Summary
Arealization of the neocortex is controlled by a regulatory hierarchy beginning with morphogens secreted from patterning centers positioned at the perimeter of the dorsal telencephalon. These morphogens establish within cortical progenitors the differential expression of transcription factors that specify their area identity, which is inherited by their neuronal progeny, providing the genetic framework for area patterning. The two patterning centers most directly implicated in arealization are the commissural plate, which expresses Fibroblast growth factors, and the cortical hem, which expresses Bone morphogenetic proteins and vertebrate orthologs of Drosophila wingless, the Wnts. A third, albeit putative, patterning center is the antihem, identified by its expression of multiple signaling molecules. We describe recent findings on roles for these patterning centers in arealization. We also present the most recent evidence on functions of the four transcription factors, Emx2, COUP-TFI, Pax6, and Sp8, thus far implicated in arealization. We also describe screens for candidate target genes of these transcription factors, or other genes potentially involved in arealization. We conclude with an assessment of a forward genetics approach for identifying genes involved in area patterning, based in part on quantitative trait locus mapping and the implications for significant differences between individuals in area size on behavioral performance.
Keywords: area specification, cortical development, cortical hem, Emx2, Coup-TFI, morphogens, neuronal specification, quantitative trait locus mapping, transcription factors
Introduction
The cerebral cortex, a brain component unique to mammals, arises from the dorsal telencephalon (dTel). The cerebral cortex is divided into regions, with the largest region, the neocortex positioned between two other regions, the archicortex (midline cortex and hippocampus) and paleocortex (olfactory piriform cortex). Among the many features that distinguish the neocortex from other regions is its laminar patterning into six major, radially organized, layers that are morphologically and connectionally distinct. In its tangential dimension, the neocortex is organized into “areas” that are functionally unique subdivisions distinguished by differences in cytoarchitecture and chemoarchitecture, input and output connections, and patterns of gene expression.
Determining the mechanisms that control the development of cortical areas, a process termed arealization, is a major issue in neurobiology that has attracted the attention and imagination of many investigators, particularly in the past decade [1–5]. Proper area patterning of the neocortex is a critical developmental event, because neocortical areas form the basis for sensory perception, control of our movements, and mediate our behavior. Many features must be properly specified during arealization—not only the unique properties that determine an area’s function and interaction with other neural structures, but also the appropriate size.
The specification and differentiation of neocortical areas is controlled by an interplay between genetic regulation intrinsic to the neocortex --characterized by transcription factors (TFs) expressed by cortical progenitors and morphogens expressed by telencephalic patterning centers --and extrinsic influences such as thalamocortical axon (TCA) input that relays in an area-specific fashion sensory information from the principal sensory nuclei of dorsal thalamus to the primary cortical areas (Figure 1). Although of undeniable importance, surprisingly little is known about the mechanisms that control arealization, and most of what we know is recent. For instance, direct evidence for the intrinsic genetic control of the area identities of cortical progenitors was first reported early in this decade [6,7]. Here we describe recent major findings most directly relevant to neocortical arealization, focusing on genetic regulation intrinsic to the neocortex. Findings in the past year have substantially expanded our understanding of this process, but at the same time they have called into question the precise role of some players.
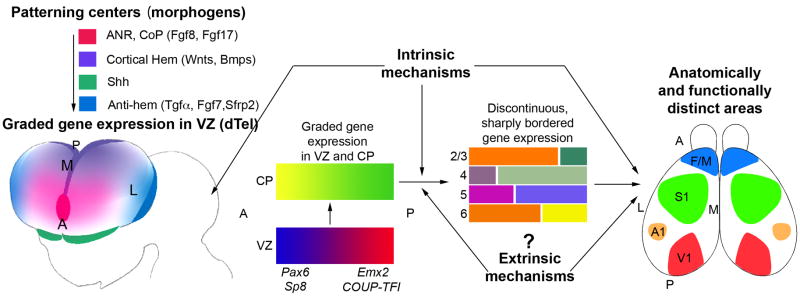
Figure 1. Patterning centers and graded transcription factors drive arealization of the neocortex.
The initial, tangential gradients of transcription factors (TFs) in the ventricular zone (VZ) are established by signaling molecules/morphogens secreted from telencephalic patterning centers, such as Fgf8 and Fgf17 from anterior neural ridge (ANR), which later becomes the commissural plate (CoP), and Wnts and BMPs from the cortical hem. The antihem is a putative patterning center identified based on its expression of secreted signaling molecules (e.g. Tgfα, Fgf7, Sfrp2, as well as Neurogulin 1 and 3) with known patterning functions. A fourth telencephalic patterning center is defined by the expression domains of sonic hedgehog (Shh) in ventral telencephalon, but it does not have defined roles in dorsal telencephalic (dTel) patterning. The graded expression of certain TFs, such as Pax6, Emx2, COUP-TFI and Sp8, imparts positional or area identities to cortical progenitors which is imparted to their neuronal progeny that form the cortical plate (CP). The CP also initially exhibits gradients of gene expression that are gradually converted to distinct patterns with sharp borders. Coincident with this process, distinct cortical layers (2–6), and the anatomically and functionally distinct areas seen in the adult (M1, S1, A1, V1), differentiate from the CP. Genes that are differentially expressed across the cortex are often expressed in different patterns in different layers, suggesting that area-specific regulation of such genes is modulated by layer-specific properties, and questions the definition of area identity. Although the initial establishment of the graded gene expression in the embryonic CP is controlled by mechanisms intrinsic to the telencephalon, the more complex differentiation patterns established postnatally might be controlled in part by extrinsic mechanisms, for example, TCA input and the sensory activity that it relays from the periphery to the cortex. The figure is modified from [64].
Neocortex primer
The neocortex has four “primary” areas; each is the cornerstone of clusters of functionally related areas that include scores of higher order areas that are prominently interconnected. Three of the primary areas are sensory: the primary visual (V1), somatosensory (S1) and auditory (A1) areas, which process primary information received from the eye/retina (vision), body (somatosensation), and inner ear/cochlea (audition), respectively. The fourth primary area is motor (M1), which controls voluntary movements. Each primary cortical area receives TCA input from a specific principal dorsal thalamic nucleus. These nuclei receive modality specific sensory information directly or indirectly from peripheral sense organs or receptors, and in turn define the functional modality of their target primary area.
In mice, the predominant model for genetic studies of cortical development, neocortical neurons are generated predominantly between embryonic days 10 and 17. Most neocortical neurons are glutamategic, including all projection neurons, and are generated by progenitors in the ventricular zone (VZ) of dTel, and later, a second germinal zone, the subventricular zone (SVZ) positioned immediately above the VZ. The VZ generates deep layer neurons, including subplate and layer 5 and 6 projection neurons, whereas the SVZ is a prominent source of neurons that form the superficial layers 2, 3 and 4 [8]. In primates, relative to the VZ, the SVZ is substantially larger, and locally enhanced proliferation in posterior occipital cortex has been reported to contribute to the major increase in the numbers of superficial layer neurons in V1 compared to adjacent higher order visual areas (e.g. V2), thereby contributing to arealization [9]. Approximately 20% of all cortical neurons are GABAergic interneurons that are generated primarily in the medial and caudal ganglionic eminences of ventral telencepahlon (vTel) and migrate along multiple pathways to reach the cortex [10,11]. Cajal-Retzius neurons, a third general category of cortical neurons, populate the MZ (layer 1) and express Reelin, a large secreted protein long thought to be required to establish appropriate cortical layering by influencing the radial migration and settling patterns of cortical neurons [12]. Cajal-Retzius neurons are also generated external to the cortical VZ, primarily within the cortical hem and also in the subpallium and septum [13,14].
