Abstract
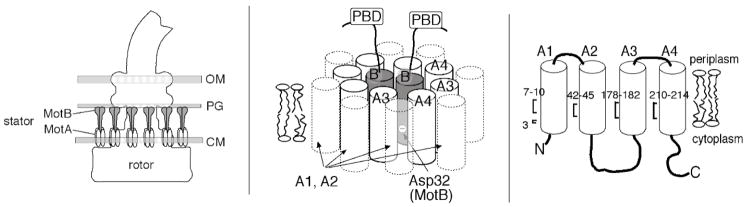
MotA and MotB are membrane proteins that form the stator of the bacterial flagellar motor. Each motor contains several MotA4MotB2 complexes, which function independently to conduct protons across the membrane and couple proton flow to rotation. The mechanism of rotation is not understood in detail but is thought to involve conformational changes in the stator complexes driven by proton association/dissociation at a critical Asp residue of MotB (Asp 32 in the protein of Escherichia coli). MotA has four membrane segments and MotB has one. Previous studies using targeted disulfide crosslinking showed that the membrane segments of the two MotB subunits are together at the center of the complex, surrounded by the TM3 and TM4 segments of the four MotA subunits. Here, the cross-linking studies are extended to TM1 and TM2 of MotA, using Cys residues introduced in several positions in the segments. The observed patterns of disulfide cross-linking indicate that the TM2 segment is positioned between segments TM3 and TM4 of the same subunit, where it could contribute to the proton-channel-forming part of the structure. TM1 is at the interface between TM4 of its own subunit and the TM3 segment of another subunit, where it could stabilize the complex. A structural model based on the cross-linking results shows unobstructed pathways reaching from the periplasm to the Asp 32 residues near the inner ends of the MotB segments. The model indicates a close proximity for certain conserved, functionally important residues. The results are used to develop an explicit model for the proton-induced conformational change in the stator.
The bacterial flagellar motor is an ion-fueled machine capable of turning at hundreds of revolutions per second (1–3). In most species, the motor can turn either CW or CCW, and reversals in direction are the basis of regulated movement such as chemotaxis (4, 5). Although more than two-dozen proteins are needed for assembly and operation of the flagellum, only five are thought to function closely in rotation. FliG, FliM, and FliN form the “switch complex” on the rotor that determines the direction of rotation (6, 7). MotA and MotB are membrane proteins that form the stator, functioning to conduct protons across the membrane and couple proton flow to rotation (8–12) (Figure 1, left). While structural studies of the rotor proteins are fairly advanced (13–16), less is known about the structure of the stator. The stator complexes have subunit composition MotA4MotB2 (17, 18). MotA has four TM segments, relatively large domains in the cytoplasm, and only short segments in the periplasm (9, 19). MotB has a short segment in the cytoplasm, a single TM segment, and a large periplasmic domain that includes features believed to bind peptidoglycan (10, 20, 21). The MotB membrane segment contains an invariant, functionally critical aspartate residue (Asp 32) that has been proposed to function directly in ion conduction (22, 23) (Figure 1, middle).
Figure 1.
Left: Cartoon of the flagellar basal body of Escherichia coli showing the approximate location of the stator complexes. Layers of the cell envelope are indicated: CM, cytoplasmic membrane; PG, peptidoglycan; OM, outer membrane. Each motor contains about 11 stator complexes (35) that function independently to generate torque (36, 37). Middle: Arrangement of membrane segments in the MotA4MotB2 complex, as deduced from previous crosslinking studies on the MotB segment (TM-B) and segments TM3 and TM4 of MotA (26, 27). Segments TM1 and TM2 of MotA are around the outside of the complex but the details of their arrangement are not known. Right: Positions of Cys residues used in the present cross-linking study of the TM segments of MotA.
In a current model for the rotation mechanism, torque is produced as the stator undergoes conformational changes driven by proton association/dissociation at Asp 32 (23). In support of this model, mutations that neutralized the charge of Asp 32 (e.g., replacement by Asn or Ala) were shown to alter the conformation of MotA in an assay based on protease susceptibility (24). This conformational change affected the cytoplasmic domain of MotA, in a segment that was also shown to interact with the rotor (25). These findings suggest that the rotor is not the actuator of movement but is responsible for setting the direction of rotation. Presumably, the switch in direction between CW and CCW rotation must involve changes in the topography and/or chemical properties at the rotor-stator interface.
