Abstract
Described herein is a general approach to identify novel compounds using the biodiversity of a megadiverse group of animals; specifically, the phylogenetic lineage of the venomous gastropods that belong to the genus Conus (“cone snails”). Cone snail biodiversity was exploited to identify three new μ-conotoxins, BuIIIA, BuIIIB and BuIIIC, encoded by the fish-hunting species Conus bullatus. BuIIIA, BuIIIB and BuIIIC are strikingly divergent in their amino acid composition compared to previous μ-conotoxins known to target the voltage-gated Na channel skeletal muscle subtype Nav1.4. Our preliminary results indicate that BuIIIB and BuIIIC are potent inhibitors of Nav1.4 (average block ~96%, at a 1 μM concentration of peptide), displaying a very slow off-rate not seen in previously characterized μ-conotoxins that block Nav1.4. In addition, the three new Conus bullatus μ-conopeptides help to define a new branch of the M-superfamily of conotoxins, namely M-5. The exogene strategy used to discover these Na channel-inhibiting peptides was based on both understanding the phylogeny of Conus, as well as the molecular genetics of venom μ-conotoxin peptides previously shown to generally target voltage-gated Na channels. The discovery of BuIIIA, BuIIIB and BuIIIC Na channel blockers expands the diversity of ligands useful in determining the structure-activity relationship of voltage-gated sodium channels.
Keywords: Biodiversity-derived compounds, Sodium channel ligands, exogenes
Introduction
A significant accomplishment of molecular neuroscience has been identifying membrane macromolecules that underlie nervous system function. An extraordinarily conserved set of such proteins, including ion channels and neurotransmitter receptors, are present in all nervous systems from C. elegans to humans. These molecules are encoded by a few gene superfamilies (such as the voltage-gated and ligand-gated ion channel superfamilies), with each superfamily comprising several families (e.g., Na channels responsible for action potentials are in a family that belongs to the voltage-gated ion channel superfamily) (Goldin, et al. 2000) (Hille 2001; Ogata 2002).
Every ion channel/receptor family, in turn, comprises multiple subtypes or isoforms. In mammals, nine isoforms of the major α-subunit of voltage-gated Na channels, each encoded by a different gene, have been characterized (Catterall 1992) (Catterall 2000). Despite the breakthroughs in identifying the molecular components of the nervous system, delineating the functions of each has been a challenge. The standard technology for investigating function is knockout mice; thus, knockouts of individual Na channel subtypes have been made (Planells-Cases, et.al. 2000; Amaya 2006; Woodruff-Pak, et al. 2006). However, the difficulty and expense of maintaining the mice, the fact that some knockouts do not thrive and the unpredictability of whether knocking out of one subtype will result in a compensatory, and possibly ectopic, over expression of another makes the development of alternative methods for investigating function highly desirable.
An attractive alternative would be to use pharmacological methods; the limiting factor is that highly subtype-selective ligands to study function are seldom available, particularly for ion channels. For the voltage-gated sodium channels, the most widely used pharmacological tool is tetrodotoxin, an alkaloid whose best known source is puffer fish (Yotsu, et al. 1987). Of the nine Na channel subtypes in mammals, six are tetrodotoxin-sensitive and three are insensitive (Catterall, et al. 2005). Thus, at the present time, pharmacological characterization using tetrodotoxin clearly does not have the resolution provided by knockout mice. More selective ligands for voltage-gated Na channels need to be developed.
One biological system being broadly exploited to generate ligands with greater subtype selectivity for ion channels are the small peptides found in cone snail venoms (Norton and Pallaghy 1998). There is remarkable molecular diversity in each venom; the 700 species of cone snails each have ~100 different peptide toxins, and rapid interspecific divergence has generated ~70,000 different peptides. Like their ion channel targets, cone snail peptides are encoded by gene superfamilies that in turn comprise families, and each family of conopeptides generally targets a corresponding ion channel or receptor family (Terlau and Olivera 2004). For example, the M superfamily includes the μ-conopeptide family, delineated with the Cys framework –CC-C-C-CC-, each member of which targets one or more of the nine subtypes of voltage-gated sodium channels. Hypermutation of Conus peptide genes occurred as cone snails speciated, and they are examples of “exogenes,” those genes responsible for mediating biotic interactions between organisms. Exogenes are characteristically extremely rapidly diverging; the accelerated evolution of exogenes expressed in Conus venom ducts has generated a biochemical and pharmacological diversity that can be exploited to develop highly subtype-selective ligands for ion channels and receptors (Olivera 2006; Ellison and Olivera 2007; Olivera and Teichert 2007).
Cone snails can be grouped into discrete clades. Our laboratory has analyzed the molecular phylogeny of cone snails; the resultant phylogenetic information has been incorporated into a general strategy for the discovery of novel classes of peptides. This “exogenome” strategy was used effectively to obtain highly subtype selective Conus peptides that discriminate between nicotinic acetylcholine receptor subtypes (Olivera 2006). The analysis of Conus peptide exogene families, when combined with informed phylogenetics, makes screening the enormous pharmacological resource afforded by cone snails venom peptides far more efficient.
In this paper, exogene analysis and phylogenetics were used to identify three new μ-conotoxins, BuIIIA, BuIIIB and BuIIIC. BuIIIA, BuIIIB and BuIIIC have a significantly different amino acid composition from previous μ-conotoxins known to target the voltage-gated Na channel subtype Nav1.4, yet are extremely potent inhibitors of this subtype. BuIIIA, BuIIIB and BuIIIC from Conus bullatus, are apart of a novel class of μ-conopeptides discovered from a newly defined clade of fish-hunting cone snails, the Textilia clade. In addition, C. bullatus peptides help define a new branch of the M-superfamily of conotoxins, namely M-5. The discovery of these three novel Na channel blockers expands the diversity of ligands useful in determining the structure-activity relationship of voltage-gated sodium channels.
Materials and Methods
Cloning and identification of C. bullatus μ-conotoxins from cDNA
Dissection of C. bullatus venom ducts, construction of cDNA, and PCR amplification and cloning of μ-conotoxins were as previously described (West, et al. 2002; Azam, et al. 2005). Briefly, venom ducts were dissected from living snails and stored at −80 °C. Venom duct tissue was homogenized and RNA extracted using Trizol reagent according to the manufacturer’s standard protocol (TRIzol Total RNA Isolation - Life Technologies/Gibco BRL, Grand Island, NY). cDNA was prepared by reverse transcription of RNA isolated from Conus venom ducts as previously described (Jacobsen, Jimenez et al. 1998). The resulting cDNA served as a template for PCR amplification using primers based on conserved 5′- and 3′-UTR regions of previously isolated μ-conotoxin genes:
Forward primer: 5′ CAA GA(AG) GGA TCG ATA GCA GTT C 3′
Reverse primer: 5′ ACT GCA ATC (AG)TT TTA CTT ATT C 3′
Analysis of the cDNA sequences identified the open reading frames encoding the complete precursor protein for μ-conotoxins. Aligned nucleotide sequences of BuIIIA, BuIIIB and BuIIIC, with primer sequence region highlighted are shown in supplementary Figure 1 (S1). DNA Sequences for BuIIIA, BuIIIB., and BuIIIC were deposited into GenBank, accession numbers FJ240165–167.