Telencephalic Patterning Centers in arealization
Arealization is controlled by a regulatory hierarchy beginning with morphogens secreted from patterning centers positioned at the perimeter of dTel, which establish within cortical progenitors the differential expression of TFs that determine their area identity and that inherited by their neuronal progeny that form the CP (Figure 1). Four telencephalic patterning centers appear to be involved directly or indirectly in cortical patterning, as well as in regionalization of the telencepahlon and/or internal patterning within other regions of the telencephalon. The two patterning centers most directly implicated in arealization are the commissural plate (CoP), which expresses Fibroblast growth factors (Fgfs), and the cortical hem, which expresses Bone morphogenetic proteins (Bmps) and vertebrate orthologs of Drosophila wingless referred to as Wnts.
A third, albeit putative, patterning center is the antihem, identified by its expression of multiple signaling molecules, including Tgfα, Neuregulin1, Neuregulin3, Fgf7 and the Wnt antagonist, secreted frizzled related protein Sfrp2 [15]. The antihem is located in the neuroepithelium near the boundary between ventro-lateral neocortex and the LGE of vTel, and forms a narrow stripe of expression extending along the entire anterior-posterior (A–P) axis of the telencephalon. The cortical hem and antihem have been suggested to cooperate with the CoP to establish identities along the A-P and medial-lateral (M–L) axes of the developing cortex. Although no function has been defined for the antihem, it is essentially absent in small eye mutant mice, which lack functional Pax6 protein, and therefore some of the major defects in telencephalic patterning observed in small eye mutants might be due to the loss of antihem function [15].
Finally, large contiguous domains of Sonic hedgehog (Shh) expression are located in vTel and the hypothalamus of ventral diencephalon [16]. Shh secreted by this patterning center has been implicated in regional patterning of the forebrain [17–21]. However, new studies have led to the proposal that Shh is not involved in dTel patterning, and that the telencephalic phenotypes in mice with a targeted deletion of Gli3, which encodes a zinc finger TF that mediates Shh signaling, occur through a Shh-independent mechanism [21].
In the following sections, we summarize recent findings of roles for these patterning centers in arealization, as well as the four TFs, Emx2, Pax6, COUP-Tf1, and Sp8, which are expressed by cortical progenitors and have been directly implicated in arealization.
Commissural Plate: an anterior patterning center
The anterior neural ridge (ANR), which is the anterior junction between neural and nonneural ectoderm, and later through morphogenesis becomes the CoP, formed by fusion of the neural plate folds at the anterior margin of the forebrain, is an anterior patterning center for arealization (Figure 1) [22]. The ANR/CoP is prominently defined by the overlapping expression domains of Fgf8, 17, and 18. Of these, Fgf8, and to a lesser degree Fgf17, have been most studied in arealization. They locally induce members of the ETS family of TFs and establish the gradients of Emx2 and COUP-TFI within cortical progenitors by repressing their expression anteriorly in a dose-dependent fashion [23,24]. Altering levels of Fgf8 or 17 has substantial affects on area patterning, presumably indirectly through their repression of Emx2, COUP-TFI and other TFs expressed by cortical progenitors [23–25]. Recent studies though show that Fgf8 and Fgf17 have distinct roles in the patterning of dorsal versus ventral frontal cortical areas: whereas Fgf8 controls the size of both dorsal frontal cortex and ventral/orbital frontal cortex, Fgf17 selectively controls the size of dorsal frontal cortex [26].
Cortical hem: a dorsal/caudal patterning center
The cortical hem is neuroepithelial tissue adjacent to the dorsal midline in the medial cortical wall, defined by its expression of multiple Bmps and Wnts [17,27] (Figure 1). The distribution and timing of Bmp/Wnt expression in the cortical hem and their receptors in the cortex suggest that the cortical hem is involved in cortical patterning (e.g. [28]). However, by comparison to the CoP, the function of the cortical hem in neocortical arealization has not been clearly defined. Genetic ablation of the cortical hem has been done using the Wnt3a locus to drive expression of the diptheria toxin A chain [13]. This ablation results in a substantial loss of Cajal-Retzius neurons, but surprisingly neither the loss of these neurons, and thereby the predominant source of Reelin in the cortical MZ, or the morphogens associated with the hem, has a significant effect on arealization or other aspects of cortical patterning including the development of cortical lamination [13], believed to be controlled by Reelin [12].
The Lhx class of Lim homedomain proteins has been implicated in controlling development of the cortical hem. Targeted deletion of Lhx5, which is expressed in the cortical hem, leads to loss of choroid plexus and cortical hem, and impaired development of the hippocampal formation [29]. Lhx2 is expressed in the cortical VZ in a high-to-low posterior-medial to anterior-lateral gradient, and exhibits an abrupt decline in its expression posterior-medially, excluding it from the cortical hem, through a repression by Bmp2 and Bmp4 expressed in the roof plate [30]. In Lhx2 knockout mice, the lack of the normally high expression of Lhx2 in medial cortex adjacent to the cortical hem results in a dramatic expansion of the hem, whereas in contrast the neocortex is dramatically reduced in size and proliferation prematurely ceases [30–32]. These findings show that establishing the boundary between the cortical hem and the adjacent cortical VZ, and their respective fates, requires the action of Lhx2. However, addressing roles for Lhx2 in arealization was not possible because the constitutive Lhx2 knockout mice die early in embryonic cortical development, and cortical development is suppressed.
However, roles for Lhx2 in dTel patterning have been substantially advanced by recent elegant use of a conditional knockout of Lhx2 and genetic mosaics in chimeric mice comprised of Lhx2 null and wild type cells [33]. These studies provide further evidence that Lhx2 specifies in a cell-autonomous fashion cortical identity and acts to suppress hem fates in medial cortex, and in a complementary fashion, to suppress antihem fates in lateral cortex. These studies demonstrate that Lhx2 is classic selector gene in regional fate determination within dTel, being required to define the regional fates of dTel, and further, that the cortical hem is a hippocampal organizer [33].
Transcription factors that specify area identities of cortical progenitors
The telencephalic patterning centers described above in principle have the capacity to interact; for example, morphogens secreted by one patterning center can repress the expression of those expressed by another center (for review see [3,4]). In addition, morphogens secreted by the CoP and cortical hem have prominent roles in establishing the graded expression of TFs in progenitors in the cortical VZ. These TFs meet the basic criteria required for candidate genes that specify area identities of cortical progenitors in that they are regulatory genes that are differentially expressed across the A–P and M–L cortical axes by progenitors. These properties suggest that these TFs also function in a differential manner across the cortical axes, which is required to impart area identities, but in addition to differential expression, this property could be achieved by the expression of co-factors or other mechanisms that differentially influence TF function. To date, four TFs, Emx2, Pax6, COUP-TFI and Sp8, have been reported to be expressed by cortical progenitors and have a direct role in arealization. The expression patterns for these four TFs and summaries of their phenotypes in genetically-engineered mice are shown in Figure 2. Below we summarize these data, and in Figure 3 we present our current view of the roles and interactions between these TFs in regulating area patterning of the A–P cortical axis.
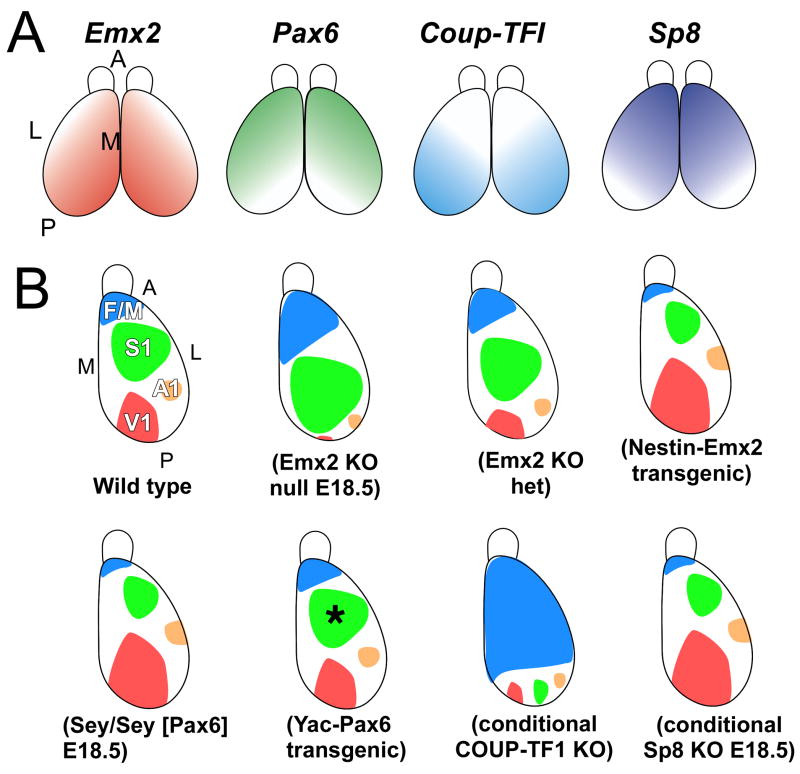
Figure 2. Summary of graded expression of transcription factors implicated in arealization and findings in mouse mutants.
