct deposition of Cnp1 and H4 in order to establish centromeric chromatin in a state required for kinetochore attachment and equal chromosome segregation. Based on results from previous work (Fujita et al., 2007), we favor a model in which Mis18 is first required for ‘priming’ of centromeric chromatin during late mitosis through establishment of acetylated H4. In human cells, treatment with the histone deacetylase (HDAC) inhibitor Trichostatin A (TSA) suppressed the loss of CENP-A centromere localization in cells treated with an RNAi against hMis18α (Fujita et al., 2007). It therefore appears that centromeric histones may need to be acetylated prior to CenH3 deposition. Once this specific chromatin modification has occurred, Mis18 may recruit Mis16 and Scm3 to centromeres in late mitosis, thereby facilitating incorporation of Cnp1 into chromatin through its direct association with Scm3. Successful deposition of Cnp1 at centromeres then results in reduced acetylation of histone H4 (Figure 7A)(Hayashi et al., 2004; Fujita et al., 2007). This model favors the idea that Scm3 and elements of the Mis16-Mis18 pathway act primarily as recruitment factors for Cnp1, as suggested by the studies of Mis18 in mammalian cells (Fujita et al., 2007), but it is also possible that in fission yeast they are required for maintenance of Cnp1-containing nucleosomes at centromeres. However, this possible role in maintenance does not involve regulation of Cnp1 protein stability, as Cnp1-GFP levels are not affected in the scm3-ts strains at the restrictive temperature (Figure 3B).
As histones are extremely basic in nature, it follows that acidic stretches are often found in histone chaperones; one such example is the H3/H4 chaperone Asf1 (Mousson et al., 2007). The region that we mapped as the Cnp1-interaction domain of Scm3 (aa1-194) has an isoelectric point of 4.89, consistent with a role in enhancement of interaction with Cnp1 through charge neutralization. In budding yeast, it is the histone fold domain (HFD) of Cnp1 that interacts with Scm3 (Stoler et al., 2007). Considering that Cnp1 and other histones share a high level of conservation across the HFD, understanding the selectivity of Scm3 for Cnp1 using structural analyses will provide important insight into the specificity of Scm3 for Cnp1 in its role as a factor required for stable incorporation of Cnp1 at the centromere.
Experimental Procedures
Strains, Media, and Genetic Analysis
Strain genotypes are listed in Table S1. Growth media and methods for S. pombe were performed as described (Moreno et al., 1991). Ectopic expression of pRep41-N-GFP-Scm3/Cnp1 for plating and pRep1-N-GST-Scm3/Cds11-190 for co-immunoprecipitation was under control of the thiamine-repressible nmt promoter. Induction of plasmid expression in selective medium lacking thiamine was for 18-21 h. The scm3 gene was deleted by replacement of the entire genomic locus with the KanMx6 cassette in a diploid strain (Bahler et al., 1998). This heterozygous diploid was sporulated and tetrads were dissected. Survival assays were performed following growth of the scm3+ and scm3-ts strains at the semi-permissive temperature of 30°C. Approximately 1000 cells were plated onto yeast extract with supplements (YES) and quantitation was performed following incubation at 25°C for 4 days. Spot assays were performed by plating 10-fold serial dilutions of exponentially growing cells onto YES or selective minimal media. Plates were incubated at the indicated temperatures and scanned after 3-5 days.
Isolation of scm3-ts Mutants
The scm3-HA:KanMx6 locus from strain JW380 was amplified by PCR using standard conditions and then re-amplified in 8 parallel PCR reactions containing excess dNTPs and polymerase. The resulting products were pooled and transformed into S. pombe. Geneticin (G418)-resistant clones were selected at 25°C and then tested for growth at 35.5°C. Temperature sensitive clones were selected, backcrossed, and sequenced. Of 24 strains, 15 contained the N100S mutation (scm3-6), and 3 contained the N50S mutation (scm3-19). Transformation with an episomal plasmid containing scm3+ confirmed that the growth defect was rescued by scm3+.
Microscopy
Cells were photographed using a Nikon Eclipse E800 microscope, Photometrics Quantix CCD camera, and IPLab Spectrum software. For visualization of chromosome I, LacI-GFP-NLS was expressed in a strain containing the LacI binding sequence at the lys1+ gene, which is pericentromeric to cen1 (Nabeshima et al., 1997; Straight et al., 1996). Wild type and scm3-ts strains were grown to mid-log phase at 25°C and then shifted to 30°C for 6 h. Methanol fixed cells were used to observe GFP and RFP-tagged proteins (Hayashi et al., 2004). Mutants were grown in YES at 20°C and shifted to 36°C for 6 h before fixation and imaging.