While high-resolution structural information is not yet available for the stator, features of the membrane-segment arrangement have been deduced from targeted disulfide- cross-linking studies using Cys residues introduced by mutagenesis (26, 27). Patterns of disulfide cross-linking showed that the TM segments of the two MotB subunits are together at the center of the complex, surrounded by the TM3 and TM4 segments of the four MotA subunits (Figure 1, middle). Segments TM1 and TM2 would then be in more outward, lipid-exposed positions in the complex, and consistent with this, TM1 and TM2 were found to be more tolerant of Trp replacements than TM3 or TM4 (28). Details of the TM1 and TM2 organization, such as their locations relative to each other, or to the ion-conducting parts of the complex, are not known. Here, we describe cross-linking studies that were undertaken to examine the organization of the TM1 and TM2 segments. The results indicate that TM2 contributes to the proton-conducting part of the complex, whereas TM1 appears to engage in inter-subunit contacts that stabilize the complex. The new constraints obtained from cross-linking experiments on TM1 and TM2 are used together with the results from previous studies to develop a model for the organization of the 18 TM segments in the MotA4MotB2 complex. The model shows clear pathways, large enough to contain water, reaching from the periplasm to the critical Asp 32 residues in the channel. The model also predicts close spatial relationships among certain residues, including Asp 32, the invariant residue Pro 173 in MotA, and a previously unremarked residue, Tyr 217. These relationships are used to develop a specific, chemically explicit model for the protonation-induced conformational change in the E. coli stator complex.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Mutagenesis
Cys-replacement mutations were made in plasmid pRF5, a derivative of pAlter-1 (Promega) that carries the motA and motB genes. The motA gene in pRF5 was mutated to replace the native Cys 240 residue with Ser. This replacement was shown previously to have only a minor effect on motility (19). The Cys replacements in MotA segments TM3 and TM4 were described previously (26). Replacements in TM1 and TM2 were made using the Altered Sites procedure (Promega). Most double-Cys replacements were made by direct mutagenesis using two oligonucleotides, and the Altered Sites procedure. Double mutants involving position 44 were constructed by transferring an MluI-NsiI fragment encoding the TM3 or TM4 mutations into the vector containing the position-44 Cys replacement. For most cross-linking experiments, proteins were expressed in strain BL21(De3) (29), without induction by IPTG. Under this condition, MotA and MotB were moderately overexpressed (at about 10-fold the wild-type level). All mutations and ligation steps were confirmed by DNA sequencing. Plasmids were isolated using the QIAprep miniprep system (Qiagen).
Measurements of swarming motility
Function of the single- and double-Cys mutant MotA proteins was assayed by tranforming the pRF5 variants into the motAB-deletion strain RP6894, a gift from J.S. Parkinson (University of Utah). In this host, pRF5 expresses MotA and MotB at levels comparable to wild-type, and conferred motility comparable to the wild type (when the proteins contained no Cys replacements). Motility was assayed by measured rates of colony expansion in soft tryptone agar, as described previously (30). Two-μl aliquots of a saturated culture in TB-Amp broth (per liter: 10 g tryptone, 5 g NaCl, 100 mg Amp) were spotted onto soft-agar plates containing tryptone and 0.27% bacto-agar, and incubated at 32° C. Once colonies began to expand, their diameters were measured hourly for at least four hours. Rates of expansion were computed by linear fits to plots of diameter vs time; the reported rates are relative to wild-type controls spotted on a subset of the same plates. In the helical-wheel diagrams (Figure 2), the degree of motility impairment in the mutants is represented by grayscale (wild-type function = white; full immotility = black). To identify helix faces most susceptible to mutation, “defect moments” were computed by multiplying the magnitude of the defect at each position (loss of motility relative to the wild-type) by a unit vector pointing toward that position, and summing over all positions. Standard helical geometry was assumed, with successive positions differing by 100°.
Figure 2.
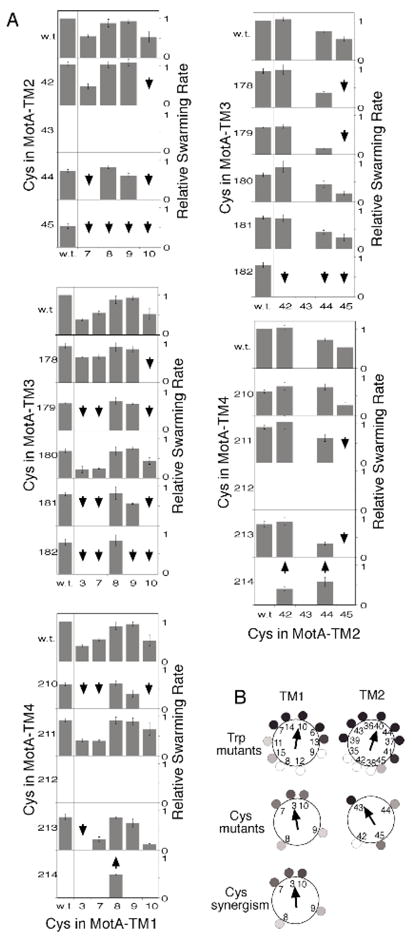
A) Effects of the Cys replacements on MotA function. A motAB-deletion strain (RP6894) was transformed with plasmids encoding the indicated motA variants, and wild-type motB, and motility was assayed by rates of colony expansion on soft-agar plates. Downward-pointing arrows indicate double mutants that functioned more poorly than expected on the basis of the single-Cys phenotypes (i.e., synergistic defects). Upward-pointing arrows indicate three instances of mutational suppression in which the motility defect in the L214C mutant (in TM4) was rescued by a second Cys replacement in TM1 or TM2. B) Effects of TM1 and TM2 mutations on MotA function, plotted on helical wheels. Top: Results of Trp-scanning mutagenesis (28). The magnitude of the motility impairment at each position is indicated in grayscale. Arrows are vector sums of the impairments (pointing toward the side most sensitive to mutation). Middle: Effects of Cys replacements made for the present study. Bottom: Relative frequency of synergistic defects in TM1. Grayscale indicates the fraction of double mutants (involving that position) that gave a synergistic defect. (The analogous plot for TM2 is not shown because sampling in that case was sparse, owing to immotility of the G43C mutant.)
