Abstract
Lutzomyia longipalpis are vectors of medically important visceral leishmaniasis in South America. Bloodfed adult females digest large amounts of protein, and xanthine dehydrogenase is thought to be a key enzyme involved in protein catabolism through the production of urate. Large amounts of heme are also released during digestion with potentially damaging consequences, as heme can generate oxygen radicals that damage to lipids, proteins and nucleic acids. However, urate is an anti-oxidant that may prevent such oxidative damage produced by heme. We investigated xanthine dehydrogenase by developing the RNAi technique for sand flies and used this technique to knock down the Lu. longipalpis xanthine dehydrogenase gene to evaluate its role in survival of adult females after blood feeding.
The gene sequence of Lu. longipalpis xanthine dehydrogenase is described together with expression in different life cycle stages and RNAi knock down. Semi quantitative RT-PCR of xanthine dehydrogenase expression showed a significant increase in expression after bloodmeal ingestion. Micro-injection of dsRNA via the thorax of 1 day old adult female sand flies resulted in approximately 40% reduction of xanthine dehydrogenase gene expression in comparison to flies injected with a control dsRNA. A significant reduction of urate in the whole body and excretions of Lu. longipalpis was observed after dsRNA xanthine dehydrogenase microinjection and feeding 96h later on rabbit blood. Sand flies injected with XDH dsRNA also exhibit significantly reduced life span in comparison with the mock-injected group when fed on sucrose or when rabbit blood fed, showing that urate could be indeed an important free radical scavenger in Lu. Longipalpis.
The demonstration of xanthine dehydrogenase knock down by dsRNA microinjection, low mortality of micro-injected insects and the successful bloodfeeding of injected insects demonstrated the utility of RNAi as a tool for functional analysis of genes in phlebotomine sand flies.
Keywords: Lutzomyia longipalpis, RNAi, urate, heme, xanthine dehydrogenase, oxidative stress
1. Introduction
Phlebotomine sand flies are the vectors of Leishmania species, parasitic protozoans responsible for the leishmaniases, a group of diseases affecting human and various animal populations throughout much of the tropics and subtropics (Bates, 2007). Leishmaniases have several diverse clinical manifestations ranging from ulcerative skin lesions, destructive mucosal inflammation to the potentially fatal disseminated visceral infection (Banuls et al., 2007). During the last 20 years, zoonotic visceral leishmaniasis due to Leishmania (Leishmania) infantum and transmitted by Lutzomyia longipalpis (Lutz & Neiva 1912) has become a serious public health problem in several Brazilian cities (Alexander et al., 2002; Michalsky et al., 2007).
The female sand fly ingests blood meals to supply protein for egg production. When feeding on vertebrate blood, haematophagous insects are adapted to overcome the pro-oxidant action of the large amounts of heme generated during digestion of haemoglobin inside their midguts (Paiva-Silva et al., 2006; Graça-Souza, 2006). Urate is the main end-product of nitrogen metabolism, including protein and purine break down (Glantzounis et al., 2005). It is released into the haemolymph and absorbed by the Malpighian tubules via a pH gradient to constitute a major component of the insect’s urine (Souza et al., 1997). Xanthine dehydrogenase (XDH) is a key enzyme as it catalyses the oxidation of xanthine to uric acid.
Mosquito microarray experiments suggest that XDH mRNA levels increase upon blood feeding in Aedes aegypti (Sanders et al., 2003). XDH enzyme activities increase in bloodfed Aedes aegypti and maximal levels precede digestive protease activities (von Dungern and Breigel, 2001), with XDH and urate synthesis correlating directly with bloodmeal size. In insects, urate has been shown to have important anti-oxidant properties that prevent lipid peroxidation (Souza et al., 1997). In Drosophila melanogaster, urate-null mutants defective for XDH/XOD are more sensitive to paraquat, ionizing radiation and hyperoxia, suggesting this gene plays a major role against oxidative stress in this insect species (Hilliker et al., 1992). Kim et al. (2001), also working with D. melanogaster rosy mutants, suggested that XDH plays an important role in the innate response to infection and that age-associated deterioration of the innate immune response may be associated to some extent with loss of XDH activity.
In organisms as diverse as nematodes, trypanosomes, plants, fungi, insects and humans, introduction of double-stranded RNA triggers the destruction of homologous mRNA, producing knockdown of the corresponding protein (Hutvagner and Zamore 2002; Tomari and Zamore, 2006). The technique of RNA interference has emerged as a powerful tool to study gene function in a wide variety of experimental systems, particularly in non-model organisms for which other methods of investigation are not available (Fraser et al., 2000; Gonczy et al., 2000). Gene silencing using dsRNA has been used to knock down a variety of genes in arthropods including Anopheles gambiae, (Vlachou et al., 2005; Boisson et al., 2006) the honeybee Apis mellifera (Maleszka et al., 2007), Bemisia tabaci (Ghanim et al., 2007), Drosophila melanogaster (Kennerdell and Carthew, 1998), Manduca sexta (Eleftherianos et al., 2006) and Ixodes scapularis (Narasimhan et al., 2004). Recently, Araújo et al (2006) described a nitrophorin 2 knock down in a triatomine species (Rhodnius proxilus) by both dsRNA injection and ingestion, the latter proving to be a non invasive way to deliver dsRNA into a biological organism. Jaubert-Possamai et al (2007) described the development of RNAi by micro-injection of dsRNA in the pea aphid Acyrthosiphon pisum, showing that specific gene knockdown is feasible for small insects, even when minor volumes of dsRNA are injected. For Lu. longipalpis, a viral vector driven RNA silencing method was tested on cultured embryonic cells (Pitaluga et al, 2007). Silencing of a luciferase gene was achieved but unrelated dsRNAs also reduced expression of the gene.
The recent completion of a Lu. longipalpis mass sequencing EST programme (Dillon et al., 2006) and cDNA micro-array analysis of bloodfed and Leishmania-infected Lu. longipalpis (unpublished data) provided an extensive list of differentially expressed sand fly genes. The development of a reverse genetics technique for phlebotomine sand flies is of crucial importance as a functional genomic tool to study genes and proteins that might have an effect on Leishmania development inside the gut of the sand fly.
