Abstract
Mucociliary clearance is a critical innate defense system responsible for clearing up invading pathogens including bacteria and virus. Although the right amount of mucus is good, excessive mucus causes airway obstruction and tends to precipitate disease symptoms. Rhinovirus (RV) is a common cold virus that causes asthma and chronic obstructive pulmonary disease exacerbation. Mucus overproduction has been linked to the pathogenesis of RV-induced diseases and disease exacerbations. However, the molecular mechanism is not clear. In this study, using one of the major airway mucin-MUC5AC as marker, we found that both major and minor groups of RV induced mucin production in primary human epithelial cells and cell line. RV1A (a minor group of RV) could induce mucous cell metaplasia in vivo. Viral replication was needed for RV-induced mucin expression, and this induction was also dependent on TLR3, suggesting the involvement of double-stranded (ds) RNA signaling. Indeed, dsRNA alone could also induce mucin expression. TLR3-mediated mucin induction was negatively regulated by MyD88, and only partially dependent on TRIF, which suggests a departure from well-documented TLR3 signaling paradigm that mediates inflammatory and other innate defense gene inductions. In addition, TLR3 signaling activated epidermal growth factor receptor (EGFR) through inductions of the expression of EGFR ligands (transforming growth factor-α and amphiregulin), which in turn activated EGFR-ERK signaling and mucin expression through an autocrine/paracrine loop. This novel coupling of antiviral defense machinery (i.e., TLR3) and major epithelial proliferation/repair pathway (i.e., EGFR) might play an important role in viral-induced airway remodeling and airway disease exacerbation.
Keywords: mucin, airway epithelium, TLR3, rhinovirus
CLINICAL RELEVANCE
The mechanism of viral infection on airway disease exacerbation is poorly understood. The present study suggests that viral-induced epidermal growth factor receptor activation and mucus production may contribute to the pathogenesis of virus-induced airway disease exacerbation.
Mucociliary clearance is a powerful innate defense system that is responsible for clearing up most of the invading pathogens, including bacteria and virus. Although the right amount of mucus is beneficial, excessive mucus production causes airway obstruction, which may lead to enhanced inflammation and exacerbations of the existing diseases. In chronic airway diseases, mucous cell metaplasia and concomitantly persistent mucus overproduction significantly increase the morbidity and mortality of those diseases (1–3).
Rhinovirus (RV) is a small (30 nm in diameter), nonenveloped, positive-stranded RNA virus (4). More than 100 serotypes have currently been identified. Based on the cellular receptor being used for viral entry, RV can be divided into two main groups: major group that uses intercellular adhesion molecule 1 (ICAM-1), and a minor group that uses low-density lipoprotein receptor (LDLR) family (4). Due to the structural difference of ICAM-1, the major group of human RV cannot infect rodents. Only the minor group can infect rodent cells, and most recently, it has been shown that RV1B, a minor group RV, can directly infect mouse lung in a live animal (5, 6).
Airway RV infection is the major cause of common cold (7). Although viral cold only causes minor symptoms, the financial burden resulting from loss of working days is very substantial. Recently, RV infection has been causally linked to the exacerbation of various airway diseases (8), particularly asthma (9). Accumulating data indicate that RV infection causes asthma exacerbation both in children (10) and in adults (11). Mucus overproduction is one of the major symptoms and significantly contributes to the morbidity of those diseases. However, the underlying mechanism is unclear. In a previous study, we have demonstrated that interferon-inducible RNA-dependent protein kinase (PKR)-mediated double-stranded (ds) RNA-dependent pathway is responsible for epithelial antiviral defense induced by both major and minor groups of RV infection (12), which suggests the importance of viral dsRNA-induced signaling. It has also been reported before that dsRNA could induce MUC2, a major colonic but a minor airway mucin, in both intestinal and airway epithelial cells through a G protein–coupled receptor and P38-mitogen-activated protein kinase (MAPK)-dependent pathways (13). Thus, we hypothesize that RV-induced mucin production may also be mediated through a dsRNA-dependent pathway. In the present study, using MUC5AC (one of the major airway mucins) as a marker, we have demonstrated that both major and minor groups of RV could induce mucin production in epithelial cells, which depends on viral replication and at least partly on TLR3 (a pattern recognition receptor [PRR] that specifically recognizes viral dsRNA). To further explore TLR3-dependent signaling, we used dsRNA as a surrogate for viral infection. By this approach, we found that TLR3-mediated MUC5AC expression was negatively regulated by the common TLR adaptor-MYD88 (14), and only partially dependent on the TLR3/4-specific adaptor-TRIF (14), suggesting a novel pathway that is not quite the same as the well-established TLR3 signaling paradigm (14). Further study indicates that TLR3-mediated MUC5AC expression was dependent on the transcriptional activation of multiple epidermal growth factor receptor (EGFR) ligands, and the subsequent activation of EGFR through an autocrine/paracrine loop. Our finding suggests a novel link between the epithelial antiviral signaling (i.e., TLR3) and the airway remodeling machinery (i.e., EGFR and mucin), which may play an important role in airway disease exacerbation.
MATERIALS AND METHODS
Viruses, Chemicals, Inhibitors, Antibodies
RV16 and RV1A stocks were amplified and purified based on the previous published protocol (15). Briefly, HeLa cell suspension was infected at room temperature for 1 hour with a multiplicity of infection (MOI) of 10 to 15 plaque-forming units (PFU) per cell. Infected cell suspensions were diluted 10-fold in prewarmed medium B and incubated at 35°C for 7 to 8 hours. The cells were then pelleted and resuspended in PBS. Virus was released from cells by three cycles of freezing and thawing and then harvested as the supernatant after centrifugation to pellet cell debris. Sucrose gradient was used to purify the virus particles released from cells. Viral titers were determined by plaque assay as described previously (15). To make replication-deficient RV16 (UV-RV16), stocks of RV16 were ultraviolet irradiated, as described previously (16), with a slight modification. Briefly, 1 ml RV stock solution, containing 5 × 108 RV16, was exposed to 200 μW cm−2 ultraviolet light for 10 minutes on ice. Synthetic dsRNA was purchased from InvivoGen (San Diego, CA). Chemical inhibitors (AG1478, BIBX1382, U0126, TAPI, GM6001) were purchased from Calbiochem (EMD Biosciences, Inc., San Diego, CA). Anti-MUC5AC monoclonal antibody was purchased from Labvision Corp. (now acquired by Thermo Scientific, Fremont, CA). Anti-TLR3 antibody was purchased from Abcam (Cambridge, MA). Neutralizing antibody anti-EGFR was purchased from Calbiochem (EMD Biosciences). Other neutralizing antibodies (anti—transforming growth factor [TGF]-α, anti-HBEGF, anti-amphiregulin) as well as their species- and isotype- matched control antibodies (anti-goat IgG, anti-mouse IgG2A) were purchased from R&D Systems, Inc. (Minneapolis, MN). Actin antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies against phospho-proteins (pEGFR, pERK) were purchased from Cell signaling technology (Danvers, MA).