ChIP
ChIP assays were performed as described (Saitoh et al., 1997). Immunoprecipitation was performed using antibodies to FLAG (M2, Sigma-Aldrich), GFP (Roche), Pk (anti-V5 tag, Serotech), H2B (Maruyama et al., 2006), H3 (GeneTex), H4 (05-858, Upstate) or AcH4 (06-598, Upstate). The nda3-KM311 mitotic-arrest experiment was performed using cells expressing chromosomally integrated Cnp1-FLAG and Scm3-Pk under their native promoters. Cells were fixed in formaldehyde after growth at 20°C for 8 h in YPD.
Immunoprecipitation
Soluble extracts were prepared from exponentially growing cells by bead-beating in lysis buffer (50 mM Tris pH 8, 150 mM NaCl, 2.5 mM EDTA, 10% glycerol, 0.3% NP-40 and protease inhibitors, 1 mM PMSF) followed by centrifugation to remove insoluble material. Immunoprecipitation was performed using glutathione-sepharose (Amersham) for 2 h at 4°C, and proteins were resolved in a 10% Tris-glycine gel. Following transfer to nitrocellulose membrane and blocking with 5% milk in TBS plus 0.1% Tween-20, western blotting was performed using anti-GST (Amersham) and anti-GFP (Roche) antibodies.
Minichromosome Loss Assay
This assay was performed as described (Niwa et al., 1986). Two independent Ade+ isolates of scm3+, scm3-6 or scm3-19 strains were analyzed for stability of minichromosome 16 (Ch16) containing an ade6-M216 mutation (Prudden et al., 2003). Host ade6-M210 cells are rendered Ade+ by allelic complementation. Following growth at the semi-permissive temperature of 30°C for 16 h, ∼1000 cells from each strain were plated on adenine-limiting (YEA) plates and incubated at 25°C for 4 days. Total colonies and red colonies (Ade- colonies accumulate a red pigment) were counted.
Yeast Two-hybrid Assay
The Gal4 activation (pGADT7) or DNA binding domain (pGBKT7) was fused to the indicated proteins and expressed in S. cerevisiae strain AH109 (Clontech Matchmaker system). Isolates containing both plasmids were identified by growth on minimal glucose medium lacking leucine and tryptophan (Dex-LW). Positive interactions were determined by growth on high stringency glucose medium lacking leucine, tryptophan, histidine and adenine (Dex-LWHA).
Supplementary Material
Acknowledgments
We thank Charly Chahwan for help with the identification of scm3+ and for discussions, and Robin Allshire for sharing unpublished data. Strains were provided by Shigeaki Saitoh and Kohta Takahashi (SP38, SP148), John Prudden and Tim Humphrey (JP970), and Hiroaki Murakami (HM1B111, HM1D21). Thanks to members of the Russell lab and the Scripps Cell Cycle Group for discussions. Work in PR's lab was supported by NIH grants GM59447 and CA77325.
Footnotes
Supplemental Data: Supplemental Data including Table S1, Figures S1-S10, and Supplemental References can be found with this article online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aravind L, Iyer LM, Wu C. Domain architectures of the Scm3p protein provide insights into centromere function and evolution. Cell Cycle. 2007;6:2511–2515. doi: 10.4161/cc.6.20.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Jr, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- Boddy MN, Lopez-Girona A, Shanahan P, Interthal H, Heyer WD, Russell P. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol Cell Biol. 2000;20:8758–8766. doi: 10.1128/mcb.20.23.8758-8766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Chen Y, Baker RE, Keith KC, Harris K, Stoler S, Fitzgerald-Hayes M. The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol Cell Biol. 2000;20:7037–7048. doi: 10.1128/mcb.20.18.7037-7048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Koning L, Corpet A, Haber JE, Almouzni G. Histone chaperones: an escort network regulating histone traffic. Nat Struct Mol Biol. 2007;14:997–1007. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- Dunleavy EM, Pidoux AL, Monet M, Bonilla C, Richardson W, Hamilton GL, Ekwall K, McLaughlin PJ, Allshire RC. A NASP (N1/N2)-related protein, Sim3, binds CENP-A and is required for its deposition at fission yeast centromeres. Mol Cell. 2007;28:1029–1044. doi: 10.1016/j.molcel.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espelin CW, Kaplan KB, Sorger PK. Probing the architecture of a simple kinetochore using DNA-protein crosslinking. J Cell Biol. 1997;139:1383–1396. doi: 10.1083/jcb.139.