(A) Graded expression in cortical progenitors of the transcription factors directly implicated in arealization, Emx2, Pax6, Coup-TFI, and Sp8, along the anterior-posterior (A–P) and lateral-medial (L–M) axes of the cortex. (B) Summary of reports of loss- or gain-of-function mutant mice of TFs that exhibit changes in area patterning. Mice with a targeted deletion of Emx2 die at birth, but late embryonic analyses suggest substantial changes in arealization as indicated in the cartoon, with a reduction in posterior areas and an expansion and posterior shift of anterior areas. Reducing Emx2 levels in the cortex of the heterozygote mutant mice (Emx2 KO het) results in posterior shifts of areas with shrinkage of V1, while overexpression of Emx2 under the control of nestin promoter (Nestin-Emx2 Transgenic) shifts areas anteriorly. Small eye mutant mice, which lack functional Pax6 protein, die at birth, but marker analyses suggest a reduction in anterior areas and an expansion and anterior shift of posterior areas. However, YAC transgenic mice of Pax6 do not show area changes other than a slight, but significant, reduction in the size of S1 (asterisk). Selective deletion of COUP-TFI in conditional knockout mice crossed with an Emx1-Cre line results in a massive expansion of frontal/motor areas and a substantial reduction of the primary sensory areas that shift posteriorly to the posterior cortical margin. Analyses of conditional knockout mice of Sp8 crossed to a BF1 (Foxg1) Cre line shows at late embryonic ages anterior shifts of gene markers, a phenotype similar to that reported for Fgf8 hypomorphic mice. The BF1-Cre line deletes Sp8 not only from cortical progenitors but also from the CoP, resulting in diminished expression of Fgf8 in the CoP. See text for details and references.
Figure 3. Roles and interactions between transcription factors that control arealization of the neocortex.
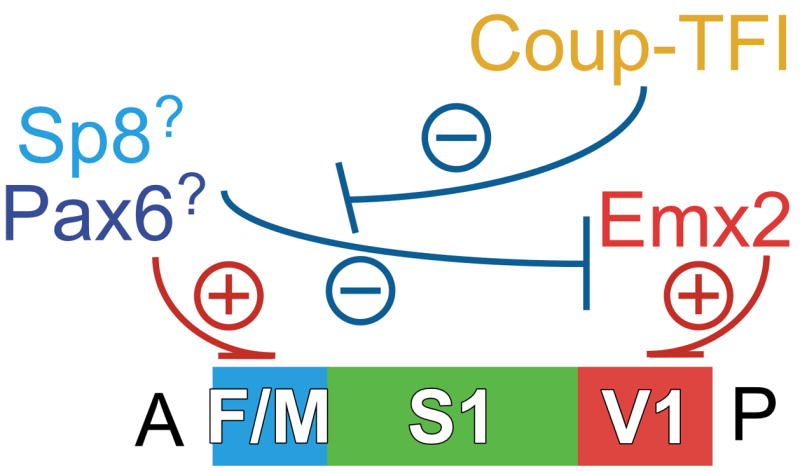
Sp8 and Pax6 have been implicated in preferentially specifying in cortical progenitors and their progeny the identities of frontal/motor (F/M) areas, although as discussed in the text, their roles require further validation. Emx2 preferentially specifies in cortical progenitors the identities of posterior (P)/sensory (e.g. V1) areas. Coup-Tf1 represses within its more robust expression domain, the phenotypic function of any TF that may specify F/M area identities, e.g. Pax6 and Sp8 and any other TF to be identified, thereby limiting their action to anterior (A) cortical progenitors that specify F/M area identities. We also suggest based on current evidence that TFs that specify F/M area identities are dominant over the TFs that specify caudal/sensory areas and can phenotypically repress their function.
Roles for Emx2 in arealization have been the most studied for any TF. Emx2, a homeodomain TF related to Drosophila empty spiracles (ems), is expressed highest in progenitors that generate posterior-medial areas of neocortex, such as V1, and lowest in progenitors that generate anterior-lateral areas, such as frontal and motor [34]. The initial studies, and the first to show a role for TFs in area patterning, were loss-of-function performed on Emx2 constitutive knockout mice [6,7]. Emx2 knockout mice die at birth, well before cortical areas differentiate, limiting these studies to marker analyses and patterning of area-specific TCA projections. However, subsequent analyses of nestin-Emx2 transgenic mice, which use nestin promoter elements to drive elevated levels of Emx2 expression limited to progenitors, and of heterozygous Emx2 constitutive knockout mice, at postnatal ages after areas emerge provide a more complete picture of roles for Emx2 in arealization [35]. These genetic manipulations that change the levels of Emx2 expression in cortical progenitors result in disproportionate changes in the sizes of the primary sensory and frontal/motor cortical areas, but have no effect on overall cortical size [35]. They also show that Emx2 operates by a concentration-dependent mechanism in cortical progenitors to specify disproportionately the sizes and positioning of the primary cortical areas, and that higher levels of Emx2 preferentially impart posterior-medial area identities, such as those associated with V1. These findings led to the “Cooperative Concentration Model” that the same set of TFs is expressed by progenitors across the entire cortex and cooperate to control arealization, and importantly, the level of expression of an individual TF such as Emx2, is a defining parameter that specifies the area identity of a cortical progenitor and its progeny [35].
Recent genetic rescue studies done by crossing the nestin-Emx2 mice, which have about a 50% increase in Emx2 expression in cortical progenitors, with Emx2 heterozygous knockout mice, which have about a 50% reduction in Emx2 expression, have validated that Emx2 controls arealization and that the levels of Emx2 expression are a critical parameter [36]. In the progeny from this cross, both Emx2 expression in cortical progenitors, as well as the size and positioning of cortical areas, are restored to wild type.
COUP-TFI is an orphan nuclear receptor expressed in a high posterior-lateral to low anterior-medial expression gradient by both progenitors and CP neurons. The initial evidence of a role for COUP-TFI in arealization came from studies of constitutive null mice, but again analyses were limited because most of the mice die within a few days after birth, and the majority of TCAs fail to reach the cortex [37]. However, these complications have been overcome by the recent analyses of conditional COUP-TFI knockout mice in which COUP-TF1 is selectively deleted from cortex at E10 by crosses to an Emx1-Cre line [38]. Cortical deletion of COUP-TFI results in a massive expansion of frontal/motor areas to occupy most of parietal and occipital cortex, which in wild type mice are occupied by somatosensory and visual areas, [39] respectively (Figure 4). This expansion of frontal/motor areas is paralleled by a substantial reduction in the sizes of the three primary sensory areas, which are compressed to the caudal pole of the cortical hemisphere. Thus, COUP-TFI is required to balance the patterning of neocortex into frontal/motor areas and sensory areas [38]. These findings suggest that COUP-TFI functions predominantly by repressing the identities of frontal/motor cortical areas within its expression domain in parietal and occipital cortex, allowing for the appropriate specification of the sensory cortical areas and limiting frontal/motor areas to their anterior domain that has very low levels of COUP-TFI expression.