The aim of this study was to develop the double stranded RNA knock down approach for the diminutive Lu. longipalpis and to characterise the XDH gene that is thought to play a key role in bloodmeal digestion and response to oxidative stress.
2. Materials and Methods
2.1. Insects
A laboratory colony of Lu. longipalpis established from flies caught in Jacobina (Bahia-Brazil) and kept at the Liverpool School of Tropical Medicine was used in all experiments and maintained using standard methods (Modi, 1997). Insects were reared under controlled conditions of temperature (28 ± 1 °C) and humidity (80-95%).
2.2. RNA extractions and RT-PCR
Total RNA was extracted from whole bodies of Lu. longipalpis using the RNaqueous 4-PCR Kit (Ambion®). RT-PCR was performed using the AccessQuick RT-PCR System (Promega®) according to the manufacturer’s instructions. The cDNA was constructed from 10ng of total RNA using 25 cycles.
2.3. Double stranded RNA synthesis
The XDH template was PCR amplified from the XDH gene plasmid preparation obtained from a normalized Lu. longipalpis cDNA Library (Dillon et al., 2006) and the ampicillin resistance gene (AMP) was PCR amplified from the pBluescript SK plasmid (Stratagene). PCR was carried out using specific primers containing the T7 site at their 5 prime ends (Table 1). The PCR products of 600bp for XDH and 534 bp for AMP were used as templates for transcription reactions using the Megascript RNAi Kit (Ambion®) according to the manufacturer’s instructions. After synthesis, column purification and elution with RNAse-free water at 65°C, dsRNA purity was determined by 1.5 % agarose gel electrophoresis quantified using a Nanodrop ND-1000 Spectrophotometer. Double stranded RNA samples were concentrated to 4.5 μg/μL prior to use using a Christ® rotational vacuum concentrator RVC 2-25 and stored at - 80°C until needed.
Table 1.
Primer sequences for dsRNA construction and RT-PCR
| Primer | 5′-3′ sequences | Product size |
|---|---|---|
| Ds-XDH forward | TAATACGACTCACTATAGGGAGATTCCAGAAGAGGCGCAAAGATG | 600bp |
| Ds-XDH reverse | TAATACGACTCACTATAGGGAGACCAGCGCCAAAGGTGAAGTAGTT | - |
| Lulong-XDH-RT-F | CGTTGAGGGGTGGGTTTGTAAGAC | 400bp |
| Lulong-XDH-RT-R | AGAGCGCCGGATTGATTGAGAA | - |
| Ds-AMP forward | TAATACGACTCACTATAGGGATGCCGGGAAGCTAGAGTAAGTA | 534bp |
| Ds-AMP reverse | TAATACGACTCACTATAGGGAAACGCTGGTGAAAGTAAAAGATG | - |
| Lulong-60S-F | TCTCATCGGAAGTTTTCTGC | 850bp |
| Lulong-60S-R | GGCTTGTGACACCCTTGAAT | - |
2.4. Double stranded RNA micro injections
Long sharp microinjection needles made from borosilicate 3.5 inch hematocrit capillary tubes (Drummond® USA) were drawn into fine tips using a PC-10 Narishige Puller. Before use the needles were bevelled using an EG-44 Narishige® microgrinder. One day old, non-fed Lu. longipalpis were briefly chilled on ice placed on an ice pack under a dissecting microscope and injected with 32 nanolitres of a 4.5 μg/μL stock dsRNA solution (144 ng per fly) for XDH (knock down group) and dsAMP (mock-injected group) in the thorax, using a Drummond® Nanoject II Auto-Nanolitre injector. After injections, flies were kept in a cage (>90% relative humidity) and supplied with 70% (w/v) sucrose solution ad libitum. Sand flies were fed with fresh rabbit blood heated to 35°C, through a chick skin feeder 96h after injection. Uninjected sand flies of the same age were used as a second control group.
2.5. Uric acid assay
To measure urate in the whole body of Lu. longipalpis, an Amplex Red Uric Acid Assay kit (Invitrogen®) was used. Double stranded RNA-injected and uninjected flies were fed on rabbit blood and whole insect bodies (fully engorged insects) were homogenised individually in 130 μL of 0.1 M Tris-HCl pH 7.5 using a mini pestle pellet rotor, 48 hours after the feed. Following centrifugation at 13,500 rpm for 15 minutes, 50 μL of the homogenate was mixed with 50 μL of the Amplex Red Reagent (0.4 U/mL horseradish peroxidase and 0.4 U/mL uricase, made up in 0.1 M Tris-HCl pH 7.5). After 30 minutes incubation at 37°C, absorbance was measured at 490 nm. For urate measurements in the excretions of Lu. longipalpis, flies were fed on rabbit blood and individually kept inside Eppendorf microfuge tubes with pierced lids to allow ventilation. Tubes were placed inside sealed plastic boxes over humidified filter paper and after removing the adult females, the insect’s excretions were collected by dissolving with 120 μL of 0.1 M Tris-HCl pH 7.5. Amount of urate was measured 48 hours after the feed as described for the whole body. Control blanks consisting of 50 μL 0.1 M Tris-HCl pH 7,5 and 50 μL of the sand fly homogenates were also read at 490 nm and subtracted from the experimental results to minimise influence of the blood colour on the absorbance.
2.6. Protein assay
To certify that all insects used in different experimental groups had taken up similar amounts of blood, the total amount of protein in the whole body was quantified (Bradford, 1976). Briefly, 4 μL of Lu. longipalpis whole body homogenate in 0.1 M Tris-HCl pH 7.5 was mixed with 196 μL of the BIORAD® Protein assay reagent, diluted 1x in distilled water and absorbance was measured in a 96 well plate at 595 nm. Bovine serum albumin was used as control.
