SUMMARY
The σ factors are the key regulators of bacterial transcription initiation. Through direct read-out of promoter DNA sequence, they recruit the core RNA polymerase to sites of initiation, thereby dictating the RNA polymerase promoter-specificity. The group 1 σ factors, which direct the vast majority of transcription initiation during log phase growth and are essential for viability, are auto-regulated by an N-terminal sequence known as σ1.1. Here we report the solution structure of Thermatoga maritima σA σ1.1. We additionally demonstrate by using chemical crosslinking strategies that σ1.1 is in close proximity to the promoter recognition domains of σA. We therefore propose that σ1.1 auto-inhibits promoter DNA binding of free σA by stabilizing a compact organization of the σ factor domains that is unable to bind DNA.
INTRODUCTION
The catalytically competent core bacterial RNA Polymerase (RNAP, subunit composition α2ββ’ω) is unable to initiate promoter specific transcription (Borukhov and Nudler, 2003; Browning and Busby, 2004; Darst, 2001; Ebright, 2000; Gross et al., 1998; Murakami and Darst, 2003). Transcription initiation requires an additional subunit, the σ factor, to form the RNAP holoenzyme (Burgess et al., 1969; Travers and Burgess, 1969). The σ factor confers upon the RNAP holoenzyme the ability to recognize and bind promoter DNA, and also plays a key role in dsDNA melting to form the transcription bubble, allowing transcription initiation.
Bacterial species have anywhere from a single σ factor to more than 60 which, together with various transcription factors, regulate different transcriptional responses and programs (Borukhov and Nudler, 2003; Browning and Busby, 2004; Gruber and Gross, 2003). σ factors fall into two broad classes, the σ70 family and the σ54 family (Gruber and Gross, 2003). Only the σ70 family members are present in all bacterial species. The σ70 family is further subdivided into 4 groups. The group 1, or primary, σ factors are responsible for the bulk of transcription during log phase growth and are essential for viability, while group 2–4 σ factors, or alternate σ factors, fulfill more specialized functions such as stress responses (Gruber and Gross, 2003).
σ factors need to be regulated in order to prevent DNA binding at inappropriate times. Specialized σ factors are often regulated through binding to anti-σ factors that prevent them from interacting with core RNAP and contacting DNA (reviewed in (Campbell and Darst, 2005; Campbell et al., 2008)). The group 1 σ factors do not bind promoter DNA prior to the formation of the RNAP holoenzyme, due in large part to an N-terminal extension that is unique to group 1 σ factors called σ1.1 (Camarero et al., 2002; Dombroski et al., 1993; Dombroski et al., 1992).
The group 1 σ factors contain 4 conserved domains, known as σ1.1, σ2, σ3 and σ4, connected by flexible linkers. A wealth of structural and biochemical information is available on σ2, σ3 and σ4 (Campbell et al., 2002a; Campbell et al., 2002b; Campbell et al., 2003; Lane and Darst, 2006; Malhotra et al., 1996; Murakami et al., 2002a; Murakami et al., 2002b; Sorenson et al., 2004; Vassylyev et al., 2002). σ2 recognizes the promoter −10 element and plays an essential role in the melting of the double stranded DNA to form the transcription bubble (Gross et al., 1998). σ3 recognizes the extended −10 element (Barne et al., 1997). σ4 recognizes the promoter −35 element (Gardella et al., 1989; Siegele et al., 1989). These three domains also form extensive contacts with the core RNAP (Murakami et al., 2002a; Murakami et al., 2002b; Vassylyev et al., 2002). There is no structural information available on σ1.1. However, biochemical information shows that it plays a role in the transition from a closed to an open promoter complex (Vuthoori et al., 2001; Wilson and Dombroski, 1997) and is responsible for preventing DNA binding prior to formation of the holoenzyme (Camarero et al., 2002; Dombroski et al., 1993; Dombroski et al., 1992).
The mechanism by which σ1.1 prevents DNA binding by the group 1 σ factors is unknown. σ constructs lacking σ1.1 are able to bind promoter DNA while full length group 1 σ factors exhibit only very weak DNA binding (Camarero et al., 2002; Dombroski et al., 1993; Dombroski et al., 1992). Additionally, σ1.1 can interfere with DNA binding by σ4 in trans (Dombroski et al., 1993). This led to the proposal that σ1.1 binds to σ4 to prevent −35 element binding. However, an NMR based study was unable to detect a direct interaction between σ1.1 and the C-terminal portion of σ4, σ4.2, which harbors the −35 element DNA binding determinants (Camarero et al., 2002). In addition, no interaction between σ1.1 and σ4 could be detected using a bacterial two-hybrid screen (Hinton et al., 2006). Thus, it is also possible that σ1.1 prevents DNA binding through an allosteric mechanism, or that σ1.1 does not interact with other domains of the σ factor and DNA binding is prevented simply by electrostatic repulsion between the highly negatively charged σ1.1 and negatively charged DNA.
Here we report the solution structure of σ1.1 of T. maritima σA. We also demonstrate, through intramolecular crosslinking, that σ1.1 is in close proximity to the DNA binding domains σ2 and σ4 in the autoinhibited state. We propose that σ1.1 interdomain contacts stabilize a compacted σ factor structure that is sterically incompatible with DNA binding.
RESULTS
σ1.1 adopts a novel helical fold
