Abstract
Abnormal expression of major histocompatibility complex (MHC) class I and class II in various tissues is associated with autoimmune disease. Autoimmune responses can be triggered by viral infections or tissue injuries. We show that the ability of a virus or a tissue injury to increase MHC gene expression is duplicated by any fragment of double-stranded (ds) DNA or dsRNA introduced into the cytoplasm of nonimmune cells. Activation is sequence-independent, is induced by ds polynucleotides as small as 25 bp in length, and is not duplicated by single-stranded polynucleotides. In addition to causing abnormal MHC expression, the ds nucleic acids increase the expression of genes necessary for antigen processing and presentation: proteasome proteins (e.g., LMP2), transporters of antigen peptides; invariant chain, HLA-DM, and the costimulatory molecule B7.1. The mechanism is different from and additive to that of γ-interferon (γIFN), i.e., ds polynucleotides increase class I much more than class II, whereas γIFN increases class II more than class I. The ds nucleic acids also induce or activate Stat1, Stat3, mitogen-activated protein kinase, NF-κB, the class II transactivator, RFX5, and the IFN regulatory factor 1 differently from γIFN. CpG residues are not responsible for this effect, and the action of the ds polynucleotides could be shown in a variety of cell types in addition to thyrocytes. We suggest that this phenomenon is a plausible mechanism that might explain how viral infection of tissues or tissue injury triggers autoimmune disease; it is potentially relevant to host immune responses induced during gene therapy.
Organ-specific autoimmune diseases are associated with abnormal expression of major histocompatibility complex (MHC) class I and aberrant expression of MHC class II antigens on the surface of cells in the affected tissue (1–4). Abnormal expression of MHC molecules on normal, nonimmune cells can present antigens to T cells (5), which leads to T cell activation, a loss in self-tolerance, and the development of autoimmune disease (e.g., Graves’ disease; ref. 6). There is no comprehensive explanation as to how abnormal MHC expression might develop in the target tissue or how this expression might contribute to the ensuing immune cell response.
Viral infections can ablate self-tolerance, mimic immune responses to self antigens, and induce autoimmune disease (3, 7, 8). Recent work has suggested that viral triggering of diverse autoimmune diseases, including rheumatoid arthritis and insulin-dependent diabetes, is caused by local viral infection of the tissue and not molecular mimicry (8–10). It is suggested that this viral triggering involves MHC genes, results in the presentation of self antigens, and induces bystander activation of the T cells. The mechanism for this viral triggering of autoimmune disease is obscure, as is its relation to the response of immune cells to cytokine/interferon (IFN; refs. 8–10). Thus, γIFN certainly can increase MHC gene expression in the target tissue (3, 4, 11); however, the mechanism by which a viral infection of nonimmune cells in a target tissue recruits and activates immune cells to produce γIFN is unclear. Additionally, it is unlikely that γIFN alone causes autoimmunity, because its administration does not induce typical autoimmune disease (12). Moreover, generalized γIFN production by immune cells cannot account for cell-specific autoimmunity (e.g., the destruction of pancreatic β cells but not α cells in insulin-dependent diabetes mellitus or the involvement of only thyroid follicular cells but not parafollicular C cells in autoimmune Graves’ disease). Finally, increased MHC class I and class II expression was reported after tissue damage in vivo, even in γIFN or γIFN-receptor knockout mice (13), also suggesting that IFN is not the sole factor for abnormal MHC induction.
We wondered whether viral or self DNA introduced into the cell by infection or tissue injury, respectively, might increase MHC gene expression. We show that any fragment of double-stranded (ds) DNA or dsRNA introduced into the cytoplasm can induce abnormal MHC expression, as well as the expression or activation of other genes or gene products, respectively, that are essential for antigen presentation by nonimmune cells. These data suggest that ds nucleic acids introduced into the cytoplasm may drive normal cells to become antigen-presenting cells (APCs) and induce an autoimmune response by bystander activation of the immune cells. We suggest that the present report also has widespread biologic significance, because transfection procedures are used widely in experimental and clinical applications (e.g., gene therapy).
MATERIALS AND METHODS
Cells.
The rat FRTL-5 thyroid cells used were fresh subclones (F1). All of their properties and the method by which they were grown have been described (14–16).
Transfection.
Unless otherwise noted, 5 μg of DNA was mixed with 30 μl of Lipofectamine Plus reagent (GIBCO/BRL) and 750 μl of serum-free medium and then incubated for 15 min at room temperature. A duplicate mixture without DNA also was incubated for 15 min at room temperature. Cells were washed with serum-free medium, and the combined mixtures were added. After 3 h, medium was replaced with normal culture medium containing serum.
Nucleic Acids.
Synthetic polynucleotides were from Amersham Pharmacia; salmon-sperm DNA was from Stratagene; calf-thymus DNA was from Sigma. Genomic DNA was purified with a Wizard Genomic DNA purification Kit (Promega). Plasmid DNAs were purified with EndoFree Plasmid Maxi Kits (Qiagen, Valencia, CA); lipopolysaccharide concentrations in plasmid DNA samples were measured with the Limulus Amebocyte Lysate test (BioWhittaker). CpG oligonucleotides are described (17, 18); methylation was performed with SssI methylase (New England Biolabs) at 37°C for 2 h. Methylation of CpG motifs was confirmed by resistance to BstUI (New England Biolabs). DNA was digested by DNase I at 37°C for 30 min, followed by phenol-chloroform extraction and ethanol precipitation. Digestion was confirmed by agarose gel electrophoresis.