2.7. Verification of XDH knock down
RT-PCR was carried out using either individual flies or pools of 20 flies from each group in triplicate. The expression of the XDH gene in Lu. longipalpis injected with XDH or AMP dsRNAs were evaluated alongside expression of a house keeping gene (AM088777, putative 60S ribosomal protein L3). RT-PCR products were analysed by 1.5% agarose gels electrophoresis and reduction in XDH gene expression was determined by densitometric measurement of bands using the software GeneSnap/GeneTools (Syngene, UK). Change in gene expression in dsXDH and dsAMP-injected flies was determined by the ratio between band intensity of the XDH gene and its correspondent rRNA (loading controls).
2.8. Time course XDH expression in Lu. longipalpis developmental stages
Three independent experiments were performed and XDH gene expression was performed by semi-quantitative RT-PCR in different life cycle stages after total RNA extraction. Individual experiments consisted of three replicates, each one containing either 5 fourth instar Lu. longipalpis larvae, 5 pupae, 5 unfed females (1 day old) or 5 hamster-fed females at 24, 48, 72 and 96h after the feed. Ribosomal RNA was used as loading control. RT-PCR products were analysed by 1.5% agarose gel electrophoresis and XDH gene expression was determined by densitometric measures of bands using the software GeneSnap/GeneTools. Relative XDH expression was determined by the ratio between band intensity of the XDH gene and its correspondent rRNA (loading controls).
2.9. Xanthine dehydrogenase gene sequencing
A partial Lu. longipalpis XDH gene sequence was previously reported (Dillon et al., 2006). A primer walking approach was used to obtain the full-length sequence of this gene. An initial fragment was obtained using degenerate primers targeting a conserved region in one of the ion/sulphur motifs, based on multiple alignments of other insect xanthine dehydrogenases using Clustal W. Based on this first sequence, new specific primers were designed and used in new sequencing reactions to obtain further sequences. The 5 prime end was obtained using the 5 prime RACE Kit (Invitrogen) according to the manufacturer’s instructions. Sequences corresponding to part of the XDH gene were aligned with the partial XDH clone to generate the full length sequence.
2.10. Sand fly survival
One day old, non-fed Lu. longipalpis were injected with 32 nanolitres of a 4.5 μg/μL stock dsRNA solution (144 ng per fly) for XDH and dsAMP in the thorax. After injections, flies were kept in a cage (>90% relative humidity) and supplied with 70% (w/v) sucrose solution ad libitum. Sand flies were fed with fresh rabbit blood heated to 35°C, through a chick skin feeder 96h after injection. Mortality was assessed in each cage every day to assess sand fly survival in sugar-fed and bloodfed dsRNA-injected sand flies compared to injected controls.
2.11.. Statistical analysis
Statistical analysis was carried out using Minitab 15. The Anderson-Darling test was used to verify normality of the data. When data followed normal distribution, ANOVA was used to verify difference between groups and a pair-wise t-test was used to test differences between the mean of two independent groups. When data distribution was non-normal, Kruskal-Wallis ANOVA was used and the Wilcoxon rank sum was used to test differences between medians of two independent groups. For sand fly survival experiments, statistical analysis was performed using the log rank test, as showed in Rogers and Bates (2007). P< 0.05 was considered statistically significant.
3. Results
3.1. Lu. longipalpis XDH cDNA
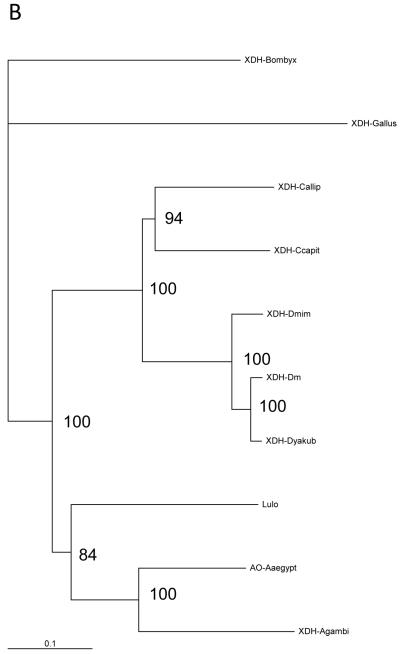
The full-length LuloXDH cDNA sequence contains 4,023 base pairs (EMBL Accession No. AM887864 and encodes a peptide of 1331 amino acids and a predicted mass of 146.4kDa when putatively translated. A partial alignment of eight available XDH proteins of insect species and one aldehyde oxidase from Aedes aegypti shows that this enzyme is highly conserved within the regions thought to bind iron-sulfur (2Fe/2S), the NAD-binding domain conferring substrate specificity to XDH and Molybdenum cofactor (MoCo) domain at the C-terminal region (Sato et al., 1995; Doyle et al., 1996; Komoto et al., 1999) (Fig. 1A). Lu. longipalpis XDH is very similar in size and sequence to other insect aldehyde oxidases (AO) and XDH, being 68% identical to Aedes aegypti AO (Accession No. EAT46105), 63% identical to three Drosophila XDH, 62% similar to the medfly Ceratitis capitata (AAG47345) Calliphora vicina (P08793) and A. gambiae XDH (AF515734) and 60% identical to a B. mori XDH (AB005911).
Fig. 1.
(a) Partial structure-based alignment of the amino acid sequence of Lulo-XDH (Accession no AM887864) and some other insect xanthine dehydrogenases/aldehyde oxidade: Anopheles gambiae XDH (Accession no AAO14865); Aedes aegypti aldehyde oxidase (Accession no EAT46105); Drosophila melanogaster XDH (Accession no CAA68409); Drosophila mimetica XDH (Accession no AAQ17532); Drosophila yakuba XDH (Accession no AAQ17528); Calliphora vicina XDH (Accession no CAA30281); Ceratitis capitata XDH (Accession no AAG47345); Bombyx mori XDH1 (Accession no BAA21640). Conserved functional areas of the enzyme are 2Fe/2S iron-sulfur domains (boxed), NAD+-binding site (black shade) and the carboxy terminal MoCo molybdenum cofactor (grey shade). (b) Phylogenetic tree of XDH sequences among insect species and Gallus gallus (domestic chicken) XDH obtained by the Neighbour Joining method. The scale bar represents 0.1% divergence. The number in the tree branches indicates the bootstrap value from 1000 interactions.