Cell Culture, RV Infection, and dsRNA Treatment
Differentiated primary cell culture.
Human tracheobronchial tissues were obtained from National Disease Research Interchange, with an approved protocol. The Hamner Institute Health and Safety Committee approved all procedures involved in tissue procurement. We have, in the past, successfully established primary airway epithelial cultures from these tissues (17, 18). Normally, primary cells were plated on a Transwell (Corning Costar, Corning, NY) chamber (25 mm) at 1–2 × 104 cells/cm2, in a Ham's F12:Dulbecco's modified Eagle's medium (1:1) supplemented with eight factors, including: insulin (5 μg/ml), transferrin (5 μg/ml), epidermal growth factor (10 ng/ml), dexamethasone (0.1 μM), cholera toxin (10 ng/ml), bovine hypothalamus extract (15 μg/ml), bovine serum albumin (0.5 mg/ml), and all-trans-retinoic acid (30 nM). After a week in immersed culture condition, cultured cells were shifted to an air–liquid interface culture condition. Under the biphasic culture condition, high transepithelial resistance (> 500 Ω · cm2), multiple cell layers, cilia beating, and the formation of mucus-secreting granules were observed (17, 18). Normally, experiments were performed at Day 21 or 2 weeks after the change of the culture condition from immersed to air–liquid interface. Medium was routinely changed once every other day. To account for the donor variation, primary cell data were repeated on at least three independent donors.
NCI-H292 cells.
Cells were obtained from ATCC, and cultivated on regular tissue culture dish in RPMI media plus 10% fetal bovine serum (FBS).
RV infection and dsRNA treatment.
For differentiated primary cell culture, 100 μl media containing desired concentration of RV or dsRNA was only added on the top of cells, which mimicked apical side of airway epithelia. For the monolayer primary cells and NCI-H292 cells, RV or dsRNA was diluted into culture media with desired concentration.
Real-Time PCR
Real-time PCR was performed as described previously (19). cDNA was prepared from 3 μg of total RNA with Moloney murine leukemia virus (MoMLV)–reverse transcriptase (Promega, Inc., Madison, WI) by oligo-dT primers for 90 minutes at 42°C in a 20-μl reaction solution, and was then further diluted to 100 μl with water for the following procedures. Two microliters of diluted cDNA was analyzed using 2× SYBR Green PCR Master Mix by an ABI 5700 or ABI Prism 7900HT Sequence Detection System (Applied Biosystems Inc., Foster City, CA), following the manufacturer's protocol. Primers (Table 1) were used at 0.2 μM. The PCR reaction was performed in 96-well optical reaction plates, and each well contained a 50-μl reaction mixture. The SYBR green dye was measured at 530 nm during the extension phase. The relative mRNA amount in each sample was calculated based on the ΔΔCt method using housekeeping gene GAPDH. The purity of amplified product was determined from a single peak of a dissociation curve. Efficiency curves were performed for each gene of interest relative to the housekeeping gene, based on the manufacturer's instructions. Results were calculated as fold induction over control, as described previously (19).
TABLE 1.
REAL-TIME PRIMERS
| Gene | Primers | |
|---|---|---|
| MUC5AC | Forward | GCCTTCACTGTACTGGCTGAG |
| Reverse | TGGGTGTAGATCTGGTTCAGG | |
| AREG | Forward | GCCTGGAAGACACCCTAATGTG |
| Reverse | GGCCGTGTCAACAAGGATACTT | |
| HBEGF | Forward | CCATTCTGAAAGGCTGGTTTG |
| Reverse | TACTCCGGAAGGGTCCTTTGT | |
| TGFa | Forward | TGATGCCACCAGATTTGACTG |
| Reverse | GTGCGCTTGAATGTCAGGAAT | |
| TLR3 | Forward | ATTGGGCAAGAACTCACAGG |
| Reverse | AGCATCAGTCGTTGAAGGCT | |
| TRIF | Forward | ACTGAACGCAGCCTACTCAGC |
| Reverse | ATGACATGTGGCTCCCAAAAG | |
| MYD88 | Forward | TGGTGGTGGTTGTCTCTGATG |
| Reverse | GGATGCTGGGGAACTCTTTCT | |
| GAPDH | Forward | CAATGACCCCTTCATTGACC |
| Reverse | GACAAGCTTCCCGTTCTCAG | |
Immunofluorescence
For immunofluorescence, the filter where the differentiated epithelial cells were grown was cut out and fixed in 4% paraformaldehyde and then embedded in paraffin. Sections were prepared in The Hamner Histology Core and incubated with 1:100 diluted anti-MUC5AC antibody overnight at 4°C. Anti-mouse IgG secondary antibody was conjugated with Alexa488 (Invitrogen, Carlsbad, CA). Fluorescence images were acquired by confocal microscope (LSM 510 meta; Carl Zeiss, Thornwood, NY).
Mouse Model of RV Infection and Immunohistochemistry
Six- to eight-week-old female Balb/C mice were anesthetized and administered 50 μl PBS containing 106 PFU RV1A as described elsewhere (5). The control mice were administered with PBS only. As an additional control, ultraviolet-crosslinked RV (UV-RV1A) was made by the same protocol as UV-RV16 and administered at the same amount to examine the role of RV replication in goblet cell induction. Twenty-four hours later mice were killed, and lungs were perfused, fixed in paraformaldehyde, and embedded in paraffin. For immunohistochemistry, sections were incubated with 1:100 diluted anti-MUC5AC antibody overnight at 4°C. An anti-mouse IgG antibody conjugated with alkaline phosphatase was used for detection. The final slides were counterstained with hematoxylin. The images were acquired by a microscope (AxioObserver Z1; Carl Zeiss).
Western Blot
Total cellular protein was collected based on the methods described previously (20). The sources of antibodies have been described in Viruses, Chemicals, Inhibitors, Antibodies. Equal protein load for both total and nuclear proteins was confirmed using the staining of anti-actin antibody.
Small Interference RNA and Transient Transfection
Control small interference (si)RNA was purchased from Ambion (Austin, TX). siRNA against TLR3 (GGTATAGCCAGCTAACTAGAA) (21), TRIF (GACCAGACGCCACTCCAAC) (22), and MYD88 (CTGGAACAGACAAACTATC) (22) were synthesized by Ambion. siRNA was transfected into cells using lipofectamine 2000 (Invitrogen) based on manufacturer's instructions. Successful knockdown of the target was confirmed by real-time RT-PCR and Western blot.
