Abstract
The Pharmacogenetics and Pharmacogenomics Knowledge Base (PharmGKB: http://www.pharmgkb.org) is devoted to disseminating primary data and knowledge in pharmacogenetics and pharmacogenomics. We are annotating the genes that are most important for drug response and present this information in the form of Very Important Pharmacogene (VIP) summaries, pathway diagrams, and curated literature. The PharmGKB currently contains information on over 500 drugs, 500 diseases, and 700 genes with genotyped variants. New features focus on capturing the phenotypic consequences of individual genetic variants. These features link variant genotypes to phenotypes, increase the breadth of pharmacogenomics literature curated, and visualize single-nucleotide polymorphisms on a gene’s three-dimensional protein structure.
Keywords: Polymorphism, Variant, Drug, Tamoxifen, Warfarin, PharmGKB, Disease, Gene, Pharmacogenetics, Pharmacogenomics
INTRODUCTION
Pharmacogenetics studies the influence of genetic variations on drug response and disease predisposition. An important goal within the field is to collect and condense the knowledge and data to catalyze research and support hypothesis generation. Therefore, we created the Pharmacogenetics and Pharmacogenomics Knowledge Base (PharmGKB: http://www.pharmgkb.org) in order to aggregate data and knowledge from the primary research literature, while creating distilled and curated information in multiple forms. These forms include: (1) Very Important Pharmacogene (VIP) summaries, (2) pathways summarizing the pharmacokinetics and pharmacodynamics for particular drugs, and (3) annotated information about specific genetic variants that affect drug-response phenotypes (Hodge et al., 2007). PharmGKB works with the National Institutes of Health (NIH)-sponsored Pharmacogenetics Research Network (PGRN) to collect, analyze, and archive primary experimental data, while providing search and display capabilities (Giacomini et al., 2007). Based upon its position as a central resource devoted entirely to pharmacogenomics, PharmGKB has recently taken an active role in the formation of pharmacogenomic data-sharing consortia (Owen et al., 2008).
PharmGKB currently captures information on more than 500 drugs, 500 diseases, and 700 variant genes with primary experimental genotype data. PharmGKB has 53 drug-related pharmacokinetic and pharmacodynamic pathways, 36 VIP gene summaries, and over 2,500 curated literature annotations.
With the availability of modern high-throughput techniques, studies investigating the influence of multigenic variants on drug response have led to a rapid increase of newly detected polymorphisms. Unfortunately, schemes for naming genetic variants and their location has not been consistent. With the establishment of dbSNP rsIDs (reference single-nucleotide polymorphism [SNP] IDs), polymorphisms are precisely positioned at an exact genomic location (Sherry et al., 2001). PharmGKB is committed to mapping previous genetic variants to a common framework in order to provide uniform access to all polymorphisms and support search and sorting operations.
PharmGKB began in April 2000 as a free public online resource. Although there are many features of the site available to anonymous users, registration is required for viewing individual subject data, such as the individual genotypes or phenotypes associated with a single person. Knowledge and data are updated on a continual basis through frequent data submissions and ongoing curation efforts of the PharmGKB team. PharmGKB has an updated user interface to serve diverse user groups and meet their interests (Hernandez-Boussard et al., 2008). The latest features include a variant-mining project to broaden functional annotation of polymorphisms, automated literature annotations, and mapping of coding SNPs onto three-dimensional (3D) protein structures.
THE PharmGKB HOMEPAGE
PharmGKB provides a broad spectrum of pharmacogenomic knowledge to the scientific community. The PharmGKB user interface serves the interests of the PharmGKB user community and aims to provide easy access to information. Recent additions include: (1) buttons for direct access to variants of interest, SNP arrays, and downloads and services; (2) a system of tabs that separate different classes of information for gene, drug, and disease pages in order to reduce visual data overload; (3) “active” menu bars providing faster loading of Web pages; and (4) help resources organized by user type (e.g., clinicians, pharmacologists, bioinformaticians, or geneticists).