To examine these relationships more closely a phylogenetic analysis was performed which grouped Lutzomyia longipalpis XDH within a clade formed with XDH/AO protein sequences from other dipterans belonging to the Suborder Nematocera (Anopheles gambiae, Aedes aegypti) (Fig.1B). Another clade closely related to the nematoceran flies was formed with three Drosophila species (D. melanogaster XDH- CAA68409; D. mimetica XDH-AAQ17532; D. yakuba XDH AAQ17528), the Medfly Ceratitis capitata XDH and Calliphora vicina XDH with the lepidopteran Bombix mori XDH sequence forming a separate clade.
3.2. XDH expression in different life cycle stages
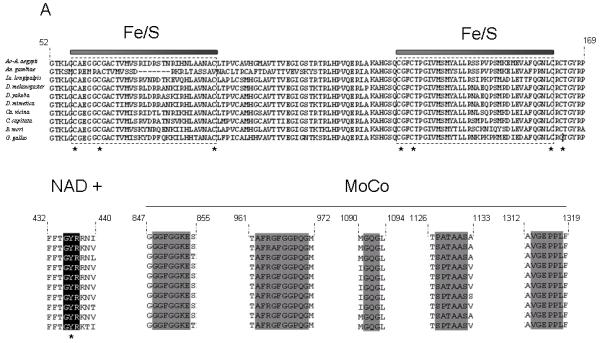
In order to identify XDH expression in Lu. longipalpis life cycle stages, 3 independent RT-PCR experiments were performed on RNA extracted from pools of 5 individuals for each of 4th instar larvae, pupae, unfed adult females and blood fed females at 24, 48, 72 and 96 hours after an artificial rabbit blood meal. There was evidence of induction of XDH expression related to bloodmeal ingestion with significantly higher expression in blood fed females 96h after the blood meal (P < 0.05) compared to all other time points and significantly higher at 72h post blood feed when compared to unfed sand flies (P<0.05). XDH expression was also significantly reduced during larval and pupal stage when compared to unfed and blood-fed adult females (P<0.05) (Fig 2A and B).
Fig. 2.
(a) Relative XDH gene expression in larvae, pupae and blood fed and unfed adult Lu. longipalpis. Three experiments (3×5 insects per data point) were performed and relative XDH gene expression was expressed by the ratio between band densitometry values in RT-PCR reactions of XDH gene and the control ribosomal 60S gene for larvae, pupae, unfed and rabbit-fed sand flies at 24, 48, 72 and 96h after blood feed. Larval and pupal expression was significantly lower than all adult samples; expression in 96h bloodfed insects was significantly higher than all other samples; 72h bloodfed insect expression was significantly higher than unfed adults. Statistical significance at p< 0.05.
(b) Time-course of RT-PCR analysis of the Lu longipalpis xanthine dehydrogenase gene (XDH) expression in unfed females and rabbit-fed sand flies at 24, 48, 72 and 96 hours after feeding, as well as larvae and pupae. RT-PCR of ribosomal 60S gene was used as loading control.
3.3. XDH knock down
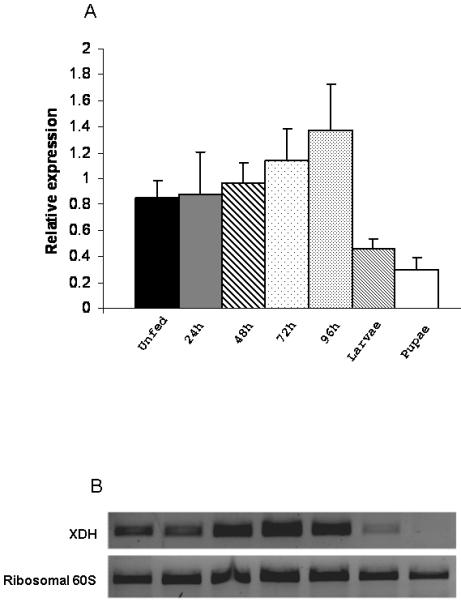
The effect of microinjection of dsRNA on XDH gene knock down was investigated by semi-quantitative RT-PCR. Reduction of XDH expression could be observed in dsXDH-injected flies (144 ng per fly) when compared to dsAMP-injected flies (Fig 3A). Relative expression of the XDH gene was calculated for each sand fly injected either with dsXDH or dsAMP and was obtained by calculating the ratio between XDH band intensity and the ribosomal 60S band intensity for both dsXDH-injected and dsAMP-injected groups. Triplicate experiments showed that double stranded XDH injections significantly reduced XDH gene expression by 40 ± 9% (P=0.035) when compared to the mock-injected group (Fig 3B). XDH expression levels were also measured for dsXDH-injected and dsAMP-injected flies by RT-PCR from day 2 to day 10 after dsRNA injection. For the dsXDH-injected group, XDH showed no significant change in gene expression 48h after the injection. However, XDH expression dropped below control levels (dsAMP-injected flies) by day 3 after injection, remaining low until day 6. From day 7 onwards, XDH expression levels increased but still remained lower than the dsAMP-injected control group (Fig 3C).
Fig. 3.
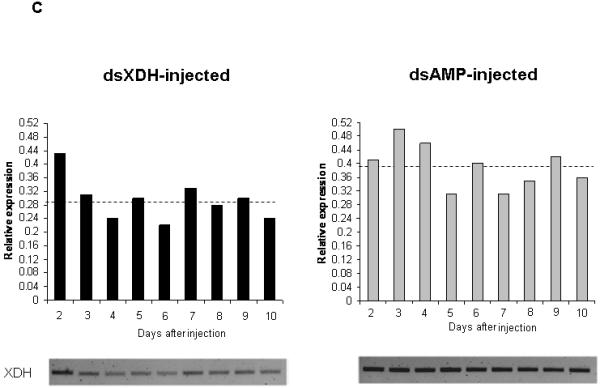
RT-PCR verification of xanthine dehydrogenase (XDH) expression from Lu. longipalpis injected with XDH and ampicillin (AMP) dsRNAs. RT-PCR of the ribosomal 60S gene was used as loading control. (a) Reduction in XDH expression in sand flies injected with XDH dsRNA (Lanes 1, 2 and 3) compared to flies injected with AMP dsRNA (lines 4, 5 and 6). Lanes 1, 2, 4 and 5 represent XDH expression from 1 sand fly and lanes 3 and 6 represent XDH expression of a pool of 20 sand flies. (b) 3 independent experiments were done and relative XDH gene expression was expressed by the ratio between band densitometry values in an RT-PCR product of the XDH gene and the control Ribosomal 60S gene for dsXDH and dsAMP-injected sand flies. Individual insects, as well as pools of 20 insects were used in each experimental groups. Asterisk represents significant statistical significance at p< 0.05. (c) XDH relative expression levels for dsXDH-injected and dsAMP-injected flies measured by semi-quantitative RT-PCR from day 2 (D2) until day 10 (D10) after dsRNA injection. Dotted lines represent mean relative expression levels for dsXDH and dsAMP-injected flies. Each bar represents a pool of 3 sand flies.