Northern Analysis.
The preparation of total RNA, the probes for MHC class I and class II, and the Northern analysis have been described (15, 16, 19). The probe for rat class II transactivator (CIITA) is a cloned rat type III CIITA cDNA fragment in pcDNA3 (K.S., A.M., L.D.K., and M. Pietrarelli, unpublished data) digested with EcoRI to release a 4098-bp fragment. The probe for IFN regulatory factor 1 (IRF-1; GenBank accession no. X14454) was a 2.1-kb fragment cut with HindIII/BamHI from pUCIRF-1 (kindly provided by T. Taniguchi, Osaka University, Osaka, Japan, and K. Sugiyama, Boehringer Ingelheim, Hyogo, Japan). Other probes were made by reverse transcription–PCR by using published cDNA sequences as primers: LMP2, TACCGTGAGGACTTGTTAGCG and ATGACTCGATGGTCCACACC (296 bp); transporter of antigen peptides 1 (TAP1), GGAACAGTCGCTTAGATGCC and CACTAATGGACTCGCACACG (504 bp); invariant chain (Ii), AATTGCAACCGTGGAGTCC and AACACACACCAGCAGTAGCC (635 bp); HLA-DMB, ATCCTCAACAAGGAAGAAGGC and GTTCTTCATCCACACCACGG (222 bp); B7.1, CCATACACCGAATCTACTGGC and TTGACTGCATCAGATCCTGC (589 bp); RFX5, AAGCTGTATCTCTACCTTCAG and TTTCAGGATCCGCTCTGCCCA (470 bp); PKR, ACAAGGTGGATAGTCACACGG and CCAGATGCTGACTGAGAAGC (352 bp); and βIFN, AAGATCATTCTCACTGCAGCC and TGAAGACTTCTGCTCGGACC (586 bp).
SDS/PAGE and Western Blotting.
Preparation of total cell protein and Western blotting were performed as described (20). Antibodies used were directed against phosphospecific Stat1 (Tyr-701), phosphospecific Stat3 (Tyr-705), phosphospecific p44/42 mitogen-activated protein kinase (MAPK; ERK-1 and ERK-2), and Stat1 (New England Biolabs).
Electrophoretic Mobility-Shift Assay.
Purification of nuclear protein, probe labeling, and the method for the electrophoretic mobility-shift assay have been described (15, 16, 21).
Flow Cytometry.
Single cell suspensions were washed with PBS (pH 7.4), and 106 cells were pelleted, suspended in 100 μl of PBS, and placed in a 96-well flat-bottomed plate. After 30 min on ice, cells were incubated for 30 min with 100 μl of fluorescein isothiocyanate-labeled human-, rat-, or mouse-specific monoclonal antibodies against MHC class I or class II antigens (Serotec). Cells were incubated for 30 min, washed three times, and kept in the dark at 4°C until they were analyzed with a fluorescence-activated cell sorter (FACSort instrument and cellquest software; Becton Dickinson). The optimal dilution of antibodies was predetermined. Leu-4 was the background control, and a subclass-matched Ig was the negative control.
RESULTS
Viral DNA Induces MHC Expression.
Herpes simplex virus (HSV) infection increased MHC class I and MHC class II RNA levels in FRTL-5 cells within 48 h of infection (Fig. 1a, lanes 1–4). The mean increase ± SD for MHC class I was (4.1 ± 0.7)-fold at 48 h, when normalized to GAPDH values. The increase in MHC class II was reproducible at 48 h. Given the absence of constitutive levels of class II RNA (Fig. 1a, lanes 1 and 5), its quantitative increase was calculated relative to the maximal effect of 100 units/ml γIFN (see Fig. 1 d and e).
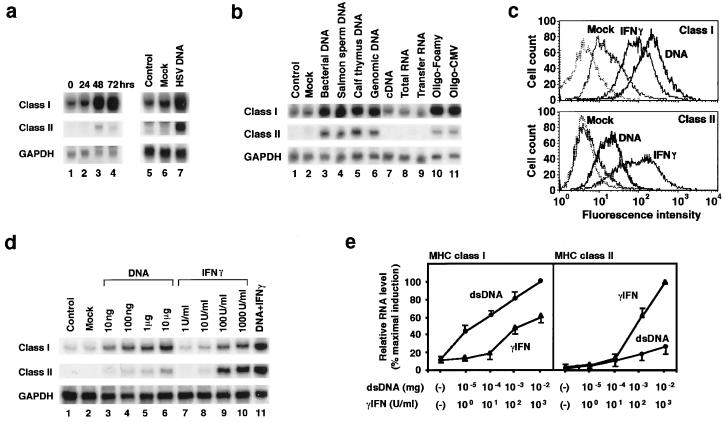
Figure 1.
