Abstract
Self-assembling peptides and peptide derivatives bearing cell-binding ligands are increasingly being investigated as defined cell culture matrices and as scaffolds for regenerative medicine. In order to systematically refine such scaffolds to elicit specific desired cell behaviors, ligand display should ideally be achieved without inadvertently altering other physicochemical properties such as viscoelasticity. Moreover, for in vivo applications, self-assembled biomaterials must exhibit low immunogenicity. In the present study, multi-peptide co-assembling hydrogels based on the beta-sheet fibrillizing peptide Q11 (QQKFQFQFEQQ) were designed such that they presented RGDS or IKVAV ligands on their fibril surfaces. In co-assemblies of the ligand-bearing peptides with Q11, ligand incorporation levels capable of influencing the attachment, spreading, morphology, and growth of human umbilical vein endothelial cells (HUVECs) did not significantly alter the materials’ fibrillization, beta-turn secondary structure, or stiffness. RGDS-Q11 specifically increased HUVEC attachment, spreading, and growth when co-assembled into Q11 gels, whereas IKVAV-Q11 exerted a more subtle influence on attachment and morphology. Additionally, Q11 and RGDS-Q11 were minimally immunogenic in mice, making Q11-based biomaterials attractive candidates for further investigation as defined, modular extracellular matrices for applications in vitro and in vivo.
INTRODUCTION
Hydrogels constructed through self-assembly are receiving increasing attention for a variety of biomedical and biotechnological applications, including scaffolds for regenerative medicine [1, 2], the controlled release of therapeutics [3–6], and defined cell culture matrices [7, 8]. Self-assembled materials are advantageous for biological applications because of their precise chemical definition, their ability to form complex fibrillar or network structures from easily synthesized precursors, and the possibility of combining more than one molecule into co-assemblies having multiple different functions. Given that cells integrate and respond to an extraordinarily complex mixture of physical, chemical, and biological signals, these advantages can be further exploited if the individual molecular constituents can be adjusted independently, without changing other properties such as the viscoelasticity of the matrix as a whole. Such modularity enables a systematic tuning of the physical, chemical, and biological milieu that ultimately dictates how cells behave [9, 10].
Beta-sheet fibrillizing peptides [7, 8, 11–19] and peptide amphiphiles [2, 20–22] have received particular attention recently as defined matrices for cells owing to their stimulus-sensitive fibril formation, their ability to form hydrogels, their ease of synthesis, and the availability of many amino acid sequences known to influence cell behavior through the binding of integrins and other receptors. Self-assembling fibril-forming peptides terminally modified with such cell-binding ligands have recently been utilized to influence the attachment of fibroblasts [18, 19], the proliferation and gene expression of mouse neural stem cells [14], the differentiation behavior of pre-osteoblasts [8], and the proliferation of endothelial cells [15]. These previous studies have been performed using the RAD16 peptide [8, 14, 15] self-assembling sequences from laminin [18], or self-assembling sequences from transthyretin [19], demonstrating that several different β-sheet fibril-forming peptides are capable of presenting bioavailable ligands on their surfaces. However, previous studies have also found that the incorporation of ligands within β-sheet fibrillar hydrogels can significantly alter their mechanical properties [15, 23], complicating the interpretation of cell behavior on these materials. Mechanical variability in ligand-bearing fibrillar assemblies can be mitigated by adsorbing the peptide fibrils onto stiff surfaces such as polystyrene culture dishes [18, 19], but this strategy is not applicable for 3-D fibrillar gels or for cases where modulating matrix stiffness is desirable. In gels, previous efforts have sought to produce uniformly stiff matrices by co-assembling ligand-bearing peptides into a background of unmodified peptides [8, 15], but the mechanical variability of such matrices has to our knowledge not been reported. Controlling the mechanical properties of these materials is important given the growing awareness of how significantly a material’s stiffness can dictate cell adhesion, proliferation, and differentiation [13, 24–26]. Although most previous work regarding substrate mechanics has been conducted on polymeric substrates, recent examples using self-assembled peptides have reiterated the impact of gel stiffness on cell behavior [13, 17]. Without the ability to adjust ligand presentation in self-assembled matrices while at the same time controlling mechanical properties, it will be difficult to systematically clarify the contributions of both factors. Additionally, as self-assembled peptide biomaterials move towards clinical applications, their potential for engaging immune responses needs to be clarified. Both their non-native amino acid sequences and their highly oligomerized structures could in principle contribute to immunogenicity [27–29]. Previous reports have described non-immunogenic unfunctionalized fibrillar peptide assemblies [16] and non-inflammatory ligand-bearing assemblies [2, 30, 31], but antibody production against ligand-bearing β-sheet assemblies has not been directly investigated.
In the work described here, we investigated the co-assembly, gelation, ligand presentation, mechanical properties, and immunogenicity of peptide hydrogels based on the sequence QQKFQFQFEQQ (Q11), which has previously been utilized by our laboratory for producing substrates for endothelial cells with adjustable stiffness [13]. In the present work, we investigated co-assemblies of Q11 with peptides having Q11 at their C-termini and one of two different ligands, RGDS or IKVAV, at their N-termini. The RGDS sequence found in fibronectin, laminin, vitronectin and many other extracellular matrix proteins is well known to enable cell attachment for many cell types by binding multiple integrins including α3β1, α5β1, αvβ3 and several others [32, 33]. The peptide IKVAV, from a cryptic sequence near the carboxy-terminal end of the 1 chain of laminin-111, is perhaps most widely known in the field of biomaterials as a modulator of neuronal cell attachment and neurite outgrowth [2, 34, 35]. It has also been found to influence the attachment, migration, morphology, and matrix remodeling of endothelial cells as well [36–40], though these mechanisms are not fully understood and appear to be context-dependent, illustrating the need to investigate the effect of this peptide in highly defined systems such as the one described here.