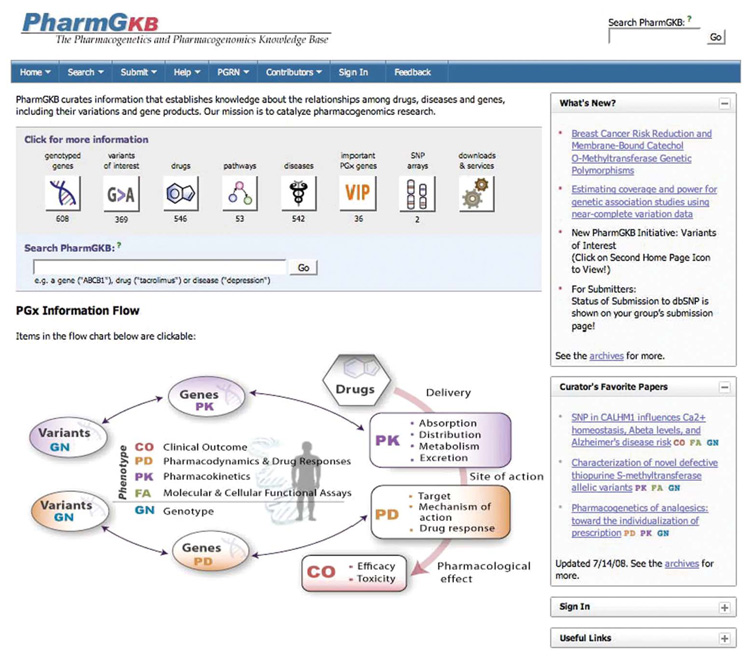
The PharmGKB homepage is designed to meet two goals: (1) to provide distinct entry points to curated pharmacogenomics-related data and knowledge for experienced users and (2) to be a pharmacogenetics educational portal for all users. In the center of the homepage, clickable icons guide the user to information about genotyped genes, variants of interest, drugs, drug-centered pathways, diseases, important PGx genes (VIP annotations), SNP arrays, and downloads and services (Fig. 1). PharmGKB holds primary experimental data annotated with tags for related genes, drugs, diseases, and literature. Data and knowledge content are also available for download. PharmGKB provide a text search box for general queries.
Figure 1.
The PharmGKB homepage (retrieved July 21, 2008, from http://www.pharmgkb.org/). The homepage highlights easily accessible pharmacogenetics and pharmacogenomics knowledge and educational information.
The contents of the PharmGKB are classified according to categories of evidence (COE) of pharmacogenetic knowledge: clinical outcome (CO), pharmacodynamics (PD), pharmacokinetics (PK), molecular and cellular functional assays (FA), and genotype (GN). These five categories of knowledge are the essential components of the diagram under the search box (Fig. 1). The image is intended to inform researchers new to pharmacogenetics about fundamental concepts of PK, PD, and how they relate to genetics. Under the “Help” tab, located in the menu bar at the top of the page, additional educational resources are available as lecture materials, tutorials, and useful links. The menu bar further provides access to general PharmGKB information (“Home” tab, “PGRN” tab, and “Contributors” tab), more specific search functions (“Search” tab), and data depositing guidelines and templates (“Submit” tab). A scientific curator will respond to questions and comments (“Feedback” tab) within 48 hours.
As shown in Figure 1, the right-hand side of the homepage provides up-to-date pharmacogenetics and pharmacogenomics information. The “Curator’s Favorite Papers” panel consists of recent publications selected from prominent journals and is updated biweekly. These papers are archived, thus providing a useful resource for teaching and for researchers new to the pharmacogenetics and pharmacogenomics fields. The “What’s New” box and the PharmGKB Blog help users to stay informed about current relevant topics and new important data sets and features on PharmGKB.