All of our studies were carried out using derivatives of σA from Thermotoga maritima (Tm). This σ factor has a high degree of sequence similarity to σ70 from E. coli, including throughout σ1.1 (Figure 1A), but is thermostable and lacks a 240 residue non-conserved insertion between σ1.2 and σ2 present in Ec σ70 so is considerably smaller (47 kDa vs. 70 kDa), making it better suited for biochemical and structural studies (Camarero et al., 2002). Based on sequence alignments, σ1.1 of Tm σA extends approximately from residue 25 to residue 120 (Figure 1A). Preliminary NMR experiments revealed that the non-conserved N-terminus of σA (residues ~1–30) and the C-terminus of σ1.1 (~95–120) were unstructured (data not shown). Thus, the construct we ultimately used for structural determination contained residues 29–95. Uniformly 15N and 13C labeled σA(29–95) was used to obtain the main- and side-chain assignments of the 1H, 15N, and 13C resonances using the standard set of double- and triple-resonance 2D and 3D experiments (Cavanagh et al., 1996). An initial assessment of the σ1.1 secondary structure was made using the deviations of assigned chemical shifts from random coil values (Wishart and Sykes, 1994). This analysis suggested that the domain was composed entirely of α helical secondary structure elements.
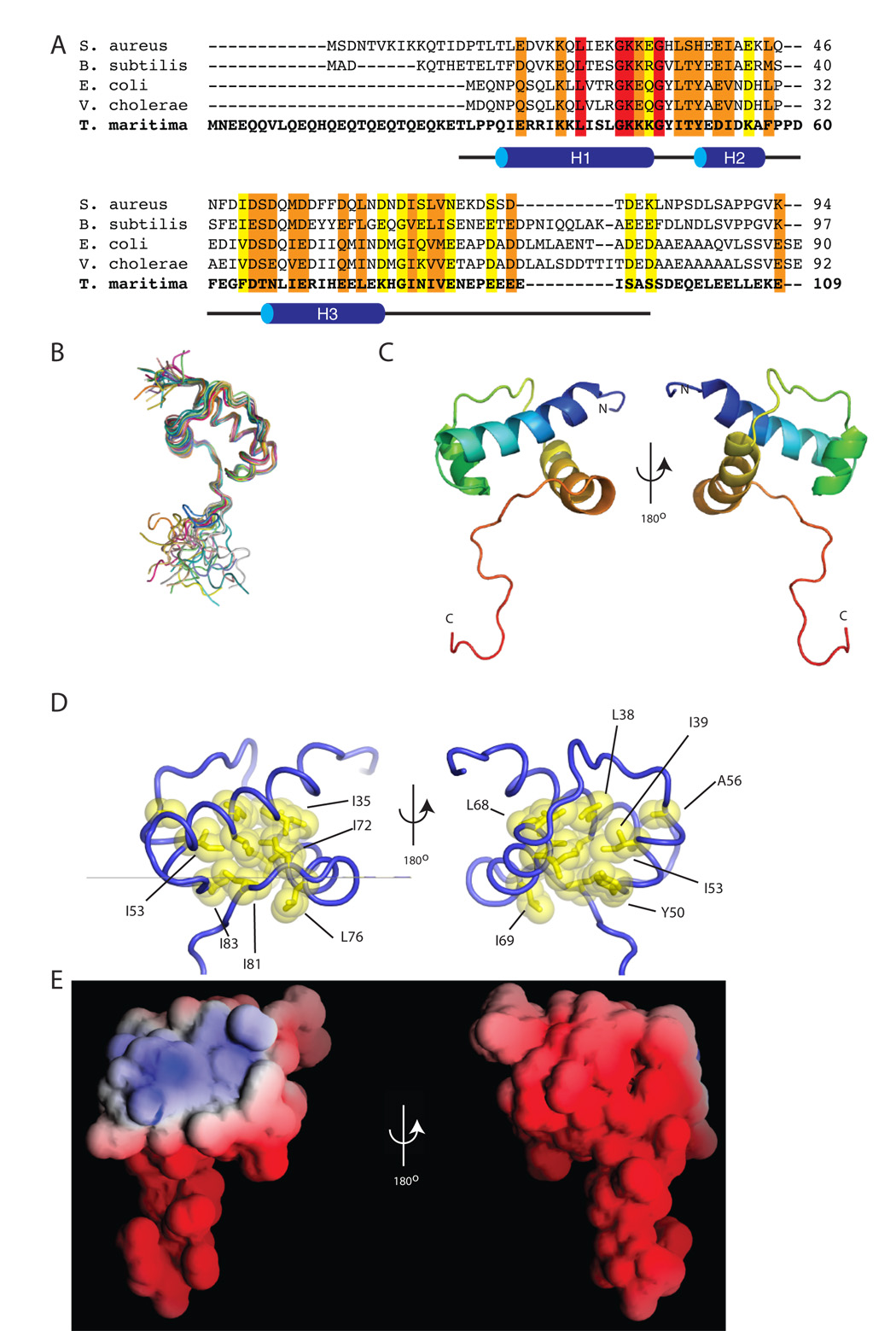
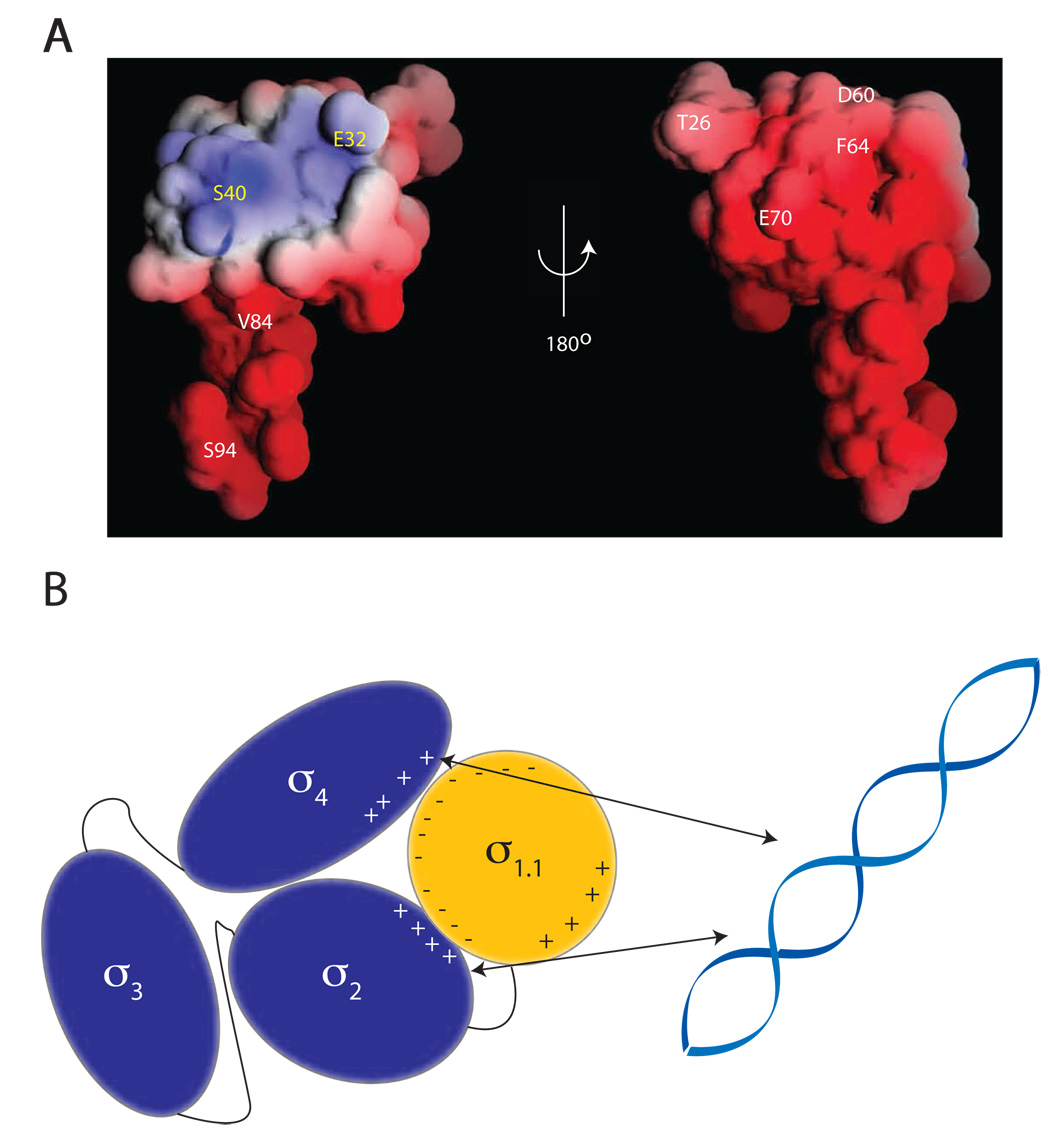
Figure 1. The structure of σ1.1.
(A) An alignment of σ1.1 from five different bacterial species generated using the ClustalW program. Highlighted in red are identical residues, in orange are conserved residues and in yellow are semi-conserved residues. The black line below the alignment represents the construct used for structural determination, with secondary structural elements (H1–H3) indicated. (B) A superimposition of the backbone of the 20 lowest energy structures of σ1.1 obtained from calculations using NMR-derived restraints. Each structure is individually colored. The N-termini are in the upper left and the C-termini are in the bottom half of the figure. (C) Cartoon representation of the secondary structure elements of σ1.1. The structure is rainbow spectrum colored from N-terminus (blue) to C-terminus (red). (D) Cartoon representation of σ1.1. Residues that form the hydrophobic core are shown in yellow. Images generated using MacPYMOL (Delano Scientific). (E) The electrostatic surface of σ1.1. Red is negative, blue is positive, white is neutral. Image generated using GRASP (Nicholls et al., 1991).
The solution structure of σ1.1 (pdb accession 2K6X) was solved using restraints generated from a series of multi-dimensional NMR experiments. Structure calculations were performed using a total of 877 unambiguous distance restraints (245 intraresidue, 314 sequential, 185 short range, 40 medium range and 93 long range). In addition, 107 dihedral (φ and ψ) angle restraints and 44 hydrogen bond restraints were used in the structure calculations. Structural statistics and analysis are given in Table 1 and the 20 lowest energy structures are superimposed in Figure 1B. For the structured region, (residues 29–86) the average pairwise root-mean-square deviation (rmsd) is 0.56 ± 0.11 Å for backbone residues and 1.25 ± 0.14 Å for all heavy atoms. Most of the residues are in the most favored (85.5%) or allowed (12.7%) conformations. Proline 58, which is largely but not universally conserved between σ factors, is in the cis conformation based on Hα-Hδ/Hd’ NOE data.
Table 1.
NMR restraints and structural statistics for the best 20 structures.
| Restraints and statistics | |
|---|---|
| Total number of restraints | 1028 |
| NOE restraints | 877 |
| Intraresidue | 245 |
| Sequential | 314 |
| Short-range | 185 |
| Medium-range | 40 |
| Long-range | 93 |
| Dihedral angle restraints | 107 |
| Hydrogen bond restraintsa | 44 |
| Structure Statisticsb | |
| NOE violations > 0.5 Å | 0 |
| Dihedral violations > 5° | 0 |
| Total Energy (kcal/mol) | −2498.83 ± 48.09 |
| NOE constraints energy (kcal/mol) | 76.07 ± 10.59 |
| RMSD from average structure c, d | |
| Backbone (N, Cα, C) (Å) | 0.56 ± 0.11 (2.07 ± 0.54) |
| Heavy atoms (Å) | 1.25 ± 0.14 (2.45 ± 0.46) |
| Ramachandran Statisticse | |
| Most favored region (%) | 96.4 (85.5) |
| Additionally allowed (%) | 3.6 (12.7) |
| Generously allowed (%) | 0.0 (1.0) |
| Disallowed (%) | 0.0 (0.8) |
Hydrogen bond restraints were HN-O distance of 1.8–2.3 Å and an N-O distance of 2.8–3.3 Å.
Structural characteristics for the final ensemble of 20 water-refined structures.
RMSD of the mean structure from individual structures in the ensemble.
RMSD for residues 29–86 shown. The numbers in the parenthesis indicate the RMSD for residues 29–95.
Ramachandran plot data shown for residues 29–86. The numbers in the parenthesis indicates the statistics for residues 29–95.
σ1.1 possesses a novel fold comprising three α-helices connected by two loops (Figure 1C). Helix 1 (H1) is the longest helix and contains residues Q30-K45. H1 is connected by a four-residue loop to the smallest helix (H2) comprising residues Y50-A56. H2 is connected via an eight-residue loop to the final helix (H3), which stretches from residues T66-K78. H2 and H3 are roughly anti-parallel to one another and pack perpendicularly against H1. σ1.1 contains a compact hydrophobic core formed by highly conserved residues from all three helices and part of the C-terminal tail (Figure 1D). Indeed, the side-chains of I81 and I83, from the tail, project directly into the hydrophobic core where they pack against residues from all three helices. These interactions help to hold residues 79–83 of the tail in an extended conformation that runs roughly parallel with H1. The apparent structural role of I81 is particularly interesting since the corresponding residue in E. coli σ70 (I53) has been shown to be required for its function (Bowers and Dombroski, 1999). As expected from the amino acid composition, σ1.1 has a mostly negative electrostatic surface potential (Figure 1E). An intriguing exception to this is a positively charged patch centered on H1. The functional relevance of this will be discussed in later sections.