Oxidative cross-linking
Most cross-linking experiments were performed on whole cells of strain BL21(De3) expressing the Cys-mutant Mot proteins. Cells were grown overnight at 32° C with aeration in 3 mL of LB-broth (per liter: 5 g yeast extract, 10 g tryptone, 5 g NaCl) containing 100 μg/mL Amp. Final OD600 was typically about 1.4. Cells were collected by centrifugation and resuspended in 50 mM Tris pH 8.0, 20 mM EDTA, to a volume giving an OD600 of 10. Two 100-μL aliquots were taken from each sample, one for oxidative cross-linking by iodine and one for use as a non-oxidized control.
Crystalline iodine (Sigma) was dissolved in 95% ethanol to make a 500 mM stock solution which was stored at −20° C. Experiments used a 100 mM working solution in 95% ethanol. Most cross-linking experiments were conducted at room temperature. Some single-Cys mutants were cross-linked at 54° C to increase the yield of single-position crosslinking (for use in making band assignments). Samples were treated with 2 mM iodine (2 μl from the 100 mM stock) for 2–3 min., then quenched with 20 mM NEM (Sigma) for 2–3 mins. The NEM was added from a 1 M stock, prepared in 95% ethanol and stored at −20° C. Samples were denatured by addition of SDS to 5%, followed by heating to 95° C for 10 min. Non-oxidized control samples were exposed to just the NEM. Experiments to examine reduction of disulfide bonds used the iodine treatment described above, followed by treatment with 2-mercaptoethanol (10%) for 10 min, followed by quenching with NEM. Samples were either frozen for later use or were prepared for SDS-PAGE by adding 1/9.5 volumes of 10X loading buffer (35% Glycerol, 8.5% Sucrose, 7% SDS, 0.5 M Tris, pH 6.8, and 0.05% Bromophenol Blue).
SDS-PAGE and Immunoblotting
Protein samples were electrophoresed on non-reducing 8.5% SDS-PAGE mini-gels (Bio-Rad MiniProtean II system). Proteins were transferred from gels onto nitrocellulose paper using a semi-dry transfer apparatus (Bio-Rad Transblot SD) for 48 min. Rabbit polyclonal antibody against the larger cytoplasmic domain of MotA (residues 48–174) was prepared as described before (30), and was used at a dilution of 3:2000 in a 4% milk/TBS solution, pH 8, containing 0.2% Tween-20 (Sigma). HRP-conjugated secondary antibody (Pierce Biotechnology) was used at a dilution of 1/5,000. Band intensities were quantified using a video densitometer and the software package NIH-Image.
MotA sequence alignment
A set of 70 phylogenetically diverse MotA sequences was collected from the NCBI web-site (ncbi.nlm.nih.gov). Sequences were aligned using Clustal-X. (MotA sequences are provided in Supporting Information.)
Structural Model
As an aid in visualizing the TM-segment relationships determined from cross-linking, and the relative sizes and dispositions of side-chains, an idealized model of the trans-membrane portion of the stator complex was constructed. A straight alpha-helix was identified in the crystal structure of the membrane protein cytochrome c oxidase of Paracoccus denitrificans (residues 177-199, chain A). Using Swiss-pdb viewer, the segment was mutated to generate a model for each of the MotA and MotB TM segments (each about 20 residues in length). The model helixes were brought together using Swiss-pdb viewer, in the relative orientations deduced from cross-linking. Helices were brought into contact, then energy minimization in Swiss-pdb Viewer was used, in combination with manual adjustment of some side-chain conformations, to give a closely packed structure. Coordinates for the model structure are provided as a pdb file in Supporting Information.
RESULTS
The arrangement of the TM1 and TM2 segments of MotA was examined by targeted disulfide cross-linking (26, 31–33), using Cys residues introduced in five TM1 positions and four TM2 positions, as indicated in Figure 1 (right). The Cys replacements in TM1 and TM2 were paired with various Cys replacements in the other TM segments, to make a set of 106 double-Cys mutant proteins for use in the cross-linking experiments. To examine the effects of the Cys replacements on protein function, plasmids expressing the Cys-replacement proteins were transformed into a motAB-deletion strain and complementation of the motility defect was assayed by measuring rates of colony expansion on soft-agar tryptone plates. The set of Cys mutants made for this study, and results of the motility assays, are summarized in Figure 2, panel A.
All of the single-Cys replacements in TM1 and TM2 were tolerated (i.e., permitted motility) except the replacement of residue 43, a Gly that is conserved in about half of the known MotA sequences. (An alignment of MotA sequences is given in Supporting Information.) As reported previously, single Cys replacements were also tolerated at most positions in TM3 and TM4, exceptions in that case being Gly 212 and Leu 214. Synergistic defects were common: about one-fourth of the double-Cys mutants were unable to swim even though the individual mutations were tolerated. A few cases of suppression were observed, all involving rescue of the position-214 mutant. When plotted on helical wheels, the functional defects in the Cys mutants followed a pattern consistent with previous results from Trp-scanning mutagenesis (28). Positions that showed severe motility impairments or a tendency toward synergistic defects occurred in regions that were also least tolerant of Trp substitutions (Figure 2, panelB). The helix faces most tolerant of mutation were centered on residues 8 and 9 in TM1, and residues 42 and 45 in TM2. On the basis of the Trp and Cys mutational results, we propose that these faces are oriented toward the lipid phase.
The first set of cross-linking experiments examined the relationship between TM1 and TM2. Mutant proteins with Cys replacements in TM1 and TM2, and corresponding single-Cys controls, were expressed in cells and cross-linking was induced using iodine. Products of cross-linking were examined on MotA immunoblots, giving the results summarized in Figure 3 (panels A and B).