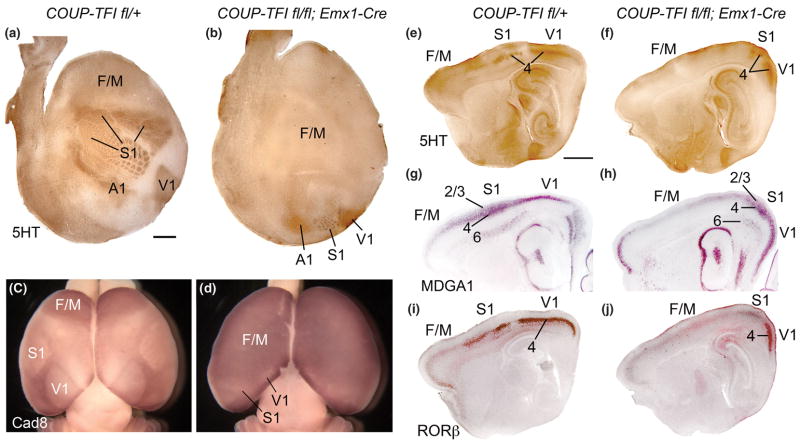
Figure 4. Selective deletion of COUP-TFI from cortex results in massive expansion of frontal/motor areas and posterior compression of primary sensory areas.
Findings from [38] showing a prominent role for COUP-TFI in arealization. (A,B) Serotonin (5HT) immunostaining on tangential sections through layer IV of flattened cortices of P7 control (COUP-TFIfl/+) and conditional mutant (fl/fl; Emx1-Cre) cortices. Anterior is to left, and medial to the top. (A) Serotonin staining reveals primary sensory areas, including primary somatosensory (S1), visual (V1) and auditory (A1) areas, by marking area-specific TCA axon terminations. (B) In COUP-TFI fl/fl; Emx1-Cre conditional mutant brains, the primary sensory areas are much smaller than in controls and are compressed to ectopic positions at the posterior pole of the cortical hemisphere. The barrelfield of the ectopic S1 retains its characteristic patterning but is substantially reduced in size and caudally shifted, while a reduced V1 is located medial and a reduced A1 lateral to the miniature S1 barrelfield. (C,D) In situ hybridization for Cad8 on whole mounts of P7 wild-type (+/+; Emx1-Cre) and homozygous conditional mutant (COUP-TFIfl/fl; Emx1-Cre) brains uniquely marks the frontal/motor areas (F/M). The F/M areas substantially expand following selective deletion of COUP-TFI from cortex. The reduced ectopic primary sensory areas (V1, S1) can be identified by small domains of diminished cad8 expression in posterior cortex. (E–J) Serotonin (5HT) immunostaining (E,F) MDGA1 (G,H) and RORβ (I,J) in situ hybridization on serial sagittal sections of P7 control (COUP-TFIfl/+) and conditional mutant (fl/fl; Emx1-Cre) cortices. Anterior is to the left, dorsal to the top. Serotonin immunostaining reveals area-specific TCA terminations in layer 4 of S1 and V1. In conditional mutant cortex, both S1 and V1 are reduced in size and are ectopically positioned at the posterior pole of the cortical hemisphere (F). (G,H) MDGA1 selectively marks layers 4 and 6 of S1, and layer 2/3 more broadly in cortex. The S1 specific expression of MDGA1 in layers 4 and 6 confirms the reduced size and posterior shift of S1 in the COUP-TFI deficient cortex, and that these changes occur in parallel across cortical layers. (I,J) RORβ is expressed predominantly in layer 4 of the primary sensory areas (e.g. S1, V1) in wild type cortex (I). RORβ expression in the COUP-TFI deficient cortex is altered to parallel the changes in area patterning in mutant cortex (J). The majority of the cortex in the conditional mutants, including all of the neocortex anterior to the reduced, caudally-shifted primary sensory areas, exhibit serotonin staining and expression of MDGA1 and RORβ that are characteristic of wild type Frontal/Motor cortex (F/M). Scale bars: 1mm. Figure is modified from [38].
Pax6 is a paired box domain TF expressed by cortical progenitors in a low posterior-medial to high anterior-lateral gradient that opposes the pattern of Emx2 expression [6]. Thus, Pax6 is most highly expressed in frontal/motor areas, consistent with the conclusion from marker analyses of small eye (sey) mutant mice, which are deficient for functional Pax6 protein, that implicated Pax6 in specifying anterior area identities associated with frontal/motor areas [6,40,41]. Again, analyses of the sey mutants are limited because they die at birth, and have other major defects that challenge the studies. However, the reported role for Pax6 in arealization has been questioned by a recent gain of function study of Pax6 that used a YAC transgenic approach to overexpress Pax6 [42]. Even in lines in which Pax6 is overexpressed in cortical progenitors by up to 300%, the authors observe no changes in area patterning other than a small but significant decrease in S1 size. Additional studies will be required to sort out these discrepancies and define the role, if any, for Pax6 in arealization.
Sp8, a zinc-finger TF related to Drosophila buttonhead, is expressed in a high anterior-medial to low posterior-lateral gradient by cortical progenitors; Sp8 is also transiently expressed coincident with the Fgf8 domain in the CoP and is a direct transcriptional activator of Fgf8 expression [43]. In the past year, two studies using complementary genetic approaches have reported roles for Sp8 in arealization. One study employed in utero electroporation of expression constructs for gain-and loss-of function analyses of Sp8 function in arealization [43], and the other analyzed a conditional knockout of Sp8 crossed to a BF1 (Foxg1)-Cre line, a “pan-telencephalic” Cre line [44]. Analyses of the conditional Sp8 knockout mice at late embryonic ages show an anterior shift of cortical markers, suggesting that Sp8 preferentially specifies identities associated with frontal/motor areas [44].
However, the use of the BF1-Cre line complicates analyses of roles for Sp8 in arealization because it results in the deletion of Sp8 from progenitors in the cortical VZ as well as from the ANR/CoP. As described above, Sp8 is a direct transcriptional activator of Fgf8 [43] and in addition is required for its maintained expression in the CoP [43, 44]. Therefore, because Fgf8 helps establish through repression the graded expression of Emx2 and COUP-TFI in cortical progenitors, and altering Fgf8 expression has prominent effects on area patterning, the marker shifts observed in the BF1-Cre mediated conditional deletion of Sp8 is consistent with either the diminished expression of Fgf8 in the CoP, or a direct role for Sp8 in specifying area identities of cortical progenitors.
A question relevant for arealization is why does Sp8 not induce Fgf8 within cortical progenitors? In vitro assays show that Emx2, which is co-expressed with Sp8 in cortical progenitors but not in the CoP, represses the ability of Sp8 to bind regulatory elements of Fgf8 and induce its expression [43]. Thus, in vivo, Emx2 likely suppresses the Sp8 transcriptional activation of Fgf8 in cortical progenitors, thereby restricting Fgf8 expression to the CoP.
Finally, analyses of mice with a targeted deletion of the homeodomain TF Otx1 have revealed an intriguing phenotype related to area patterning. Otx1 is expressed by layer 5 projection neurons –the predominant output projection of the cortex, and earlier by their progenitors in the VZ. In adults, layer 5 neurons that project to the spinal cord are limited to sensorimotor areas, but during development they are much more broadly distributed and are even found within visual areas. They acquire their area-specific adult distribution through a process of selective axon elimination [45]. Otx1 null mice have an aberrant areal distribution of layer 5 corticospinal neurons that extends more caudomedial than in wild type mice [46]. Thus, Otx1 is involved in some manner in determining the areal identity of layer 5 projection neurons and/or the process of axon elimination, but the details are presently unclear.