σ1.1 makes interdomain contacts
We next turned to the question of whether region 1.1 makes contacts with other domains in the σ factor. Our initial approach to this problem was to use NMR spectroscopy employing segmental isotopic labeled σA constructs prepared using expressed protein ligation (Muralidharan and Muir, 2006; Ottesen et al., 2003). Although the NMR spectra obtained using these constructs were consistent with there being inter-domain interactions involving σ-1.1, the data were insufficient to determine the details of these interactions (Supplemental Figures 1 and 2). Despite considerable efforts to overcome these technical issues, we concluded that an alternative methodology was needed to unambiguously characterize the putative inter-domain interactions in the sigma factor.
We decided that a site-directed crosslinking strategy might be a more productive approach to this problem. Our approach exploits the fact the Tm σA contains no native cysteines, a feature which should allow selective attachment of a chemical crosslinker to a mutant cysteine introduced into σ1.1. With this in mind, we synthesized a novel crosslinker containing a fluorophenylazide photo-reactive warhead, a 3-nitro-2-pyridinesulfenyl group for attachment to the protein through disulfide exchange, and a biotin handle for downstream detection (Figure 2A, Supplemental Figure 3). Once attached to the cysteine sulfhydryl, the reactive azide within the crosslinker is separated from the side-chain by a distance of ~14 Å. The crosslinking strategy we devised is based on transfer of the biotin tag from σ1.1 to another domain upon irradiation and reduction (Figure 2B). In principle, the tagged domain could then be identified by electrophoretic or mass spectrometry resolution of the fragments produced by proteolytic or chemical cleavage methods.
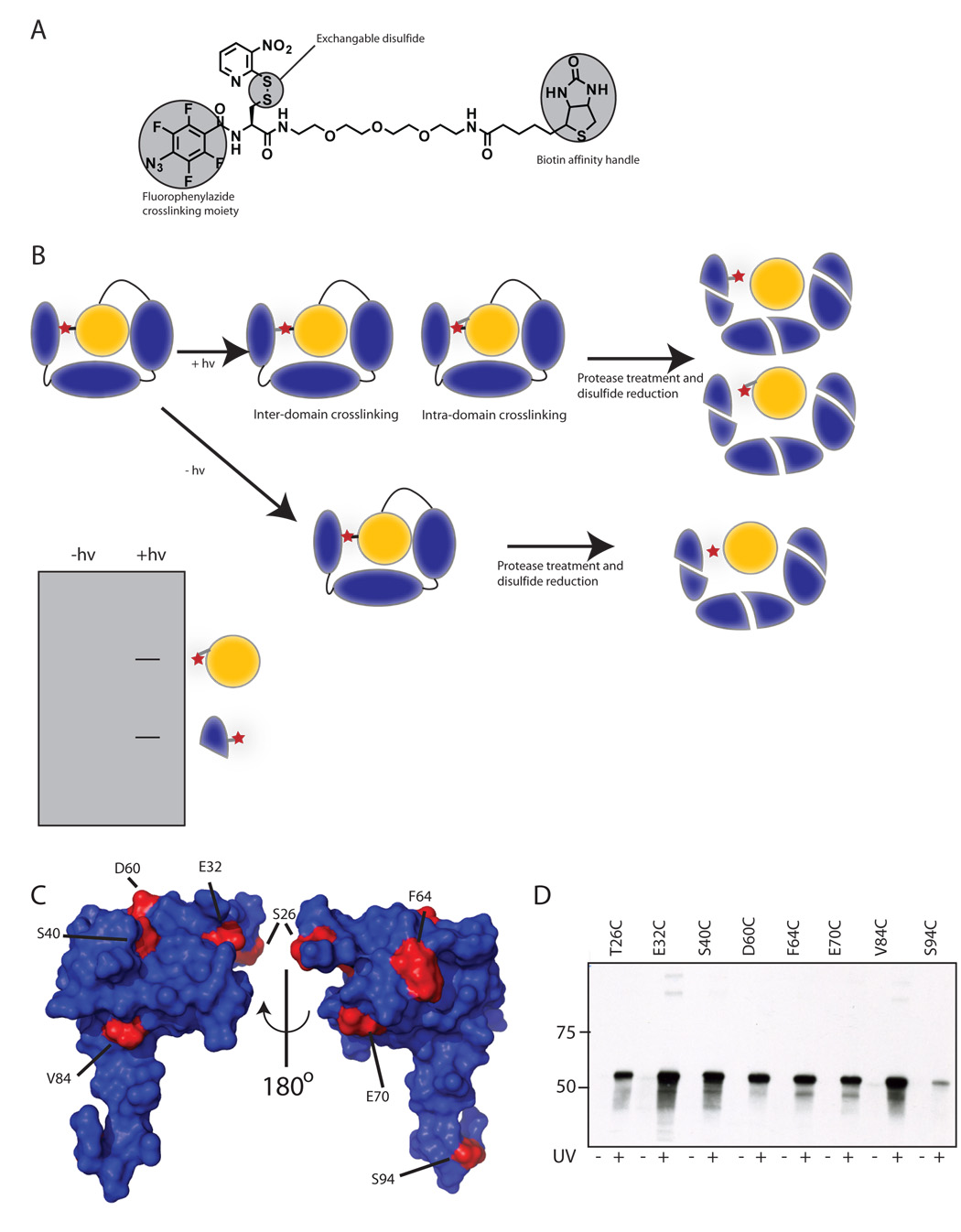
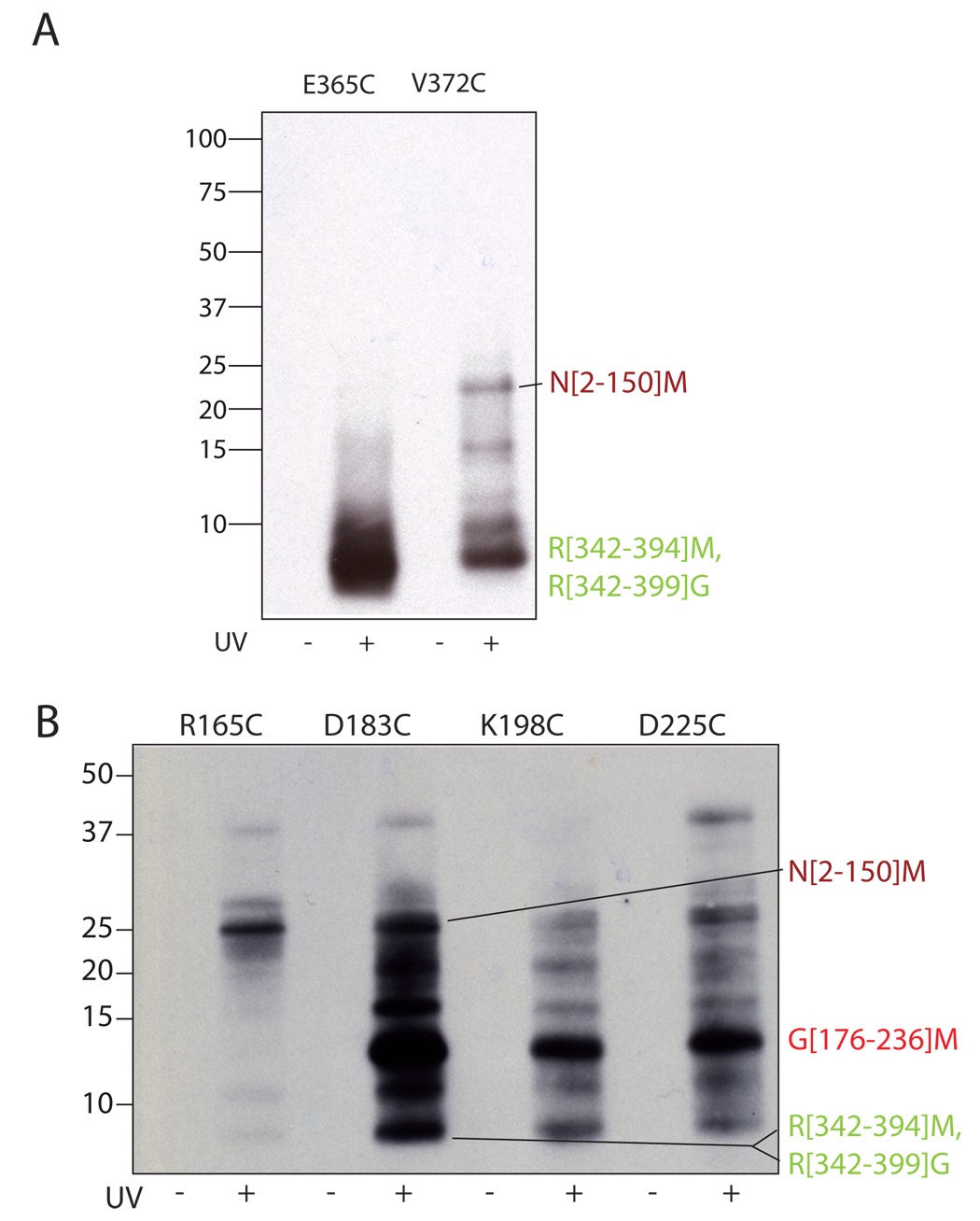
Figure 2. Crosslinking to detect intramolecular interactions.