Figure 3.
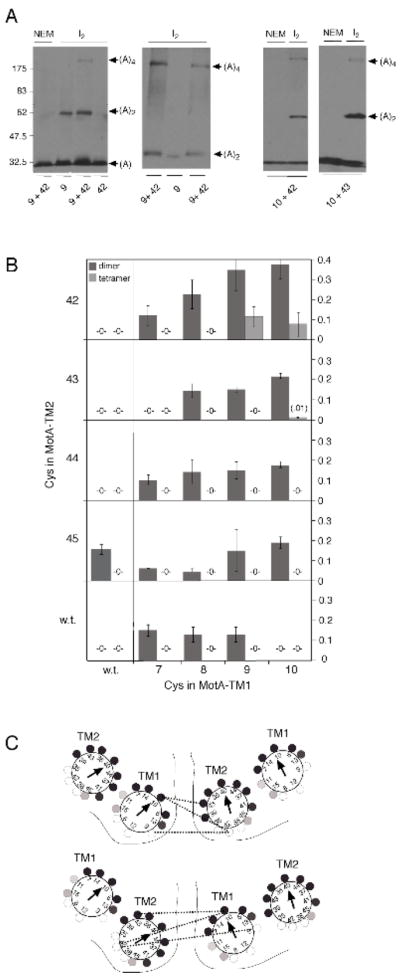
Crosslinking results with TM1 and TM2. A) Representative blots illustrating dimer and tetramer formation in the three double-Cys mutants that gave evidence of TM1-TM2 cross-linking. Electrophoresis was carried out for twice the normal time in the second panel to increase the separation between the 9+42 and 9-only bands. B) Summary of dimer and tetramer yields in the TM1/TM2 double-Cys mutants and the single-Cys controls. C) Two alternative models for the relative arrangement of the TM1 and TM2 segments in MotA. Dotted lines indicate the observed TM1-TM2 cross-links and solid lines indicate boundaries between subunits.
Some of the TM1 and TM2 single-Cys controls gave readily detectable levels of cross-linked dimer upon oxidation with iodine. Such single-position crosslinking was not seen in the previous study of the TM3 and TM4 segments (26). Its occurrence in the TM1 and TM2 mutants might indicate that these segments are relatively mobile, or that ome cross-linking can occur between different MotA4MotB2 complexes in the membrane. Whatever the cause, the cross-linking in single-Cys mutants has the potential to complicate results with the double mutants. As shown below, the various products exhibited distinctive mobilities in most cases, facilitating interpretation.
Most of the TM1/TM2 double mutants showed only a single dimer band, at the same position and with comparable yield as that seen in the constituent single-Cys mutants. Three of the double mutants (9/42, 10/42, and 10/43) gave an additional product at the position expected for a MotA tetramer (Figure 3, panel B). They also gave the highest yields of cross-linked dimer, even though dimer yields were negligible in most of the corresponding single-Cys mutants. Both the high dimer yield and the formation of tetramer could be explained by the occurrence of TM1-TM2 cross-linking. Consistent with this, the mobility of the dimer band formed by the 9/42 double mutant was slightly different from that of the 9-only single-Cys product (Figure 3, panel A, middle). The double-Cys mutation that gave highest tetramer yield (9/42) allowed good function in the motility assays (Figure 2), and thus does not appear to disrupt or destabilize the complex. By identifying positions at the subunit interface, these TM1-TM2 cross-links favor one of two possible models for the relative positions of TM1 and TM2 in the complex (Figure 3, panel C).
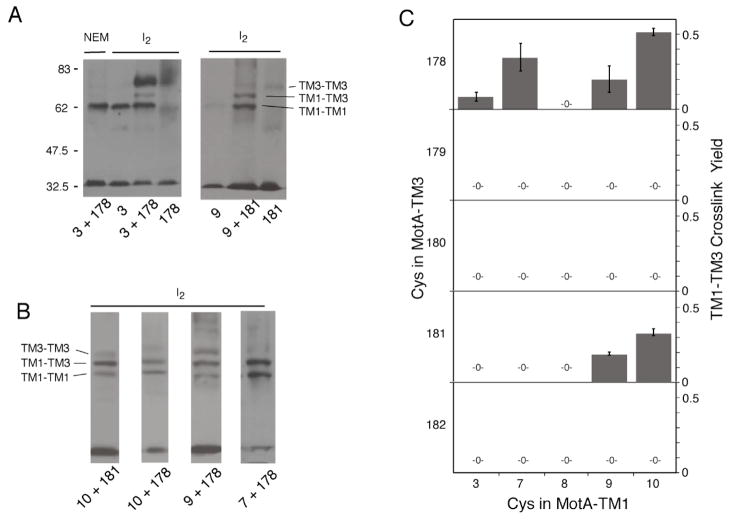
We next examined the relationship between the TM1 and TM2 segments and the more interior segments TM3 and TM4. The Cys replacements in TM1 and TM2 were paired with Cys replacements in TM3 and TM4, and iodine-induced cross-linking was examined as before. Several positions in TM1 showed cross-linking to positions in TM3, as evidenced by the appearance of new bands in the dimer region (Figure 4). Most such double mutants showed three bands in this region, which were assigned by comparison with single-Cys mutants on the same gels. In all cases, the lower band could be assigned to a TM1-TM1 single-Cys product and the upper band to a TM3-TM3 single-Cys product, leaving the intermediate-position band as the TM1-TM3 product. Yields of TM1-TM3 cross-linking are summarized in Figure 4. In TM1, cross-linking to TM3 was greatest at the inward-facing positions 7, 9, and 10. In TM3, the strongest cross-linking to TM1 was through positions 178 and 181, which are near the MotA/MotA subunit interface but are predicted to be somewhat exposed to the outside (26). Most of the cross-linkable TM1-TM3 double mutants retained strong function in the motility assays. The only exceptions involved position 10, which tolerates Cys by itself but not in combination with most other Cys replacements (Figure 2).