Screens for genes differentially expressed along cortical axes and candidate target genes of TFs and morphogens that control cortical arealization
Defining the target genes of TFs that control arealization and determining how they function to generate area specializations is one of many major challenges for the future. An initial step in this process is to do large scale screens to define candidate target genes. Some screens have been designed to identify additional genes that are differentially expressed within the cortex and therefore might be involved in arealization. The first reported screen of this type was a differential display PCR screen that compared RNAs derived from frontal and occipital embryonic cortex, and identified scores of known and novel genes, including for example, the graded cortical expression of COUP-TFI and Close Homolog of L1 (CHL1) [39], both of which have been subsequently shown to have significant functions in cortical development. More recently, others have used microarray technology to do similar searches for genes differentially expressed along the axes of developing mouse cortex [47–49]. A distinct series of recent screens have used a different approach, and were designed to identify genes that are candidate targets of TFs or morphogens implicated in arealization, such as Emx2 and Pax6 [50–53], or Fgfs [54]. Each of these screens identified hundreds of candidate targets with increased or decreased expression, and therefore potentially involved in cortical arealization as well as functions relevant to other prominent phenotypes exhibited by Emx2 and Pax6 (sey) mutants, as well as Fgfr1 mutants, including proliferation, neuronal differentiation, migration, axon guidance, and regional patterning of the telencephalon.
One screen used a Representational Display Analysis that compared Emx2 null cortex to wild type, and vice versa, and among the many genes identified was Odz4/Ten_m4, which, along with the other 3 members of this gene family, was analyzed [52]. The vertebrate Odz genes (also referred to as the Ten_m family in mouse) are orthologs of the Drosophila pair-rule patterning gene, Odd Oz (Odz), which encodes a transmembrane protein with structural domains similar to tenascin and is involved in segmental patterning in Drosophila. In embryonic mice, Odz4 has an expression pattern that parallels the graded expression of Emx2, but rather than being expressed in the VZ, Odz4 is expressed in the CP throughout its development. Odz2 and Odz3 have similar gradients of expression as Odz4 in the CP, whereas Odz1 has an opposing expression gradient [52]. Postnatally, these graded expression patterns refine into more restricted patterns, with Odz2, 3 and 4 having patterns that relate to the posterior-medial positioned visual areas, and Odz1 to the more anterior sensorimotor areas. The Odz genes also have distinct laminar expression patterns [52]. Each Odz family member exhibits an anterior shift in cortical expression in Emx2 mutants and a posterior shift in Pax6 (sey) mutants, consistent with the opposing area patterning functions of Emx2 and Pax6 and potential roles for the Odz genes in arealization as targets of Emx2 and Pax6 [52].Odz3/Ten_m3 was also independently identified in a microarray screen designed to identify genes differentially expressed in somatosensory versus visual areas of developing mouse cortex [49]. These investigators also find the preferential expression of Odz3 within visual cortical areas and provide evidence that Odz3 promotes homophilic adhesion and neurite outgrowth by neurons that express it [49].
Primary cortical areas exhibit significant variation in size between normal individuals
The general spatial relationship between the primary areas is largely conserved across mammals, although some animals with unusual or large and atypical peripheral appendages/sense organs (e.g. the platypus’ bill or the echo-location system in bats) have modifications on this general geometrical scheme of area patterning to reflect their sensory specializations [55]. A straightforward example of this concept comes from a comparison of area patterning in the mouse, ghost bat, and short-tailed opossum. Overall cortical size in these species is similar, but the sizes of the three primary sensory cortical areas (S1, V1 and A1) differ substantially between them reflecting their unique sensory specializations and needs [56].
Area patterning also varies substantially across individuals of the same species. For example, the sizes of primary areas in human neocortex vary by two- to three-fold within the normal population, despite overall cortical volume varying only by about 30% [57,58]. Mice that are essentially genetically identical, i.e. isogenic inbred strains of mice, such as C57Bl/6J and DBA/2J mice, do not have significant variation in overall cortical surface area or in the sizes of specific cortical areas whereas comparisons between the inbred strains that are genetically distinct, show significant differences in sizes [59]. These studies have focused primarily on size differences of S1, particularly on the posteromedial barrel subfield (PMBSF) of S1, and V1, delineated in adult mice of the isogenic inbred strains C57Bl/6J and DBA/2J. The overall surface area of the neocortex is 7% larger in the C57Bl/6J strain than in the DBA/2J strain of mice. However, after normalizing for this overall size difference, V1 is 12% larger in the C57Bl/6J strain than in the DBA/2J strain whereas PMBSF is 10% larger in the C57Bl/6J strain than in the DBA/2J strain [59]. Interestingly, these size differences alone are 90% effective as a blind predictor of the strain. As described below, such area size differences can result in differences in modality-specific behavioral performance [36].
These differences between adult C57BL/6J and DBA/2J mice in their cortical area patterning has led some groups to employ a forward genetic approach to define the genetic contributions to these phenotypic variations, and in particular the use of Quantitative trait locus (QTL) mapping. A few groups, especially by Waters, Williams and colleagues, have recently championed this tool. In particular when used for analysis of recombinant inbred (RI) strains of mice, this approach can be used to map and characterize genes responsible for heritable variation in complex phenotypes. A significant advantage of using RI strains derived from parental strains with mapped genomes is that they facilitate determining the genetic contributions to complex traits, including area size, which can be readily mapped to specific genetic loci and even to specific genes using QTL mapping.
The size difference in PMBSF between the C57BL/6J and DBA/2J strains and the heritability of this trait has been investigated further by analyzing 42 RI strains derived from C57Bl/6J and DBA/2J mice (referred to as BXD lines), generated by crosses between the two parental strains [60]. Using this approach, a difference of up to 33% is found in size between the largest and smallest PMBSF in the BXD RI strains, with a continuous size distribution, suggesting a polygenic trait. Using QTL linkage analysis and other criteria, the identified candidate genes responsible for the size differences include carbonic anhydrase-related protein VIII and Rab2, which belongs to the Rab subfamily of small GTP-binding proteins. Both of these genes have properties and functions that make them intriguing candidates for further study. In addition, mRNA expression profiles obtained with GeneNetwork indicate a strong correlation between total PMBSF area and two genes, Adcy1 and Gap43, important in S1 development. However, because many factors unrelated to genetic patterning mechanisms of arealization likely contribute to differences in area sizes between adults, many of the genes identified using QTL mapping of adult traits, including those defined in this study, are unlikely to be directly involved in arealization. Nonetheless, these forward genetic approaches complement reverse genetics and may well yield important insights into the genetic regulation of area patterning.
Recent studies have defined “Expression Level Polymorphisms” (ELP) characterized by differences between individuals in the expression level of genes [61]; these differences in expression levels approximate those that have been genetically created for Emx2 in mice and result in significant changes in area sizes [35]. Therefore, modest differences in naturally occurring gene expression due to ELP could underlie the naturally occurring differences in area size in humans and mice. Indeed, polymorphisms in the regulatory region of the human serotonin 5-HT2A receptor gene have been recently shown to significantly influence the level of expression of this gene, which has potential implications for neurological disorders [62].
Behavioral implications of variation in area size
Recent studies in mice indicate that variations in area size within the ranges found between inbred mouse strains, and well below the ranges reported for normal humans, can have dramatic, modality-specific effects on behavior [36]. For example, alterations of the levels of Emx2 in cortical progenitors that result in either relatively modest decreases or increases in the sizes of somatosensory and motor areas in adult mice result in significant, and specific, deficiencies at tests of tactile and motor performance. These findings indicate that area size can be a critical parameter in determining performance at modality-specific behaviors [36]. They also underscore the importance of establishing during development the appropriate expression levels of TFs that specify area identities, as changes in them can result in a change in area sizes. Thereby relatively subtle changes in early developmental events can have a prominent influence on behavior later in life, affecting performance and likely underlying many forms of cognitive dysfunction and neurological disorders. Further, they support the hypothesis that cortical areas have evolved an optimal size defined and tuned by their relationships with other components of their neural system to maximize functional efficiency and behavioral performance [55].