(A) Chemical structure of the photo-crosslinker used in this study. A synthetic scheme is provided in supplemental material. (B) The crosslinker (red star) is attached by disulfide exchange to a surface cysteine in the σ factor. Following photo-crosslinking and digestion, the disulfide is reduced, transferring the tag to regions in close proximity to σ1.1. The transferred-tag is detected by electrophoretic methods. (C) σ1.1 residues mutated for crosslinking studies. The surface of σ1.1 is rendered in blue with residues that were mutated to cysteine for attachment of the crosslinker highlighted in red. Images were generated using MacPYMOL (Delano Scientific). (D) Crosslinking can be induced by uv irradiation. Indicated σA mutants were labeled with crosslinker by disulfide exchange. Samples that had either been or had not been irradiated at 325 nm light were then reduced with DTT. Samples were then resolved by SDS-PAGE and analyzed by Western blot (Streptavidin-HRP).
Using the structure of σ1.1 as a guide, we selected ten residues to mutate to cysteine for crosslinker attachment (Figure 2C). These were designed to give broad coverage of the surface of σ1.1. Of the ten point mutations made, eight could be expressed and purified (Supplemental Figure 4 and Supplemental Table 1). The E51C and E77C mutants did not express. The mutant proteins were then labeled with crosslinker using a disulfide exchange process (Figure 2D). In all cases, the crosslinker could be removed by reduction with DTT prior to uv irradiation at 325 nm but not following, indicating that crosslinking to the protein had occurred. Moreover, we observed no evidence of the formation of crosslinked σA dimers or higher oligomers (Supplemental Figure 5), indicating that under the conditions of all our experiments, crosslinking is intramolecular. The presence of the label did not interfere with DNA binding autoinhibition by σ1.1 in any of the mutants (Supplemental Figure 6A). Furthermore, labeled σ factors behaved as wild type in abortive transcription initiation assays (Supplemental Figure 6B).
Having established that crosslinks from σ1.1 could be formed, we next set about determining what portions of the σ factor were being contacted by σ1.1. Numerous attempts were made to directly identify crosslinked regions by mass spectrometry analysis of tryptic fragments, but in no case could the unambiguous identification of a crosslinked peptide be made. We thus turned to gel based methods to map the sites of crosslinking. Analysis of the σA sequence suggested that cyanogen bromide (CNBr) digestion of the protein would be informative. The expected CNBr digestion pattern is shown in Figure 3A. The majority of crosslinking is predicted to occur locally, to nearby portions of σ1.1. Fortunately, σ1.1 is entirely contained within a single CNBr fragment that is more than twice as large as the next largest. Thus, σ1.1 should be easily resolved from other CNBr fragments by SDS-PAGE, in principle, allowing one to distinguish between local and inter-domain crosslinking.
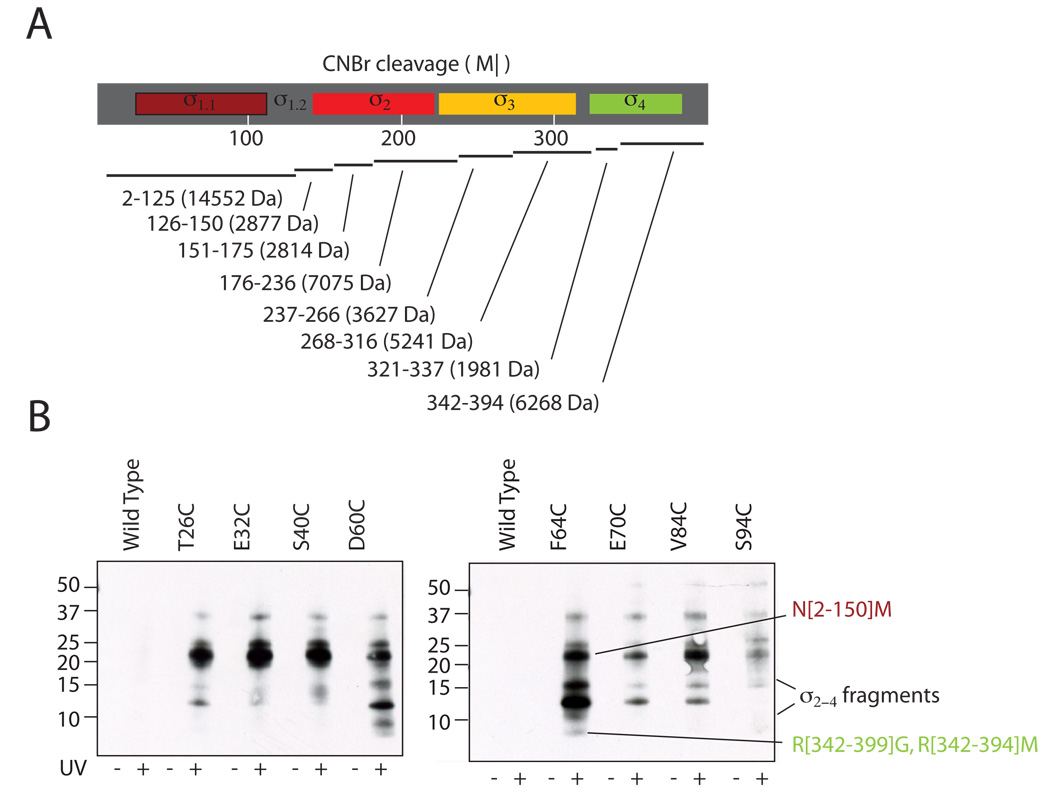
Figure 3. Interdomain crosslinking from σ1.1.
(A) Predicted CNBr digestion of Tm σA. The domain structure of σA (top) is shown with the predicted CNBr digestion fragments below. Residue numbers of cleavage products are shown with molecular weights in parenthesis. (B) Photo-crosslinking from σ1.1 analyzed by label transfer and CNBr digestion. Crosslinker was attached at the indicated residues within σ1.1 through a disulfide. Wild-type σA, which lacks a native cysteine, was included as a control. Irradiated (UV+) and non-irradiated (UV−) samples were digested with CNBr, separated by reducing SDS-PAGE (10–20% acrylamide, tris-tricine buffer system) and probed for biotin by Western blotting (HRP-streptavidin). Note, only the irradiated cysteine mutants retained the biotin. The identity of the labeled bands is shown at right. Bands that have been identified by mass spectrometry are labeled with residue numbers. A full account of the assignment protocol is given in supplemental methods.
In order to map protein crosslinking site(s), the irradiated samples were digested with CNBr followed by reduction to transfer the biotin tag. In all cases, western blotting revealed the presence of biotin-probe on fragments that contain σ1.1 (Figure 3B). Note, several of the CNBr digestion fragments of σA, including σ1.1, migrate slower than predicted based on molecular weight (a detailed description of the procedure used to identify the CNBr digestion products is provided in the Supplemental Information). Labeling of σ1.1 is consistent with local crosslinking, which was expected. The presence of biotin in bands that were smaller than the σ1.1 was interpreted to indicate inter-domain crosslinks. By this criterion, the E32C and S40C attachment sites did not display strong crosslinking to fragments from any other region. By contrast, the other mutants all showed crosslinking to fragments that were smaller than σ1.1, and must therefore come from domains 2–4. Crosslinking from the D60C and F64C attachment sites led to labeling of a ~6 kDa band that was unambiguously assigned to σ4.2 (R[342–399]G and R[342–394]M) by mass spectrometry. Additionally, crosslinking from the T26C, D60C, F64C, E70C and V84C attachment sites led to robust labeling of a fragment with an apparent mobility between 10 and 15 kDa. This mobility is consistent with fragments from either σ2 or σ3. However, it is also possible that partial digestion products containing σ4 could account for some of the bands observed between 10 and 15 kDa.
To help resolve this ambiguity, we turned to BNPS-skatole (2-(2'-nitrophenylsulfenyl)-3-methyl-3-bromoinolenine) digestion, which cleaves following tryptophan residues. There are three tryptophan residues in σA: W201, W213 and W214. Thus, BNPS-skatole digestion effectively cuts σA in half (Figure 4A). Crosslinked protein samples were digested with BNPS-skatole, separated by reducing SDS-PAGE and blotted with streptavidin-HRP (Figure 4B). As expected, when the crosslinker was attached to residues 32 or 40, very little labeling was seen on the band representing the C-terminal half of the protein (202/215–399). In contrast, label-transfer to the C-terminal fragment clearly occurred when the crosslinker was attached to the other 6 sites, indicating interdomain crosslinking. In all cases, crosslinking was observed to the N-terminal fragment of the protein, comprising residues 1-201/214 (Figure 4B). This is consistent with the formation of local crosslinks to σ1.1, but could also represent crosslinking to σ2.