Statistical Analysis
Experimental groups were compared using a two-sided Student's t test, with significance level set as P < 0.05. When data were not distributed normally, significance was assessed with the Wilcoxon matched-pairs signed-ranks test, and P < 0.05 was considered to be significant. Matlab 6.0 with statistics toolbox (MathWorks, Inc., Natick, MA) was used for analyses of the data.
RESULTS
Both Major and Minor Groups of RV Induced Mucin Gene Expression
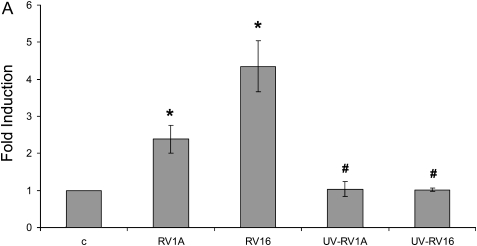
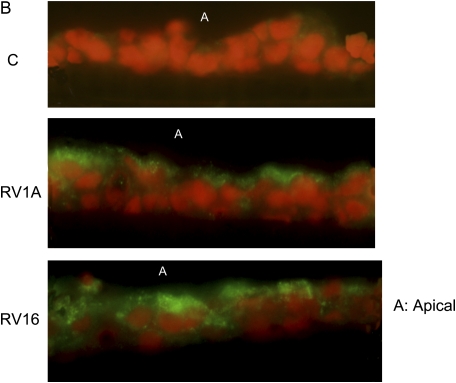
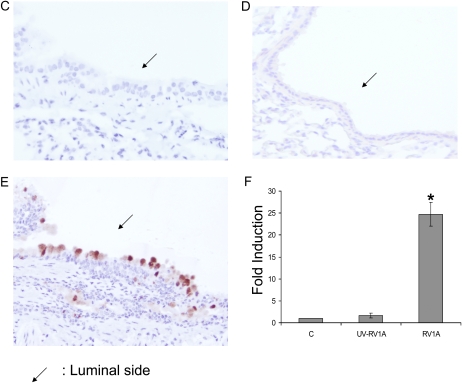
To test if RV could induce mucin production in airway epithelial cells, we infected differentiated primary human tracheobronchial epithelial cells (TBE) with either RV16 (a major group RV) or RV1A (a minor group RV) based on the protocol described previously (12). Both RVs induced MUC5AC expression at both RNA (Figure 1A) and protein level (Figure 1B). And these inductions appeared to depend on RV replication because ultraviolet-crosslinked RVs failed to induce any MUC5AC expression (Figure 1A). Consistently, RV1A infection on mouse trachea also induced goblet cell metaplasia (Figure 1E) and Muc5ac expression (Figure 1F), which agrees with the recent report on mucin inducing effect of RV1B (another minor group of RV) infection of a mouse model of asthma (5). In contrast, PBS control and ultraviolet-crosslinked RV1A did not have any effect (Figures 1C, 1D, and 1F), suggesting that RV replication is important in the induction of goblet cell metaplasia and mucin expression. Due to the difference between human and mouse ICAM-1, RV16 could not infect mouse directly. But the recent report has demonstrated that RV16 could induce mucin in a human ICAM-1 transgenic mouse model (5). Thus, both the major and the minor group of RV could induce mucin expression in vitro and in vivo.
Figure 1.
Both major and minor groups of rhinovirus (RV) induced mucin gene expression. Animals, culture condition, RV infection protocol, and other related methods have been described in Materials and Methods. (A) RV1A, RV16, and their nonreplicating counterparts induced by ultraviolet radiation (UV-RV1A, UV-RV16) were used to infect differentiated human primary epithelial cells at 5 × 106 plaque-forming units at the apical side. Forty-eight hours later, total cellular RNA was collected and MUC5AC expression was detected by real-time PCR. *P < 0.05 when comparing with control (n = 4); #P < 0.05 compared with non–UV-radiated counterparts (i.e., UV-RV1A versus RV1A; UV-RV16 versus RV16) (n = 4). (B) Differentiated primary cells were infected by RV for 48 hours as described above. The filters were cut and fixed as described in Materials and Methods. The sections made from those filters were then incubated with anti-MUC5AC monoclonal antibody and detected by immunofluorescence. A: Apical side. Green: anti-MUC5AC antibody; red: nucleus staining by propidium iodide. This is the representative image from three independent experiments. (C–E) Mice were infected by (C) PBS, (D) 106UV-RV1A, (E) 106 RV1A as described in Materials and Methods. Twenty-four hours later, their lungs were perfused and fixed. The tissue sections were incubated with anti-MUC5AC antibody and detected by colorimetric method. The brown color shows the MUC5AC protein staining, and those were all surface goblet cells. Five mice were used for each group (i.e., control and RV-infected). These are the representative images from those lungs. (F) Real-time PCR result of Muc5ac expression from whole lung. C: PBS control, UV-RV1A:UV-crosslined RV1A, RV1A: RV1A treatment. *P < 0.05, n = 5.
RV-Induced Mucin Expression Was Mediated at Least Partly through TLR3
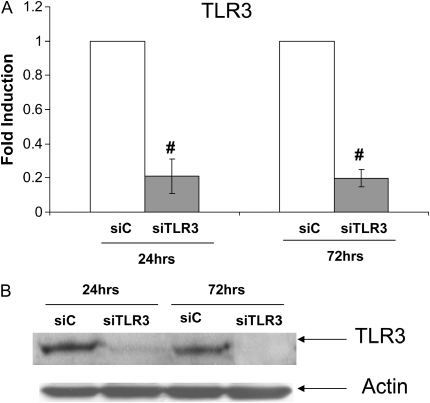
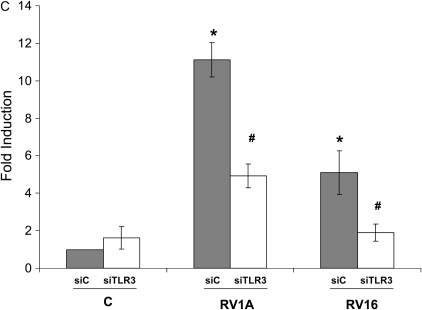
Since RV16 and RV1A used different receptor, RV-induced mucin expression did not appear to depend on the viral entry. Considering its dependence on viral replication (Figure 1A) and our previous report of dsRNA-mediated signaling in epithelial antiviral defense (12), we then tested the hypothesis that RV-induced MUC5AC expression was mediated by viral dsRNA-induced signaling. siRNA interference approach was used for this discovery. To facilitate the transfection study, we used an epithelial cell line, NCI-H292, which has been a common model cell line for mucin study and showed similar RV-induced mucin expression (Figure 2C). We first tested PKR and got no effect (data not shown). We then tested another well-known dsRNA receptor-TLR3. By transfecting cells with TLR3-specific siRNA (siTLR3), we observed more than 80% reduction in its mRNA level by real-time PCR (Figure 2A). This reduction of TLR3 expression was started at 24 hours after transfection and persisted for at least 72 hours later (Figure 2A). Because the widely used TLR3 monoclonal antibody (clone 3.7) cannot be used for Western blot, we screened many commercial vendors and finally identified one antibody from Abcam that recognized the right TLR3 protein in Western blot. As shown in Figure 2B, siTLR3 could significantly knock down TLR3 protein production at both 24 and 72 hours, consistent with mRNA knockdown result (Figure 2A). This observation suggests that TLR3 siRNA could persistently reduce TLR3 expression at least until 72 hours later. Interestingly, knocking down TLR3 significantly blocked both RV1A- and RV16-induced MUC5AC expression (Figure 2C), suggesting that TLR3-mediated signaling is at least partly responsible for RV-induced mucin expression. Therefore, we focused on TLR3-mediated MUC5AC expression in the following study.