Conclusion
The coming years look very promising for significant advances in understanding the mechanisms that control area patterning. The study of cortical arealization has captured the attention of a rapidly increasing number of investigators bringing to bear on the issue a diverse range of backgrounds and talents. In addition, the tools required for these studies, ranging from genetically engineered mice to data bases, are expanding rapidly. The availability of fully sequenced genomes for strains of mice, and the data bases of gene and protein expression patterns and even quantitative data on expression levels, will speed progress, as will the forward genetics approach being advanced that complements the reverse genetics that have thus far yielded most of our knowledge. Finally, a particularly intriguing issue is the use of gene expression data bases, such as the Allen Brain Atlas, the Gensat Project, or numerous other databases (for review and list of URLs see [63]), to correlate the expression patterns of thousands of genes to area maps based on anatomy, mainly cytoarchitecture. Among the goals of these types of approaches is to re-define area patterns and even the relationships between areas based on gene expression profiles. These types of analyses should provide greater insight into the definition of a cortical area and hopefully provide the markers to facilitate the important extension of studies of arealization from primary areas to higher order areas.
Acknowledgments
Work in the authors’ lab on this topic is supported by NIH grants R37 NS31558 and R01 NS 050646. We thank Chuck Stevens and members of the O’Leary lab, particularly Shen-ju Chou, Todd Kroll, Axel Leingartner, Scott May, Carlos Perez-Garcia, Andreas Zembrzycki, for their contributions and helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rakic P. Specification of cerebral cortical areas. Science. 1988;241:170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- 2.O’Leary DD. Do cortical areas emerge from a protocortex? Trends Neurosci. 1989;12:400–406. doi: 10.1016/0166-2236(89)90080-5. [DOI] [PubMed] [Google Scholar]
- 3.O’Leary DD, Chou SJ, Sahara S. Area patterning of the mammalian cortex. Neuron. 2007;56:252–269. doi: 10.1016/j.neuron.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 4.Rash BG, Grove EA. Area and layer patterning in the developing cerebral cortex. Curr Opin Neurobiol. 2006;16:25–34. doi: 10.1016/j.conb.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Sur M, Rubenstein JL. Patterning and plasticity of the cerebral cortex. Science. 2005;310:805–810. doi: 10.1126/science.1112070. [DOI] [PubMed] [Google Scholar]
- 6.Bishop KM, Goudreau G, O’Leary DD. Regulation of area identity in the mammalian neocortex by Emx2 and Pax6. Science. 2000;288:344–349. doi: 10.1126/science.288.5464.344. [DOI] [PubMed] [Google Scholar]
- 7.Mallamaci A, Muzio L, Chan CH, Parnavelas J, Boncinelli E. Area identity shifts in the early cerebral cortex of Emx2−/− mutant mice. Nat Neurosci. 2000;3:679–686. doi: 10.1038/76630. [DOI] [PubMed] [Google Scholar]
- 8.Kriegstein AR, Noctor SC. Patterns of neuronal migration in the embryonic cortex. Trends Neurosci. 2004;27:392–399. doi: 10.1016/j.tins.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Dehay C, Kennedy H. Cell-cycle control and cortical development. Nat Rev Neurosci. 2007;8:438–450. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- 10.Marin O, Rubenstein JL. Cell migration in the forebrain. Annu Rev Neurosci. 2003;26:441–483. doi: 10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- 11.Nery S, Fishell G, Corbin JG. The caudal ganglionic eminence is a source of distinct cortical and subcortical cell populations. Nat Neurosci. 2002;5:1279–1287. doi: 10.1038/nn971. [DOI] [PubMed] [Google Scholar]
- 12.Tissir F, Goffinet AM. Reelin and brain development. Nat Rev Neurosci. 2003;4:496–505. doi: 10.1038/nrn1113. [DOI] [PubMed] [Google Scholar]
- 13.Yoshida M, Assimacopoulos S, Jones KR, Grove EA. Massive loss of Cajal-Retzius cells does not disrupt neocortical layer order. Development. 2006;133:537–545. doi: 10.1242/dev.02209. [DOI] [PubMed] [Google Scholar]
- 14.Bielle F, Griveau A, Narboux-Neme N, Vigneau S, Sigrist M, Arber S, Wassef M, Pierani A. Multiple origins of Cajal-Retzius cells at the borders of the developing pallium. Nat Neurosci. 2005;8:1002–1012. doi: 10.1038/nn1511. [DOI] [PubMed] [Google Scholar]
- 15.Assimacopoulos S, Grove EA, Ragsdale CW. Identification of a Pax6-dependent epidermal growth factor family signaling source at the lateral edge of the embryonic cerebral cortex. J Neurosci. 2003;23:6399–6403. doi: 10.1523/JNEUROSCI.23-16-06399.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crossley PH, Martinez S, Ohkubo Y, Rubenstein JL. Coordinate expression of Fgf8, Otx2, Bmp4, and Shh in the rostral prosencephalon during development of the telencephalic and optic vesicles. Neuroscience. 2001;108:183–206. doi: 10.1016/s0306-4522(01)00411-0. [DOI] [PubMed] [Google Scholar]
- 17.Grove EA, Tole S, Limon J, Yip L, Ragsdale CW. The hem of the embryonic cerebral cortex is defined by the expression of multiple Wnt genes and is compromised in Gli3-deficient mice. Development. 1998;125:2315–2325. doi: 10.1242/dev.125.12.2315. [DOI] [PubMed] [Google Scholar]
- 18.Ohkubo Y, Chiang C, Rubenstein JL. Coordinate regulation and synergistic actions of BMP4, SHH and FGF8 in the rostral prosencephalon regulate morphogenesis of the telencephalic and optic vesicles. Neuroscience. 2002;111:1–17. doi: 10.1016/s0306-4522(01)00616-9. [DOI] [PubMed] [Google Scholar]
- 19.Shimogori T, Banuchi V, Ng HY, Strauss JB, Grove EA. Embryonic signaling centers expressing BMP, WNT and FGF proteins interact to pattern the cerebral cortex. Development. 2004;131:5639–5647. doi: 10.1242/dev.01428. [DOI] [PubMed] [Google Scholar]
- 20.Kuschel S, Ruther U, Theil T. A disrupted balance between Bmp/Wnt and Fgf signaling underlies the ventralization of the Gli3 mutant telencephalon. Dev Biol. 2003;260:484–495. doi: 10.1016/s0012-1606(03)00252-5. [DOI] [PubMed] [Google Scholar]
- 21.Rash BG, Grove EA. Patterning the dorsal telencephalon: a role for sonic hedgehog? J Neurosci. 2007;27:11595–11603. doi: 10.1523/JNEUROSCI.3204-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crossley PH, Martin GR. The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development. 1995;121:439–451. doi: 10.1242/dev.121.2.439. [DOI] [PubMed] [Google Scholar]
- 23.Garel S, Huffman KJ, Rubenstein JL. Molecular regionalization of the neocortex is disrupted in Fgf8 hypomorphic mutants. Development. 2003;130:1903–1914. doi: 10.1242/dev.00416. [DOI] [PubMed] [Google Scholar]
- 24.Storm EE, Garel S, Borello U, Hebert JM, Martinez S, McConnell SK, Martin GR, Rubenstein JL. Dose-dependent functions of Fgf8 in regulating telencephalic patterning centers. Development. 2006;133:1831–1844. doi: 10.1242/dev.02324. [DOI] [PubMed] [Google Scholar]
- 25.Fukuchi-Shimogori T, Grove EA. Neocortex patterning by the secreted signaling molecule FGF8. Science. 2001;294:1071–1074. doi: 10.1126/science.1064252. [DOI] [PubMed] [Google Scholar]