DNA induces MHC expression in cells. FRTL-5 cells (2 × 106 cells in 10-cm dishes) were infected with HSV (a, lanes 1–4; ref, 29), or, alternatively, they were transfected with 5 μg of HSV DNA fragments (a, lane 7), the indicated DNAs (b, lanes 3–7), RNA (b, lanes 8 and 9), or the following 54-bp ds oligodeoxynucleotides (ODNs) from Foamy virus or cytomegalovirus (CMV; b, lanes 10 and 11): CATCTTCCAGTTCATCTCTAGTCATTTGGGCTGTTTCGGCCATTGTTACTGGTC and CTATTTGCACCACGTTCGCAGCCATACCAATCTACCTGATCCCATCTCCAGGCT, respectively. GAPDH = glyceraldehyde-3-phosphate dehydrogenase. (c) Fluorescence-activated cell sorter analysis of cell-surface MHC class I and class II expression induced by DNA or 100 units/ml rat γIFN 48 h after treatment. Cells were transfected with 5 μg of pcDNA3 (Invitrogen), exactly as they were for all dsDNAs in a and b. The dashed line represents control staining with fluorescein isothiocyanate-labeled normal mouse IgG1. The data shown in a, b, and c are typical results from three experiments performed on different batches of cells. (d and e) Cells were transfected with the indicated amounts of dsDNA (d, lanes 3–6), treated with γIFN (d, lanes 7–10), or exposed to both (d, lane 11). d depicts Northern blotting from one representative experiment; e presents the mean ± SD from four separate experiments expressed relative to the maximum induction of class I RNA (Left) or the maximum induction of class II RNA by γIFN (Right). (a–e) The total RNA was prepared, and Northern analysis was performed at the times noted or 48 h after treatment. Lipofectamine treatment alone served as a control transfection procedure (Mock).
Transfected HSV DNA fragments similarly increased MHC RNA levels (Fig. 1a, lanes 5–7), and this increase was not a viral DNA-specific phenomenon. Rather, all types of dsDNA, but not single-stranded (ss) DNA, increased MHC RNA levels (Fig. 1b). Regardless of the DNA used, the mean increase ± SD for MHC class I was (3.9 ± 0.5)-fold at 48 h; the quantitative increase in MHC class II relative to the effect of γIFN is evident in Fig. 1 d and e.
tRNA, which has a clover-leaf structure with linear ds segments of fewer than 10 bp, did not experience an increase in MHC RNA levels. The effect of dsDNA was concluded to be specific, because there was no increase in control genes, such as GAPDH (Fig. 1 a and b). Control transfections without DNA had no effect (Fig. 1 a, lane 6, and b, lane 2). We found less than 0.02 endotoxin units per mg of plasmid DNA in preparations that we purified with the Qiagen Endo-free kits; moreover, 15 μg/ml lipopolysaccharide from Sigma (7,500 endotoxin units), when used as a control, had no effect on MHC induction. Additionally, more rigorously defined DNAs without lipopolysaccharide contamination were active, (e.g., synthetic polynucleotides; Fig. 2).
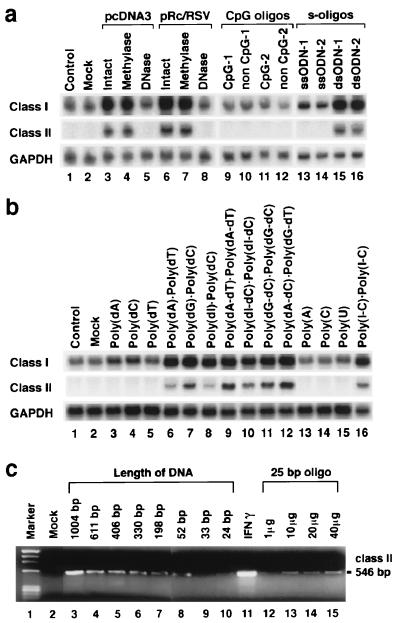
Figure 2.
Properties of the nucleic acid needed to induce MHC expression in cells. Transfection and Northern analysis were performed exactly as described for Fig. 1. (a) FRTL-5 cells were transfected with intact, methylated, or DNase-treated plasmids, pcDNA3 or pRc/RSV (lanes 3–8), ss CpG ODNs or control ODNs (lanes 9–12), or ss or ds phosphorothioate oligonucleotides (s-oligos; lanes 13–16). Lane 1 contains RNA from nontreated cells, and lane 2 contains only RNA from cells subjected to the transfection procedure. ODN-1 is the Foamy virus oligonucleotide. ODN-2 is the cytomegalovirus oligonucleotide. CpG-1 is GCTAGACGTTAGCGT; non CpG-1 is GCTAGATGTTAGCGT; CpG-2 is TCAACGTTGA; and non CpG-2 is TCAAGCTTGA. (b) Various synthetic polynucleotides and their duplexes were transfected similarly and analyzed (lanes 3–16). (c) Cells were transfected with 5 μg of dsDNA fragments from 24 bp to 1,004 bp in length (lanes 3–10) or with the indicated amount of 25-bp dsODNs (lanes 12–15). MHC class II expression was measured 48 h later by reverse transcription–PCR (15, 16). Cells treated with 100 units/ml γIFN for 48 h were the positive control. These results are typical of at least five different experiments performed on different batches of cells.
The effect was independent of the method of introducing the ds nucleic acids into the cytoplasm, i.e., different transfection procedures (with Lipofectamine, electroporation, or DEAE dextran) similarly increased MHC RNA levels (data not shown).