PCR amplification of 16S rRNA gene segment of mtDNA for C. bullatus species and 16S sequence analysis
16S rRNA was amplified as previously described (Espiritu, et al. 2001). Nucleic acid sequences were aligned manually using MEGA version 3.1 (Kumar, et al. 2004). The phylogenetic tree was created from two independent runs using the software program MrBayes (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003). 8,000,000 trees were made in each run, 80,000 of which were saved. 1,500 of each of those 80,000 were also discarded as burn-in. Each run had four chains (one cold and three heated). The two independent runs were combined into a single tree where branches were retained if they were found in 50% or more of those trees not discarded. The standard deviation after 8,000,000 generations was 2.836 × 10−3. A general time reversible (GTR) model was used, with the rate variation of some sites kept invariable and the remaining rates drawn from a gamma distribution.
Chemical synthesis and two-step oxidative folding of μ-conotoxins
Synthetic conotoxins were produced using methods identical to those described previously (Bulaj, et al. 2005; Bulaj, et al. 2006). Briefly, the peptides were synthesized on solid support using standard Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry. The peptides were cleaved from the resin by a 3- to 4-hr treatment with reagent K (trifluoroacetic acid (TFA)/water/ethanedithiol/phenol/thioanisole; 82.5/5/2.5/7.5/5 by volume). The peptides were subsequently filtered and precipitated with cold methyl-tert butyl ether (MTBE). The linear peptides were purified by reversed-phase HPLC using a semi-preparative C18 Vydac column (218TP510, 10 mm × 250 mm), over a linear gradient of 10–30% ACN in 40 min and the fractions were detected at 220 nm. Peptide was further characterized using ESI-MS (Micromass Quattro II mass spectrometer) at Mass Spectrometry and Proteomic Core Facility of the University of Utah.
To facilitate the folding of C. bullatus peptides with the native Cys connectivity (Cys1-Cys4, Cys2-Cys5 and Cys3-Cys6), peptides containing acetamidomethyl (Acm)-protection of Cys3 and Cys6 were constructed. A two-step folding protocol was used to produce the correctly folded species of BuIIIA, BuIIIB, and BuIIIC. HPLC comparison of traditional oxidation and folding protocols, in which none of the Cys are protected, and the two-step approach indicated the two-step approach garnered a better yield of the final product (see Supplementary Figure 2). Formation of the first two-disulfide bridges (Cys1-Cys4 and Cys2-Cys5) was accomplished by oxidation of linear peptides in buffered solution (0.1 M Tris-HCl, pH 7.5) containing 1 mM EDTA, 1 mM oxidized glutathione, 1 mM reduced glutathione, and 1 M NaCl. Folding was quenched by acidification with formic acid (8% final concentration). The main oxidation product was purified from the folding mixture by semi-preparative HPLC, and further characterized using ESI-MS. Formation of the final disulfide bridge was accomplished by a simultaneous removal of Acm-protecting groups, and subsequent oxidation with 1 mM iodine containing 25% ACN in H2O. Iodine oxidation was quenched after 10 minutes at room temperature by drop wise addition of 1 M ascorbic acid. Final purification of fully folded peptide was accomplished by semi-preparative HPLC using conditions described above and also characterized using mass spectrometry.
Electrophysiology of cloned rat NaV1.4 expressed in Xenopus oocytes
The cDNA clone for NaV1.4 was kindly provided by Dr. Al Goldin. The cDNA was linearized with NotI and transcribed with T7 RNA polymerase using the mMessage mMachine RNA transcription kit from Ambion (Austin, TX) to generate capped cRNA. The cRNA was then purified using the Qiagen RNeasy kit (Qiagen, Valencia, CA).
Oocytes were each injected with about 15 ng of cRNA in about 30 nl of distilled water and incubated at 16 °C in ND96 (composition in mM: 96 NaCl, 2 KCl, 1.8 CaCl2, 1 MgCl2, 5 HEPES, pH ~7.3) supplemented with Pen/Strep, Septra and Amikacin for 1 to 5 days. To record sodium currents from a given oocyte, it was placed in a cylindrical well (4 mm diameter, volume ~ 50 μl) in a wafer made of Sylgard (Dow Corning, Midland, MI). The oocyte was impaled with glass microelectrodes filled with 3 M KCl (0.1 to 0.5 MΩ) and two-electrode voltage clamped using a Warner OC-725C amplifier (Warner Instruments, Hamden, CT). The membrane potential was held at −80 mV, and sodium channels were activated by a 50 ms step to -10 mV applied every 30 s. The current signals were low-pass filtered at 3 kHz, digitized at a sampling frequency of 10 kHz, and leak-subtracted by a P/6 protocol using in-house software written in LabVIEW (National Instruments, Austin, Texas). The oocyte chamber was intermittently perfused with ND96 using a motorized syringe (Cavro XL3000, Tecan Systems, San Jose, CA) or peristaltic pump (Rainin, Woburn, MA). To apply toxin, the perfusion was halted, 5 μL of toxin solution (at ten times of the final concentration) was applied to the 50 μL bath, and the bath manually stirred for about 5 seconds by gently aspirating and expelling ~ 5 μL of bath fluid several times with a micropipette. Recordings were conducted at room temperature.