3.4. Urate production in whole body and excretory material of Lu. longipalpis
To determine whether the knockdown in XDH mRNA led to a decrease in XDH enzyme activity the amount of urate in flies was estimated. Reduction in urate production after RNAi for XDH expression was confirmed by a colorimetric assay on insect whole body and excretions of individual sand flies. In the sand fly whole body, the amount of urate produced after a blood meal in dsXDH-injected Lu. longipalpis was considerably lower when compared to dsAMP-injected sand flies (P< 0.001), this reduction being also statistically significant when comparing urate production in dsXDH-injected to uninjected sand flies (Fig. 4A). dsXDH injections also significantly reduced urate production in the excretory material of sand flies when compared to the dsAMP-injected and uninjected groups (Fig 4B). Protein quantification in both dsXDH and dsAMP-injected sand fly whole bodies after the blood meal showed that insects in both groups had fed on similar amounts of rabbit blood, indicating that differences in urate production were not due to differences in blood intake (Fig 4C).
Fig. 4.
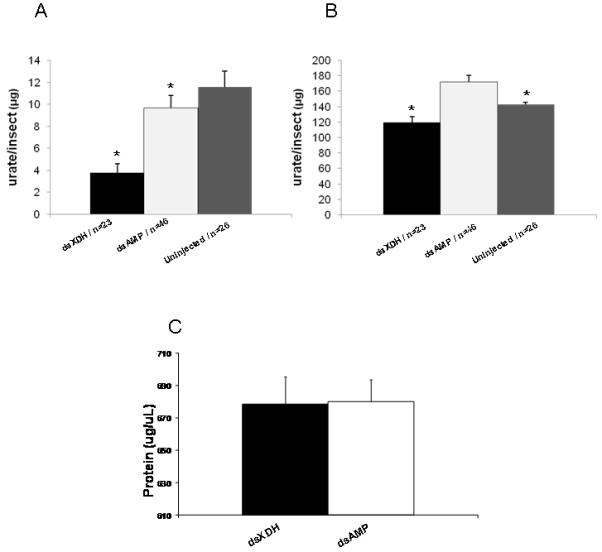
Amount of urate in Lutzomyia longipalpis (1 day old) injected with dsXDH or dsAMP. (a) Urate in the whole body 48 hours after feeding on rabbit blood (4 days after injection). Insects injected with dsXDH had significantly lower urate than dsAMP or uninjected insects; urate in dsAMP injected insects was significantly lower than uninjected insects. (b) Urate in the excretory material 48 hours after feeding on rabbit blood. dsXDH injected insects had significantly lower urate in the excretory material compared to the dsAMP and uninjected controls. Uninjected controls contained significantly less urate in the excretory material. (c) Protein content of sand fly whole bodies 48 h after feeding on rabbit blood. Asterisk indicates statistical significance at P< 0.05. Results (a) and (b) are presented as mean ± SEM, (c) mean ± SD.
3.5.Effect of XDH knockdown on the lifespan of sugar-fed and blood fed Lu. longipalpis
Unfed female Lu. longipalpis were injected with XDH and AMP dsRNA and subsequent sand fly survival in sugar-fed and rabbit blood fed sand flies was recorded. Double stranded XDH injections were found to significantly reduce sand fly longevity in sugar-fed (Fig 5A; log rank test: Chi-squared: 8.396, P<0.0038, 3 experiments) and bloodfed sand flies (Fig 5B; log rank test: Chi-squared: 17.71, P <0.0001, 2 experiments), when compared to dsAMP-injected sand flies.
Fig.5.
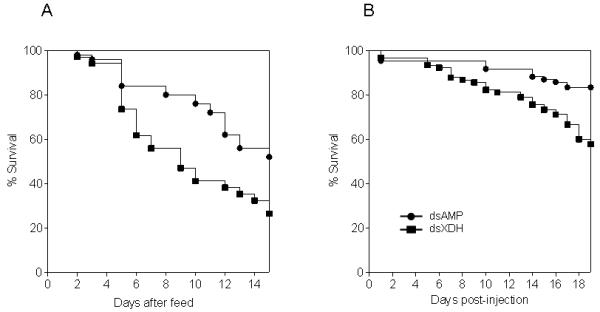
Kaplan-Meier plots showing survival of Lu. longipalpis under different nutritional conditions (a) Representative plot of sand flies bloodfed 96h after injection with XDH or AMP dsRNA where the mortality of an initial group of 34 (dsXDH) and 50 (dsAMP) sand flies was monitored daily. (b) Representative plot of flies injected with XDH and AMP dsRNA and sugar fed after injections. An initial group of 90 (dsXDH) and 84 (dsAMP) sand flies were monitored daily for mortality. Statistical analysis was based on at least two independent experiments.
4. Discussion
This study showed for the first time that specific gene knock down using RNAi is feasible for phlebotomine sand flies using an injected volume of 140 ng dsRNA per fly. Microinjection of dsRNA via the thorax of 1 day old adult female sand flies resulted in approximately 40% reduction of XDH gene expression in comparison to flies injected with a control dsRNA. Examination of the expression profile post-injection suggested that expression of this gene was not reduced until 48h after injection. This effect on mRNA was reflected in the reduction of urate production in the whole bodies and excretory material of blood fed insects. Incomplete knock down may be due to the timing of injection; gene expression may occur during the first day post-emergence from the pupae. Attempts to inject the pupae to obtain more complete knock down were unsuccessful.