Figure 4.
Inter-molecular crosslinking between positions in TM1 and TM3. A) Representative gels showing results with the 3/178 and 9/181 mutants, illustrating the method of band assignment by comparison with single-Cys mutants. Double mutants were oxidized using iodine at room temperature (approximately 20°); single mutants were oxidized at a higher temperature (54°) to increase yield. B) Additional examples of TM1-TM3 crosslinking. The corresponding single-Cys mutants (not shown) gave results similar to those in panel A, with TM1-TM1 products at the lowest position and TM3-TM3 products at the highest position. C) Summary of yields of TM1-TM3 cross-linking (n.d., none detected).
Besides crosslinking to TM3 of another subunit, TM1 also exhibited intra-molecular crosslinking to TM4, evidenced by the appearance of a fast-migrating band below the monomer (Figure 5). The band was eliminated by reduction with 2-mercaptoethanol and is thus the product of disulfide cross-linking. Intra-molecular crosslinking to TM4 occurred through positions 8 and 10 in TM1, thus identifying a surface somewhat distinct from that involved in inter-molecular crosslinking to TM3. On the TM4 side, crosslinking to TM1 occurred through positions 210, 211, and 212.
Figure 5.
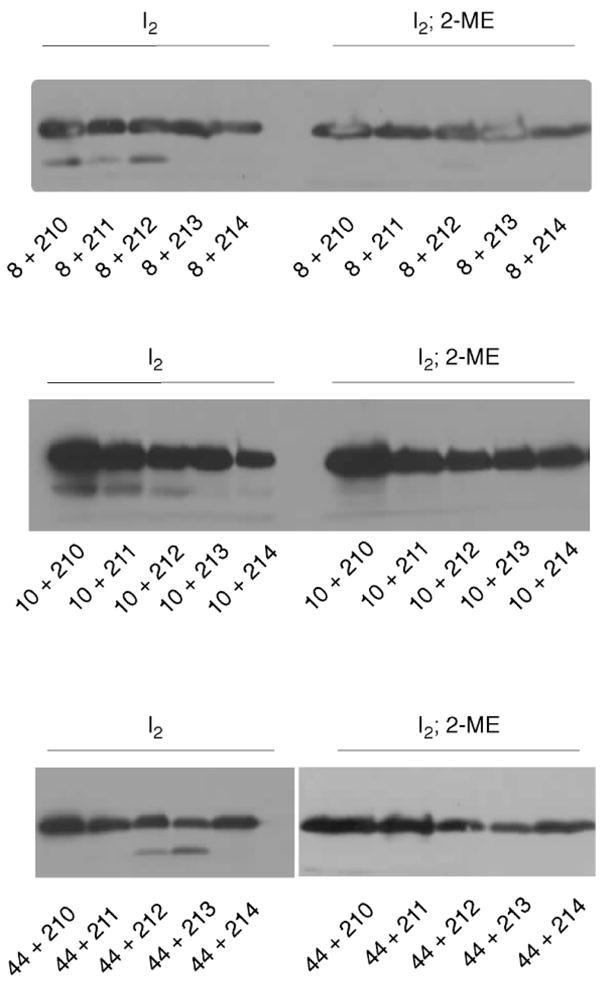
Intra-molecular crosslinking between TM1 and TM4 (top two blots), and between TM2 and TM4 (bottom blot). The monomer region of the gels is shown; the dimer region showed only products of single-position (TM1-TM1 and TM4-TM4) cross-linking. Intra-molecular crosslinking gives rise to the fast-migrating band below the MotA monomer, which disappears on treatment with 10% 2-mercaptoethanol (right). All observed instances of intra-molecular cross-linking are shown.
The double mutants involving TM2 did not show any dimer bands other than those assignable to single-position (TM2-TM2) crosslinking, which suggests that TM2 is not adjacent to the TM3 or TM4 segments of an adjacent subunit. Instead, TM2 showed intra-molecular crosslinking to TM4, through position 44. On the TM4 side, intra-molecular crosslinking to TM2 was through position 213 and (more weakly) position 212.
The frequent occurrence of synergistic defects in the swarming assays (Figure 2) suggests that two Cys replacements can act together to alter the structure or stability of the complex. The cross-linking experiments give further evidence of destabilization in some double-Cys mutants. The proteins with single Cys replacements in TM4 did not show any cross-linking into dimers unless the experiment was done at relatively high temperature (54°), but TM4-TM4 crosslinking occurred at a readily detectable level when a second Cys replacement was present in TM1 (Figure S1 in Supporting Information). (Products of single-position crosslinking were identified by comparison with the products of high-temperature crosslinking in single-Cys mutants.) Likewise, TM1-TM1 crosslinking was increased by the presence of a Cys replacement in TM4; i.e., the effect is reciprocal. These results indicate that Cys replacements can act together to make the complex more dynamic, and that the observed single-position crosslinking is most likely the result of this dynamic character.