Results
Using phylogeny for μ-conotoxin discovery
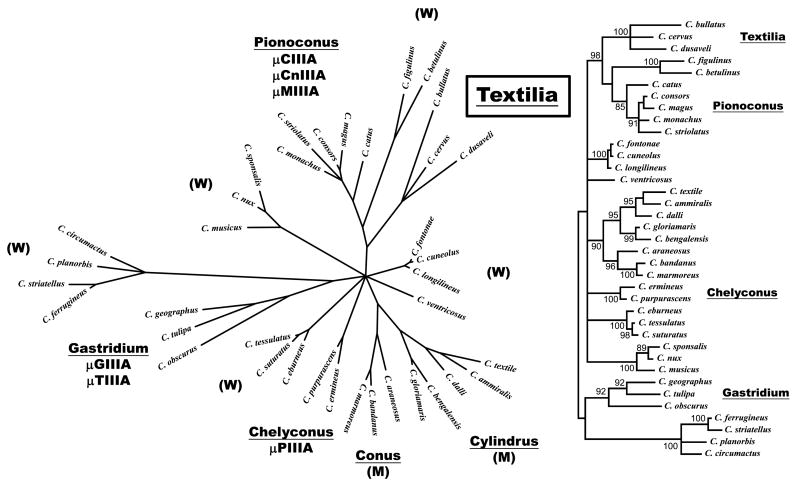
μ-conotoxins are a family of Na channel antagonists widely distributed across Conus. Several groups have reported a molecular phylogeny for the cone snails (Duda Jr and Palumbi 1999; Espiritu, et al. 2001) (Duda Jr and Kohn 2005) a section of the phylogenetic tree is shown in Figure 1. New Conus included in the tree are: C. bullatus, C. cervus and C. dusaveli, all fish-hunting species from relatively deep water (see Fig. 2). This species group is referred to as the Textilia clade; the type species of Textilia (Swainson, 1840) is the bubble cone C. bullatus. As shown in Figure 1, previously characterized μ-conotoxins belong to three different clades of fish-hunting Conus (Gastridium, Chelyconus and Pionoconus), each represented by a discrete branch on the phylogenetic tree. The skeletal muscle (NaV1.4)-specific peptides, μ-GIIIA (Cruz, et al. 1985) and μ-PIIIA (Shon, et al. 1998), came from snails in two different clades, Gastridium and Chelyconus. Notably, the μ-conopeptides from species in the Pionoconus clade were previously shown to target neuronal Na channel subtypes (Zhang, et al. 2006).
Figure 1. Phylogenetic tree of Conus. The phylogenetic tree was constructed based on 16S mitochondrial DNA sequences as described in Materials and Methods.
Not all clades of Conus are shown; however, all lineages with previously characterized μ-conotoxins are indicated, including the Gastridium, Chelyconus and Pionoconus clades. μ-Conotoxins from the species shown in the phylogenetic tree are indicated below the name of the clade. Three species not previously included in an analysis of the molecular phylogeny of Conus, C. bullatus, C. cervus and C. dusaveli, form a distinct branch defining the Textilia clade. There are two clades that comprise all known mollusk-hunting species, the subgenera Conus and Cylindrus, and their branches are labeled “(M)”. The branches labeled “(W)” are various worm-hunting clades, the taxonomic status of these is presently being evaluated by A. Kohn and co-workers. The same tree is shown on the right, but with confidence values above 80% indicated; note that the four clades that are labeled are all branches on the tree with high confidence values.
Figure 2. Fish-hunting Conus species belonging to the Textilia clade.
Shown are shells of three different Conus species (top, C. bullatus; middle, C. cervus; bottom, C. dusaveli) that form a well-defined branch on the phylogentic tree (See Figure 1); the taxonomic designation for this group is Textilia (Swainson, 1840) with the type species being the bubble cone, C. bullatus. These species have not been extensively analyzed previously largely because of their deep-water habitat; it has been a challenge to collect species such as C. cervus and C. dusaveli, considered by collectors as among the 50 rarest seashells (a good shell specimen of C. cervus sell for $400 – $1,2 00).
Representative species from ten clades of Conus from which μ-conotoxins had not been characterized, including the newly defined Textilia clade were chosen for analysis. We took advantage of the fact that μ-conotoxins belong to the M-superfamily (Corpuz, et al. 2005). Potential μ-conotoxin sequences were obtained by PCR using primers based on conserved sequence elements of M-superfamily genes. A large number of M-superfamily sequences (>100) were obtained; the analysis of these sequences revealed putative new sequences that belong to the μ-conotoxin family. Of these, those from the representative species of the Textilia clade, C. bullatus, seemed the most distinctive and promising.
Three putative μ-conotoxin cDNA clones with unique structural features were identified from the cDNA library from C. bullatus venom ducts (see Materials and Methods). The predicted prepropeptide sequences are shown in Table 1; they are typical of precursors in the μ-conotoxin family. The striking signal sequence conservation of the μ-conopeptide precursors is evident by comparison of C. bullatus sequences with previously determined μ-conopeptide precursor sequences derived from Conus species in other clades, which are also shown in Table 1. The post-translational processing of a μ-conopeptide (prepropeptide) is well defined and predictable (Corpuz, et al. 2005); the precursors are proteolytically converted to the mature toxin at defined sites (---KR in the specific prepropeptide sequences shown).
Table 1.
Conus bullatus μ-conotoxins precursors

|
It should be noted that of the total number, 21, of C. bullatus cDNA M-superfamily clones analyzed, 10 encoded μ-BuIIIA and a polymorphic variant, two encoded μ-BuIIIB and one encoded μ-BuIIIC.
Analysis of the translated μ-conotoxin sequences from C. bullatus
The predicted mature toxins from the Conus bullatus cDNA sequences are compared to those of previously characterized μ-conotoxins in Tables 2 and 3. The three C. bullatus peptides, BuIIIA, BuIIIB and BuIIIC clearly define a distinctive class of μ-conotoxins, different from those discovered so far from the other phylogenetic lineages of Conus. The three mature μ-conotoxin sequences obtained from C. bullatus differ from the overall consensus sequence of all the μ-conotoxins from Pionoconus, Gastridium and Chelyconus shown in Table 2, which is: X3CCX5CX4CX4–5CC, where X is an amino acid other than Cys. In contrast, the consensus for the three C. bullatus peptides is: X4CCX5–7CX3CX5CC. Thus, in addition to AA substitutions, Conus bullatus peptides as a class have unique features: all three peptides have 4 AA before the first cysteine, a variable length of from 5–7 AA in the first loop (between the second and third Cys residues), and only three amino acids in the second loop (between the third and fourth Cys residues), instead of the 4 AA present in the other μ-conotoxins (Tables 2 and 3). In addition, neither BuIIIA, BuIIIB, nor BuIIIC have a glutamine residue on the amino terminus that is often postranslationally modified to pyroglumate, unlike several of the μ-conotoxins in the Gastridium and Chelyconus clades (Table 2).
Table 2.