A colorimetric assay for urate revealed significant reduction in both whole body and excretions of the female insect subjected to dsXDH injection. Reduced urate production in sand fly excretions after dsXDH injection was less obvious when compared to XDH knock down in the whole body. This might be due to urate production and accumulation in the hind gut previous to dsXDH introduction. Increased urate production in the excretions of dsAMP-injected flies was observed when compared to uninjected flies. As uric acid is a potent antioxidant molecule, it may be that the injury caused by the injection itself might have increased the oxidative stress in injected flies, therefore increasing uric acid production in mock-injected sand flies. Moreover, the introduction of a non-sterile substance in the sand fly haemolymph might have contributed to increase the oxidative stress. Such difference could not be observed in the whole body, perhaps because most of the urate produced during blood digestion is excreted, urate from excretory material being from 12 to 25 fold higher than urate in the whole body in our experiments.
The role of XDH and urate production in protection against oxidative stress has been studied in other insects. In Drosophila melanogaster, urate-null rosy mutants defective for XDH are sensitive to oxygen-derived stress (Hilliker et al., 1992). Moreover, previous studies using D. melanogaster rosy mutant suggested that XDH might play an important role in the innate immune response and age-associated deterioration of the innate immune system (Kim et al., 2001). In Rhodnius prolixus, very high urate levels were measured in the haemolymph after a blood meal and hemin injection, suggesting this may reflect an adaptation to overcome high oxidative stress generated by heme (Souza et al.,1997). Sand flies injected with XDH dsRNA exhibit significantly reduced life span in comparison with the mock-injected group fed on rabbit blood through chick skin (Fig 5A). This suggests, in line with previous studies, that urate may be operating as an important radical scavenger in Lu. longipalpis. Urate also seems to have an impact on sand fly survival even when blood is absent from the diet as survival of sugar fed flies injected with XDH dsRNA was reduced compared to controls (Fig 5B). In our experiments, sand flies were microinjected with considerable amounts of dsRNA, considering the sand flies diminutive body size. The injections “per se “ probably cause an important injury and physiological stress to the fly and considering the lack of sterile conditions, it is also possible that bacterial infections were associated with the injections, suggesting some degree of association between XDH activity and innate immune responses in the sand fly. However, experiments with XDH knockdown followed by injection/ingestion of gram positive and gram negative bacteria would be required to confirm any involvement of XDH with immune responses in Lu. longipalpis.
Up-regulation of the XDH gene was detected towards the end of blood meal digestion (72-96h post feeding) of adult female Lu. longipalpis. Transcript data from bloodfed mosquito microarray experiments suggests that up-regulation of XDH occurred earlier in the bloodfeeding process (Aedes aegypti; Sanders et al., 2003; Anopheles gambiae, Ribeiro, 2003). A detailed study of enzyme activity profiles of XDH in bloodfed Aedes aegypti (von Dungern and Briegel, 2001) showed that maximal XDH activity precedes digestive proteolytic activities, with the bulk of the enzyme being confined to the fat body. It was suggested that this reflects the importance of catabolic activities to ensure prompt clearance of excess protein ingested during the blood meals through a uricotelic pathway. It is unclear whether the up-regulation of XDH during the latter part of bloodmeal digestion in Lu longipalpis is a response to the increased protein catabolism or simply reflects the restoration of XDH levels in this insect in preparation for the next bloodmeal.
Xanthine dehydrogenases are members of the molybdenum hydrolase family of enzymes, catalyzing the oxidation of hypoxanthine to xanthine and xanthine to uric acid (Pitts and Zwiebel, 2001). Lu. longipalpis XDH sequence was similar to previously described XDH peptides from other insects, there is a high degree of conservancy within the two N-terminal ion-sulfur centres, a NAD-binding site and a C-terminal domain required for MoCo binding and dimerization (Mendel 2007) (Fig 1A). A phylogenetic analysis showed that Lu. longipalpis XDH grouped closely to other nematoceran XDH/AO sequences, being more distantly related, but forming a broader clade with three Drosophila sequences and two other brachyceran flies (Fig 1B). These results are in accordance with previous analyses where dipteran XDH sequences formed a monophyletic group within a dipteran clade, with the lepidopteran sequences being grouped in a separate clade (Komoto et al., 1999).
Double-stranded RNA is now used in a wide range of eukaryotes to suppress a variety of genes, allowing rapid analyses of gene function (Tomari and Zamore, 2005). The injection of dsRNA to disrupt gene expression in insects vectors of diseases has been performed before to study gene function in mosquitoes and triatomine bugs (eg. Blandin et al., 2002; Vlachou et al., 2005; Paskewitz et al., 2006; Boisson et al., 2006; Araújo et al., 2007). Until recently, molecular studies on sand fly-Leishmania relationships have focussed on the parasite-derived determinants of vector specificity. Considering that the parasites are confined to the gut lumen, Leishmania is expected to have a wide impact on gene regulation in the insects’ gut, as well as in other tissues such as fat body and ovaries. The production of Lu. longipalpis cDNA libraries (Dillon et al., 2006; Jochim et al., 2008) and a sand fly full genome sequencing programme (http://www.vectorbase.org/Other/AdditionalOrganisms/?organism=sandflies) together with production of a cDNA microarray means that the development of reverse genetic techniques for sand flies will provide a new approach to the study of sand fly - Leishmania interactions.
Studies addressing the effect of antioxidants in promoting trypanosome survival in tsetse flies (McLeod et al., 2007) and production of nitric oxide metabolites in mosquitoes during Plasmodium infection (Peterson et al., 2007) showed that supplementation of the blood meal with urate increased parasite prevalence in both trypanosome-infected tsetse flies and midgut oocyst production in Plasmodium berghei-infected Anopheles stephensi. Therefore, xanthine dehydrogenase knock down by RNAi followed by infection of the sand fly with Leishmania infantum, as well as infective blood meal supplementation with urate would help establish the importance of urate in Leishmania survival in the sand fly midgut.