- 26*.Cholfin JA, Rubenstein JL. Patterning of frontal cortex subdivisions by Fgf17. Proc Natl Acad Sci U S A. 2007;104:7652–7657. doi: 10.1073/pnas.0702225104. The authors show novel distinct functions for Fgf8 and Fgf17, two of many Fgf family members secreted by the Commissural plate, in patterning subdivisions of frontal cortex (FC). Using a set of gene expression markers and BAC-GFP mice that distinguish subdivisions of the newborn mouse FC, they show by analyzing Fgf17 and Fgf8 mutant mice that Fgf17 selectively controls the size of dorsal frontal cortex, whereas Fgf8 controls the size of both dorsal FC and ventral/orbital FC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furuta Y, Piston DW, Hogan BL. Bone morphogenetic proteins (BMPs) as regulators of dorsal forebrain development. Development. 1997;124:2203–2212. doi: 10.1242/dev.124.11.2203. [DOI] [PubMed] [Google Scholar]
- 28.Kim AS, Pleasure SJ. Expression of the BMP antagonist Dan during murine forebrain development. Brain Res Dev Brain Res. 2003;145:159–162. doi: 10.1016/s0165-3806(03)00213-x. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Y, Sheng HZ, Amini R, Grinberg A, Lee E, Huang S, Taira M, Westphal H. Control of hippocampal morphogenesis and neuronal differentiation by the LIM homeobox gene Lhx5. Science. 1999;284:1155–1158. doi: 10.1126/science.284.5417.1155. [DOI] [PubMed] [Google Scholar]
- 30.Monuki ES, Porter FD, Walsh CA. Patterning of the dorsal telencephalon and cerebral cortex by a roof plate-Lhx2 pathway. Neuron. 2001;32:591–604. doi: 10.1016/s0896-6273(01)00504-9. [DOI] [PubMed] [Google Scholar]
- 31.Bulchand S, Grove EA, Porter FD, Tole S. LIM-homeodomain gene Lhx2 regulates the formation of the cortical hem. Mech Dev. 2001;100:165–175. doi: 10.1016/s0925-4773(00)00515-3. [DOI] [PubMed] [Google Scholar]
- 32.Porter FD, Drago J, Xu Y, Cheema SS, Wassif C, Huang SP, Lee E, Grinberg A, Massalas JS, Bodine D, et al. Lhx2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development. 1997;124:2935–2944. doi: 10.1242/dev.124.15.2935. [DOI] [PubMed] [Google Scholar]
- 33**.Mangale VS, Hirokawa KE, Satyaki PR, Gokulchandran N, Chikbire S, Subramanian L, Shetty AS, Martynoga B, Paul J, Mai MV, et al. Lhx2 selector activity specifies cortical identity and suppresses hippocampal organizer fate. Science. 2008;319:304–309. doi: 10.1126/science.1151695. This elegant study generated and used a conditional knockout of the Lim homeodomain gene Lhx2 and mouse genetic mosaics to provide evidence that Lhx2 acts as a classic selector gene and an intrinsic determinant of cortical identity. Lhx2 selector activity is restricted to an early critical period and acts cell-autonomously to specify cortical identity and suppress the fates of cortical hem medially and antihem laterally. They also show that the cortical hem is a hippocampal organizer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simeone A, Gulisano M, Acampora D, Stornaiuolo A, Rambaldi M, Boncinelli E. Two vertebrate homeobox genes related to the Drosophila empty spiracles gene are expressed in the embryonic cerebral cortex. Embo J. 1992;11:2541–2550. doi: 10.1002/j.1460-2075.1992.tb05319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamasaki T, Leingartner A, Ringstedt T, O’Leary DD. EMX2 regulates sizes and positioning of the primary sensory and motor areas in neocortex by direct specification of cortical progenitors. Neuron. 2004;43:359–372. doi: 10.1016/j.neuron.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 36*.Leingartner A, Thuret S, Kroll TT, Chou SJ, Leasure JL, Gage FH, O’Leary DD. Cortical area size dictates performance at modality-specific behaviors. Proc Natl Acad Sci U S A. 2007;104:4153–4158. doi: 10.1073/pnas.0611723104. The sizes of primary cortical areas can vary between normal individuals raising the question of whether area size dictates behavioral performance. The authors show that adult mice genetically engineered to overexpress the homeodomain transcription factor EMX2 in embryonic cortical stem cells, resulting in reductions in sizes of somatosensory and motor areas, exhibit substantial deficiencies at tactile and motor behaviors. Even increasing the size of sensorimotor areas by decreasing cortical EMX2 levels can lead to diminished sensorimotor behaviors. Genetic crosses that retain ectopic Emx2 transgene expression subcortically, and restore cortical Emx2 expression to wild type levels also restore cortical areas to wild type sizes and in parallel restore the deficient tactile and motor behaviors to wild type performance. These findings demonstrate a direct link between area size and modality-specific behavioral performance, and suggest that cortical areas have an optimal size defined within the context of other components of their neural system to maximize functional efficiency and behavioral performance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou C, Qiu Y, Pereira FA, Crair MC, Tsai SY, Tsai MJ. The nuclear orphan receptor COUP-TFI is required for differentiation of subplate neurons and guidance of thalamocortical axons. Neuron. 1999;24:847–859. doi: 10.1016/s0896-6273(00)81032-6. [DOI] [PubMed] [Google Scholar]
- 38**.Armentano M, Chou SJ, Srubek Tomassy G, Leingartner A, O’Leary DD, Studer M. COUP-TFI regulates the balance of cortical patterning between frontal/motor and sensory areas. Nat Neurosci. 2007;10:1277–1286. doi: 10.1038/nn1958. The transcription factor COUP-TFI (also known as Nr2f1) has graded expression in developing cortex, with highest expression in the cortical progenitors and progeny in parietal and occipital cortex that form sensory areas, and the lowest expression was observed in frontal cortex that includes motor areas. Cortical deletion of COUP-TFI results in massive expansion of frontal/motor areas to occupy most of neocortex, paralleled by substantial compression of sensory areas to caudal occipital cortex. These area patterning changes are preceded and paralleled by corresponding changes in molecular markers of area identity and altered thalamocortical inputs and layer 5 and 6 output projections. The authors conclude that COUP-TFI is required for balancing area patterning of neocortex by repressing in its expression domain frontal/motor area identities allowing fro proper specification of sensory areas. [DOI] [PubMed] [Google Scholar]
- 39.Liu Q, Dwyer ND, O’Leary DD. Differential expression of COUP-TFI, CHL1, and two novel genes in developing neocortex identified by differential display PCR. J Neurosci. 2000;20:7682–7690. doi: 10.1523/JNEUROSCI.20-20-07682.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bishop KM, Rubenstein JL, O’Leary DD. Distinct actions of Emx1, Emx2, and Pax6 in regulating the specification of areas in the developing neocortex. J Neurosci. 2002;22:7627–7638. doi: 10.1523/JNEUROSCI.22-17-07627.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muzio L, DiBenedetto B, Stoykova A, Boncinelli E, Gruss P, Mallamaci A. Emx2 and Pax6 control regionalization of the pre-neuronogenic cortical primordium. Cereb Cortex. 2002;12:129–139. doi: 10.1093/cercor/12.2.129. [DOI] [PubMed] [Google Scholar]