Predicted mature toxins from μ-Conus cDNA sequences
| Species | μ–Conotoxin | Mature toxin Sequence | Reference | |
|---|---|---|---|---|
| Textilia | ||||
| C. bullatus | BuIIIA | VTDRCCKGKRECGRWCRDHSRCC# | This work | |
| C. bullatus | BuIIIB | VGERCCKNGKRGCGRWCRDHSRCC# | This work | |
| C. bullatus | BuIIIC | IVDRCCNKGNGKRGCSRWCRDHSRCC# | This work | |
|
| ||||
| Gastridium | ||||
| C. geographus | GIIIA | RDCCTOOKKCKDRQCKOQRCCA# | Cruz, et al,1989 | |
| C. geographus | GIIIB | RDCCTOORKCKDRRCKOMKCCA# | Cruz, et al,1989 | |
| C. tulipa | TIIIA | RHGCCKGOKGCSSRECROQHCC# | Lewis, et al., 2007 | |
|
| ||||
| Chelyconus | ||||
| C. purpurascens | PIIIA | ZRLCCGFOKSCRSRQCKOHRCCG# | Shon, et al., 1998 | |
|
| ||||
| Pionoconus | ||||
| C. consors | CnIIIA | GRCCDVPNACSGRWCRDHAQCC# | Zhang, et al. 2006 | |
| C. consors | CnIIIB | ZGCCGEPNLCFTRWCRNNARCCRQQ^ | Zhang, et al. 2006 | |
| C. magus | MIIIA | ZGCCNVPNGCSGRWCRDHAQCC# | Zhang, et al. 2006 | |
| C. catus | CIIIA | GRCCEGPNGCSSRWCKDHARCC# | Zhang, et al. 2006 | |
Z and O represent post-translationally modified residues pyroglutamate and hydroxyproline, respectively.
C-terminal amidation,
C-terminal free acid.
Table 3.
New M-5 superfamily of μ-conotoxins
| Species | μ-Conotoxin | Mature toxin Sequence |
|---|---|---|
| C. bullatus | BuIIIA | VTDRCCKGKRECGRWCRDHSRCC# |
| C. bullatus | BuIIIB | VGERCCKNGKRGCGRWCRDHSRCC# |
| C. bullatus | BuIIIC | IVDRCCNKGNGKRGCSRWCRDHSRCC# |
|
| ||
| C. consors | CnIIIA | GRCCDVPNACSGRWCRDHAQCC# |
| C. consors | CnIIIB | ZGCCGEPNLCFTRWCRNNARCCRQQ^ |
| C. magus | MIIIA | ZGCCNVPNGCSGRWCRDHAQCC# |
| C. catus | CIIIA | GRCCEGPNGCSSRWCKDHARCC# |
|
| ||
| C. kinoshitai | KIIIA | CCNCSSKWCRDHSRCC# |
| C. striatus | SIIIA | ZNCCNGGCSSKWCRDHARCC# |
| C. stercusmuscarum | SmIIIA | ZRCCNGRRGCSSRWCRDHSRCC# |
Chemical synthesis of C. bullatus μ-conotoxins
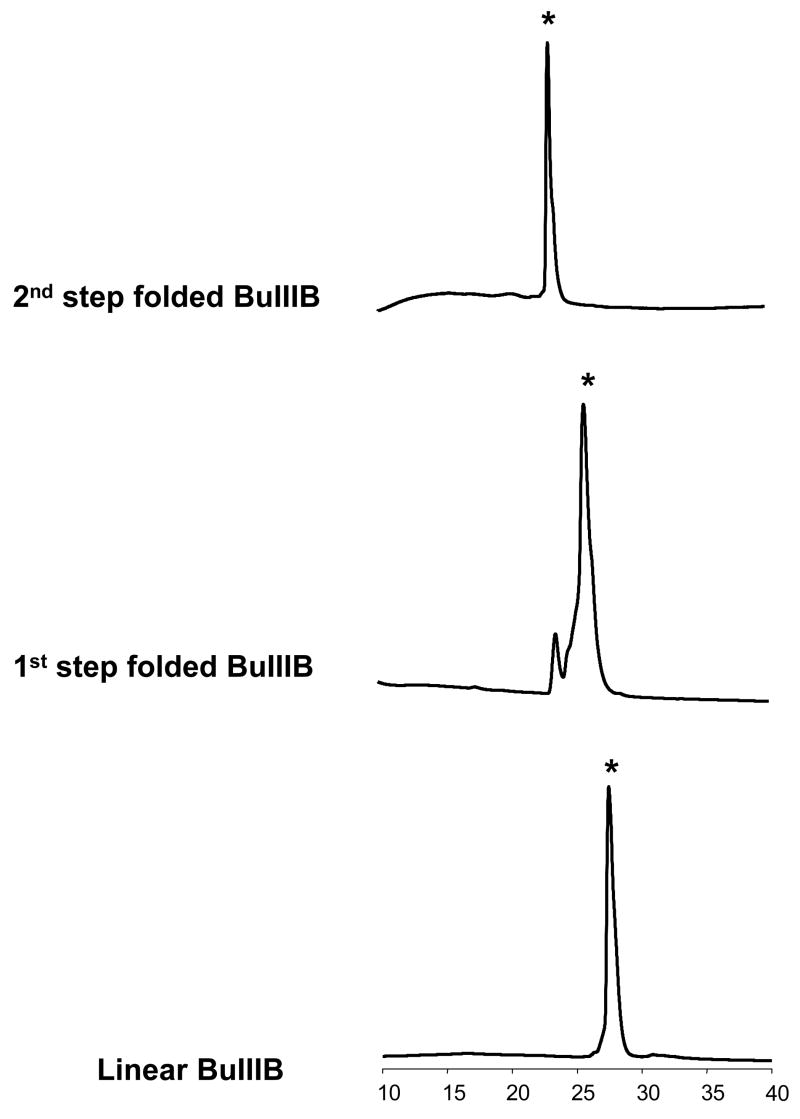
We chemically synthesized and folded all C. bullatus peptides, μ-conotoxins BuIIIA, BuIIIB, and BuIIIC, and tested their activities. The chemical synthesis and the folding protocols are described in Materials and Methods. A two-step protocol using analogues containing acetamidomethyl (Acm)-protection of Cys3 and Cys6 was designed to facilitate folding of C. bullatus μ-conotoxins into the form with native Cys connectivity (Cys1-Cys4, Cys2-Cys5 and Cys3-Cys6). Figure 3 depicts a representative HPLC profile of the two-step oxidative folding of BuIIIB. The first folding step with glutathione produced a major species that was purified by preparative HPLC, and subsequently oxidized with iodine. The final oxidation step yielded a single major product. The same methodology was used to produce BuIIIA and BuIIIC. In each case, the chemical identity of the final product was verified by mass spectrometry.
Figure 3. Two-step oxidative folding of C. bullatus μ-conotoxin BuIIIB.
RP-HPLC elution profiles of all intermediates obtained during the folding of linear BuIIIB. The peptide was folded by the two-step procedure described in Materials and Methods. BuIIIA and BuIIIC were prepared similarly. The correctly folded products are indicated with an asterisk (*).