DISCUSSION
The present data provide constraints on the organization of the TM1 and TM2 segments of MotA that can be combined with previous cross-linking results to generate a model for the arrangement of all the membrane segments in the stator complex (Figure 6, panel A) The model is based on the subunit composition MotA4MotB2 (18), and has the TM1 and TM2 segments in outward (i.e., lipid-facing) locations. Within each MotA subunit, TM2 is positioned between the TM3 and TM4 segments where it forms the outer wall of the channel in the MotA subunits that form the proton pathways. The TM1 segments do not contribute to the proton channels directly but are at the interfaces between adjacent MotA subunits, where they could function to stabilize the complex.
Figure 6.
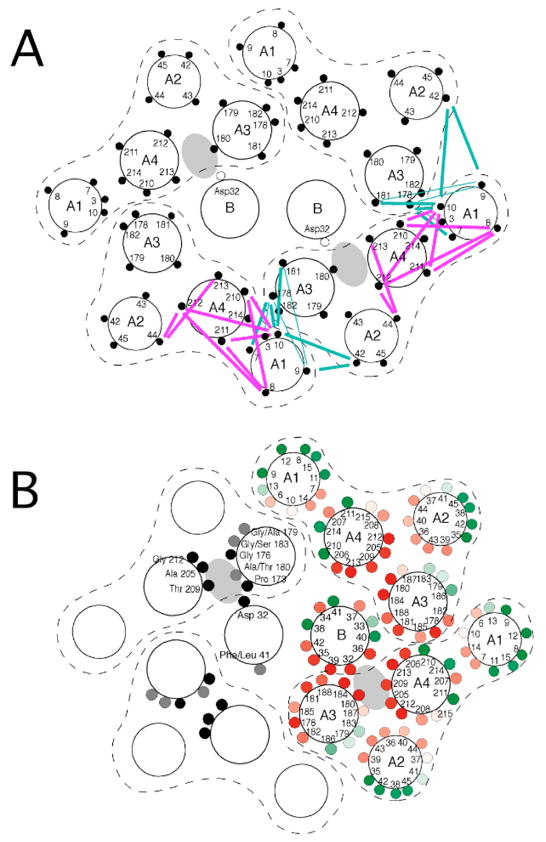
A) Model for the arrangement of membrane segments in the MotA4MotB2 complex. The arrangement of interior segments (TM3, TM4, and TM-B) is based on previous crosslinking studies (26, 27). The arrangement of the TM1 and TM2 segments is based on cross-links detected in the present study, which are indicated by the colored lines (cyan for inter-molecular, and magenta for intra-molecular). B) Effects of Trp substitutions, and patterns of residue conservation, in the membrane segments of the stator complex. In the subunits on the right, positions are colored red if function was eliminated by a Trp replacement, with the intensity of color indicating the degree of negative dominance (the magnitude of the motility impairment caused by expressing the mutant protein in a wild-type cell). Positions are colored green if function was retained in the Trp mutant, with intensity indicating the relative swarming rate. (data from (22, 28)). Patterns of sequence conservation are shown in the subunits on the left, with invariant positions colored black and positions having just two fairly closely-related residues colored gray. (Position 180 is very rarely Ser but is included because Ser has properties intermediate to those of Ala and Thr). (See Supporting Information for an alignment of MotA sequences.)
As discussed previously (26), the core membrane segments (TM-B, TM3, and TM4) contain several well conserved residues, all of which occur at positions in or near the proton channels (Figure 6). In the peripheral segments TM1 and TM2, only the residues Gly 6 and Gly 39 are well conserved. These are not in the channel but are at positions between membrane segments where they could be important for allowing close, stable contacts, particularly between TM1 of one subunit and TM3 of the adjacent subunit. Also as noted previously, with the exception of Thr209, residues predicted to line the channel are non-polar, and in most cases small. The model accommodates the Trp-scanning mutational results on TM1 and TM2 by orienting the positions that tolerate Trp toward the outer, lipid-facing surface (Figure 6 panel B).
To help in visualizing features of the proposed TM arrangement, we constructed a computer model of the complex based on standard α-helical conformation. The model shows unobstructed pathways extending from the periplasmic side of the complex to the Asp 32 residues near the inner end, owing to the presence of the aforementioned small residues (Supplemental Figure). Below Asp 32, the residues are larger (including Ile 54, Leu165, Leu 223, MotB Ile 28) and a clear path is not evident. The exit of protons from Asp 32 to the cell interior might therefore require a conformational change to open a pathway through this region.
Pro 173 of MotA is strictly conserved and a mutational study showed that it is important for motor function (30). Pro residues have been found to be essential in the function of several membrane proteins, including ion transporters, and are believed to act by facilitating bending in α-helices (34). If TM3 of MotA is helical as the cross-linking and mutational data suggest, then the presence of Pro 173 will prevent the hydrogen bonding that would otherwise occur to the backbone carbonyl of residue 169. In the model developed here, this carbonyl group of MotA is near (~ 5 Å from) the side-chain of Asp 32. This suggests a simple mechanism for linking protonation to a conformational change: Upon protonation, Asp 32 might hydrogen bond to carbonyl 169, inducing a bend in this part of TM3 (Supplemental Figure).