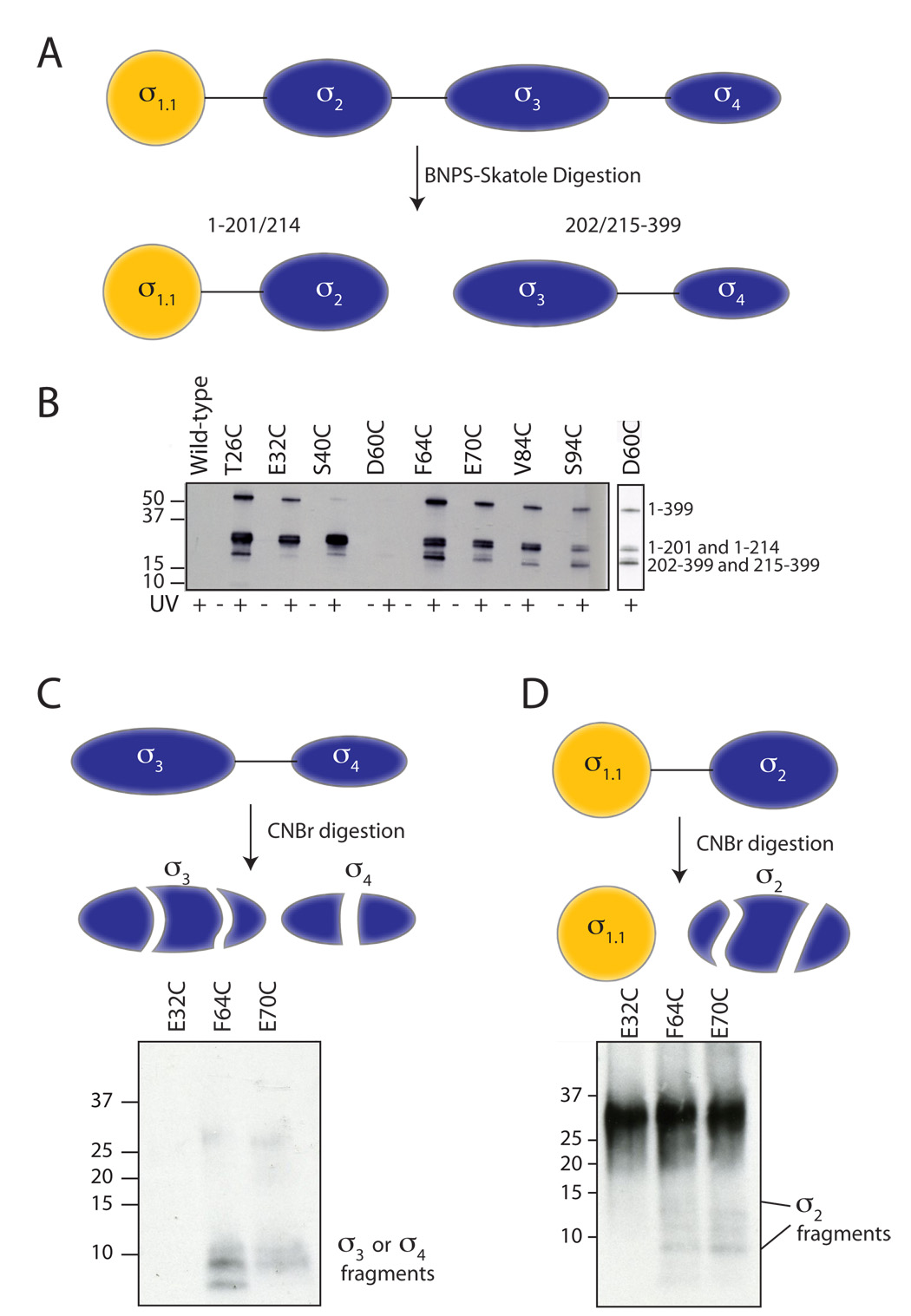
Figure 4. BNPS-Skatole and two-stage digestion of crosslinked products.
(A) BNPS-skatole cleaves after Trp residues and, as a consequence, conveniently cuts σA between σ2 and σ3. (B) Photo-crosslinking from σ1.1 analyzed by label transfer and BNPS digestion. Crosslinker was attached at the indicated residues within σ1.1 through a disulfide. Wild-type σA, which lacks a native cysteine, was included as a control. Irradiated (UV+) and non-irradiated (UV−) samples were digested with BNPS, separated by reducing SDS-PAGE (4–12% acrylamide) and probed for biotin by Western blotting (HRP-streptavidin). Note, the D60C sample did not label well, so a longer exposure is shown to the right. The identity of the labeled bands (shown at right) was determined by in-gel trypsinolysis followed by mass spectrometry (data not shown). (C,D) Two-step digestion protocol. Irradiated crosslinker samples of the indicated attachment sites were digested with BNPS-skatole, separated by SDS-PAGE (4–12% acrylamide). Following excision and extraction from the gel, the C-terminal BNPS-skatole fragment (panel C) and N-terminal BNPS-skatole fragment (panel D) were further digested with CNBr and analyzed by SDS-PAGE (10–20% acrylamide, tris-tricine buffer system) and Western blot (HRP-streptavidin).
To further map the interactions, a subset of the crosslinked samples was subjected to a two-stage digestion procedure. We chose E32C, F64C and E70C as representatives of what appears to be three distinct crosslinking patterns (Figure 3B). Following BNPS-skatole digestion, the bands corresponding to the N- and C-fragments were cut out of the coomassie stained gel, extracted and digested with CNBr before being analyzed by blotting with HRP-streptavidin. For the F64C and E70C crosslinker attachment sites, CNBr digestion of the C-terminal BNPS-skatole fragment led to the generation of smaller biotin-containing fragments (Figure 4C). From this we can conclude that σ1.1 is situated close to σ3 and/or σ4 in the autoinhibited state. In the case of F64C, the labeling pattern includes the σ4.2 fragment previously seen in the CNBr digestion of the intact protein. No biotin-labeled fragments were detected in the extracted and digested C-fragment from the E32C mutant, consistent with no inter-domain crosslinking occurring from this attachment site. CNBr digestion of the BNPS-skatole N-fragment was more revealing. While the E32C attachment site showed no crosslinking to smaller digestion fragments, the F64C and E70C sites both displayed faint, but reproducible, biotin-labeled fragments with greater gel mobility than σ1.1 (Figure 4D). These can only represent digestion fragments containing σ2, thus suggesting that σ1.1 and σ2 are close in space.
σ1.1 and σ4 are close in space
To confirm that σ1.1 is proximal to σ4, we performed crosslinking experiments in the reverse direction, i.e. from σ4 back to σ1.1. Using the structure of the highly homologous σ4 from Thermus aquaticus as a guide (Campbell et al., 2002b), we designed two surface exposed sites for crosslinker attachment, E365 and V372, both of which are located in σ4.2. The corresponding cysteine mutants (E365C and V372C) were generated (Supplemental Figure 4) and crosslinking/label-transfer experiments performed as with the σ1.1 mutants. As expected, local crosslinking to σ4 was observed in both cases (Figure 5A). More importantly, we also observed substantial crosslinking to σ1.1 from the V372C attachment site, based on the appearance of a biotin-labeled σ1.1 fragment in the CNBr digest. There was no evidence of crosslinks from the E365C site back to σ1.1. This difference in behavior may reflect the fact that E365C, but not the V372C, is highly conserved in group 1 sigma factors and so the structure and/or function of σ4 may be more sensitive to perturbations at this site. The key result, however, is the observation of crosslinks to σ1.1 from position 372, which serves to confirm that σ1.1 is situated near σ4 in the autoinhibited state
Figure 5. Interdomain crosslinking from σ4 and σ2.
Crosslinker was attached at the indicated residues within σ4 (A) and σ2 (B) through a disulfide. Irradiated (UV+) and non-irradiated (UV−) samples were digested with CNBr, separated by reducing SDS-PAGE (10–20% acrylamide, tris-tricine buffer system) and probed for biotin by Western blotting (HRP-streptavidin). The identity of the labeled bands is shown at right. The N[2–150]M, R[342–394]M and R[342–399]G bands were identified by mass spectrometry, the G[176–236]M band was identified as described in the text.
σ1.1, σ2 and σ4 are situated close in space
Since the crosslinks between σ1.1 and σ2 were identified following double digestion and were only faintly detected, we felt it was particularly important to confirm these by showing crosslinking in the reverse direction. Accordingly, we generated four cysteine mutants of σ2 for crosslinker attachment (Supplemental Figure 4). These were designed to give broad surface coverage based on the structure of the homologous σ2 from Thermus aquaticus (Campbell et al., 2002b). These samples were crosslinked and analyzed as with the σ1.1 and σ4 mutants (Figure 5B). For the D183C, K198C and D225C attachment sites, the predominant labeled band migrated between 10 and 15 kDa. We reasoned that this fragment likely results from local crosslinking and as such corresponds to the CNBr fragment of σ2 containing the crosslinker attachment sites, i.e. G[176–236]M. Consistent with this interpretation, this band was completely absent in the digest from the R165C attachment site from which local crosslinking affords a CNBr fragment of only 2.8 kDa which would not be seen in this figure. More importantly, we observed crosslinking from σ2 back to σ1.1 for all four crosslinker attachment sites (Figure 5B). This is clearly illustrated for the R165C attachment site where the principle labeled band in the CNBr digest migrates at the position that corresponds to σ1.1. Additionally, we observed crosslinking from σ2 to σ4.2. Such crosslinks could not be unambiguously assigned when performing crosslinking from the σ4 mutants, due to the possibility of detecting partial digestion products containing σ4. However, since the σ4.2 bands display unique gel mobility, they could be easily identified in this experiment.