Evaluation of BuIIIA, BuIIIB, and BuIIIC activity on Nav1.4 channels
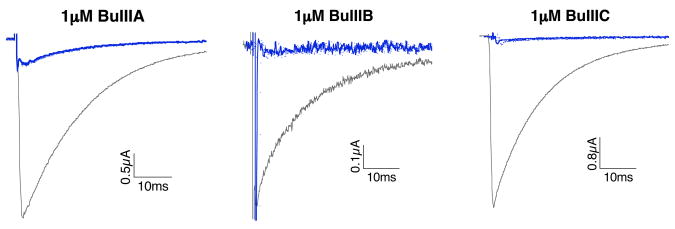
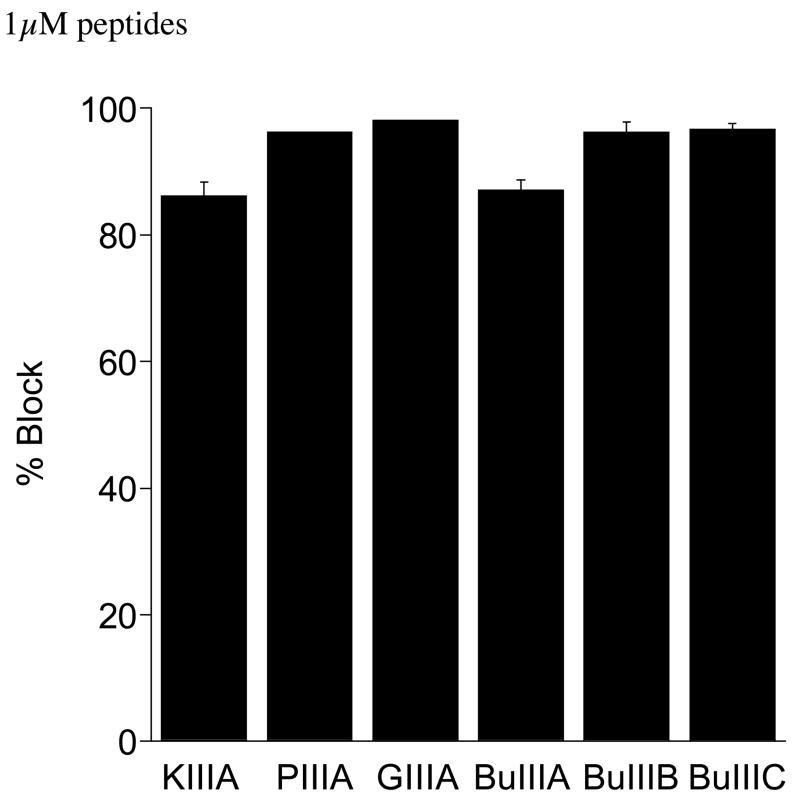
The activities of the three C. bullatus peptides were assessed on Nav1.4 expressed in Xenopus oocytes (see Materials and Methods) and compared to the activities of a representative set of μ-conotoxins, PIIIA, GIIIA, and KIIIA (33). PIIIA and GIIIA belong to the Chelyconus and Gastridium clades, and were selected for their remarkable divergence in amino acid sequence compared with BuIIIA, BuIIIB and BuIIIC; PIIIA and GIIIA contain postranslationally modified residues on the amino-terminal and in the first and third Cys-loop (Table 2). KIIIA is similar to the C. bullatus μ-conotoxins; however it has significant structural differences at the N-terminus and first Cys loop (Table 3). All C. bullatus μ-conotoxins were tested at the same concentration (1 μM) and under identical conditions, representative recordings are shown in Figure 4. BuIIIA, BuIIIB and BuIIIC inhibited most, if not all, of Nav1.4 sodium current. As shown in the bar graph of Figure 5, C. bullatus peptides BuIIIB and BuIIIC are potent antagonists of Nav1.4 with an average block of ~96%. KIIIA and BuIIIA inhibit Nav1.4 similarly (~87% block). PIIIA and GIIIA activity on Nav1.4 were determined from IC50 values published by Safo and colleagues (Safo, Rosenbaum et al. 2000). The calculated block of Nav1.4 by PIIIA (96% block) and GIIIA (98% block) is similar to that by BuIIIB and BuIIIC.
Figure 4. Block of Nav1.4 by μ-conotoxins BuIIIA, BuIIIB, BuIIIC.
Voltage-clamp recordings were performed on Xenopus oocytes expressing NaV1.4 as describe in Materials and Methods. Representative sodium currents recorded in the absence of toxin (control traces, gray) and following ~30 min exposure to 1μM BuIIIA, BuIIIB, and BuIIIC (black traces). Each trace represents the average of 5 responses.
Figure 5. Comparison of the block of Nav1.4 produced by 1 μM μ-conotoxins from C. bullatus and other Conus species.
PIIIA is from C. purpurascens (Chelyconus clade), GIIIA is from C. geographus (Gastridium clade), and KIIIA is from C. kinoshitai, whose phylogeny has not been determined. Values for PIIIA and GIIIA were calculated from IC50 values given in the Safo, et al. (31).
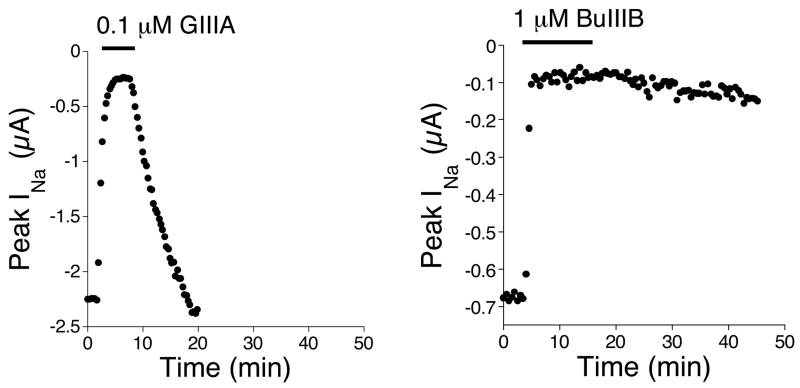
The block of Nav1.4 by BuIIIA was reversible (koff = 0.021 ± 0.006 min−1, N = 3 oocytes), and the current largely recovered in an hour. In contrast, BuIIIB is very slowly reversible (Figure 6) as is BuIIIC (not shown), and recovery from block in each case was too slow to determine koff values in the time frame of the experiments. GIIIA is the best studied μ-conotoxin, and its reversibility (koff = 0.14 ± 0.015 min−1, N = 3 oocytes) is also illustrated in Figure 6 for comparison. The detailed electrophysciological characterization of BuIIIA, BuIIIB and BuIIIC will be described elsewhere.
Figure 6. Time course of block and recovery of NaV1.4 following exposure to GIIIA and BuIIIB.
The bar at the top of each graph represents when the peptide was present at the indicated concentration. GIIIA is readily reversible whereas BuIIIB is relatively irreversible.