These results were not limited to rat FRTL-5 thyroid cells and were duplicated in primary and continuous cultures of human and mouse fibroblasts, NIH 3T3 cells, SkMC human muscle cells, HUVEC human endothelial cells, C2C12 mouse smooth-muscle cells, WEHI231 pre B cells, and P381D1 macrophages, as well as primary cultures of mouse spleen dendritic cells, mouse peritoneal macrophages, and mouse spleen macrophages (data not shown). In each case there was an increase in class I and class II RNA levels as well as in antigen expression, as determined by fluorescence-activated cell sorter analyses, albeit this increase was less dramatic in the immune cells where constitutively high levels of MHC class I, MHC class II, or both exist. Therefore, induction of MHC expression by naked ds nucleic acid seemed to be a universal phenomenon.
The effect of dsDNA transfection on MHC gene expression differed from that of γIFN. The dsDNA increased class I levels more than class II, both with respect to cell-surface expression (Fig. 1c) and RNA (Fig. 1d, lanes 1–10), whereas γIFN increased class II more than class I. Quantitative changes in class I RNA levels, relative to the maximum induction of class I RNA by 10 μg of dsDNA, are presented in Fig. 1e (Left). Changes in class II RNA levels relative to the maximum induction of class II RNA by 100 units/ml γIFN, are presented in Fig. 1e (Right). The mean increase in MHC class I fluorescence intensity (channel number), induced by dsDNA relative to basal values (29), was 284 vs. 131 for γIFN. Relative to a basal value of 8, the mean increase in MHC class II induced by dsDNA was 23 vs. 156 for γIFN. Relative fluorescence-intensity values varied by less than 10% in three experiments, and changes were evident in 100% of cells. The effect of dsDNA transfection and γIFN on MHC gene expression was additive (Fig. 1d, lane 11).
CpG Motifs Are Not Responsible for the Effect.
Unmethylated CpG motifs within bacterial and viral DNA sequences have been shown to activate immune cells by inducing the production of various cytokines or Igs (17, 18). We evaluated the possible role of CpG motifs by transfecting FRTL-5 cells with intact or methylated dsDNA or known CpG ODNs and their non-CpG controls (Fig. 2a). Both methylated and unmethylated plasmid DNA had similar effects on MHC class I and II induction (Fig. 2a, lanes 3 vs. 4 and 5 vs. 6). Also, neither the ODNs having one or more CpG motifs (CpG-1 and/or CpG-2), which were confirmed to induce interleukin 6 and 12 or γIFN in lymphocytes (18), nor their non-CpG controls induced MHC expression (Fig. 2a, lanes 9–12).
However, the induction of MHC was abolished when the dsDNA was pretreated with DNase (Fig. 2a, lanes 5 vs. 3 and 8 vs. 6) but not RNase (data not shown). DNase digests the dsDNA into short (<10-bp) fragments or individual nucleotides, suggesting that dsDNA that has a certain length—and not a contaminant—is the active moiety. The effect of DNase digestion was confirmed by agarose gel electrophoresis. Additionally, ss phosphorothioate ODNs had no effect, whereas ds phosphorothioate ODNs induced MHC expression (Fig. 2a, lanes 13–16). The dsDNA effect on MHC gene expression, therefore, seemed to be dsDNA-specific and not to involve CpG motifs.
The Effect Is Sequence Independent.
To determine whether there was any sequence specificity, we transfected FRTL-5 cells with various synthetic polynucleotides (Fig. 2b). dsDNA copolymers (Fig. 2b, lanes 9–12) or duplexes (Fig. 2b, lanes 6–8) induced MHC expression, whereas ssDNA polymers had no effect (Fig. 2b, lanes 3–5). Regardless of the polynucleotide used, the mean increase ± SD for MHC class I, relative to constitutive values, was (4.2 ± 0.9)-fold at 48 h, when normalized to GAPDH values. The increase in MHC class II—again clearly significant and reproducible at 48 h—was 36 ± 8% of the maximal effect of 100 units/ml γIFN, which was run as a control. Of interest, dsRNA, which is known to induce various antiviral reactions, also induced a similar increase in MHC gene expression, whereas ssRNA had no effect (Fig. 2b, lanes 13–16). The effect was length- and concentration-dependent (Fig. 2c); in four separate experiments, ds oligonucleotides as short as 25 bp were reproducibly effective (Fig. 2c, lanes 12–15).
The ds Nucleic Acids Induce Expression of Multiple Genes Related to Antigen Processing and Presentation.
Multiple genes and proteins are required for the multiple steps involved in antigen processing and presentation by MHC genes (22, 23). In the case of MHC class I, increases in proteasome proteins (e.g., LMP2) and activity are necessary for antigen processing to peptides (22). Also, TAP molecules are required to allow antigenic peptides to bind the class I molecule at the cell surface (22). In case of MHC class II, Ii and HLA-DMB proteins are required to regulate the binding of antigen peptides; and costimulatory molecules (B7.1 or CD80, for example) are required to activate T lymphocytes (23). Expression of all of these genes is induced reproducibly by dsDNA in multiple experiments, as well as by γIFN, concomitantly with increased MHC gene expression (Fig. 3), suggesting that the cells can acquire full capability to present antigen to immune cells.