DISCUSSION
The DNA binding functions of bacterial σ factors are regulated through a variety of mechanisms (Gruber and Gross, 2003). The group 1 σ factors, which perform the bulk of transcription during exponential growth, are converted from an autoinhibited state, which cannot interact with promoter DNA, to an active form that recognizes promoter DNA upon binding to core RNAP. Biochemical and genetic studies have established that the N-terminal domain of group 1 σ factors, σ1.1, is required for autoinhibition (Dombroski et al., 1993; Dombroski et al., 1992). The mechanistic basis for this σ1.1 function has remained unclear. Biochemical studies indicate that σ1.1 can inhibit the DNA binding properties of σ4 in trans, suggesting a direct physical interaction between the domains (Dombroski et al., 1993; Dombroski et al., 1992). However, NMR studies of segmental labeled versions of Tm σA do not support a direct interaction between σ1.1 and at least the C-terminal half of σ4 (Camarero et al., 2002). In the present study, we have shown that, in the autoinhibited state, σ1.1 of Tm σA is in close proximity to both DNA binding domains of the protein, namely σ2 and σ4. Critical to these studies was the determination of the solution structure of σ1.1, since this allowed us to rationally design point mutants of the protein for use in a site-directed chemical crosslinking strategy employing a novel tri-functional chemical crosslinking reagent.
While there is no structure available of an intact, isolated group 1 σ factor, the crystal structure of a full-length alternative sigma factor from Aquifex aeolicus, σ28, has been solved in complex with its anti-sigma factor FlgM (Sorenson et al., 2004). σ28 has a highly compacted structure in this complex, with extensive interfaces between conserved σ2–4. Biochemical studies suggest that σ28 maintains this compacted structure even in the absence of FlgM (Sorenson and Darst, 2006). This led to the proposal that autoinhibition of DNA binding in σ28 results from a combination of sub-optimal interdomain distances and steric occlusion of the promoter binding determinants. An obvious question then is whether similar interdomain interactions exist in autoinhibited group 1 σ factors? The high level of sequence homology between σ2 and σ4, and to a lesser extent σ3, argues in favor of a similar compacted structure. However, since alternative σ factors like σ28 lack σ1.1, it is quite possible that a different arrangement of the domains is present in autoinhibited group 1 σ factors. We observed direct crosslinks between σ2 and σ4 in free σA (Figure 5), which is consistent with a σ28 –like structure. However, given the low resolution of crosslinking experiments, it is unclear whether this reflects a direct physical interaction between these domains, versus their being in close apposition but not actually in contact. In the structure of σ28, the C-terminal portion of σ3 (which is highly acidic in both primary and alternative σ factors) forms a bent helix that sits between σ2 and σ4 and packs against the basic −35 element binding region of σ4. Therefore, we would expect to see crosslinks to σ3 if σA assumes a similar structure as σ28. Unfortunately, it was not possible to unambiguously assign crosslinked bands to σ3 fragments using the chemical cleavage methods we employed. Thus, while we cannot rule such crosslinks out, the question of whether part of σ3 inserts between σ2 and σ4 in σA remains unclear.
Our crosslinking data indicate that σ1.1 packs close to σ2 and σ4 in the autoinhibited state of σA and that, furthermore, surface electrostatics plays a role in this compaction. As expected, based on the highly acidic sequence of σ1.1 (calculated pI = 4.4), the majority of the protein surface is negatively charged. However, there also exists a small positively charged patch centered on H1 (Figure 1E). Of the 8 sites in σ1.1 chosen for crosslinker attachment, all but 2 exhibited interdomain crosslinking (Figure 3). What is remarkable is how this apportioning of activity correlates with surface electrostatics; all the sites from which interdomain crosslinking was observed map to the negative surface, whereas both of the sites from which no inter-domain crosslinking was observed map to the positive patch (Figure 6A). σ2 and σ4 are both positively charged DNA binding domains (calculated pIs for σ2 and σ4 are 10.5 and 10, respectively). It is therefore likely that electrostatic forces play an important role in stabilizing the close proximity of σ1.1, σ2 and σ4 revealed by our crosslinking data. Consistent with this idea, the autoinhibitory effect of σ1.1 is known to be highly sensitive to changes in ionic strength (Camarero et al., 2002).
Figure 6. Proposed model of DNA binding inhibition by σ1.1.
(A) Crosslinker attachment sites are indicated on the electrostatic surface map of σ1.1 generated using the GRASP program. Indicated in white are the sites that were shown to make interdomain crosslinks. Indicated in yellow are the sites from which no interdomain crosslinking was observed. (B) Schematic showing the compaction model of σA autoinhibition. The negative surface of σ1.1 is capable of forming crosslinks to the DNA binding domains σ2 and σ4. It is thus likely that σ1.1 organizes the σ factor into a compacted structure that is incapable of binding DNA. σ2 forms crosslinks to σ4, indicating that these two domains must also be in close proximity.
Our crosslinking data presents a picture of the free group 1 σ factor that is very different from the structure in the RNAP holoenzyme. In the latter context, the σ factor adopts an extended conformation, with each domain widely separated in space (Murakami et al., 2002a; Vassylyev et al., 2002). For instance, in the crystal structure of the Thermus thermophilus holoenzyme, the centers of σ2 and σ4 are separated by ~65 Å (versus ~25 Å in the structure of σ28), and σ1.1, which is not resolved in the structure, is believed to be separated from the rest of the holoenzyme by a flexible linker (Vassylyev et al., 2002). By contrast, our crosslinking data indicates that the domains in the free σ factor are in close proximity; σ1.1, σ2 and σ4 all crosslink to one another. We again stress that the presence of these crosslinks does not prove that all of the domains are physically interacting, particularly since the attachment chemistry results in a ~14 Å linker between the cysteine and the reactive azide group. Nonetheless, the crosslinking data is clearly incompatible with the open conformation of σA found in the holoenzyme structures. Rather, our data suggest that the free factor assumes a much more compacted state relative to the conformation in the holoenzyme. This idea is also supported by LRET and FRET studies on E. coli σ70, which indicate that the interdomain distances are considerably smaller in free σ than in the RNAP holoenzyme (Callaci et al., 1999) (Mekler et al., 2002).
The crosslinking data presented herein, together with the aforementioned biochemical and biophysical studies, point to an autoinhibition mechanism involving a significant compaction of the free σ factor relative to its more open conformation in the holoenzyme. Since interdomain crosslinks were only observed from the negative surface of σ1.1, we propose that electrostatic interactions between this surface and the positively charged surfaces of σ2 and σ4 underlie this compaction (Figure 6B). The autoregulatory role of σ1.1 is therefore fulfilled by locking the DNA binding domains in a conformation that is incompatible with promoter binding. We cannot, using the approaches we have employed, determine the strength of the interactions between σ1.1 and the DNA binding domains of the σ factor. We predict, however, that they are fairly weak, and likely transient, for a number of reasons. First, mechanistically, the interdomain interactions need to be broken in order to form the DNA binding competent RNAP holoenzyme. If the interactions between the sigma domains are of high affinity, it would be energetically expensive to form productive holoenzyme complexes due to effective molarity (Kobe and Kemp, 1999). Second, while σ constructs containing σ4 could be inhibited by the addition of σ1.1 in trans, achieving a 50% reduction in DNA binding required a 15–20 fold excess of σ1.1 over σ4, suggesting that binding between the two domains is weak (Dombroski et al., 1993). Additionally, σ2 binding by σ1.1 could not be detected in this trans inhibition assay, once again implying weak binding. Finally, the NMR spectra of σ4.2 is not significantly perturbed by σ1.1 (Camarero et al., 2002), despite the fact that our crosslinking data shows that σ1.1 and σ4.2 must be in close proximity. This points to an interaction that is both weak and somewhat fluid, which could be difficult to detect when using NMR based approaches. This highlights the sensitivity of the crosslinking approach as it can capture relatively weak and transient interactions.
SIGNIFICANCE
In this work, we have reported the first high resolution structure of a σ1.1 domain. While there is a wealth of structural data on bacterial sigma factors and fragments thereof, to date there had been no structure available on σ1.1. We have addressed a long-standing mechanistic issue in prokaryotic transcription, namely how sigma factors are autoinhibited. While it has long been known that σ1.1 is needed for autoinhibition, quite how it works has been less clear – some studies have pointed to a direct interdomain binding mechanism, others have not. Our data clearly show that in the autoinhibited state, σ1.1 is juxtaposed with the two DNA binding domains (σ2 and σ4) of the sigma factor and that this compacted state is held together via complementary electrostatic forces between the domains. For steric reasons, this conformation is incompatible with DNA binding. In performing this work, we have synthesized and successfully applied a new trifunctional chemical crosslinking reagent, which will be useful for studying transient interactions in other systems.