- 42.Manuel M, Georgala PA, Carr CB, Chanas S, Kleinjan DA, Martynoga B, Mason JO, Molinek M, Pinson J, Pratt T, et al. Controlled overexpression of Pax6 in vivo negatively autoregulates the Pax6 locus, causing cell-autonomous defects of late cortical progenitor proliferation with little effect on cortical arealization. Development. 2007;134:545–555. doi: 10.1242/dev.02764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43*.Sahara S, Kawakami Y, Izpisua Belmonte JC, O’Leary DD. Sp8 exhibits reciprocal induction with Fgf8 but has an opposing effect on anterior-posterior cortical area patterning. Neural Develop. 2007;2:10. doi: 10.1186/1749-8104-2-10. This study shows that the zinc finger transcription factors Sp5, Sp8, and Sp9, homologues of Drosophila buttonhead (btd), are expressed coincident with the three telencephalic patterning centers, the cortical hem, commissural plate (CoP) and the sonic hedgehog domain, respectively, and that Sp5 and Sp8 also have graded expression in cortical progenitors. To study function, they focused on Sp8, and show that it binds Fgf8 regulatory elements and is a direct transcriptional activator of Fgf8, a morphogen expressed in the CoP that influences area patterning. Using in utero electroporation of gain-of-function and loss-of-function expression constructs they show that Sp8 is required to maintain Fgf8 expression in the CoP and controls the anterior-posterior area patterning of the neocortex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44*.Zembrzycki A, Griesel G, Stoykova A, Mansouri A. Genetic interplay between the transcription factors Sp8 and Emx2 in the patterning of the forebrain. Neural Develop. 2007;2:8. doi: 10.1186/1749-8104-2-8. This study complements that of Sahara et al. [43]. The authors generate mice with floxed alleles of Sp8 and perform a selective telencephalic deletion using BF1 (Foxg1) cre line, which deletes Sp8 from the CoP, resulting in diminished Fgf8 expression, and from cortical progenitors. They limit their analyses to embryonic stages (E18.5), before areas differentiate. The cortex shows many defects, including aberrant preplate splitting and aberrant laminar identities. The cortex is also reduced in size and the cortical wall is thinner—phenotypes are due to a reduced progenitor pool and exaggerated apoptosis. Emx2 expression is increased and Pax6 expression is diminished, and consistent with these findings, A–P positional markers exhibit an anterior shift. These arealization changes are similar to those observed when Fgf8 expression in the CoP is diminished, which they find. Sp8 is also essential for the maintenance of ventral cell identity in the septum and medial ganglionic eminence (MGE). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Leary DD, Koester SE. Development of projection neuron types, axon pathways, and patterned connections of the mammalian cortex. Neuron. 1993;10:991–1006. doi: 10.1016/0896-6273(93)90049-w. [DOI] [PubMed] [Google Scholar]
- 46.Ando K, Yagi H, Suda Y, Aizawa S, Sakashita M, Nagano T, Terashima T, Sato M. Establishment of framework of the cortical area is influenced by Otx1. Neurosci Res. 2008;60:457–459. doi: 10.1016/j.neures.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 47.Funatsu N, Inoue T, Nakamura S. Gene expression analysis of the late embryonic mouse cerebral cortex using DNA microarray: identification of several region- and layer-specific genes. Cereb Cortex. 2004;14:1031–1044. doi: 10.1093/cercor/bhh063. [DOI] [PubMed] [Google Scholar]
- 48.Muhlfriedel S, Kirsch F, Gruss P, Chowdhury K, Stoykova A. Novel genes differentially expressed in cortical regions during late neurogenesis. Eur J Neurosci. 2007;26:33–50. doi: 10.1111/j.1460-9568.2007.05639.x. [DOI] [PubMed] [Google Scholar]
- 49.Leamey CA, Glendining KA, Kreiman G, Kang ND, Wang KH, Fassler R, Sawatari A, Tonegawa S, Sur M. Differential gene expression between sensory neocortical areas: potential roles for Ten_m3 and Bcl6 in patterning visual and somatosensory pathways. Cereb Cortex. 2008;18:53–66. doi: 10.1093/cercor/bhm031. [DOI] [PubMed] [Google Scholar]
- 50.Arai Y, Funatsu N, Numayama-Tsuruta K, Nomura T, Nakamura S, Osumi N. Role of Fabp7, a downstream gene of Pax6, in the maintenance of neuroepithelial cells during early embryonic development of the rat cortex. J Neurosci. 2005;25:9752–9761. doi: 10.1523/JNEUROSCI.2512-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gangemi RM, Daga A, Muzio L, Marubbi D, Cocozza S, Perera M, Verardo S, Bordo D, Griffero F, Capra MC, et al. Effects of Emx2 inactivation on the gene expression profile of neural precursors. Eur J Neurosci. 2006;23:325–334. doi: 10.1111/j.1460-9568.2005.04559.x. [DOI] [PubMed] [Google Scholar]
- 52.Li H, Bishop KM, O’Leary DD. Potential target genes of EMX2 include Odz/Ten-M and other gene families with implications for cortical patterning. Mol Cell Neurosci. 2006;33:136–149. doi: 10.1016/j.mcn.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 53.Holm PC, Mader MT, Haubst N, Wizenmann A, Sigvardsson M, Gotz M. Loss- and gain-of-function analyses reveal targets of Pax6 in the developing mouse telencephalon. Mol Cell Neurosci. 2007;34:99–119. doi: 10.1016/j.mcn.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 54.Sansom SN, Hebert JM, Thammongkol U, Smith J, Nisbet G, Surani MA, McConnell SK, Livesey FJ. Genomic characterisation of a Fgf-regulated gradient-based neocortical protomap. Development. 2005;132:3947–3961. doi: 10.1242/dev.01968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krubitzer L. The magnificent compromise: cortical field evolution in mammals. Neuron. 2007;56:201–208. doi: 10.1016/j.neuron.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 56.Krubitzer L, Kahn DM. Nature versus nurture revisited: an old idea with a new twist. Prog Neurobiol. 2003;70:33–52. doi: 10.1016/s0301-0082(03)00088-1. [DOI] [PubMed] [Google Scholar]
- 57.Dougherty RF, Koch VM, Brewer AA, Fischer B, Modersitzki J, Wandell BA. Visual field representations and locations of visual areas V1/2/3 in human visual cortex. J Vis. 2003;3:586–598. doi: 10.1167/3.10.1. [DOI] [PubMed] [Google Scholar]
- 58.White LE, Andrews TJ, Hulette C, Richards A, Groelle M, Paydarfar J, Purves D. Structure of the human sensorimotor system. I: Morphology and cytoarchitecture of the central sulcus. Cereb Cortex. 1997;7:18–30. doi: 10.1093/cercor/7.1.18. [DOI] [PubMed] [Google Scholar]
- 59.Airey DC, Robbins AI, Enzinger KM, Wu F, Collins CE. Variation in the cortical area map of C57BL/6J and DBA/2J inbred mice predicts strain identity. BMC Neurosci. 2005;6:18. doi: 10.1186/1471-2202-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60*.Jan TA, Lu L, Li CX, Williams RW, Waters RS. Genetic analysis of posterior medial barrel subfield (PMBSF) size in somatosensory cortex (SI) in recombinant inbred strains of mice. BMC Neurosci. 2008;9:3. doi: 10.1186/1471-2202-9-3. This study takes a forward genetics approach using quantitative trait locus (QTL) mapping to identify candidate genes that control area size and other complex traits. They find a 33% difference in size of the posterior medial barrel subfield, a part of the mouse primary somatosensory area, between 42 recombinant inbred (RI) strains of mice generated from C57BL/6J and DBA/2J parental strains. QTL mapping and other criteria identified 4 genes potentially responsible for the size differences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cowles CR, Hirschhorn JN, Altshuler D, Lander ES. Detection of regulatory variation in mouse genes. Nat Genet. 2002;32:432–437. doi: 10.1038/ng992. [DOI] [PubMed] [Google Scholar]
- 62.Myers RL, Airey DC, Manier DH, Shelton RC, Sanders-Bush E. Polymorphisms in the regulatory region of the human serotonin 5-HT2A receptor gene (HTR2A) influence gene expression. Biol Psychiatry. 2007;61:167–173. doi: 10.1016/j.biopsych.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 63.Sunkin SM, Hohmann JG. Insights from spatially mapped gene expression in the mouse brain. Hum Mol Genet. 2007;16(Spec No 2):R209–219. doi: 10.1093/hmg/ddm183. [DOI] [PubMed] [Google Scholar]
- 64.O’Leary DD, Nakagawa Y. Patterning centers, regulatory genes and extrinsic mechanisms controlling arealization of the neocortex. Curr Opin Neurobiol. 2002;12:14–25. doi: 10.1016/s0959-4388(02)00285-4. [DOI] [PubMed] [Google Scholar]