DRUG, GENE, AND DISEASE RELATIONSHIP
The drug, gene, and disease summary pages are entry points to more detailed information. Entering a drug, gene, or disease in the search box displays a list of hits pertaining to the query, with the option to narrow the search to information with phenotype data, genotype data, and/or literature annotation. Because a text search can hit different types of objects (e.g., genes, drugs, diseases, or literature citations), each hit is labeled as to the type of information it contains. For example, searching for a drug produces a first hit that points to an individual drug page. To provide faster access to the user’s relevant interests, the drug page is structured through a tab system displayed under the drug name (Fig. 2). The “Overview” tab provides generic and trade names for the entered drug. Under the “Properties” tab is information about molecular weight, indications, contraindications, mechanisms of action, and other pharmacokinetics data. Phenotypic primary data from clinical and basic science studies involving the entered drug are summarized under the “Datasets” tab. The “Curated Publications” tab displays a list of related genes and diseases gathered from literature. The pharmacogenetics literature is curated by using the five categories of evidence (CO, PK, PD, FA, and GN) and standardized vocabularies of genes, drugs, and diseases. The COEs and standard vocabularies provide an indexing scheme and highlight drug-gene and drug-disease relationships easily to the user. Genes are cataloged according to the Human Genome Nomenclature Committee terms (HGNC; Wain et al., 2002), and diseases are categorized by using Medline’s “Medical Subject Headings” terms (MeSH terms; Lindberg et al., 1993). Genes and diseases are clickable, leading to similarly organized pages with the tab system for easy navigation. Interactions of genes and drugs are visualized in drug-centered pathways that contain more in-depth knowledge.
Figure 2.
Tamoxifen drug page. Drug pages are structured with a tab system (“Overview,” “Properties,” “Datasets,” “Pathways,” “Curated Publications,” “Non-curated Publications,” and “Downloads/Cross-references”) providing access to different information types. The “Datasets” tab provides curated primary phenotype data sets deposited by the research community. The “Curated Publications” tab links knowledge about drugs with genes and diseases and are similarly structured to the drug pages. The five categories of evidences are used by the curators to characterize the drug-gene and drug-disease relationship. Publication abstracts and PubMed IDs are available through the “View” link.
PharmGKB AND CONSORTIA
A primary mission of the PharmGKB has been to serve as a repository for primary genotype and phenotype data related to pharmacogenetics and pharmacogenomics research from members of the scientific community. There is an increasing interest in creating larger patient cohorts in order to detect smaller effects with greater statistical power. Therefore, PharmGKB has initiated efforts to encourage researchers to pool data (particularly clinical data) in order to create larger data sets for analysis and sharing. These data-sharing consortia then work with PharmGKB curators to focus on a specific pharmacagenomics question. The International Warfarin Pharmacogenetics Consortium (IWPC) is the first such consortium and focuses on the development of a globally relevant warfarin pharmacogenetics dosing algorithm as its first specific research question. Warfarin is an anticoagulant that is difficult to dose and is associated with a set of genes whose variants are known to affect the optimal dose for individual patients. As a result of this collaborative world-wide effort, a new dosing algorithm has been defined that appears to perform better than a clinical-only or fixed-dose approach (IWPC, submitted). Encouraged by the IWPC’s success, PharmGKB has helped form a second consortium, the International Tamoxifen Pharmacogenomics Consortium (ITPC), with a focus on collecting and sharing data sets relating to CYP2D6 genotypes and clinical outcomes in women receiving adjuvant tamoxifen therapy for early breast cancer to gain a better understanding of the relationships between CYP2D6 metabolism status and tamoxifen (http://www.pharmgkb.org/views/project.jsp?pId=63). Tamoxifen, a selective estrogen-receptor modulator, is an antihormonal treatment for breast cancer patients whose tumors express the estrogen receptors and/or the progesterone receptor. It is used in women with metastatic breast cancer and women at high risk of developing breast cancer as a prevention strategy (Jordan, 2008). The cytochrome P450 enzyme, CYP2D6, is one of the major contributors to the metabolism of tamoxifen, especially through its role in the formation of antiestrogenic metabolites endoxifen and 4-hydroxytamoxifen. There are discordant reports relating to the relationship between CYP2D6 genotype and clinical outcomes with tamoxifen. Contrary to initial studies, which found no relationship, more recent studies have demonstrated that genetic variation of CYP2D6 and inhibitors of the enzyme markedly reduce endoxifen plasma concentrations in tamoxifen-treated women. Those studies showed that impaired tamoxifen metabolism results in worse treatment outcomes (Goetz et al., 2007; Schroth et al., 2007). The ITPC is committed to aggregating data on CYP2D6 and other genetic variants involved in the metabolism and effects of tamoxifen with clinical outcomes to address this complex question. This effort will clarify the role of the CYP2D6 genotype as an independent predictor for the treatment outcome of women with breast cancer with tamoxifen treatment. Thus, a priori genetic assessment of CYP2D6 could result in a more favorable tamoxifen treatment regime.