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espelin CW, Simons KT, Harrison SC, Sorger PK. Binding of the essential Saccharomyces cerevisiae kinetochore protein Ndc10p to CDEII. Mol Biol Cell. 2003;14:4557–4568. doi: 10.1091/mbc.E02-08-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Furuyama T, Dalal Y, Henikoff S. Chaperone-mediated assembly of centromeric chromatin in vitro. Proc Natl Acad Sci U S A. 2006;103:6172–6177. doi: 10.1073/pnas.0601686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Kiyomitsu T, Yoda K, Yanagida M. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J Cell Biol. 2003;160:25–39. doi: 10.1083/jcb.200210005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Saitoh S, Yanagida M. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 1999;13:1664–1677. doi: 10.1101/gad.13.13.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Dalal Y. Centromeric chromatin: what makes it unique? Curr Opin Genet Dev. 2005;15:177–184. doi: 10.1016/j.gde.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Toda T, Yanagida M. The NDA3 gene of fission yeast encodes beta-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyola A, Almouzni G. Histone chaperones, a supporting role in the limelight. Biochim Biophys Acta. 2004;1677:3–11. doi: 10.1016/j.bbaexp.2003.09.012. [DOI] [PubMed] [Google Scholar]
- Maruyama T, Nakamura T, Hayashi T, Yanagida M. Histone H2B mutations in inner region affect ubiquitination, centromere function, silencing and chromosome segregation. EMBO J. 2006;25:2420–2431. doi: 10.1038/sj.emboj.7601110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Morris CA, Moazed D. Centromere assembly and propagation. Cell. 2007;128:647–650. doi: 10.1016/j.cell.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Mousson F, Ochsenbein F, Mann C. The histone chaperone Asf1 at the crossroads of chromatin and DNA checkpoint pathways. Chromosoma. 2007;116:79–93. doi: 10.1007/s00412-006-0087-z. [DOI] [PubMed] [Google Scholar]
- Nabeshima K, Saitoh S, Yanagida M. Use of green fluorescent protein for intracellular protein localization in living fission yeast cells. Methods Enzymol. 1997;283:459–471. doi: 10.1016/s0076-6879(97)83037-6. [DOI] [PubMed] [Google Scholar]
- Niwa O, Matsumoto T, Yanagida M. Construction of a mini-chromosome by deletion and its mitotic and meiotic behaviour in fission yeast. Mol Gen Genet. 1986;203:397–405. [Google Scholar]
- Pidoux AL, Richardson W, Allshire RC. Sim4: a novel fission yeast kinetochore protein required for centromeric silencing and chromosome segregation. J Cell Biol. 2003;161:295–307. doi: 10.1083/jcb.200212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudden J, Evans JS, Hussey SP, Deans B, O'Neill P, Thacker J, Humphrey T. Pathway utilization in response to a site-specific DNA double-strand break in fission yeast. EMBO J. 2003;22:1419–1430. doi: 10.1093/emboj/cdg119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh S, Takahashi K, Yanagida M. Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell. 1997;90:131–143. doi: 10.1016/s0092-8674(00)80320-7. [DOI] [PubMed] [Google Scholar]
- Schuh M, Lehner CF, Heidmann S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- Smith MM. Centromeres and variant histones: what, where, when and why? Curr Opin Cell Biol. 2002;14:279–285. doi: 10.1016/s0955-0674(02)00331-9. [DOI] [PubMed] [Google Scholar]
- Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, Baker RE. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci U S A. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- Sugata N, Li S, Earnshaw WC, Yen TJ, Yoda K, Masumoto H, Munekata E, Warburton PE, Todokoro K. Human CENP-H multimers colocalize with CENP-A and CENP-C at active centromere--kinetochore complexes. Hum Mol Genet. 2000;9:2919–2926. doi: 10.1093/hmg/9.19.2919. [DOI] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Chen ES, Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa O, Yanagida M. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol Biol Cell. 1992;3:819–835. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Takayama Y, Masuda F, Kobayashi Y, Saitoh S. Two distinct pathways responsible for the loading of CENP-A to centromeres in the fission yeast cell cycle. Philos Trans R Soc Lond B Biol Sci. 2005;360:595–606. doi: 10.1098/rstb.2004.1614. discussion 606-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