Two of the MotA subunits in the complex appear not to make any direct contribution to the proton channels, which raises the question of why the same protein is utilized in two such different contexts. The model of the complex reveals a previously unrecognized similarity between the channel- and non-channel subunits that may provide a clue. This involves residue Tyr 217 in TM4, which is conserved in diverse MotA sequences as either Tyr or Asn (very rarely Gln). The Tyr 217 residue arising from a channel subunit points into the adjacent, non-channel subunit, with its side-chain positioned near Pro 173 of the non-channel subunit. Thus, Tyr 217 might exert an effect analogous to that proposed for Asp 32, except in the non-channel subunits, interacting with Pro173 or nearby groups to regulate the conformation of the non-channel subunit. Such a linkage could serve to coordinate events in the channel- and non-channel subunits: The conformational change induced in the channel subunits upon protonation of Asp 32 might not only bend TM3 but could also shift TM4, and thus alter the relationship between Tyr 217 and relevant groups in the non-channel subunits. This would allow the channel- and non-channel subunits to undergo similar conformational changes, either synchronously or out-of-phase (Supplemental Figure).
In conclusion, the present study has led to a particular model for the arrangement of TM segments in the stator complex of the flagellar motor, and specific ideas regarding the conformational change that drives rotation. The proposed model for the conformational change leads naturally to a model for motor rotation and switching, described in Supporting Information.
Acknowledgments
We thank J.S. Parkinson for strains, and S. Berry, M. Sarkar, and K. Paul for helpful disussions. W. C. C. was partially supported by a minigrant from the Biosciences Undergraduate Research Program at the University of Utah.
Supported by grant 1-R01-GM64664 from the U.S. National Institutes of Health. The Protein-DNA core facility at the University of Utah receives support from the National Cancer Institute (5P30 CA42014).
Abbreviations
- CW
clockwise
- CCW
counter-clockwise
- TM
trans-membrane
- Amp
ampicillin
- TRIS
tris(hydroxymethyl)aminomethane
- EDTA
ethylenediaminetetraacetic acid
- NEM
N-ethylmaleimide
- SDS
sodium dodecylsulfate
- PAGE
polyacrylamide gel electrophoresis
- 2-ME
2-mercaptoethanol
Footnotes
Supplemental Information. Additional cross-linking data, MotA sequence alignments, a pdb file for the structural model, and illustrations of hypotheses for the stator conformational change, and for the role of the conformational change in torque generation, are provided in Supplemental Information, at http:www.biochemistry,notme.
References
- 1.Larsen SH, Adler J, Gargus JJ, Hogg RW. Chemomechanical coupling without ATP: the source of energy for motility and chemotaxis in bacteria. Proc Natl Acad Sci USA. 1974;71:1239–1243. doi: 10.1073/pnas.71.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lowe G, Meister M, Berg HC. Rapid rotation of flagellar bundles in swimming bacteria. Nature. 1987;325:637–640. [Google Scholar]
- 3.Magariyama Y, Sugiyama S, Muramoto K, Maekawa Y, Kawagishi I, Imae Y, Kudo S. Very fast flagellar rotation. Nature. 1994;371:752. doi: 10.1038/371752b0. [DOI] [PubMed] [Google Scholar]
- 4.Silverman M, Simon M. Flagellar rotation and the mechanism of bacterial motility. Nature. 1974;249:73–74. doi: 10.1038/249073a0. [DOI] [PubMed] [Google Scholar]
- 5.Larsen SH, Reader RW, Kort EN, Tso WW, Adler J. Change in direction of flagellar rotation is the basis of the chemotactic response in E. coli. Nature. 1974;249:74–77. doi: 10.1038/249074a0. [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi S, Aizawa SI, Kihara M, Isomura M, Jones CJ, Macnab RM. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J Bacteriol. 1986;168:1172–1179. doi: 10.1128/jb.168.3.1172-1179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Macnab RM. In: Two-component signal transduction. Hoch JA, Silvahy TJ, editors. ASM Press; Washington DC: 1995. pp. 181–199. [Google Scholar]
- 8.Silverman M, Simon M. Operon controlling motility and chemotaxis in E. coli. Nature. 1976;264:577–580. doi: 10.1038/264577a0. [DOI] [PubMed] [Google Scholar]
- 9.Dean GE, Macnab RM, Stader J, Matsumura P, Burke C. Gene sequence and predicted amino-acid sequence of the MotA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J Bacteriol. 1984;143:991–999. doi: 10.1128/jb.159.3.991-999.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stader J, Matsumura P, Vacante D, Dean GE, Macnab RM. Nucleotide sequence of the Escherichia coli motB gene and site-limited incorporation of its product into the cytoplasmic membrane. J Bacteriol. 1986;166:244–252. doi: 10.1128/jb.166.1.244-252.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blair DF, Berg HC. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell. 1990;60:439–449. doi: 10.1016/0092-8674(90)90595-6. [DOI] [PubMed] [Google Scholar]
- 12.Stolz B, Berg HC. Evidence for interactions between MotA and MotB, torque-generating elements of the flagellar motor of Escherichia coli. J Bacteriol. 1991;173:7033–7037. doi: 10.1128/jb.173.21.7033-7037.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lloyd SA, Whitby FG, Blair DF, Hill CP. Structure of the C-terminal domain of FliG, a component of the rotor in the bacterial flagellar motor. Nature. 1999;400:472–475. doi: 10.1038/22794. [DOI] [PubMed] [Google Scholar]