KNOWLEDGE DISPLAY IN PATHWAYS
Several studies have shown that both adverse and beneficial responses to drugs can be influenced by polymorphisms in genes encoding drug-metabolizing enzymes, drug transporters, and drug targets. There are large amounts of data and knowledge about single gene-drug interactions, but to understand the entire drug lifecycle, simultaneous analysis of multiple genetic variants in molecular pathways are important to investigate their effect on drug disposition and action.
PharmGKB pathways are comprehensive maps summarizing prominent genes involved in PK and PD. PharmGKB pathways are drug centered, connecting involved genes with highly curated knowledge and primary data. Pathways about drugs used in chemotherapy of neoplastic diseases, affecting cardiovascular and respiratory functions, acting on the blood and blood-forming organs, affecting the central nervous system and sensory organs, affecting metabolism and gastrointestinal function, and antiviral agents, are currently available. Pathways are created by PharmGKB curators or collaborators who not only help lay out the pathway, but also provide documentation supporting each element of the pathway.
All icons in the pathway diagrams are clickable, linking to the relevant gene and drug pages described above. For gene and drug groups containing more than one item, clicking on the symbol will open a window displaying all involved genes or drugs. A legend explaining the different-shaped and -colored symbols is available at the upper left side of the image. In addition to the pathway diagram, a summary is provided describing the contents of the image. To the right of the pathway diagram, related information is listed, indicating interactions with other drugs and pathways, as well as the option to download the image file and a supporting evidence file with literature references for every arrow in the graphic. The pathways are updated at least every 2 years by the incorporation of newly gained knowledge. PK pathways describe which genes coding for metabolizing enzymes and transporters are involved in drug metabolism and summarize known active and inactive metabolites, with their significantance symbolized by a golden star (Fig. 3A). PD pathways summarize genes involved in signaling cascades of physiological processes in which the drug interacts (Fig. 3B).
Figure 3.
PharmGKB pathway diagrams. (A) The antiestrogen pathway: PK describes the hepatic metabolism of the drug tamoxifen (purple rectangle) to several primary and secondary metabolites (purple-yellow rectangles) by the cytochrome P450 (CYP) enzymes. Like tamoxifen, some of these metabolites exhibit antiestrogenic effects, indicated by arrows to the breast cancer cell. Along the arrows, the genes (blue ovals) are displayed, which code for the involved CYP isoforms. In the transformation to the potent antiestrogenic metabolite, 4-hydroxy-tamoxifen, CYP2D6 plays a significant role, indicated by the star. A summary explaining the different shapes and colors used in the graphic is available through the “legend” link. (B) The antiestrogen pathway: PD displays genes (blue ovals) involved in the synthesis of estrogens (green ovals) and the effect of estrogens on downstream signals by binding to estrogen receptors. Aromatase inhibitors (purple rectangle) are a group of drugs that reduce the amount of estrogens by blocking their transformation from androgens, whereas selective estrogen-receptor modulators (SERM) decrease the cellular effects of estrogens by binding to estrogen receptors in breast cells. The two drug groups are major pharmacological strategies in breast cancer therapy of estrogen receptor and/or progesterone receptor–positive breast tumors.