Figure 2.
RV-induced mucin expression was mediated at least partly through TLR3. For the convenience of transfection, NCI-H292 cells were used for this study. (A) Control siRNA (siC) and TLR siRNA (siTLR3) were transfected into NCI-H292 cells. Twenty-four and 72 hours later, RNA was extracted and TLR3 expression was determined by real-time PCR. #P < 0.05 compared with control (n = 4). (B) Western blot was performed using the specific anti-TLR3 antibody. Anti-actin antibody was used as a control for loading. This is the representative image from three independent experiments. (C) TLR3-dependent induction of MUC5AC expression by RV1A or RV16 infection on NCI-H292 cells. The cells were transfected with control siRNA (siC) or TLR3 siRNA (siTLR3), and then infected with RV1A or RV16 separately at 106 for 48 hours. Shaded bars represent siC-transfected cells, and open bars represent siTLR3-transfected cells. *P < 0.05 compared with noninfected control (C) (n = 4); #P < 0.05 when comparing siTLR3-transfected cells (open bars) with siC-transfected cells (shaded bars) (n = 4).
TLR3 Ligand–dsRNA Induced MUC5AC Expression Both in Primary Cells and in NCI-H292 Cells
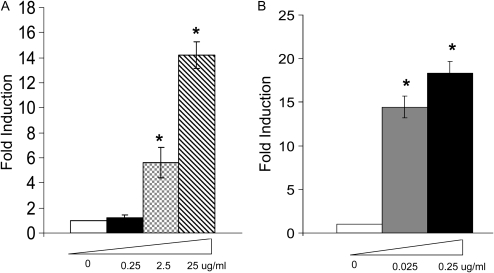
We further tested whether direct treatment of TLR3 ligand–dsRNA could induce MUC5AC expression. Indeed, dsRNA induced MUC5AC expression in both primary TBE cells (Figure 3A) and NCI-H292 cells (Figure 3B), and these inductions were dose dependent (Figure 3). NCI-H292 cells appeared to be more sensitive to dsRNA treatment than primary cells, and the reason was unclear. One possibility could be that since differentiated primary cells had multiple layers, different layers would experience different concentrations of dsRNA; thus, the overall response appeared to be weaker than monolayered NCI-H292 cells.
Figure 3.
TLR3 ligand–dsRNA induced MUC5AC expression both in primary cells and NCI-H292 cells. (A) Differentiated primary cells were treated with various doses (0.25, 2.5, 25 μg/ml) of dsRNA for 24 hours as described in Materials and Methods. Control was treated with PBS. RNA was collected and MUC5AC expression was determined by real-time PCR. *P < 0.05 compared with control. (B) NCI-H292 cells were treated with various doses (0.025, 0.25 μg/ml) of dsRNA for 24 hours as described in Materials and Methods. Control was treated with PBS. RNA was collected and MUC5AC expression was determined by real-time PCR. *P < 0.05 compared with control.
TLR3-Mediated MUC5AC Expression Was Partially Dependent on TRIF and Negatively Regulated by MYD88
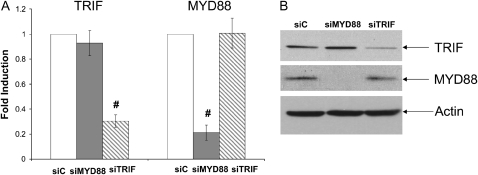
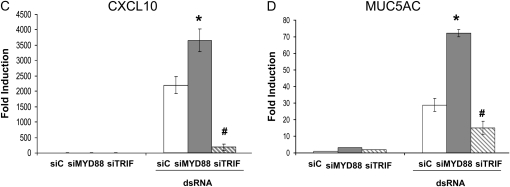
It has been well established that TLR3 uses an adaptor protein-TRIF to transduce signaling (14). We tested whether TRIF was the downstream adaptor of TLR3-mediated MUC5AC expression. Successful knockdown of both TRIF mRNA and protein were confirmed (Figures 4A and 4B). Surprisingly, siRNA knockdown of TRIF could only partially affect TLR3-mediated MUC5AC expression (Figure 4D), while under the same condition CXCL10 (a TLR3-TRIF–dependent gene) expression was almost completely repressed (Figure 4C), suggesting that TLR3-mediated MUC5AC expression might not completely depend on TRIF. Although MYD88 was not a common TLR3 adaptor, it is well involved in many other TLR signaling (14). Thus, we also tested whether the knockdown of MYD88 could affect TLR3-mediated MUC5AC expression. Interestingly, instead of reducing, knockdown of MYD88 could super-induce TLR3-mediated MUC5AC expression (Figure 4D), which might be consistent with a recent report showing a negative regulatory role of MYD88 on TLR3 signaling (23). Nonetheless, it appears that there may exist another yet unknown pathway that is very different from common TLR3 paradigm (14).
Figure 4.