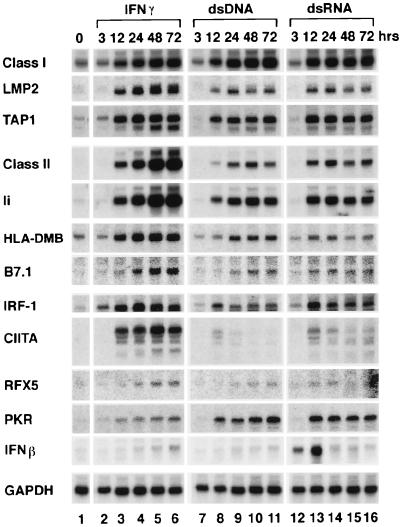
Figure 3.
Effect of ds nucleic acids on genes required for antigen processing and presentation by MHC genes. Transfection, IFN treatment, and Northern analysis were performed 3–72 h after treatment as described for Figs. 1 and 2. Transfection was with 5 μg of poly(dI-dC)⋅poly(dI-dC) (dsDNA) or poly(I-C)⋅poly(I-C) (dsRNA). These are typical results from at least four different experiments on different batches of cells.
γIFN-increased MHC gene expression is mediated by a several IFN-inducible genes, including CIITA, RFX5, and IRF-1 (24, 25). The effect of dsDNA on the RNA levels of these genes, particularly CIITA, is different from γIFN, both as a function of time and level (Fig. 3). These results were reproduced in three separate experiments.
The dsRNA behaves more like dsDNA than γIFN (Fig. 3), but dsRNA, not dsDNA, increases βIFN production by the FRTL-5 thyroid cell within 3 h. Of interest, the dsRNA-dependent protein kinase (26) PKR is also induced by dsDNA (Fig. 3).
The ds Nucleic Acids Activate Multiple Signaling Pathways.
Like γIFN (27), dsDNA induced significant phosphorylation of Stat1 and Stat3 (Fig. 4a). However, the dsDNA was very different from γIFN, whose effect on Stat1 and Stat3 phosphorylation was significantly lower and more delayed in time (Fig. 4a). In contrast, the γIFN effect on total Stat1 protein is greater and more advanced in time (Fig. 4a). Gel-shift analysis with nuclear protein from cells treated with dsDNA uncovered a marked increase in specific binding of Stat3 to its consensus DNA sequence in comparison with extracts from cells treated with IFN (Fig. 4b, Upper).
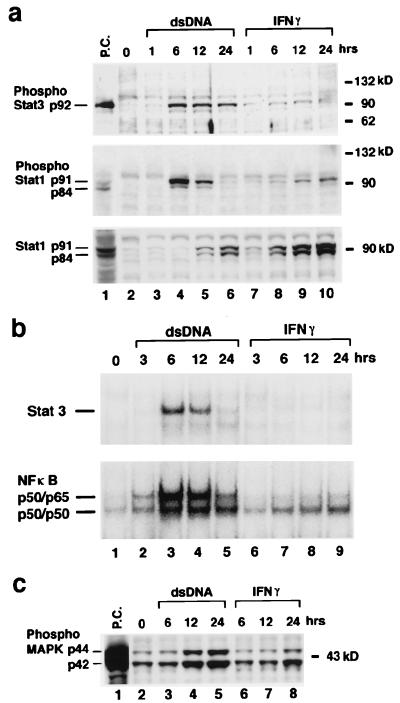
Figure 4.
dsDNA activates Stat1, Stat3, MAPK, and NF-κB. dsDNA transfection and IFN treatment of FRTL-5 cells were performed exactly as described for Figs. 1–3 by using 5 μg of poly(dI-dC)⋅poly(dI-dC). (a) Total cell lysate was prepared, and Western blot analysis was performed as described (20). Lane 1 (P.C.) contains a positive control cell lysate (New England Biolabs). (b) Nuclear protein was prepared, and gel-shift analysis was performed as described (15, 16, 19). Consensus ODNs for Stat3 and NF-κB are from Santa Cruz Biotechnology. (c) Western blot analysis with an antibody against phosphorylation-specific p44/p42 MAPK. Shown are typical results from at least four different experiments performed on different batches of cells.
NF-κB is an important transcription factor for the expression of many genes, including MHC class I. Significantly increased binding, and presumably formation, of a p50/p65 and a p50 homodimer to a consensus NF-κB oligonucleotide binding site was measurable by using nuclear extracts from cells transfected with dsDNA (Fig. 4b, Lower). γIFN treatment induced a significantly lower level of binding of both (Fig. 4b, Lower). Another difference between dsDNA and γIFN action involved phosphorylation of MAPK (Fig. 4c). Phosphorylation seemed to occur faster as a function of time and seemed to involve a quantitatively larger fraction of the protein pool.
The dsRNA induced the same changes indicated in Fig. 4 in all respects. This result is not, however, unexpected, because dsRNA activation of Stat1, NF-κB, and IRF-1 had been reported in other systems (28, 29). Of interest, PKR is implicated in this activation in those reports, possibly explaining the ability of dsDNA to induce PKR. Thus, PKR plays an important role in IFN and dsRNA-signaling pathways by modulating the transcriptional function of Stat1 and acting as a signal transducer for genes dependent on the transcription factors IRF-1 and NF-κB (28, 29).
In sum, therefore, dsDNA acts significantly differently from IFN in its effects on key components of the protein processing as well as transcriptional activation events involved in the expression of MHC and other genes important for antigen presentation. These results were reproduced in three separate experiments.
Cell Injury Can Induce MHC Expression.