Supplementary Material
Acknowledgements
The authors would like to thank Dr. Craig Wilding for helping with XDH phylogenetic analysis, Dr. Ray Wilson for his help during the Lu. longipalpis XDH cloning and sequencing and Dr. Matthew Rogers for helping with sand fly survival analysis. We gratefully acknowledge the technical assistance of Davina Moor and Jan Lewis. This work was funded by the Wellcome Trust.
References
- Alexander B, de Carvalho RL, McCallum H, Pereira MH. Role of the domestic chicken (Gallus gallus) in the epidemiology of urban visceral leishmaniasis in Brazil. Emerging Infect. Dis. 2002;8(12):1480–5. doi: 10.3201/eid0812.010485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo RN, Campos IT, Tanaka AS, Santos A, Gontijo NF, Lehane MJ, Pereira MH. Brasiliensin: A novel intestinal thrombin inhibitor from Triatoma brasiliensis (Hemiptera: Reduviidae) with an important role in blood intake. Int. J. Parasitol. 2007;37(12):1351–8. doi: 10.1016/j.ijpara.2007.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo RN, Santos A, Pinto FS, Gontijo NF, Lehane MJ, Pereira MH. RNA interference of the salivary gland nitrophorin 2 in the triatomine bug Rhodnius prolixus (Hemiptera: Reduviidae) by dsRNA ingestion or injection. Insect Biochem. Mol. Biol. 2006;36(9):683–93. doi: 10.1016/j.ibmb.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bañuls AL, Hide M, Prugnolle F. Leishmania and the leishmaniases: a parasite genetic update and advances in taxonomy, epidemiology and pathogenicity in humans. Adv. Parasitol. 2007;64:1–109. doi: 10.1016/S0065-308X(06)64001-3. [DOI] [PubMed] [Google Scholar]
- Blandin S, Moita LF, Köcher T, Wilm M, Kafatos FC, Levashina EA. Reverse genetics in the mosquito Anopheles gambiae: targeted disruption of the Defensin gene. EMBO Rep. 2002;3(9):852–6. doi: 10.1093/embo-reports/kvf180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson B, Jacques JC, Choumet V, Martin E, Xu J, Vernick K, Bourgouin C. Gene silencing in mosquito salivary glands by RNAi. FEBS Lett. 2006;580(8):1988–92. doi: 10.1016/j.febslet.2006.02.069. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–54. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Dillon RJ, Ivens AC, Churcher C, Holroyd N, Quail MA, Rogers ME, Soares MB, Bonaldo MF, Casavant TL, Lehane MJ, Bates PA. Analysis of ESTs from Lutzomyia longipalpis sand flies and their contribution toward understanding the insect-parasite relationship. Genomics. 2006;88(6):831–40. doi: 10.1016/j.ygeno.2006.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle WA, Burke JF, Chovnick A, Dutton FL, Whittle JR, Bray RC. Properties of xanthine dehydrogenase variants from rosy mutant strains of Drosophila melanogaster and their relevance to the enzyme’s structure and mechanism. Eur. J. Biochem. 1996;239(3):782–95. doi: 10.1111/j.1432-1033.1996.0782u.x. [DOI] [PubMed] [Google Scholar]
- Eleftherianos I, Marokhazi J, Millichap PJ, Hodgkinson AJ, Sriboonlert A, ffrench-Constant RH, Reynolds SE. Prior infection of Manduca sexta with non-pathogenic Escherichia coli elicits immunity to pathogenic Photorhabdus luminescens: roles of immune-related proteins shown by RNA interference. Insect Biochem. Mol. Biol. 2006;36(6):517–25. doi: 10.1016/j.ibmb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408(6810):325–30. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Ghanim M, Kontsedalov S, Czosnek H. Tissue-specific gene silencing by RNA interference in the whitefly Bemisia tabaci (Gennadius) Insect Biochem. Mol. Biol. 2007;37(7):732–8. doi: 10.1016/j.ibmb.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Glantzounis GK, Tsimoyiannis EC, Kappas AM, Galaris DA. Uric acid and oxidative stress. Curr. Pharm. Des. 2005;11(32):4145–51. doi: 10.2174/138161205774913255. [DOI] [PubMed] [Google Scholar]
- Graça-Souza AV, Maya-Monteiro C, Paiva-Silva GO, Braz GR, Paes MC, Sorgine MH, Oliveira MF, Oliveira PL. Adaptations against heme toxicity in blood-feeding arthropods. Insect Biochem. Mol. Biol. 2006;36(4):322–35. doi: 10.1016/j.ibmb.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Gönczy’ P, Echeverri C, Oegema K, Coulson A, Jones SJ, Copley RR, Duperon J, Oegema J, Brehm M, Cassin E, Hannak E, Kirkham M, Pichler S, Flohrs K, Goessen A, Leidel S, Alleaume AM, Martin C, Ozlü N, Bork P, Hyman AA. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408(6810):331–6. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- Hilliker AJ, Duyf B, Evans D, Phillips JP. Urate-null rosy mutants of Drosophila melanogaster are hypersensitive to oxygen stress. Proc. Natl. Acad. Sci. USA. 1992;89(10):4343–7. doi: 10.1073/pnas.89.10.4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G, Zamore PD. RNAi: nature abhors a double-strand. Curr. Opin. Genet. Dev. 2002;12(2):225–32. doi: 10.1016/s0959-437x(02)00290-3. [DOI] [PubMed] [Google Scholar]