TLR3-mediated MUC5AC expression was not dependent on either MYD88 or TRIF. (A) Control siRNA (siC), MYD88 siRNA (siMYD88), and TRIF siRNA (siTRIF) were transfected into NCI-H292 cells. Forty-eight hours later, RNA was extracted; MYD88 or TRIF expression was determined by real-time PCR. #P < 0.05 compared with control (n = 4). (B) Western blot was further used to confirm the knockdown of either MYD88 or TRIF. Anti-Actin antibody was used as a control for loading. This is the representative image from four independent experiments. (C) Control siRNA (siC), MYD88 siRNA (siMYD88), and TRIF siRNA (siTRIF) were transfected into NCI-H292 cells. Twenty-four hours later, cells were treated with dsRNA (0.025 μg/ml), and RNA was collected after an additional 24 hours. CXCL10 expression was determined by real-time PCR. *P < 0.05 when comparing CXCL10 expression in siMYD88-transfected and dsRNA-treated cells (shaded bar) with CXCL10 expression in siC-transfected and dsRNA-treated cells (open bar) (n = 4); #P < 0.05 when comparing CXCL10 expression in siTRIF-transfected and dsRNA-treated cells (hatched bar) with CXCL10 expression in siC-transfected and dsRNA-treated cells (open bar) (n = 4). (D) Control siRNA (siC), MYD88 siRNA (siMYD88), and TRIF siRNA (siTRIF) were transfected into NCI-H292 cells. Twenty-four hours later, cells were treated with dsRNA (0.025 μg/ml), and RNA was collected after an additional 24 hours. MUC5AC expression was determined by real-time PCR. *P < 0.05 when comparing MUC5AC expression in siMYD88-transfected and dsRNA-treated cells (shaded bar) with MUC5AC expression in siC-transfected and dsRNA-treated cells (open bar) (n = 4). #P < 0.05 when comparing MUC5AC expression in siTRIF-transfected and dsRNA-treated cells (hatched bar) with MUC5AC expression in siC-transfected and dsRNA-treated cells (open bar) (n = 4).
TLR3-Mediated MUC5AC Expression Was Dependent on EGFR-ERK Signaling
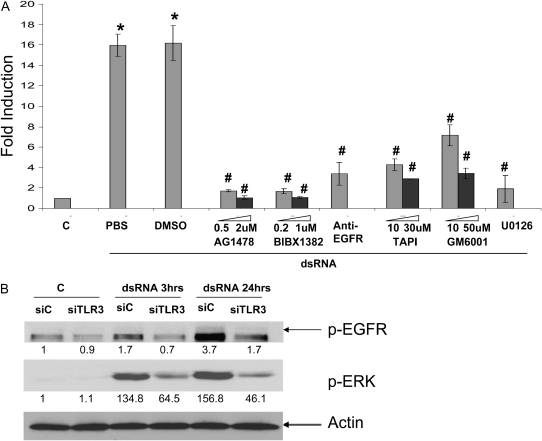
We further tested if those common mucin inducing pathways were involved. Among them, EGFR-ERK pathway is the most established one (24). Indeed, inhibition of either EGFR (AG1478 or BIBX1382) or ERK (U0126) significantly inhibited TLR3-mediated MUC5AC expression in a dose-dependent manner (Figure 5A). Since the EGFR activation could be either ligand-dependent or -independent, we tested whether the blockade of EGFR could have effect. By using a blocking antibody specifically binding to EGFR ligand binding domain (25), we found that TLR-3–mediated MUC5AC expression was significantly reduced (Figure 5A), suggesting that the extracellular ligands were required. It has been well established that EGF ligands (i.e., TGF-α, HB-EGF, amphiregulin) are synthesized as transmembrane precursors, and need a “convertase” TNF-α–converting enzyme (TACE/ADAM17) to cleave and generate the soluble growth factor (26, 27). To further test the ligand-dependent nature of TLR-3–mediated MUC5AC expression, we inhibited TACE using either specific inhibitor-TAPI or general metalloprotease inhibitor-GM6001, and found that mucin expression was reduced significantly in a dose-dependent manner (Figure 5A). All these data suggest that TLR3-mediated MUC5AC expression depended on ligand-dependent EGFR activation.
Figure 5.
TLR3-mediated MUC5AC expression was dependent on EGFR-ERK signaling. (A) Chemical inhibitors were pre-treated at different doses (when applicable, shaded bar represents lower dose, and solid bar represents high dose) on NCI-H292 cells for 1 hour; the cells were then treated with dsRNA at 0.025 μg/ml for 24 hours. RNA was collected, and MUC5AC expression was determined by real-time PCR. *P < 0.05 compared with control (C) (n = 4); #P < 0.05 compared with corresponding DMSO or PBS controls (n = 4). Only anti-EGFR antibody was dissolved in PBS. All chemical inhibitors were dissolved in DMSO. (B) Control siRNA (siC) and TLR3 siRNA (siTLR3) were transfected into NCI-H292 cells. Twenty-four hours later, cells were treated with dsRNA (0.025 μg/ml), and protein was collected after either 3 or 24 hours. Western blot analysis was used to determine the level of phosphrylated/activated EGFR (pEGFR) and ERK (p-ERK). Anti-actin antibody was used as a control for loading. Quantification was performed using IMAGE J (http://rsbweb.nih.gov/ij/). The data are presented as the fold inductions comparing with siC-transfected and nontreated control. This is the representative image from three independent experiments.
To further test the relationship between TLR3 and EGFR-ERK, we knocked down TLR3 using siRNA approach, and found that the activation of both EGFR and ERK were significantly reduced (Figure 5B). Thus, TLR3 appears to be upstream of EGFR and ERK.
TLR3-Initiated EGF Ligands Autocrine/Paracrine Loop Was Responsible for the Activation of EGFR-ERK Signaling and Mucin Gene Expression
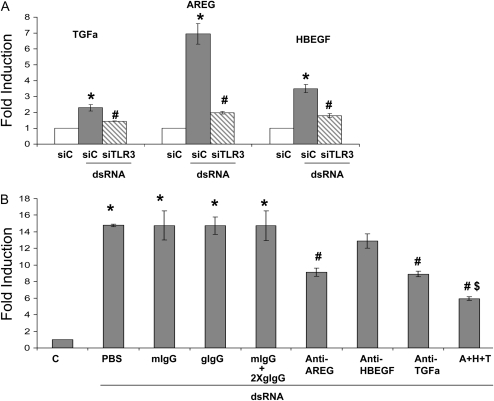
Since there are dozens of EGFR ligands, we tested three common ligands (TGF-α, amphiregulin, HB-EGF) that have been documented in epithelial mucin regulation (27–29). dsRNA appeared to elevate mRNA level of all three ligands, which was mediated by TLR3 (Figure 6A). When used individually for neutralization assay, TGF-α–neutralizing antibody (anti–TGF-α) and amphiregulin-neutralizing antibody (anti-AREG) significantly reduced TLR3-mediated MUC5AC expression by approximately 40% (Figure 6B). Anti–HB-EGF neutralizing antibody showed very slightly reduction, but failed to reach statistical significance. When three antibodies were combined (A+H+T) for treatment, they further reduced TLR3-mediated MUC5AC expression by an additional approximately 20% (Figure 6B) to approximately 60% reduction in total, suggesting that the effects of those antibodies were partly additive. To exclude the nonspecific effect of the antibody addition, we used species- and isotype-matched IgG as controls, and no effects were observed. Thus, at least both TGF-α and amphiregulin were involved in regulating TLR3-mediated MUC5AC expression.
Figure 6.