There are multiple ways for cells to be exposed to ds nucleic acids other than by viral infection. One is escape and migration of self genomic or mitochondrial DNA into the cytoplasm (30, 31). In the case of tissue injury, which can induce MHC gene expression and the escape of genomic DNA (31), the increased MHC class I and class II expression was reported after tissue damage in vivo, even in IFN or IFN-receptor knockout mice (13). This result suggested that the dsDNA phenomenon described herein might be a reasonable mechanistic explanation of how tissue injury might increase MHC expression.
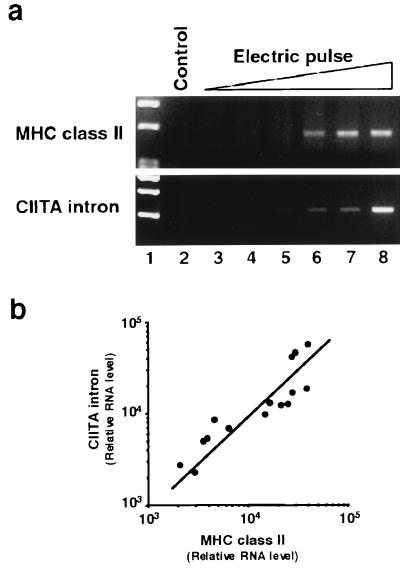
When FRTL-5 cells were exposed to progressively higher levels of electric pulsing, increased MHC RNA expression was observed (Fig. 5a, lanes 6–8). Genomic CIITA intron sequences (i.e., nuclear DNA) amplified by PCR from total RNA without first-strand synthesis, paralleled the appearance of MHC RNA and the strength of electric pulse (Fig. 5a, lanes 6–8, and b). Relative amplification of MHC class II and CIITA intron sequences was quantitatively similar in four replicate experiments on different batches of cells, despite problems inherent in the absolute quantitation of PCR results. PCR studies showed, additionally, that increases in TAP, LMP2, Ii, HLA-DMB, and B7.1 RNA levels were present along with increased MHC gene expression (data not shown).
Figure 5.
Tissue damage by electric pulsing coordinately increases MHC gene expression and genomic DNA in the cytoplasm. FRTL-5 cells (5 × 106 cells in Dulbecco’s PBS) were pulsed once with a Gene Pulser (Bio-Rad) set at 0.3 kV and at capacitances of 0.25, 25, 125, 250, and 960 μF or pulsed twice with a capacitance of 960 μF (lanes 3–8, respectively). Cells were washed with medium, returned to a 10-cm dish, and cultured for 48 h until RNA was recovered. Damage was estimated microscopically by trypan-blue exclusion and plating efficiency after pulsing. After two pulses at 960 μF, 60% of cells were fused or died. (a) Reverse transcription–PCR data compare MHC class II expression with contamination of total RNA preparations by leaked genomic DNA, measured by using PCR primers that detect an intron sequence of rat CIITA genome DNA (M. Pietrarelli, K.S., and L.D.K., unpublished results). Data are typical results from four different experiments performed on different batches of cells. (b) The correlation of MHC class II and CIITA intron levels for pulses eliciting significant increases of each (as shown in a, lanes 5–8) is presented after densitometry of the results. Data are mean values from four experiments.
These results suggest that abnormal MHC expression as a result of tissue damage is correlated directly with the leak of genomic DNA into the cytoplasm and that leaked nuclear DNA is a good candidate for the inducer of MHC gene expression after tissue damage.
DISCUSSION
We have shown that any ds nucleic acid fragment introduced in the cytoplasm by infection or leakage of self DNA can induce MHC expression directly, and, concomitantly, increase or activate other essential genes or gene products important for antigen presentation. The effect is sequence-independent, requires only small dsDNA fragments, and is different from and additive to that of γIFN. Although the precise mechanism of this effect is unknown, it will allow “nonprofessional” APCs such as thyrocytes to present self or foreign antigens to immune cells. The effect is not restricted to FRTL-5 thyroid cells, which we use as a model because of their nontransformed phenotype, their responsiveness to γIFN, and the evidence that viral infections are involved in autoimmune thyroid disease (14, 19, 32, 33). The effect is the same in the multiple cell lines we have tested.
With respect to autoimmunity, increasing evidence supports the ability of viral infection or environmentally induced damage to increase MHC gene expression in nonimmune cells of target tissues, thereby allowing them to present self antigens to immune cells in the normal repertoire, to break self-tolerance, to induce bystander activation of immune cells, and to induce the cytokine (interleukin 18/interleukin 12/IFN) cascade (8–10, 30, 31). The present report defines the plausible mechanism that makes these events occur and shows that this mechanism does not involve CpG residues.
Thus, in this study, methylation did not alter activity, whereas methylation eliminates CpG activity. This study found no sequence specificity, whereas optimal CpG stimulation depends on sequence (i.e., the ODN must contain at least one unmethylated CpG dinucleotide flanked by two 5′ purines, optimally GpA, and two 3′ pyrimidines, optimally TpC or TpT). CpG responses can be elicited with short ODNs that do not induce the dsDNA response. Most importantly, CpG motifs act directly on cells of the immune system, whereas the ds nucleic acids described herein work on nonimmune cells and convert them to APCs. In contrast, CpG motifs have no known effect on nonprofessional APCs. CpG ODNs induce γIFN by cell (18), which is not the case for ds nucleic acids. Only in the case of dsRNA can the cell be stimulated to produce IFN, and in this case, it is βIFN.