VERY IMPORTANT PHARMACOGENE (VIP) SUMMARIES
VIP genes are summaries of genes that are important for pharmacogenomics, based on their involvement in the pharmacodynamics and pharmacokinetics of one or more drugs. The PharmGKB team curates these structured summaries—often in collaboration with PGRN members and the scientific community. Currently, there are 36 VIP summaries, 26 of which are also part of the pathway diagrams. VIP summaries are accessible by using the VIP icon on the homepage. These summaries are also integrated in the gene pages. PharmGKB curators designate VIP genes based on the amount and quality of data they uncover during their review of the literature indicating the role of a gene in pharmacogenomics.
A VIP annotation consists of an introduction page, an important variants of functional significance page, and, if applicable, haplotype and splice variant pages. All pages provide direct links to cited literature and a list of relevant drugs and phenotypes associated with the pharmacogene. The Overview page displays information about the gene and its endogenous and pharmacological roles. The Important Variants page lists significant polymorphisms of the gene (Fig. 4). For each variant, a summary informs the user about the effect the variant has on the gene product and its association with diseases and drug response. For unambiguous identification, information about the nucleotide change and position for genomic DNA and the human genome University of California at Santa Cruz (UCSC) Golden Path position (Karolchik et al., 2008) are provided. This mapping allows a summarization of the different variant names found in the literature to connect key studies for further comparison. The human genome UCSC Golden Path position is used to determine the dbSNP rsID, if it is not provided in the literature source. If the polymorphism is located in an exon, the nucleotide and amino-acid change and position for mRNA and protein reference sequences are provided. If haplotypes have been defined for the gene in the literature, a haplotype page is presented. Short summaries for each haplotype detail the individual variants that characterize the haplotype and describe if the combined effect of multiple polymorphisms differs from the effect of single variants.
Figure 4.
CYP2D6 Very Important Pharmacogene (VIP) gene. The CYP2D6 VIP contains a gene summary about CYP2D6 and its most important variants and haplotypes. CYP2D6 is an enzyme of the hepatic cytochrome P450 system and metabolizes many drugs as substrates. Throughout the introductory text, links to specific research articles are available that support the claims being made in the sentences. The “Important Variants” link opens the variant information page for CYP2D6, which contains 10 important polymorphisms. Each of them has a concise section containing a summary and an unambiguous identification of the variant by position and accession number for genomic DNA, mRNA, and protein. Also included are the dbSNP rsID and the UCSC Golden Path number.
VARIANT BROWSER
Highlighting the relationship between genes, diseases, and drugs is integral to the mission of the PharmGKB. As high-throughput DNA microarray and sequencing technologies continue to improve and become more available, researchers have access to rapidly growing amounts of genotype and phenotype data. The “Variants” feature of the PharmGKB showcases information on genetic variants and presents publication-supported relationships between specific variants and associated phenotypes. Variant information is organized for both ease of use and maximum information content. The PharmGKB currently contains over 370 variant annotations in 176 genes. These numbers are increasing rapidly as PharmGKB curators continue to create new variant entries.
There are several access points to the “Variants” feature. One can click on the “variants of interest” icon found on the PharmGKB homepage to browse through a list of genes for which there exist annotated variants, or one can search for variants in a particular gene by entering that gene’s name in the “Search PharmGKB” box. Entering a dbSNP rsID as a search term will also take the user to the gene in which that variant is located.
Figure 5 depicts a typical Variants page. In this example, variants in the ABCB1 gene are graphically represented in a UCSC Genome Browser–like format. Tick marks designating the locations of SNPs submitted as genotyping data to the PharmGKB are displayed on the PharmGKB labeled track. The height of each tick mark reflects the frequency of that variant. Introns, exons, untranslated regions, promoters, and flanking regions are color-coded to facilitate visualization of these features. The Golden Path track shows the location of gene exons from the UCSC Genome Browser assembly (Karolchik et al., 2008; Kent et al., 2002). SNPs found on a custom Illumina 317 SNP array microchip are indicated on the PharmGKB Illumina track (Hernandez-Boussard et al., 2008). Finally, SNPs in this region from the dbSNP and JSNP databases are represented on the dbSNP (Sherry et al., 2001) and JSNP (Hirakawa et al., 2002) tracks, respectively.