TLR3-initiated EGF-ligands autocrine/paracrine loop was responsible for the activation of EGFR-ERK signaling and mucin gene expression. (A) Control siRNA (siC) and TLR3 siRNA (siTLR3) were transfected into NCI-H292 cells. Twenty-four hours later, cells were treated with dsRNA (0.025 μg/ml), and RNA was collected after an additional 24 hours. The expressions of TGF-α, amphiregulin (AREG), and HB-EGF were determined by real-time PCR. *P < 0.05 when comparing the gene expression in siC-transfected and dsRNA-treated cells (shaded bar) with the gene expression in siC-transfected and PBS-treated cells (open bar) (n = 4); #P < 0.05 when comparing the gene expression in siTLR3-transfected and dsRNA-treated cells (hatched bar) with the gene expression in siC-transfected and dsRNA-treated cells (shaded bar) (n = 4). (B) Effect of EGFR ligand neutralization on MUC5AC expression. NCI-H292 cells were treated with neutralization antibodies or with isotype-matched control for 24 hours; RNA was then collected and MUC5AC expression was determined by real-time PCR. One hundred percent antibody neutralization doses were initially chosen on the basis of the manufacturer's data sheet. Those doses were further tested by examining the doses five times lower or five times higher. The lowest dose that could inhibit MUC5AC expression the most was chosen. If no effect was observed, the highest dose was chosen. Anti–TGF-α: TGFa neutralizing antibody (10 μg/ml), which is a goat IgG; anti-AREG: amphiregulin-neutralizing antibody (10 μg/ml), which is a goat IgG; anti-HBEGF: HB-EGF–neutralizing antibody (50 μg/ml), which is a mouse IgG2A; A+H+T: combined treatment of anti-AREG (10 μg/ml), anti-HBEGF (50 μg/ml), and anti–TGF-α (10 μg/ml). mIgG: matching mouse IgG2A (50 μg/ml); gIgG: matching goat IgG (10 μg/ml); mIgG+2XgIgG: combined treatment of mIgG (50 μg/ml) and gIgG (20 μg/ml). *P < 0.05 when comparing with the control (C) (n = 4); #P < 0.05 when comparing the neutralizing antibody–treated cells with their corresponding IgG control–treated cells n = 4; $P < 0.05 when comparing combined antibodies–treated cells (A+H+T) with single antibody–treated cells (n = 4).
DISCUSSION
Although mucus overproduction is one of the major illnesses associated with respiratory viral infection, the underlying mechanism is largely unclear. Studies using animal models have long attributed mucus overproduction to the side-effect of airway inflammation (5, 30–32). Although dozens of mucin-inducing cytokines and other inflammatory agents can be produced in viral infected airways (33, 34), whether or not the amount of those detected in vivo is sufficient to drive mucin expression is largely untested, with the exception of IL-13, which has been extensively tested in Sendai and RSV infection models (30, 32). However, not all viral infections induce IL-13 (as in the case of RV infection, for example) (5). Recently, it has been shown that RV14, one of the major group of RV, can directly elevate mucin production though an MAPK-dependent pathway by airway epithelial cells in vitro (35). Thus, virus infection may be able to directly activate mucin expression in airway epithelial cells in the absence of inflammation. It has been shown the MAPK pathway is involved (35), but the upstream signal that activating MAPK is still unclear.
Interestingly, most common respiratory viral infections are caused by RNA viruses. And even some respiratory DNA viruses (e.g., adenovirus) generate RNA molecules during their intracellular life cycles. There are several viral RNA “sensors” expressed in airway epithelial cells, including PKR (12), TLRs (TLR3 for dsRNA and TLR7/8 for ssRNA), and recently found Caspase recruitment domain (CARD) containing molecules such as RIG-I and MDA5 (36). Considering the scope of this study, we have only tested PKR, which has been shown by our previous study to be responsible for epithelial antiviral defense (12), and TLR3, which recognizes dsRNA (36); dsRNA has been reported by Londhe and coworkers to induce MUC2 (13), a major colonic but a minor airway mucin. To further support the potential mucin-inducing effect of TLR3, our preliminary study indicated that dsRNA appears to be the most potent mucin inducer among all other TLR-ligands tested in airway epithelial cells (unpublished observation). Consistent with our previous notion that RV dsRNA is a critical signaling molecular triggering IFN-dependent antiviral defense (12), it is also responsible for mucin induction, and the difference in the present scenario is that viral dsRNA was recognized by TLR3 instead of PKR. Accordingly, these two different “sensors” led to different downstream signaling pathways and effectors. For PKR, we have shown that it induced IFN-dependent antiviral defense genes are activated and contribute to the intracellular viral clearance (12), presumable through an NF-κB–dependent pathway later reported by Edwards and colleagues (37). For TLR3, we have shown in this study that it induced mucin expression through the increase of multiple EGF ligands transcription and subsequent activation of EGFR-ERK pathway through an autocrine/paracrine mechanism. The increase of mucin expression may presumably accelerate viral clearance in vivo through enhanced mucociliary clearance, which needs to be further tested using in vivo model of RV infection described in this and other (5) studies.
Another interesting finding is that TLR3-mediated mucin expression is only partially dependent on TRIF, and appears to be negatively regulated by MYD88. The lack of sufficient knockdown can be ruled out by the confirmatory experiments using Western and real-time PCR. In addition, the expression of CXCL10, a TLR3-TRIF–dependent gene, could be almost completely knocked down by siRNA approach, which further supports the notion that TLR3-mediated mucin expression may also use a TRIF-independent pathway, which has been suggested before (38). One could still argue that the signaling pathway leading to CXCL10 expression might need more TRIF adaptor than the one leading to mucin expression, and thus the latter might be less sensitive to the reduction of TRIF, because siRNA could only knock down around 70% of TRIF expression. One approach to test this hypothesis would be using TRIF knockout mouse, because the epithelial cell derived from those mice is completely devoid of TRIF. However, this study is beyond the scope of current study, and the result could be difficult to interpret because (1) mouse and human epithelial cells have very different cell types; and (2) the complete deprivation of TRIF from embryonic stage will certainly lead to the establishment of a whole new cell signaling circuitry that may be very different from that of the wide-type cells. Nonetheless, our present study suggests that the TLR-3–mediated mucin expression may use a downstream signaling different from that of the conventional paradigm (36).