If transfection of any DNA into cells can cause such marked results, it is somewhat surprising that these results have not been observed previously, and it is important that they be considered in the future. One part of this phenomenon has been described in the context of studies showing that plasmid DNA, rather than a live virus, can increase MHC class I expression (34, 35). However, this phenomenon actually involves not only altered MHC class I expression but also increased class II, increased expression of genes important for antigen presentation, and activation of the Janus kinase/Stat, NF-κB, and MAPK systems. The regulation of many genes could be affected, and the data could be significant to the results of all transfection studies, particularly because the effect does not involve CpG. The recognition of the phenomenon and its underlying basis—i.e., the insertion of ds polynucleotides as short as 25 to 35 bp into the cytoplasm—has not been considered in other studies (8–10).
This phenomenon may contribute to the development of autoimmunity when plasmid DNA is introduced during gene therapy and may be important when dsDNA is used in plasmid DNA vaccinations. In DNA vaccination, abnormal MHC gene expression at the site of injection (e.g., smooth muscle cells) might help long-term antigen presentation. Bone marrow-derived cells can induce a cytotoxic T lymphocyte response to influenza nucleoprotein when plasmid is injected intramuscularly (36), and transfer of antigen from muscle cell to professional APCs can occur (37). Other studies have shown that there is an increase in MHC expression in the injected muscle itself (38) and that muscle cells can present antigen. Thus, transplantation of myoblasts expressing antigen can induce high titers of specific antibody (39).
ABBREVIATIONS
- APC
antigen-presenting cell
- CIITA
class II transactivator
- ds
double-stranded
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HSV
herpes simplex virus
- IFN
interferon
- Ii
invariant chain
- IRF-1
IFN regulatory factor 1
- MAPK
mitogen-activated protein kinase
- MHC
major histocompatibility complex
- ODN
oligodeoxynucleotide
- ss
single-stranded
- TAP
transporter of antigen peptides
References
- 1.Bottazzo G F, Pujol-Borrell R, Hanafusa T, Feldmann M. Lancet. 1983;ii:1115–1119. doi: 10.1016/s0140-6736(83)90629-3. [DOI] [PubMed] [Google Scholar]
- 2.Todd I, Londei M, Pujol-Borrell R, Mirakian R, Feldmann M, Bottazzo G F. Ann N Y Acad Sci. 1986;475:241–249. doi: 10.1111/j.1749-6632.1986.tb20873.x. [DOI] [PubMed] [Google Scholar]
- 3.Guardiola J, Maffei A. Crit Rev Immunol. 1993;13:247–268. [PubMed] [Google Scholar]
- 4.Singer D S, Mozes E, Kirshner S, Kohn L D. Crit Rev Immunol. 1997;17:463–468. [PubMed] [Google Scholar]
- 5.Londei M, Lamb J R, Bottazzo G F, Feldmann M. Nature (London) 1984;312:639–641. doi: 10.1038/312639a0. [DOI] [PubMed] [Google Scholar]
- 6.Shimojo N, Kohno Y, Yamaguchi K, Kikuoka S, Hoshioka A, Niimi H, Hirai A, Tamura Y, Saito Y, Kohn L D, et al. Proc Natl Acad Sci USA. 1996;93:11074–11079. doi: 10.1073/pnas.93.20.11074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gianani R, Sarvetnick N. Proc Natl Acad Sci USA. 1996;93:2257–2259. doi: 10.1073/pnas.93.6.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horwitz M S, Bradley L M, Harbertson J, Krahl T, Lee J, Sarvetnick N. Nat Med. 1998;4:781–785. doi: 10.1038/nm0798-781. [DOI] [PubMed] [Google Scholar]
- 9.Wekerle H. Nat Med. 1998;4:770–771. doi: 10.1038/nm0798-770. [DOI] [PubMed] [Google Scholar]
- 10.Benoist C, Mathis D. Nature (London) 1998;394:227–228. doi: 10.1038/28282. [DOI] [PubMed] [Google Scholar]
- 11.Ting J P-Y, Baldwin A S. Curr Opin Immunol. 1993;5:8–16. doi: 10.1016/0952-7915(93)90074-3. [DOI] [PubMed] [Google Scholar]
- 12.Schuppert F, Rambusch E, Kirchner H, Atzpodien J, Kohn L D, von zur Muhlen A. Thyroid. 1997;7:837–842. doi: 10.1089/thy.1997.7.837. [DOI] [PubMed] [Google Scholar]
- 13.Halloran P F, Goes N, Urmson J, Ramassar V, Hobart M, Sims T, Lui S L, Miller L W. Transplant Proc. 1997;29:1041–1044. doi: 10.1016/s0041-1345(96)00361-2. [DOI] [PubMed] [Google Scholar]