Discussion
A noteworthy aspect of this work is the discovery of a distinctive group of μ-conotoxins that are expressed in the venom of Conus species in the Textilia clade. One reason why the fish-hunting cone snails in the Textilia clade have not been extensively analyzed is their deep-water habitat; it has been a challenge to collect species in this clade such as Conus cervus and Conus dusaveli, considered by collectors as among the 50 rarest seashells. However, Conus bullatus is more accessible, and we were able to gradually accumulate enough venom ducts to carry out an extensive analysis of cDNA clones.
Ultimately, we were able to identify and characterize three μ-conotoxins from the Textilia clade of Conus that antagonizes Nav1.4. This particular tetrodotoxin-sensitive isoform of voltage-gated Na channels has previously been shown to be a target for μ-conotoxins, however, the strikingly divergent sequences of Conus bullatus peptides from previously characterized μ-conotoxins and their potency in inhibiting Nav1.4, make them novel tools for probing the structure activity relations of voltage gated sodium channels. A full investigation of the activity of C. bullatus peptides on other subtypes of Na channels is currently underway. The discovery of C. bullatus μ-conotoxins was accelerated by use of an exogene strategy to identify evolutionarily relevant Conus species.
It is significant that the first three peptides from the Textilia clade characterized differ greatly in their amino acid composition compared to previous antagonists of the Nav1.4 subtype. All C. bullatus peptides can be differentiated from previously characterized μ-conotoxins by their longer N-terminal extension (4 vs. 0–3 AA) and fewer residues in the second cys-flanked loop (3 instead of 4). Previously, M-superfamily conotoxins were divided into four branches based on the number of residues in the third Cys-flanked loop, namely, M-1, M-2, M-3, and M-4 (Corpuz, et al. 2005). BuIIIA, BuIIIB, and BuIIIC, together with SmIIIA (West, et al. 2002), SIIIA (Bulaj, et al. 2005), KIIIA (Bulaj, et al. 2005) and the μ-conotoxins identified in the Pionoconus clade in Table 2, comprise a new branch of the M-superfamily, M-5, in which there are 5 residues in the third Cys-loop of the mature toxin sequence (Table 3). M-5 μ-conotoxins have a significant degree of sequence homology in the third Cys loop, but differ as described above at the amino termini and in the first and second Cys-flanked loops. The ratio of residues in the second versus third loops, which we refer to as M-3/5 for BuIIIA, BuIIIB, and BuIIIC, and M-4/5 for the other μ-conotoxins in the M-5 family may account, in part, for the differences in their activity on Nav1.4. Absent from the M-5 superfamily is hydroxyproline, which are found in the second and third loops in the M-4 family members GIIIA, GIIIB, TIIIA, and PIIIA in Table 2.
Recent comparative functional analyses of the structure-activity-relationship of M-4 μ-conotoxin GIIIA (Choudhary, et al. 2007) and M-5 μ-conotoxin KIIIA (Zhang, et al. 2007) has led to the identification of specific residues involved in high affinity interactions with NaV1.4. Using pair-wise alanine mutations and double mutant cycle analysis Choudary and colleagues identified novel toxin-channel interactions involving residues K9, K11, K16, and R19 in GIIIA. Of these, only R19, the homolog of which is R24 in BuIIIC, and R13 in GIIIA, which was previously identified as critical for activity (Sato, et al. 1991; Dudley Jr, et al. 1995; French, et al. 1996; Hui, et al. 2002), the homolog of which is R17 in BuIIIC, are conserved for all M-5 μ-conotoxins except MIIIA and CnIIIA where the residue in the equivalent location of GIIIA’s R19 is glutamine (Table 2). The residues homologous to those believed to be important for KIIIA’s inhibition of NaV1.4 (W8, R10, D11, H12 and R14), are highly conserved in BuIIIA, BuIIIB, and BuIIIC (Table 3). However, our results indicate these shared features, by themselves, are not sufficient to explain the higher affinity for Nav1.4 of C. bullatus peptides BuIIIB and BuIIIC compared to KIIIA (Figure 5). BuIIIA and KIIIA have similar activities on NaV1.4 (Fig. 5), including being easily reversible (not illustrated). In contrast, BuIIIB and BuIIIC are more potent inhibitors of NaV1.4, like PIIIA and GIIIA (Figure. 5). However, PIIIA and GIIIA are readily reversible inhibitors (Safo, Rosenbaum et al. 2000), while BuIIIB and BuIIIC are almost irreversible (Figure. 6), both displaying a uniquely slow off-rate. The fact that BuIIIB and BuIIIC block Nav1.4 with about the same affinity as PIIIA and GIIIA given the significant differences in their amino acid composition is of importance. In frog neuromuscular preparations, PIIIA specifically blocks skeletal muscle Na channels irreversibly (28), and this property can be exploited to study synaptic potentials without confounding muscle action potentials. In view of their irreversibility, BuIIIB and BuIIIC may likewise be useful for studies of mammalian muscle preparations. Additionally, C. bullatus μ-conotoxins lack post-translationally modified residues such as pyroglutamate and hydroxyproline found in PIIIA and GIIIA, making them easier to chemically synthesize.
One possible hypothesis to be tested to explain the difference in activities of C. bullatus μ-conotoxins, BuIIIA, BuIIIB, and BuIIIC is that the specific combination of additional residues on the N-terminus, and the absence of a fourth residue in the second loop are critical for determining higher affinity of μ-conotoxins for Nav1.4. Experiments along these lines are currently being investigated.
In addition to the intrinsic importance of ligands specific for Nav1.4, this work pioneers a new approach for the discovery of subtype specific ligands in the Conus venom peptide system. In principle, the same general strategy can be applied to discovery from any phylogenetic lineage of animals. Some features of this discovery strategy may seem almost trivial: for example, it is critical to identify the tissue producing the compounds that the organism uses to interact with other organisms. For a venomous animal such as Conus, this is unambiguously the venom duct. Directly analyzing the source tissue makes the requisite molecular biology vastly more facile.
A second requirement of the discovery strategy is to elucidate the molecular genetics of the ligands of interest; in this case, the relevant gene family is the μ-conopeptides, which belong to the M-superfamily. This family and superfamily are sufficiently well defined to be accessible for analysis by PCR (Corpuz, et al. 2005). A third component of the discovery strategy is knowledge of the phylogenetics (as illustrated in Figure 1). Different M-superfamily sequences were collected from over 10 different clades of Conus, and the bioinformatics analysis of the sequences suggested that the Conus bullatus μ-conotoxin sequences from the Textilia clade were a sufficiently distinctive class so that further characterization, i.e., chemical synthesis and folding, was justified. Thus, a chemical synthetic capacity, and the ability to fold the linear conopeptides efficiently are additional technical requirements for the type of conopeptide discovery detailed in this paper. The final requirement is routine access to a sensitive assay – in this case, cloned Nav1.4 expressed in Xenopus oocytes.