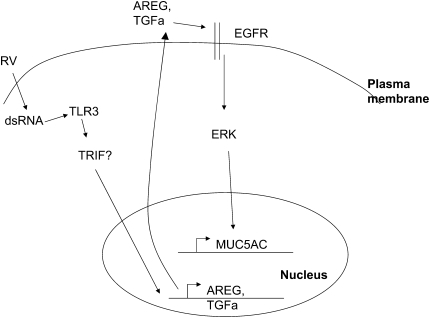
Londhe and coworkers were the first to report that dsRNA could induce epithelial mucin (13). However, their work was focused on a major colonic (39) but a minor airway mucin-MUC2, which only consists of around 2.5% of the airway mucus gel (40). Our present study, instead, focuses on the major airway mucin MUC5AC. Interestingly, these two mucins are controlled by different signaling pathways. MUC2 has been demonstrated to be induced through GPCR-PLC-PKC-P38-NFKB pathway (13), while MUC5AC has been shown in the present study to be induced by TLR3-mediated EGFR ligand transcription and subsequent activation of EGFR by an autocrine/paracrine mechanism (Figure 7). Different regulations suggest that MUC2 and MUC5AC may have different functions in the airway.
Figure 7.
Signaling pathway links RV infection and mucin expression. Question mark indicates that there may be other TRIF-independent pathways.
EGFR activation has been well established to be one of the major mucin-inducing signaling pathways (27). However, it has not been reported to mediate viral-induced mucin expression. In addition, the EGFR signaling demonstrated in this study has important different aspects from the current paradigm (27). In the current paradigm, the activity of TACE is induced by reactive oxygen species (ROS) that is generated by particular agonist (i.e., neutrophil elastase, PMA, etc.), which leads to the enhancement of TGF-α cleavage from surface membrane; the increased soluble TGF-α, in turn, activates EGFR (27). In the present study, we have shown that at least one other ligand, AREG, in addition to TGF-α, was also indispensable for EGFR activation, presumably because we used the different inducer (dsRNA instead of PMA, etc.). We have also shown the increase of EGFR ligand production, which should provide a much simpler explanation for EGFR activation. It is unclear whether or not TGF-α transcription was also induced in those previous studies (27). If yes, the increase of EGFR ligand transcription might play a more universal role in activating EGFR. Otherwise, our finding may suggest a unique mechanism triggered specifically by viral infection through TLR3. Even though the TACE is definitely required on all occasions reported by us (in the present study) and others (27), it is unclear whether or not the alteration of TACE activity is required. To our knowledge, there is no direct measurement of TACE activity in any of the previous reports (27). Thus, TACE activity might just be an essential residential enzyme responsible for EGFR ligand release, but its activity might not be altered by those mucin stimuli (27). This notion has been partly supported by the study showing TACE as a physiologic EGFR ligand convertase (26). Further study will be needed to clarify this issue.
In summary, the present study has demonstrated that both major and minor groups of RV directly induce mucin production though a novel TLR3-mediated pathway that is partly dependent on TRIF and negatively regulated by MYD88. This TLR3-mediated mucin-inducing pathway further leads to the increase of TGF-α and AREG production, and subsequent activation of EGFR through an autocrine/paracrine mechanism (Figure 7). Complete elucidation this novel pathway will advance our understanding of viral-induced airway remodeling and disease exacerbation.
This study was supported by Long Range Initiative (LRI) of the American Chemistry Council and by National Institutes of Health grant RO1AI061695 to Y.C.
Originally Published in Press as DOI: 10.1165/rcmb.2008-0223OC on October 31, 2008
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Jeffery PK. Comparison of the structural and inflammatory features of COPD and asthma. Giles F. Filley Lecture. Chest 2000;117:251S–260S. [DOI] [PubMed] [Google Scholar]
- 2.Aikawa T, Shimura S, Sasaki H, Ebina M, Takishima T. Marked goblet cell hyperplasia with mucus accumulation in the airways of patients who died of severe acute asthma attack. Chest 1992;101:916–921. [DOI] [PubMed] [Google Scholar]
- 3.Carroll N, Carello S, Cooke C, James A. Airway structure and inflammatory cells in fatal attacks of asthma. Eur Respir J 1996;9:709–715. [DOI] [PubMed] [Google Scholar]
- 4.Savolainen C, Blomqvist S, Hovi T. Human rhinoviruses. Paediatr Respir Rev 2003;4:91–98. [DOI] [PubMed] [Google Scholar]
- 5.Bartlett NW, Walton RP, Edwards MR, Aniscenko J, Caramori G, Zhu J, Glanville N, Choy KJ, Jourdan P, Burnet J, et al. Mouse models of rhinovirus-induced disease and exacerbation of allergic airway inflammation. Nat Med 2008;14:199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newcomb DC, Sajjan US, Nagarkar DR, Wang Q, Nanua S, Zhou Y, McHenry CL, Hennrick KT, Tsai WC, Bentley JK, et al. Human rhinovirus 1B exposure induces phosphatidylinositol 3-kinase-dependent airway inflammation in mice. Am J Respir Crit Care Med 2008;177:1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makela MJ, Puhakka T, Ruuskanen O, Leinonen M, Saikku P, Kimpimaki M, Blomqvist S, Hyypia T, Arstila P. Viruses and bacteria in the etiology of the common cold. J Clin Microbiol 1998;36:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Temte JL. A family physician's perspective on picornavirus infections in primary care. Arch Fam Med 2000;9:921–922. [DOI] [PubMed] [Google Scholar]
- 9.Busse WW, Gern JE. Viruses in asthma. J Allergy Clin Immunol 1997;100:147–150. [DOI] [PubMed] [Google Scholar]
- 10.Johnston SL, Pattemore PK, Sanderson G, Smith S, Lampe F, Josephs L, Symington P, O'Toole S, Myint SH, Tyrrell DA, et al. Community study of role of viral infections in exacerbations of asthma in 9–11 year old children. BMJ 1995;310:1225–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicholson KG, Kent J, Ireland DC. Respiratory viruses and exacerbations of asthma in adults. BMJ 1993;307:982–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Hamati E, Lee PK, Lee WM, Wachi S, Schnurr D, Yagi S, Dolganov G, Boushey H, Avila P, et al. Rhinovirus induces airway epithelial gene expression through double-stranded RNA and IFN-dependent pathways. Am J Respir Cell Mol Biol 2006;34:192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Londhe V, McNamara N, Lemjabbar H, Basbaum C. Viral dsRNA activates mucin transcription in airway epithelial cells. FEBS Lett 2003;553:33–38. [DOI] [PubMed] [Google Scholar]