EXPERIMENTAL PROCEDURES
NMR samples
T. maritima σA σ1.1 (residues 1–96 or 1–116, with mutations Q21L, E22V, Q23P, K24R, E25G, T26S, L27H, P28M to install a thrombin site and mutations S96G or S116G to facilitate intein thiolysis and expressed protein ligation) was cloned into the pTXB1 vector (New England Biolabs) to generate a fusion construct of σA σ1.1 – Mxe GyrA intein – chitin binding domain. BL-21(DE3) (Invitrogen) cells were transformed with this construct, grown overnight on LB plates (100 µg/mL ampicillin) then grown in 1L M9 minimal media (100 µg/mL ampicillin) with 1.2 g 15NH4SO4 and 2 g 13C glucose as the only nitrogen and carbon sources. The culture was grown at 37 °C to an OD600 of ~0.6. Expression was induced with 0.8 mM IPTG and was carried out at 30 °C for 4.5 h. Bacteria were harvested by centrifugation, resuspended in lysis buffer (500 mM NaCl, 20 mM Tris pH 8.0, 1 mM EDTA) and lysed using a french press. The lysate was centrifuged and the supernatant applied to a chitin bead column (New England Biolabs) and washed extensively with lysis buffer. Intein cleavage was induced with 50 mM DTT in lysis buffer and allowed to proceed overnight at 4 °C with gentle rocking. Cleaved protein was eluted with lysis buffer and was exchanged into 1x PBS by dialysis. Thrombin protease (Amersham) was added (50 U) and proteolysis was carried out overnight at room temperature with gentle rocking. The resulting protein was H2N-GSHM-(σ-A 29-95/115)-G-COOH. The sample was further purified by ion exchange chromatography (Hiprep Q16/10 column, AKTA FPLC, Amersham) using a 0.1–1.0 M NaCl gradient in 20 mM Tris·HCl (pH 8.0). Protein samples were characterized by electrospray mass spectrometry (ESMS, Sciex API-100 single quadropole electrospray mass spectrometer). The protein sample was treated at 65 °C for 30 min to eliminate E. coli proteases and other contaminants and then concentrated in NMR buffer (10 mM NaPi, pH 6.5, 100 mM NaCl, 1mM EDTA) to a final concentration of 300–800 µM depending on the sample. Protein concentrations were determined using Bradford assay (BioRad) using a BSA (New England Biolabs) standard curve.
NMR Spectroscopy
All NMR experiments were recorded at 25 °C using 600, 700 and 800 MHz Bruker Avance NMR spectrometers equipped with cryoprobes. NMR data were processed using Topspin 1.1 (Bruker) and analyzed by NMRView (Johnson, 2004). 1H chemical shifts were referenced to water at 4.75 ppm (at 25 °C) and 13C and 15N shifts were derived from indirect referencing. 1H-15N-HSQC, 1H-13C-HSQC, 3D-15N-NOESY-HSQC, HNCO, HNCACB, CBCA(CO)NH experiments were used to obtain the sequential assignment of the backbone (Sattler et al., 1999). Side-chain carbon and proton assignments were obtained using C(CO)NH, HC(CO)NH, HBHA(CO)NH and HCCH-TOCSY experiments. For assigning the aromatic resonances, 1H-13C HSQC, (Hβ)Cβ(CγCδ)Hδ and (Hβ)Cβ(CγCδCε)Hε (Yamazaki et al., 1993), and 3D 13C NOESY-HSQC data was utilized.
Structure Calculations
Backbone dihedral angles (φ and ψ) were calculated by analyzing the 13Cα, 13Cβ, 13C′ and 15N chemical shifts with the TALOS program that predicts the backbone torsion angles from the amino acid sequence and chemical shift information (Cornilescu et al., 1999). Hydrogen bond restraints were inferred from slow H/D exchange of backbone amides. Distance restraints were derived from 3D 13C-edited NOESY and 3D 15N-edited NOESY experiments. Using NMRView, the NOESY cross peak volumes/intensities were obtained and converted into distance restraints using the symmetry ambiguous distance restraints (ADR) protocol within the ARIA program (Linge et al., 2003). A total of 512 structures were calculated using the Cartesian dynamics simulated annealing protocol within ARIA/CNS. 100 lowest energy structures without any distance and dihedral violations were further refined in water as described by Linge et al. The NMR structures were analyzed using PROCHECK-NMR (Laskowski et al., 1996). The electrostatic potentials were calculated using GRASP (Nicholls et al., 1991). Structures were displayed using MACPymol (Delano Scientific).
Comparison of the calculated structures to existing known protein folds was made using the DALI search engine (Holm and Sander, 1996). The overall fold of three interacting helices is represented in a number of existing structures at high z-score and RMSD in the range 2.5 – 3.0 Å. However, these structures were of uniformly low sequence similarity (4–22%) and appeared to have only a topological correspondence of overlapped helices with a similar crossing angle.
Electrophoretic Analysis of Crosslinking Experiments
SDS-PAGE analysis in the crosslinking section was performed using the Criterion system (Biorad). Criterion gels used were 4–12% acrylamide Bis-Tris buffered gels and 10–20% acrylamide Tris-tricine buffered gels, as indicated. Samples for Western blotting were separated by SDS-PAGE before being transferred to PVDF (Biorad) using standard western blotting procedures. The membrane was blocked in 5% milk in TBST for 1 hour at room temperature then washed 3 times with TBST and incubated with Streptavidin-HRP (GE Health Sciences, 1:5000 in TBST) for 1 hour. The membrane was washed 3 times with TBST and developed using ECL reagents (Perkin Elmer) and Kodak Biomax MR film.
Crosslinking Samples
T. maritima σA was cloned into pET15B (Novagen) with an N-terminal hexahistidine tag and a thrombin recognition site for removal of the affinity tag (Final sequence: MGSSHHHHHHSSGLVPRGSH-σA). Cysteine mutants were generated using the Quikchange II XL kit (Stratagene). All mutations were verified by DNA sequencing. BL-21(DE3) cells were transformed with wild-type or mutant σA plasmid and grown in LB broth (100 µg/mL ampicillin) at 37 °C to an OD600 of ~0.6. Protein expression was induced with 0.8 mM IPTG at 37 °C for 4–6 h. Bacteria were harvested by centrifugation, treated at 65 °C for 20 minutes, lysed using a french press and purified using the Ni-NTA system (Novagen). Protein samples were then transferred to 1x PBS by dialysis and treated with thrombin protease (Amersham, 50 U) overnight at room temperature. Proteolyzed samples were further purified by ion exchange chromatography (HiTrap SP HP, Amersham) with a 0.1–1.0 M NaCl gradient in 20 mM Tris·HCl (pH 8.0). Fractions containing pure protein were concentrated and stored at −80 °C in 10% glycerol and at −20 °C in 50% glycerol.
Attachment of Crosslinker and Crosslinking
Crosslinker 1.5 was attached to the purified cysteine mutant of σA by disulfide exchange. Details of the crosslinker synthesis can be found in supplemental methods. Reaction mixtures contained 6 µM protein and 100 µM crosslinker in 50 mM sodium borate (pH 8.3), 8 mM Tris-HCl (pH 8.3), 80 mM KCl, 0.4 mM EDTA, 5 mM methionine, 0.6 mM tyrosine, 2% glycerol and 1% DMSO. Reactions were carried out for 20 minutes at room temperature. Reaction mixtures were then buffer exchanged to 1 x PBS using a centrifugal filter device (Vivascience, 10,000 MWCO) and concentrated to approximately 25 µM protein. Crosslinking was then induced by irradiation with a 325 nM HeCd laser for 10–15 seconds.
CNBr digestion
CNBr digestion was performed by heating protein (1 mg/mL for crosslinking samples, 5 mg/mL for WT sample used to identify CNBr digestion products) with 1.3% SDS at 42 °C for 10 minutes. Reactions were then carried out in 0.1 N HCl, 100 mM CNBr, 1% SDS overnight at room temperature. Reactions were quenched by neutralization with 1N NaOH.
BNPS-Skatole Digestion
BNPS-Skatole digestion was performed in 70% acetic acid with ~3.5 mg/mL BNPS-skatole (Sigma) overnight at room temperature. Protein concentration was 0.5–1 µg/mL depending on the experiment. Following overnight digestion, samples were treated with 67 mM DTT at room temperature for 20 minutes. Samples were extracted with ether and aqueous fractions were lyophilized and resuspended in SDS loading buffer.
Two Step Digestion
BNPS-Skatole digested samples were separated by SDS-PAGE and visualized by coomassie stain. Bands were excised with a clean razor blade and homogenized in 1 N HCl, ~0.5–1% SDS with mortar and pestle then incubated at 100 °C for 5 minutes. Samples were digested with 100 mM CNBr overnight at room temperature, as above.
Supplementary Material
Additional methods and supplemental Figures S1–S4.