- 14.Kohn L D, Giuliani C, Montani V, Napolitano G, Ohmori M, Ohta M, Saji M, Schuppert F, Shong M, Suzuki K, et al. In: Antireceptor Immunity. Rayner D, Champion B, editors. Austin, TX: R. G. Landers Biomedical; 1995. pp. 115–170. [Google Scholar]
- 15.Suzuki K, Lavaroni S, Mori A, Ohta M, Saito J, Pietrarelli M, Singer D S, Kimura S, Katoh R, Kawaoi A, et al. Proc Natl Acad Sci USA. 1998;95:8251–8256. doi: 10.1073/pnas.95.14.8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki K, Lavaroni S, Mori A, Okajima F, Kimura S, Katoh R, Kawaoi A, Kohn L D. Mol Cell Biol. 1998;18:7410–7422. doi: 10.1128/mcb.18.12.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krieg A M, Yi A K, Matson S, Waldschmidt T J, Bishop G A, Teasdale R, Koretzky G A, Klinman D M. Nature (London) 1995;374:546–549. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- 18.Klinman D M, Yi A K, Beaucage S L, Conover J, Krieg A M. Proc Natl Acad Sci USA. 1996;93:2879–2883. doi: 10.1073/pnas.93.7.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balducci-Silano P L, Suzuki K, Ohta M, Saito J, Ohmori M, Montani V, Napolitano G, Shong M, Taniguchi S I, Pietrarelli M, et al. Endocrinology. 1998;139:2300–2313. doi: 10.1210/endo.139.5.5991. [DOI] [PubMed] [Google Scholar]
- 20.Hirai A, Nakamura S, Noguchi Y, Yasuda T, Kitagawa M, Tatsuno I, Oeda T, Tahara K, Terano T, Narumiya S, et al. J Biol Chem. 1997;272:13–16. [PubMed] [Google Scholar]
- 21.Suzuki K, Kobayashi Y, Katoh R, Kohn L D, Kawaoi A. Endocrinology. 1998;139:3014–3017. doi: 10.1210/endo.139.6.6126. [DOI] [PubMed] [Google Scholar]
- 22.York I A, Rock K L. Annu Rev Immunol. 1996;14:369–396. doi: 10.1146/annurev.immunol.14.1.369. [DOI] [PubMed] [Google Scholar]
- 23.Pieters J. Curr Opin Immunol. 1997;9:89–96. doi: 10.1016/s0952-7915(97)80164-1. [DOI] [PubMed] [Google Scholar]
- 24.Mach B, Steimle V, Martubez-Soria E, Reith W. Annu Rev Immunol. 1996;14:301–331. doi: 10.1146/annurev.immunol.14.1.301. [DOI] [PubMed] [Google Scholar]
- 25.Ten R M, Blank V, Le Bail O, Kourilsky P, Israel A. C R Acad Sci Ser III. 1993;316:496–501. [PubMed] [Google Scholar]
- 26.Clemens M J, Elia A. J Interferon Cytokine Res. 1997;17:503–524. doi: 10.1089/jir.1997.17.503. [DOI] [PubMed] [Google Scholar]
- 27.Pellegrini S, Dusanter-Fourt I. Eur J Biochem. 1997;248:615–633. doi: 10.1111/j.1432-1033.1997.00615.x. [DOI] [PubMed] [Google Scholar]
- 28.Kumar A, Yang Y-L, Flati V, Der S, Kadereit S, Haque J, Reis L, Weissmann C, Williams B R G. EMBO J. 1997;16:406–416. doi: 10.1093/emboj/16.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong A H-T, Tam N W N, Yang Y-L, Cuddihy A R, Li S, Kirchoff S, Hauser H, Decker T, Koromilas A E. EMBO J. 1997;16:1291–1304. doi: 10.1093/emboj/16.6.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thorsness P E, Weber E R. Int Rev Cytol. 1996;165:207–234. doi: 10.1016/s0074-7696(08)62223-8. [DOI] [PubMed] [Google Scholar]
- 31.Moffett C W, Paden C M. J Neuroimmunol. 1994;50:139–151. doi: 10.1016/0165-5728(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 32.Saji M, Moriarty J, Ban T, Singer D S, Kohn L D. J Clin Endocrinol Metab. 1992;75:871–878. doi: 10.1210/jcem.75.3.1381373. [DOI] [PubMed] [Google Scholar]
- 33.Tomer Y, Davies T. Endocr Rev. 1993;14:107–121. doi: 10.1210/edrv-14-1-107. [DOI] [PubMed] [Google Scholar]
- 34.Park J-H, Chang S-H, Kim M-C, Shin S-Y, Youn H-J, Kim J-K, Jang Y-S, Kim C-W. FEBS Lett. 1998;436:55–60. doi: 10.1016/s0014-5793(98)01097-7. [DOI] [PubMed] [Google Scholar]
- 35.Fox B A, Drury M, Cao Z W, Huntzicker E G, Qie W X, Urba W J. Cancer Gene Ther. 1998;5:307–312. [PubMed] [Google Scholar]
- 36.Corr M, Lee D J, Carson D A, Tighe H. J Exp Med. 1998;184:1555–1560. doi: 10.1084/jem.184.4.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu T M, Ulmer J B, Caulfield M J, Deck R R, Friedman A, Wang S, Liu X, Donnelly J J, Liu M A. Mol Med. 1997;3:362–371. [PMC free article] [PubMed] [Google Scholar]
- 38.Boudinot P, Bllanco M, de Kinkelin P, Benmansour A. Virology. 1998;249:297–306. doi: 10.1006/viro.1998.9322. [DOI] [PubMed] [Google Scholar]
- 39.Ullmer J B, Deck R R, DeWitt C M, Fu T M, Donnelly J J, Caulfield M J, Liu M A. Vaccine. 1997;15:839–841. doi: 10.1016/s0264-410x(96)00256-3. [DOI] [PubMed] [Google Scholar]