- 14.O'Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol 2007;7:353–364. [DOI] [PubMed] [Google Scholar]
- 15.Wang W, Lee WM, Mosser AG, Rueckert RR. WIN 52035-dependent human rhinovirus 16: assembly deficiency caused by mutations near the canyon surface. J Virol 1998;72:1210–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duits LA, Nibbering PH, van Strijen E, Vos JB, Mannesse-Lazeroms SP, van Sterkenburg MA, Hiemstra PS. Rhinovirus increases human beta-defensin-2 and -3 mRNA expression in cultured bronchial epithelial cells. FEMS Immunol Med Microbiol 2003;38:59–64. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y, Zhao YH, Di YP, Wu R. Characterization of human mucin 5B gene expression in airway epithelium and the genomic clone of the amino-terminal and 5′-flanking region. Am J Respir Cell Mol Biol 2001;25:542–553. [DOI] [PubMed] [Google Scholar]
- 18.Wu R, Zhao YH, Chang MM. Growth and differentiation of conducting airway epithelial cells in culture. Eur Respir J 1997;10:2398–2403. [DOI] [PubMed] [Google Scholar]
- 19.Kao CY, Chen Y, Thai P, Wachi S, Huang F, Kim C, Harper RW, Wu R. IL-17 markedly up-regulates beta-defensin-2 expression in human airway epithelium via JAK and NF-kappaB signaling pathways. J Immunol 2004;173:3482–3491. [DOI] [PubMed] [Google Scholar]
- 20.Deng J, Chen Y, Wu R. Induction of cell cornification and enhanced squamous-cell marker SPRR1 gene expression by phorbol ester are regulated by different signaling pathways in human conducting airway epithelial cells. Am J Respir Cell Mol Biol 2000;22:597–603. [DOI] [PubMed] [Google Scholar]
- 21.Li K, Chen Z, Kato N, Gale M Jr, Lemon SM. Distinct poly(I-C) and virus-activated signaling pathways leading to interferon-beta production in hepatocytes. J Biol Chem 2005;280:16739–16747. [DOI] [PubMed] [Google Scholar]
- 22.Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat Immunol 2003;4:161–167. [DOI] [PubMed] [Google Scholar]
- 23.Johnson AC, Li X, Pearlman E. MyD88 functions as a negative regulator of TLR3/TRIF-induced corneal inflammation by inhibiting activation of c-Jun N-terminal kinase. J Biol Chem 2008;283:3988–3996. [DOI] [PubMed] [Google Scholar]
- 24.Burgel PR, Nadel JA. Roles of epidermal growth factor receptor activation in epithelial cell repair and mucin production in airway epithelium. Thorax 2004;59:992–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masui H, Kawamoto T, Sato JD, Wolf B, Sato G, Mendelsohn J. Growth inhibition of human tumor cells in athymic mice by anti-epidermal growth factor receptor monoclonal antibodies. Cancer Res 1984;44:1002–1007. [PubMed] [Google Scholar]
- 26.Lee DC, Sunnarborg SW, Hinkle CL, Myers TJ, Stevenson MY, Russell WE, Castner BJ, Gerhart MJ, Paxton RJ, Black RA, et al. TACE/ADAM17 processing of EGFR ligands indicates a role as a physiological convertase. Ann N Y Acad Sci 2003;995:22–38. [DOI] [PubMed] [Google Scholar]
- 27.Nadel JA. Innate immune mucin production via epithelial cell surface signaling: relationship to allergic disease. Curr Opin Allergy Clin Immunol 2007;7:57–62. [DOI] [PubMed] [Google Scholar]
- 28.Basbaum C, Li D, Gensch E, Gallup M, Lemjabbar H. Mechanisms by which gram-positive bacteria and tobacco smoke stimulate mucin induction through the epidermal growth factor receptor (EGFR). Novartis Found Symp 2002;248:171–6; discussion 176–80, 277–82. [PubMed] [Google Scholar]
- 29.Lemjabbar H, Basbaum C. Platelet-activating factor receptor and ADAM10 mediate responses to Staphylococcus aureus in epithelial cells. Nat Med 2002;8:41–46. [DOI] [PubMed] [Google Scholar]
- 30.Tyner JW, Kim EY, Ide K, Pelletier MR, Roswit WT, Morton JD, Battaile JT, Patel AC, Patterson GA, Castro M, et al. Blocking airway mucous cell metaplasia by inhibiting EGFR antiapoptosis and IL-13 transdifferentiation signals. J Clin Invest 2006;116:309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buchweitz JP, Harkema JR, Kaminski NE. Time-dependent airway epithelial and inflammatory cell responses induced by influenza virus A/PR/8/34 in C57BL/6 mice. Toxicol Pathol 2007;35:424–435. [DOI] [PubMed] [Google Scholar]
- 32.Lukacs NW, Moore ML, Rudd BD, Berlin AA, Collins RD, Olson SJ, Ho SB, Peebles RS Jr. Differential immune responses and pulmonary pathophysiology are induced by two different strains of respiratory syncytial virus. Am J Pathol 2006;169:977–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voynow JA, Gendler SJ, Rose MC. Regulation of mucin genes in chronic inflammatory airway diseases. Am J Respir Cell Mol Biol 2006;34:661–665. [DOI] [PubMed] [Google Scholar]
- 34.Rose MC, Voynow JA. Respiratory tract mucin genes and mucin glycoproteins in health and disease. Physiol Rev 2006;86:245–278. [DOI] [PubMed] [Google Scholar]
- 35.Inoue D, Yamaya M, Kubo H, Sasaki T, Hosoda M, Numasaki M, Tomioka Y, Yasuda H, Sekizawa K, Nishimura H, et al. Mechanisms of mucin production by rhinovirus infection in cultured human airway epithelial cells. Respir Physiol Neurobiol 2006;154:484–499. [DOI] [PubMed] [Google Scholar]
- 36.Kawai T, Akira S. TLR signaling. Cell Death Differ 2006;13:816–825. [DOI] [PubMed] [Google Scholar]
- 37.Edwards MR, Hewson CA, Laza-Stanca V, Lau HT, Mukaida N, Hershenson MB, Johnston SL. Protein kinase R, IkappaB kinase-beta and NF-kappaB are required for human rhinovirus induced pro-inflammatory cytokine production in bronchial epithelial cells. Mol Immunol 2007;44:1587–1597. [DOI] [PubMed] [Google Scholar]
- 38.Hoebe K, Janssen EM, Kim SO, Alexopoulou L, Flavell RA, Han J, Beutler B. Upregulation of costimulatory molecules induced by lipopolysaccharide and double-stranded RNA occurs by Trif-dependent and Trif-independent pathways. Nat Immunol 2003;4:1223–1229. [DOI] [PubMed] [Google Scholar]
- 39.Tytgat KM, Buller HA, Opdam FJ, Kim YS, Einerhand AW, Dekker J. Biosynthesis of human colonic mucin: Muc2 is the prominent secretory mucin. Gastroenterology 1994;107:1352–1363. [DOI] [PubMed] [Google Scholar]
- 40.Kirkham S, Sheehan JK, Knight D, Richardson PS, Thornton DJ. Heterogeneity of airways mucus: variations in the amounts and glycoforms of the major oligomeric mucins MUC5AC and MUC5B. Biochem J 2002;361:537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]