Acknowledgements
This work was supported by National Institutes of Health Grants GM 053759-10 (TWM, SAD), GM55843-11 (TWM) and EB-001991 (TWM) and by a Howard Hughes Medical Institute fellowship (E.C.S.). NMR resources at NYSBC are supported by P41 GM66354. We thank Lars Westblade and Andrey Feklistov for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barne KA, Bown JA, Busby SJ, Minchin SD. Region 2.5 of the Escherichia coli RNA polymerase sigma70 subunit is responsible for the recognition of the 'extended-10' motif at promoters. EMBO J. 1997;16:4034–4040. doi: 10.1093/emboj/16.13.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borukhov S, Nudler E. RNA polymerase holoenzyme: structure, function and biological implications. Curr. Opin. Microbiol. 2003;6:93–100. doi: 10.1016/s1369-5274(03)00036-5. [DOI] [PubMed] [Google Scholar]
- Bowers CW, Dombroski AJ. A mutation in region 1.1 of σ70 affects promoter DNA binding by Escherichia coli RNA polymerase holoenzyme. EMBO J. 1999;18:709–716. doi: 10.1093/emboj/18.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2004;2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- Burgess RR, Travers AA, Dunn JJ, Bautz EK. Factor stimulating transcription by RNA polymerase. Nature. 1969;221:43–46. doi: 10.1038/221043a0. [DOI] [PubMed] [Google Scholar]
- Callaci S, Heyduk E, Heyduk T. Core RNA polymerase from E. coli induces a major change in the domain arrangement of the sigma 70 subunit. Mol. Cell. 1999;3:229–238. doi: 10.1016/s1097-2765(00)80313-5. [DOI] [PubMed] [Google Scholar]
- Camarero JA, Shekhtman A, Campbell EA, Chlenov M, Gruber TM, Bryant DA, Darst SA, Cowburn D, Muir TW. Autoregulation of a bacterial σ factor explored by using segmental isotopic labeling and NMR. Proc. Natl. Acad. Sci. USA. 2002;99:8536–8541. doi: 10.1073/pnas.132033899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EA, Darst SA. Regulation of bacterial transcription by anti-s factors. In: Waksman G, Caparon M, Hultgren S, editors. Structural Biology of Bacterial Pathogenesis. Washington, DC: ASM Press; 2005. pp. 1–15. [Google Scholar]
- Campbell EA, Masuda S, Sun JL, Muzzin O, Olson CA, Wang S, Darst SA. Crystal structure of the Bacillus stearothermophilus anti-sigma factor SpoIIAB with the sporulation sigma factor sigmaF. Cell. 2002a;108:795–807. doi: 10.1016/s0092-8674(02)00662-1. [DOI] [PubMed] [Google Scholar]
- Campbell EA, Muzzin O, Chlenov M, Sun JL, Olson CA, Weinman O, Trester-Zedlitz ML, Darst SA. Structure of the Bacterial RNA Polymerase Promoter Specificity Sigma Subunit. Mol. Cell. 2002b;9:527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- Campbell EA, Tupy JL, Gruber TM, Wang S, Sharp MM, Gross CA, Darst SA. Crystal Structure of Escherichia coli σE with the Cytoplasmic Domain of Its Anti-σ RseA. Mol. Cell. 2003;11:1067–1078. doi: 10.1016/s1097-2765(03)00148-5. [DOI] [PubMed] [Google Scholar]
- Campbell EA, Westblade LF, Darst SA. Regulation of bacterial RNA polymerase sigma factor activity: a structural perspective. Curr. Op. Microbiol. 2008;11:121–127. doi: 10.1016/j.mib.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh J, Fairbrother JW, Palmer AG, III, Skelton NJ. Protein NMR Spectroscopy; Principles and Practice. 1st ed. San Diego, CA: Academic Press; 1996. [Google Scholar]
- Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Darst SA. Bacterial RNA polymerase. Curr Opin Struct Biol. 2001;11:155–162. doi: 10.1016/s0959-440x(00)00185-8. [DOI] [PubMed] [Google Scholar]
- Dombroski AJ, Walter WA, Gross CA. Amino-terminal amino acids modulate sigma-factor DNA-binding activity. Genes & Development. 1993;7:2446–2455. doi: 10.1101/gad.7.12a.2446. [DOI] [PubMed] [Google Scholar]
- Dombroski AJ, Walter WA, Record MTJ, Siegele DA, Gross CA. Polypeptides Containing Highly Conserved Regions of Transcription Initiation Factor Sigma-70 Exhibit Specificity of Binding to Promoter DNA. Cell. 1992;70:501–512. doi: 10.1016/0092-8674(92)90174-b. [DOI] [PubMed] [Google Scholar]
- Ebright RH. RNA polymerase: structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J. Mol. Biol. 2000;304:687–698. doi: 10.1006/jmbi.2000.4309. [DOI] [PubMed] [Google Scholar]
- Gardella T, Moyle H, Susskind MM. A mutant Escherichia coli sigma 70 subunit of RNA polymerase with altered promoter specificity. J Mol Biol. 1989;206:579–590. doi: 10.1016/0022-2836(89)90567-6. [DOI] [PubMed] [Google Scholar]
- Gross CA, Chan C, Dombroski A, Gruber T, Sharp M, Tupy J, Young B. The functional and regulatory roles of sigma factors in transcription. Cold Spring Harb. Symp. Quant. Biol. 1998;63:141–155. doi: 10.1101/sqb.1998.63.141. [DOI] [PubMed] [Google Scholar]
- Gruber TM, Gross CA. Multiple Sigma Subunits and the Partitioning of Bacterial Transcription Space. Ann. Rev. Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- Hinton DM, Vuthoori S, Mulamba R. The bacteriophage T4 inhibitor and coactivator AsiA inhibits Escherichia coli RNA Polymerase more rapidly in the absence of sigma70 region 1.1: evidence that region 1.1 stabilizes the interaction between sigma70 and core. J. Bacteriol. 2006;188:1279–1285. doi: 10.1128/JB.188.4.1279-1285.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Sander C. Mapping the protein universe. Science. 1996;273:595–603. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- Johnson BA. Using NMR View to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- Kobe B, Kemp BE. Active site-directed protein regulation. Nature. 1999;402:373–376. doi: 10.1038/46478. [DOI] [PubMed] [Google Scholar]
- Lane WJ, Darst SA. The structural basis for promoter −35 element recognition by the group IV sigma factors. PLoS Biol. 2006;4:e269. doi: 10.1371/journal.pbio.0040269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Linge JP, Habeck M, Rieping W, Nilges M. ARIA: automated NOE assignment and NMR structure calculation. Bioinformatics. 2003;19:315–316. doi: 10.1093/bioinformatics/19.2.315. [DOI] [PubMed] [Google Scholar]
- Malhotra A, Severinova E, Darst SA. Crystal structure of a sigma 70 subunit fragment from E. coli RNA polymerase. Cell. 1996;87:127–136. doi: 10.1016/s0092-8674(00)81329-x. [DOI] [PubMed] [Google Scholar]
- Mekler V, Kortkhonjia E, Mukhopadhyay J, Knight J, Revyakin A, Kapanidis AN, Niu W, Ebright YW, Levy R, Ebright RH. Structural organization of bacterial RNA polymerase holoenzyme and the RNA polymerase-promoter open complex. Cell. 2002;108:599–614. doi: 10.1016/s0092-8674(02)00667-0. [DOI] [PubMed] [Google Scholar]
- Murakami KS, Darst SA. Bacterial RNA polymerases: the wholo story. Curr. Op. in Struct. Biol. 2003;13:31–39. doi: 10.1016/s0959-440x(02)00005-2. [DOI] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural Basis of Transcription Initiation: An RNA Polymerase Holoenzyme-DNA Complex. Science. 2002a;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Darst SA. Structural Basis of Transcription Initiation: RNA Polymerase Holoenzyme at 4 A Resolution. Science. 2002b;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- Muralidharan V, Muir TW. Protein ligation: an enabling technology for the biophysical analysis of proteins. Nat. Meth. 2006;3:429–438. doi: 10.1038/nmeth886. [DOI] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Ottesen JJ, Blashke UK, Cowburn D, Muir TW. In: Biological Magnetic Resonance. Krishna NR, Berliner LJ, editors. New York: Kluwer Academic; 2003. pp. 35–51. [Google Scholar]
- Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulse field gradients. Prog. NMR Spectrosc. 1999;34:93–158. [Google Scholar]
- Siegele DA, Hu JC, Walter WA, Gross CA. Altered promoter recognition by mutant forms of the sigma 70 subunit of Escherichia coli RNA polymerase. J. Mol. Biol. 1989;206:591–603. doi: 10.1016/0022-2836(89)90568-8. [DOI] [PubMed] [Google Scholar]
- Sorenson MK, Darst SA. Disulfide cross-linking indicates that FlgM-bound and free σ28 adopt similar conformations. Proc. Natl. Acad. Sci. USA. 2006;103:16722–16727. doi: 10.1073/pnas.0606482103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorenson MK, Ray SS, Darst SA. Crystal Structure of the Flagellar sigma/Anti-sigma Complex Sigma-28/FlgM Reveals an Intact Sigma Factgor in an Inactive Conformation. Mol. Cell. 2004;14:127–138. doi: 10.1016/s1097-2765(04)00150-9. [DOI] [PubMed] [Google Scholar]
- Travers AA, Burgess RR. Cyclic re-use of the RNA polymerase sigma factor. Nature. 1969;222:537–540. doi: 10.1038/222537a0. [DOI] [PubMed] [Google Scholar]
- Vassylyev DG, Sekine S-i, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- Vuthoori S, Bowers CW, McCracken A, Dombroski AJ, Hinton DM. Domain 1.1 of the Sigma-70 Subunit of Escherichia coli RNA Polymerase Modulates the Formation of Stable Polymerase/Promoter Complexes. J. Mol. Biol. 2001;309:561–572. doi: 10.1006/jmbi.2001.4690. [DOI] [PubMed] [Google Scholar]
- Wilson C, Dombroski AJ. Region 1 of σ70 is Required for Efficient Isomerization and Initiation of Transcription by Escherichia coli RNA Polymerase. J. Mol. Biol. 1997;267:60–74. doi: 10.1006/jmbi.1997.0875. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Forman-Kay JD, Kay LE. Two-dimensional NMR experiments for correlating C-13b and H-1d/e chemical shifts of aromatic residues in 13C-labeled proteins via scalar couplings. J. Am. Chem. Soc. 1993;115:11054–11055. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional methods and supplemental Figures S1–S4.