This work highlights the exogene strategy as an effective method for identifying novel peptides with unique structural and functional properties. The three μ-conotoxins from C. bullatus, BuIIIA, BuIIIB, and BuIIIC, contain novel structural determinants that confer noteworthy activity for skeletal muscle subtype Nav1.4. These ligands could be used in the continuing effort to accurately define the features of the Nav1.4 pharmacophore and thereby facilitate the design of highly subtype-specific ligands that target Nav1.4.
Supplementary Material
Aligned nucleotide sequences of BuIIIA, BuIIIB and BuIIIC, with primer sequence region highlighted. Conserved residues are shaded. The similarity ranges from 90.2% between BuIIIA and BuIIIC to 94.1% between BuIIIB and BuIIIC. BuIIIA and BuIIIB have 94.0% similarity.
RP-HPLC elution profiles of reduced, one-step oxidized, two-step oxidized, and co-eluted products. The major species indicated by the asterisk (*) is the correctly folded product. When the one-step and two-step products are combined and applied to the HPLC, they co-elute as shown in the figure, indicating these two folding protocols generate the same product.
Acknowledgments
This work was supported by the NIH program project GM 48677. MH acknowledges joint support from NSF CHE and OISE 0610202 postdoctoral fellowship. We thank Professor Al Goldin for the rat NaV1.4 clone, and Kerry Matz for the Conus images.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amaya F, Wang H, Costigan M, Allchorne AJ, Egerton J, Stean J, Morisset V, Grose D, Gunthorpe MJ, Chessell IP, Tate S, Green PJ, Woolf CJ. The voltage-gated sodium channel Na(v)1.9 is an effector of peripheral inflammatory pain hypersensitivity. Journal of Neuroscience. 2006;26:12852–60. doi: 10.1523/JNEUROSCI.4015-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam L, Dowell C, Watkins M, Stitzel JA, Olivera BM, McIntosh JM. α-Conotoxin BuIA, a novel peptide from Conus bullatus distinguishes among neuronal nicotinic acetylcholine receptors. J Biol Chem. 2005;280:80–7. doi: 10.1074/jbc.M406281200. [DOI] [PubMed] [Google Scholar]
- Bulaj G, West PJ, Gareett JE, Marsh M, Zhang MM, Norton RS, Smith BJ, Yoshikami D, Olivera BM. Novel conotoxins from Conus striatus and Conus kinoshitai selectively block TTX-resistant sodium channels. Biochemistry. 2005;44:7259–7265. doi: 10.1021/bi0473408. [DOI] [PubMed] [Google Scholar]
- Bulaj G, Zhang MM, Green BR, Fiedler B, Layer RT, Wei S, Nielsen JS, Low Sj, Klein BD, Wagstaff JD, Chicoine L, Harty TP, Terlau H, Yoshikami D, Olivera BM. Synthetic m-O conotoxin MrVIB blocks TTX-resistant sodium channel Nav1.8 and has long-lasting analgesic activity. Biochemistry. 2006;45:7404–7414. doi: 10.1021/bi060159+. [DOI] [PubMed] [Google Scholar]
- Catterall W, Goldin AL, Waxman SG. International union of Pharmacology. XLVII Nomenclature and structure-function realtionships of voltage-gated sodium channels. Pharmacol Rev. 2005;57:397–409. doi: 10.1124/pr.57.4.4. [DOI] [PubMed] [Google Scholar]
- Catterall WA. Cellular and molecular biology of voltage-gated sodium channels. Physiol Rev. 1992;72:S15–S48. doi: 10.1152/physrev.1992.72.suppl_4.S15. [DOI] [PubMed] [Google Scholar]
- Catterall WA. From ionic currents to molecular mechanisms, the structure and function of voltage-gated sodium channels. Neuron. 2000;26:13–25. doi: 10.1016/s0896-6273(00)81133-2. [DOI] [PubMed] [Google Scholar]
- Choudhary G, Aliste MP, Tieleman DP, French RJ, Dudley SC., Jr Docking of _-Conotoxin GIIIA in the Sodium Channel Outer Vestibule. Channels. 2007;1(5):344–352. doi: 10.4161/chan.5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpuz GP, Jacobsen RB, Jimenez EC, Watkins M, Walker C, Colledge C, Garrett JE, McDougal O, Li W, Gray WR, Hillyard DR, Rivier J, McIntosh JM, Cruz LJ, Olivera BM. Definition of the M-conotoxin superfamily, characterization of novel peptides from molluscivorous Conus venoms. Biochemistry. 2005;44:8176–8186. doi: 10.1021/bi047541b. [DOI] [PubMed] [Google Scholar]
- Cruz LJ, Gray WR, Olivera BM, Zeikus RD, Kerr L, Yoshikami D, Moczydlowski E. Conus geographus toxins that discriminate between neuronal and muscle sodium channels. J Biol Chem. 1985;260:9280–9288. [PubMed] [Google Scholar]
- Duda T, Jr, Kohn AJ. Species-level phylogeography and evolutionary history of the hyperdiverse marine gastropod genus Conus. Molecular Phylogenetics & Evolution. 2005;34:256–272. doi: 10.1016/j.ympev.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Duda TF, Jr, Palumbi SR. Developmental shifts and species selection in gastropods. Proc Natl Acad Sci USA. 1999;96:10272–10277. doi: 10.1073/pnas.96.18.10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley SC, Jr, Todt H, Lipkind G, Fozzard HA. A μ-conotoxin-insensitive Na+ channel mutant, possible localization of a binding site at the outer vestibule. Biophys J. 1995;69:1657–1665. doi: 10.1016/S0006-3495(95)80045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison M, Olivera BM. α4/3 Conotoxins, Phylogenetic Distribution, Functional Properties, and Structure-Function Insights. The Chemical Record. 2007;7:341–353. doi: 10.1002/tcr.20131. [DOI] [PubMed] [Google Scholar]
- Espiritu DJD, Watkins M, Dia-Monje V, Cartier GE, Cruz LJ, Olivera BM. Venomous cone snails, molecular phylogeny and the generation of toxin diversity. Toxicon. 2001;39:1899–1916. doi: 10.1016/s0041-0101(01)00175-1. [DOI] [PubMed] [Google Scholar]
- French RJ, Prusak-Sochaczewski E, Zamponi GW, Becker S, Kularatna AS, Horn R. Interactions between a pore-blocking peptide and the voltage-sensor of the sodium channel, an electrostatic approach to channel geometry. Neuron. 1996;16:407–413. doi: 10.1016/s0896-6273(00)80058-6. [DOI] [PubMed] [Google Scholar]