Figure 5.
Screenshot of the ABCB1 Variants page from the PharmGKB Web site (retrieved May 30, 2008, from http://www.pharmgkb.org/do/serve?ojbCls=Gene&objId=PA267&tabType=12). The rectangular box in the upper portion of the figure shows the genomic region containing the ABCB1 gene, with single-nucleotide polymorphism (SNP) locations represented as tick marks. The lower section of the figure is a table displaying ABCB1 variants, for which genotyping information has been submitted to PharmGKB. For each variant, the Golden Path position provides unambiguous polymorphism positioning information. The dbSNP rsID, the nucleotide(s) observed at the polymorphic position, the genetic feature in which the polymorphism occurs, and amino-acid translations of codons containing polymorphic nucleotides are listed. Additionally, two- and three-star designations reflect that the variant has been curated to supply brief (two-star) or detailed (three-star) descriptions of phenotype(s) observed for that variant. (Note: To include more curated variants in this image, some noncurated variants are not shown, as indicated by the white area between the second and third variants listed.).
Importantly, the Variants display table located beneath the graphical SNP distribution image includes the Golden Path position of each variant to unambiguously pinpoint its location in the human genome relative to a reference sequence and, if known, the dbSNP rsID is listed. This standardized, precise localization of variants eliminates uncertainty in variant position that is sometimes encountered when various variant notations are employed. Additionally, the nucleotides observed at each variant location are listed, and the genomic context of each variant is indicated (e.g., exon, intron, etc.), as well as the amino acids found in variant codons. By clicking on a variant’s Golden Path position, dbSNP rsID, or polymorphic nucleotide, the user is routed to the UCSC Genome Browser, to dbSNP, or to the PharmGKB allele frequency information for that variant, respectively.
The PharmGKB has recently introduced a feature to highlight variants with phenotypic consequences. Variants with an entry in the “Variant of Interest Curation Level” column are supplemented with phenotype information from the literature and from data submissions to the PharmGKB. Three-star (***) annotations indicate in-depth variant curation, with a phenotype summary, literature references, and allele frequency data. Clicking on the “view” link adjacent to the “***” symbol brings up the three-star variant’s “Annotations” tab. One can then access detailed variant information by clicking on the URL provided. This information is the same “Important Variants Information” included in the VIP feature discussed above. Two-star (**) annotations reflect shorter phenotype summaries with accompanying literature references. To access variant annotation content, one can either click on the “view” link, or simply position the mouse pointer over the “view” link to display the variant phenotype summary. One-star (*) annotations are used for those that are generated by computer programs and have not been reviewed by curators.
The PharmGKB Variant Browser serves users in multiple ways. Variants are easy to locate in a gene, and the nucleotides observed at the polymorphic position by data submitters—as well as the amino acids encoded by polymorphic codons—are readily identifiable. This data, used in conjunction with the two- and three-star variant annotations, can serve as the basis for selecting variants to genotype in pharmacogenetics and pharmacogenomics studies.
AUTOMATED LITERATURE CURATION
The PharmGKB initially provided only human-curated and annotated literature on pharmacogenetics and pharmacogenomics. “Non-curated Publications” is a new feature consisting of pharmacogenetics literature collected through automated computer algorithms. In particular, MScanner is a supervised learning-based classifier that retrieves relevant literature automatically using journal titles, and Medical Subject Headings (MeSH; Poulter et al., 2008). If one clicks on “Non-curated Publications,” and then on the “Details” link position after the article title, followed by “Marked-up Text,” the screen displays sentences from automatically retrieved articles with drug names, variant names, genes and gene products, and disease names highlighted and color-coded for each category, using a method based upon Pharmspresso (Fig. 6; Garten and Altman, submitted). The combination of automated literature curation and marked-up text serves as an additional pipeline to provide users more rapid access to the literature. Of course, the computer algorithms can introduce noise, including incorrect annotations, and the curators work to review automated annotations and “upgrade” them to two stars (**).