- Jaubert-Possamai S, Le Trionnaire G, Bonhomme J, Christophides GK, Rispe C, Tagu D. Gene knockdown by RNAi in the pea aphid Acyrthosiphon pisum. BMC Biotechnol. 2007;7:63. doi: 10.1186/1472-6750-7-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochim RC, Teixeira CR, Laughinghouse A, Mu J, Oliveira F, Gomes RB, Elnaiem DE, Valenzuela JG. The midgut transcriptome of Lutzomyia longipalpis: comparative analysis of cDNA libraries from sugar-fed, blood-fed, post-digested and Leishmania infantum chagasi-infected sand flies. BMC Genomics. 2008;9:15. doi: 10.1186/1471-2164-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell. 1998;95(7):1017–26. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- Kim YS, Nam HJ, Chung HY, Kim ND, Ryu JH, Lee WJ, Arking R, Yoo MA. Role of xanthine dehydrogenase and aging on innate immune response of Drosophila. J. Amer. Aging Assoc. 2001;24:187–194. doi: 10.1007/s11357-001-0020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kômoto N, Yukuhiro K, Tamura T. Structure and expression of tandemly duplicated xanthine dehydrogenase genes of the silkworm (Bombyx mori) Insect Mol. Biol. 1999;8(1):73–83. doi: 10.1046/j.1365-2583.1999.810073.x. [DOI] [PubMed] [Google Scholar]
- MacLeod ET, Maudlin I, Darby AC, Welburn SC. Antioxidants promote establishment of trypanosome infections in tsetse. Parasitology. 2007;134(Pt 6):827–31. doi: 10.1017/S0031182007002247. [DOI] [PubMed] [Google Scholar]
- Maleszka J, Forêt S, Saint R, Maleszka R. RNAi-induced phenotypes suggest a novel role for a chemosensory protein CSP5 in the development of embryonic integument in the honeybee (Apis mellifera) Dev. Genes. Evol. 2007;217(3):189–96. doi: 10.1007/s00427-006-0127-y. [DOI] [PubMed] [Google Scholar]
- Mendel RR. Biology of the molybdenum cofactor. J. Exp. Bot. 2007;58(9):2289–96. doi: 10.1093/jxb/erm024. [DOI] [PubMed] [Google Scholar]
- Michalsky EM, Rocha MF, da Rocha Lima AC, França-Silva JC, Pires MQ, Oliveira FS, Pacheco RS, Dos Santos SL, Barata RA, Romanha AJ, Fortes-Dias CL, Dias ES. Infectivity of seropositive dogs, showing different clinical forms of leishmaniasis, to Lutzomyia longipalpis phlebotomine sand flies. Vet. Parasitol. 2007;147(1-2):67–76. doi: 10.1016/j.vetpar.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Modi GB. Care and maintenance of phlebotomine sandfly colonies. In: Crampton JM, Beard CB, Louis C, editors. The Molecular Biology of Insect Disease Vectors. Chapman & Hall; London: 1997. pp. 21–30. [Google Scholar]
- Narasimhan S, Montgomery RR, DePonte K, Tschudi C, Marcantonio N, Anderson JF, Sauer JR, Cappello M, Kantor FS, Fikrig E. Disruption of Ixodes scapularis anticoagulation by using RNA interference. Proc. Natl. Acad. Sci. USA. 2004;101(5):1141–6. doi: 10.1073/pnas.0307669100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva-Silva GO, Cruz-Oliveira C, Nakayasu ES, Maya-Monteiro CM, Dunkov BC, Masuda H, Almeida IC, Oliveira PL. A heme-degradation pathway in a blood-sucking insect. Proc. Natl. Acad. Sci. USA. 2006;103(21):8030–5. doi: 10.1073/pnas.0602224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paskewitz SM, Andreev O, Shi L. Gene silencing of serine proteases affects melanization of Sephadex beads in Anopheles gambiae. Insect Biochem. Mol. Biol. 2006;36(9):701–11. doi: 10.1016/j.ibmb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Peterson TM, Gow AJ, Luckhart S. Nitric oxide metabolites induced in Anopheles stephensi control malaria parasite infection. Free Radic. Biol. Med. 2007;42(1):132–42. doi: 10.1016/j.freeradbiomed.2006.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitaluga AN, Mason PW, Traub-Cseko YM. Non-specific antiviral response detected in RNA-treated cultured cells of the sandfly, Lutzomyia longipalpis. Dev. Comp. Immunol. 2007 doi: 10.1016/j.dci.2007.06.008. doi:10.1016/j.dci.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Pitts RJ, Zwiebel LJ. Isolation and characterization of the Xanthine dehydrogenase gene of the Mediterranean fruit fly, Ceratitis capitata. Genetics. 2001;158(4):1645–55. doi: 10.1093/genetics/158.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JM. A catalogue of Anopheles gambiae transcripts significantly more or less expressed following a blood meal. Insect Biochem. Mol. Biol. 2003;33(9):865–82. doi: 10.1016/s0965-1748(03)00080-8. [DOI] [PubMed] [Google Scholar]
- Rogers ME, Bates PA. Leishmania manipulation of sand fly feeding behavior results in enhanced transmission. PLoS Pathog. 2007;3(6):e91. doi: 10.1371/journal.ppat.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders HR, Evans AM, Ross LS, Gill SS. Blood meal induces global changes in midgut gene expression in the disease vector, Aedes aegypti. Insect Biochem. Mol. Biol. 2003;33(11):1105–22. doi: 10.1016/s0965-1748(03)00124-3. [DOI] [PubMed] [Google Scholar]
- Sato A, Nishino T, Noda K, Amaya Y, Nishino T. The structure of chicken liver xanthine dehydrogenase. cDNA cloning and the domain structure. J. Biol. Chem. 1995;270(6):2818–26. doi: 10.1074/jbc.270.6.2818. [DOI] [PubMed] [Google Scholar]
- Souza AV, Petretski JH, Demasi M, Bechara EJ, Oliveira PL. Urate protects a blood-sucking insect against hemin-induced oxidative stress. Free Radic. Biol. Med. 1997;22(1-2):209–14. doi: 10.1016/s0891-5849(96)00293-6. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19(5):517–29. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- Vlachou D, Schlegelmilch T, Christophides GK, Kafatos FC. Functional genomic analysis of midgut epithelial responses in Anopheles during Plasmodium invasion. Curr. Biol. 2005;15(13):1185–95. doi: 10.1016/j.cub.2005.06.044. [DOI] [PubMed] [Google Scholar]
- von Dungern P, Briegel H. Protein catabolism in mosquitoes: ureotely and uricotely in larval and imaginal Aedes aegypti. J. Insect Physiol. 2001;47(2):131–141. doi: 10.1016/s0022-1910(00)00096-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.