- Goldin AL, Barchi RL, Caldwell JH, Hofmann F, Howe JR, Hunter JC, Kallen RG, Mandel G, Meisler MH, Berwald Netter Y, Noda M, Tamkun MM, Waxman SG, Wood JN, Catterall WA. Nomenclature of voltage-gated sodium channels. Neuron. 2000;28:365–368. doi: 10.1016/s0896-6273(00)00116-1. [DOI] [PubMed] [Google Scholar]
- Hille B. Ion Channels of Excitable Membranes. 3. Sunderland, MA: Sinauer Associates, Inc; 2001. [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES, Bayesian inference of phylogeny. Bioinformatics. 2001;17:753–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Hui K, Lipkind G, Fozzard HA, French RJ. Electrostatic and steric contributions to block of the skeletal muscle sodium channel by μ-conotoxin. J Gen Physiol. 2002;119:45–54. doi: 10.1085/jgp.119.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen R, Jimenez EC, Grilley M, Watkins M, Hillyard D, Cruz LJ, Olivera BM. The contryphans, a D-tryptophan-containing family of Conus peptides, interconversion between conformers. J Peptide Res. 1998;51:173–179. doi: 10.1111/j.1399-3011.1998.tb01213.x. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3, Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Norton RS, Pallaghy PK. The cystine knot structure of ion channel toxins and related polypeptides. Toxicon. 1998;36:1573–1583. doi: 10.1016/s0041-0101(98)00149-4. [DOI] [PubMed] [Google Scholar]
- Ogata N, Ohishi Y. Molecular diversity of structure and function of the voltage-gated Na+ channels. Japanese Journal of Pharmacology. 2002;88:365–377. doi: 10.1254/jjp.88.365. [DOI] [PubMed] [Google Scholar]
- Olivera BM. Conus peptides, biodiversity-based discovery and exogenomics. Journal of Biological Chemistry. 2006;281:31173–31177. doi: 10.1074/jbc.R600020200. [DOI] [PubMed] [Google Scholar]
- Olivera BM, Teichert RW. Diversity of the Neurotoxic Conus peptides, A Model for Concerted Pharmacological Discovery. Molecular Interventions. 2007;7(5):253–262. doi: 10.1124/mi.7.5.7. [DOI] [PubMed] [Google Scholar]
- Planells-Cases R, Caprini M, Zhang J, Rockenstein EM, Rivera RR, Murre C, Masliah E, Montal M. Neuronal death and perinatal lethality in voltage-gated sodium channel alpha(II)-deficient mice. Biophys J. 2000;78:2878–91. doi: 10.1016/S0006-3495(00)76829-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MRBAYES 3, Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Safo P, Rosenbaum T, Shcherbatko A, Choi D, Han E, Toledo-Aral J, Olivera BM, Brehm P, Mandel G. Distinction among neuronal subtypes of voltage-activated sodium channels by μ-conotoxin PIIIA. J Neurosci. 2000;20:76–80. doi: 10.1523/JNEUROSCI.20-01-00076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Ishida Y, Wakamatsu K, Kato R, Honda H, Ohizumi Y, Nakamura H, Ohya M, Lancelin JM, Kohda D, Inagaki F. Active site μ-conotoxin GIIIA, a peptide blocker of muscle sodium channels. J Biol Chem. 1991;266:16989–16991. [PubMed] [Google Scholar]
- Shon K, Olivera BM, Watkins M, Jacobsen RB, Gray WR, Floresca CZ, Cruz LJ, Hillyard DR, Bring A, Terlau H, Yoshikami D. μ-Conotoxin PIIIA, a new peptide for discriminating among tetrodotoxin-sensitive Na channel subtypes. J Neurosci. 1998;18:4473–4481. doi: 10.1523/JNEUROSCI.18-12-04473.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terlau H, Olivera BM. Conus venoms, a rich source of novel ion channel-targeted peptides. Physiol Rev. 2004;84:41–68. doi: 10.1152/physrev.00020.2003. [DOI] [PubMed] [Google Scholar]
- West PJ, Bulaj G, Garrett JE, Olivera BM, Yoshikami D. μ-Conotoxin SmIIIA, a potent inhibitor of TTX-resistant sodium channels in amphibian sympathetic and sensory neurons. Biochemistry. 2002;41:15388–15393. doi: 10.1021/bi0265628. [DOI] [PubMed] [Google Scholar]
- Woodruff-Pak D, Grenn TJ, Levin SI, Meisler MH. Inactivation of sodium channel Scn8A (Na-sub(v)1.6) in Purkinje neurons impairs learning in Morris water maze and delay but not trace eyelink classical conditioning. Behav Neurosci. 2006;120:229–240. doi: 10.1037/0735-7044.120.2.229. [DOI] [PubMed] [Google Scholar]
- Yotsu M, Yamazaki T, Meguro Y, Endo A, Murata M, Naoki H, Yasmuoto T. Production of tetrodotoxin and its derivatives by Pseudomonas sp. isolated from the skin of a pufferfish. Toxicon. 1987;25:225–228. doi: 10.1016/0041-0101(87)90245-5. [DOI] [PubMed] [Google Scholar]
- Zhang M, Fiedler B, Green BR, Catlin P, Watkins M, Garett JE, Smith BJ, Yoshikami D, Olivera BM, Bulaj G. Structural and functional diversities among _-conotoxins targeting TTX-resistant sodium channels. Biochemistry. 2006;45:3723–3732. doi: 10.1021/bi052162j. [DOI] [PubMed] [Google Scholar]
- Zhang M-M, Green BR, Catlin P, Fiedler B, Azam L, Chadwick A, Terlau H, McArthur JR, French RJ, Gulyas J, Rivier JE, Smith BJ, Norton RS, Olivera BM, Yoshikami D, Bulaj G. Structure/Function Characterization of _-conotoxin KIIIA, an Analgesic, Nearly Irreversible Blocker of Mammalian Neuronal Sodium Channels. The Journal of Biological Chemistry. 2007;282(42):30699–30706. doi: 10.1074/jbc.M704616200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Aligned nucleotide sequences of BuIIIA, BuIIIB and BuIIIC, with primer sequence region highlighted. Conserved residues are shaded. The similarity ranges from 90.2% between BuIIIA and BuIIIC to 94.1% between BuIIIB and BuIIIC. BuIIIA and BuIIIB have 94.0% similarity.
RP-HPLC elution profiles of reduced, one-step oxidized, two-step oxidized, and co-eluted products. The major species indicated by the asterisk (*) is the correctly folded product. When the one-step and two-step products are combined and applied to the HPLC, they co-elute as shown in the figure, indicating these two folding protocols generate the same product.