Figure 6.
Screenshot of marked-up text generated through an automated literature curation (retrieved July 21, 2008, from http://www.pharmgkb.org/do/serve?objId=PA161796011&objCls=AutomaticAnnotation#tabview=tab1). Genes are highlighted blue, diseases are highlighted green, drugs are highlighted red, variants are highlighted orange, and indeterminate terms are highlighted black.
MAPPING OF CODING SNPS ONTO 3D PROTEIN STRUCTURES
Nonsynonymous polymorphisms in coding exons may occur that affect structure-function relationships. With the development of high-throughput sequencing technology, researchers are able to identify new SNPs before characterizing their functional relevance. Mapping coding SNPs onto 3D protein structures enables researchers to visualize the macromolecular context of the variants. Initial efforts at mapping coding SNPs onto 3D protein structures use X-ray and nuclear magnetic resonance (NMR) structures stored in the Protein Data Bank (RCSB PDB; Berman et al., 2003). PharmGKB uses the MBT Protein Workshop software from the PDB for structure viewing over the Web. Structures mapped with variant location, when available, can be accessed from the “Variants” tab. Annotated structures are displayed as they are curated. Figure 7 shows the PDB structure for angiotensin I converting enzyme (ACE) with the locations of coding SNPs highlighted in red.
Figure 7.
Display of coding single-nucleotide polymorphisms (SNPs) mapped onto three-dimensional (3D) protein structure: (retrieved August 13, 2008, from “View Coding Variants on Protein Structure” link on https://preview.pharmgkb.org/do/serve?objId=PA139&objC1s=Gene#tabview=tab2). The coding SNPs for the angiotensin I converting enzyme (ACE) stored in PharmGKB are mapped onto the 3D protein structure of the ACE, somatic isoform stored in the Protein Data Bank (RCSB PDB; http://www.rcsb.org/pdb/explore/explore.do?structureId=2C6F). PharmGKB uses the MBT Protein Workshop software from the PDB for structure viewing. Variant locations are displayed in red.
CONCLUSIONS
Bridging the fields of pharmacology, genetics, and medicine, the PharmGKB provides diverse researchers access to pharmacogenomics research results and discoveries. VIP summaries, pathway diagrams, variant annotations, literature annotations, and other features provide high-level summaries of knowledge that can be used to design new studies. The PharmGKB can also be used as an educational tool to teach clinical and basic science students about pharmacogenetics (Owen et al., 2007). All primary data submitted to PharmGKB by researchers are available to view and download. Recently, PharmGKB has taken on an exciting new role as an independent broker for data-sharing consortia focused on answering important pharmacogenomics questions. Results of the first consortium focused on warfarin has yielded a warfarin-dosing algorithm, based on studies of thousands of patients (IWPC, submitted). A new consortium is addressing the pharmacogenetics of tamoxifen, thus continuing PharmGKB’s involvement in multicenter, international studies.
ACKNOWLEDGMENTS
This work was supported by the NIH/NIGMS Pharmacogenetics Research Network (PGRN) and Database (U01GM61374). The authors thank the PGRN Publications Committee for their critical reading and helpful comments on this manuscript.
Footnotes
Full terms and conditions of use: http://www.informaworld.com/terms-and-conditions-of-access.pdf
This article may be used for research, teaching and private study purposes. Any substantial or systematic reproduction, re-distribution, re-selling, loan or sub-licensing, systematic supply or distribution in any form to anyone is expressly forbidden.
Publisher's Disclaimer: The publisher does not give any warranty express or implied or make any representation that the contents will be complete or accurate or up to date. The accuracy of any instructions, formulae and drug doses should be independently verified with primary sources. The publisher shall not be liable for any loss, actions, claims, proceedings, demand or costs or damages whatsoever or howsoever caused arising directly or indirectly in connection with or arising out of the use of this material.