- 14.Brown PN, Hill CP, Blair DF. Crystal structure of the middle and C-terminal domains of the flagellar rotor protein FliG. EMBO J. 2002;21:3225–3234. doi: 10.1093/emboj/cdf332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown PN, Mathews MAA, Joss LA, Hill CP, Blair DF. Crystal structure of the flagellar rotor protein FliN from Thermotoga maritima. J Bacteriol. 2005;187:2890–2902. doi: 10.1128/JB.187.8.2890-2902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park SY, Lowder B, Bilwes AM, Blair DF, Crane BR. Structure of FliM provides insight into assembly of the switch complex in the bacterial flagella motor. Proc Natl Acad Sci U S A. 2006;103:11886–11891. doi: 10.1073/pnas.0602811103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato K, Homma M. Functional reconstitution of the Na(+)-driven polar flagellar motor component of Vibrio alginolyticus. J Biol Chem. 2000;275:5718–5722. doi: 10.1074/jbc.275.8.5718. [DOI] [PubMed] [Google Scholar]
- 18.Kojima S, Blair DF. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry. 2004;43:26–34. doi: 10.1021/bi035405l. [DOI] [PubMed] [Google Scholar]
- 19.Zhou J, Fazzio RT, Blair DF. Membrane topology of the MotA protein of Escherichia coli. J Mol Biol. 1995;251:237–242. doi: 10.1006/jmbi.1995.0431. [DOI] [PubMed] [Google Scholar]
- 20.Chun SY, Parkinson JS. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science. 1988;239:276–278. doi: 10.1126/science.2447650. [DOI] [PubMed] [Google Scholar]
- 21.DeMot R, Vanderleyden J. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both Gram-positive and Gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol Microbiol. 1994;12:333–334. doi: 10.1111/j.1365-2958.1994.tb01021.x. [DOI] [PubMed] [Google Scholar]
- 22.Sharp LL, Zhou J, Blair DF. Tryptophan-scanning mutagenesis of MotB, an integral membrane protein essential for flagellar rotation in Escherichia coli. Biochemistry. 1995;34:9166–9171. doi: 10.1021/bi00028a028. [DOI] [PubMed] [Google Scholar]
- 23.Zhou J, Sharp LL, Tang HL, Lloyd SA, Billings S, Braun TF, Blair DF. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J Bacteriol. 1998;180:2729–2735. doi: 10.1128/jb.180.10.2729-2735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kojima S, Blair DF. Conformational change in the stator of the bacterial flagellar motor. Biochemistry. 2001;40:13041–13050. doi: 10.1021/bi011263o. [DOI] [PubMed] [Google Scholar]
- 25.Zhou J, Lloyd SA, Blair DF. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc Natl Acad Sci USA. 1998;95:6436–6441. doi: 10.1073/pnas.95.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braun TF, al-Mawsawi LQ, Kojima S, Blair DF. Arrangement of core membrane segments in the MotA/MotB proton-channel complex of Escherichia coli. Biochemistry. 2004;43:35–45. doi: 10.1021/bi035406d. [DOI] [PubMed] [Google Scholar]
- 27.Braun TF, Blair DF. Targeted disulfide cross-linking of the MotB protein of Escherichia coli: Evidence for two H+ channels in the stator complex. Biochemistry. 2001;40:13051–13059. doi: 10.1021/bi011264g. [DOI] [PubMed] [Google Scholar]
- 28.Sharp LL, Zhou J, Blair DF. Features of MotA proton channel structure revealed by tryptophan-scanning mutagenesis. Proc Natl Acad Sci USA. 1995;92:7946–7950. doi: 10.1073/pnas.92.17.7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 30.Braun TF, Poulson S, Gully JB, Empey JC, Van Way S, Putnam A, Blair DF. Function of proline residues of MotA in torque generation by the flagellar motor of Escherichia coli. J Bacteriol. 1999;181:3542–3551. doi: 10.1128/jb.181.11.3542-3551.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Falke JJ, Dernburg AF, Sternberg DA, Zalkin N, Milligan DL, Koshland DEJ. Structure of a bacterial sensory receptor: a site-directed sulfhydryl study. J Biol Chem. 1988;263:14850–58. [PubMed] [Google Scholar]
- 32.Pakula A, Simon MI. Determination of transmembrane protein structure by disulfide cross-linking: the Escherichia coli Tar receptor. Proc Natl Acad Sci USA. 1992;89:4144–48. doi: 10.1073/pnas.89.9.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee GF, Burrows GG, Lebert MR, Dutton DP. Deducing the organization of a transmembrane domain by disulfide cross-linking. J Biol Chem. 1994;269:29920–29927. [PubMed] [Google Scholar]
- 34.Brandl CJ, Deber CM. Hypothesis about the function of membrane-buried proline residues in transport proteins. Proc Natl Acad Sci U S A. 1986;83:917–921. doi: 10.1073/pnas.83.4.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reid SW, Leake MC, Chandler JH, Lo CJ, Armitage JP, Berry RM. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc Natl Acad Sci U S A. 2006;103:8066–8071. doi: 10.1073/pnas.0509932103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Block SM, Berg HC. Successive incorporation of force-generating units in the bacterial rotary motor. Nature. 1984;309:470–472. doi: 10.1038/309470a0. [DOI] [PubMed] [Google Scholar]
- 37.Blair DF, Berg HC. Restoration of torque in defective flagellar motors. Science. 1988;242:1678–1681. doi: 10.1126/science.2849208. [DOI] [PubMed] [Google Scholar]