Declaration of Interest: The authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Contributor Information
Katrin Sangkuhl, Department of Genetics, Stanford University, Stanford, California, USA.
Dorit S. Berlin, Department of Genetics, Stanford University, Stanford, California, USA
Russ B. Altman, Department of Genetics, Stanford University, Stanford, California, USA Department of Bioengineering, Stanford University, Stanford, California, USA.
Teri E. Klein, Department of Genetics, Stanford University, Stanford, California, USA
REFERENCES
- Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003;10(12):980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- Garten Y, Altman RB. Pharmspresso: a text mining tool for extraction of pharmacogenetics concepts and relationships from full text. 2008 AMIA Translational Bioinformatics Summit. 2008 doi: 10.1186/1471-2105-10-S2-S6. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomini KM, Brett CM, Altman RB, Benowitz NL, Dolan ME, Flockhart DA, et al. The pharmacogenetics research network: from SNP discovery to clinical drug response. Clin. Pharmacol. Ther. 2007;81(3):328–345. doi: 10.1038/sj.clpt.6100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz MP, Knox SK, Suman VJ, Rae JM, Safgren SL, Ames MM, et al. The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res. Treat. 2007;101(1):113–121. doi: 10.1007/s10549-006-9428-0. [DOI] [PubMed] [Google Scholar]
- Hernandez-Boussard T, Whirl-Carrillo M, Hebert JM, Gong L, Owen R, Gong M, et al. The pharmacogenetics and pharmacogenomics knowledge base: accentuating the knowledge. Nucleic Acids Res. 2008;36:D913–D918. doi: 10.1093/nar/gkm1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa M, Tanaka T, Hashimoto Y, Kuroda M, Takagi T, Nakamura Y. Nucleic Acids Res. 2002;30(1):158–162. doi: 10.1093/nar/30.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge AE, Altman RB, Klein TE. The PharmGKB: integration, aggregation, and annotation of pharmacogenomic data and knowledge. Clin. Pharmacol. Ther. 2007;81(1):21–24. doi: 10.1038/sj.clpt.6100048. [DOI] [PubMed] [Google Scholar]
- Jordan VC. Tamoxifen: catalyst for the change to targeted therapy. Eur. J. Cancer. 2008;44(1):30–38. doi: 10.1016/j.ejca.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, et al. The UCSC genome browser database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Gen. Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Wafarin Pharmacogenetics Consortium. Global Warfarin Pharmacogenetic Dosing Algorithm. 2008 Submitted. [Google Scholar]
- Lindberg DA, Humphreys BL, McCray AT. The Unified Medical Language System. Meth. Inf. Med. 1993;32:281–291. doi: 10.1055/s-0038-1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen RP, Altman RB, Klein TE. PharmGKB and the International Warfarin Pharmacogenetics Consortium: the changing role for pharmacogenomic databases and single-drug pharmacogenetics. Hum. Mutat. 2008;29(4):456–460. doi: 10.1002/humu.20731. [DOI] [PubMed] [Google Scholar]
- Owen RP, Klein TE, Altman RB. The education potential of the pharmacogenetics and pharmacogenomics knowledge base (PharmGKB) Clin. Pharmacol. Ther. 2007;82(4):472–475. doi: 10.1038/sj.clpt.6100332. [DOI] [PubMed] [Google Scholar]
- Poulter GM, Rubin DL, Altman RB, Seoighe C. MScanner: a classifier for retrieving Medline citations. BMC Bioinform. 2008;9:108. doi: 10.1186/1471-2105-9-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroth W, Antoniadou L, Fritz P, Schwab M, Muerdter T, Zanger UM, et al. Breast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypes. J. Clin. Oncol. 2007;25(33):5187–5193. doi: 10.1200/JCO.2007.12.2705. [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wain HM, Bruford EA, Lovering RC, Lush MJ, Wright MW, Povey S. Guidelines for human gene nomenclature. Genomics. 2002;79(4):464–470. doi: 10.1006/geno.2002.6748. [DOI] [PubMed] [Google Scholar]