MATERIALS AND METHODS
Peptide design and synthesis
All ligand-bearing peptides possessed the same basic construction, consisting of two N-terminal Gly residues, the ligand sequence, a triglycine spacer, and a C-terminal Q11 sequence (Table 1). Biotinylated versions of these peptides included an additional N-terminal biotin and a Ser-Gly-Ser-Gly spacer. Peptide synthesis reagents were purchased from NovaBiochem. All peptides were synthesized on a CS Bio 136 automated peptide synthesizer using standard Fmoc-based solid phase chemistry, HOBt/HBTU activation, and Rink amide AM resin (NovaBiochem cat# 01-64-0038) or Wang resin (NovaBiochem cat# 01-64-0014). All peptides were N-terminally acetylated or N-terminally biotinylated with biotin-ONp (NovaBiochem cat# 01-63-0116), cleaved using conventional TFA/TIS/H2O cocktails, and collected by precipitation in diethyl ether. For purification and peptide entrapment experiments, a Varian ProStar HPLC system was utilized with a Vydac 214TP C4 reverse-phase column and water-acetonitrile gradients. Peptides were stored as lyophilized powders at −20°C until experimentation. Similar to other β-sheet fibril-forming peptides, diminished solubility was periodically observed for Q11 and ligand-bearing Q11 peptides that had been stored for longer than several months, but solubility could be completely restored by dissolving them in a small amount of neat TFA for 15 min, re-precipitating in diethyl ether, and re-dissolving in water.
Table 1.
Peptides synthesized.
| Name | Sequence | m/z(calc’d) | m/z(found) |
|---|---|---|---|
| Q11 | Ac-QQKFQFQFEQQ-CONH2 | 1526.7 | 1527.7 |
| RGDS-Q11 | Ac-GGRGDSGGG-(Q11)-CONH2 | 2227.3 | 2227.6 |
| RDGS-Q11 (scrambled) | Ac-GGRDGSGGG-(Q11)-CONH2 | 2227.3 | 2227.0 |
| IKVAV-Q11 | Ac-GGIKVAVGGG-(Q11)-CONH2 | 2322.6 | 2322.4 |
| VAKVI-Q11 (scrambled) | Ac-GGVAKVIGGG-(Q11)-CONH2 | 2322.6 | 2321.9 |
| OVA | H2N-ISQAVHAAHAEINEAGR-COOH | 1773.9 | 1774.9 |
| Biotin-RGDS-Q11 | Biotin-SGSGRGDSGGG-(Q11)-CONH2 | 2585.8 | 2586.2 |
| Biotin-IKVAV-Q11 | Biotin-SGSGIKVAVGGG-(Q11)-CONH2 | 2681.0 | 2681.3 |
| Biotin-Q11 | Biotin-SGSG-(Q11)-CONH2 | 1999.2 | 2000.4 |
| Biotin-OVA | Biotin-SGSGISQAVHAAHAEINEAGR-COOH | 2288.5 | 2289.4 |
Gel formation and bulk measurements of peptide entrapment
Peptides were dissolved in water at specified concentrations and sonicated with a probe-type ultrasonicator to ensure complete dissolution. Overlaying these peptide solutions with Dulbecco’s phosphate buffered saline (PBS) for 40 min induced complete gelation in the peptide layer without mixing of the two layers. Gels were cast in culture inserts for cell experiments, microcentrifuge tubes for bulk analysis of peptide entrapment, and on the lower plate of an oscillating rheometer for viscoelasticity measurements. For cell culture experiments, which required exceptionally smooth surfaces in order to minimize any contributions of gel topography to cell behavior, prior to PBS addition the peptide solutions were incubated at 4°C overnight in culture inserts in sealed multi-well culture plates, which increased the viscosity of the peptide layer so that it was completely undisturbed by the addition of PBS. To measure the entrapment of ligand-bearing peptides, peptide solutions were prepared containing 20 mM total peptide, 5–50% ligand-bearing peptide, and the balance Q11. Mixed peptide solutions were overlaid with PBS, incubated for 1 h, washed three times with PBS for 30 min each, dissolved by mixing 1:2.3 with TFA, and analyzed on a C4 HPLC column. Relative peptide concentrations were determined by measuring peak areas at 215 nm, and gelled samples were compared with ungelled solutions to determine if any peptide leaching or unpredictable incorporation occurred. Gels were formed and measured in triplicate.
Transmission electron microscopy
Solutions of 1.5 mM aqueous peptide were mixed 1:2 with PBS, vortexed, sonicated, and allowed to fibrillize overnight at room temperature. Fibrils were then applied to 400 mesh carbon grids, stained with 1% uranyl acetate, and analyzed immediately using TEM (FEI Tecnai F30). For colloidal gold staining, grids with adsorbed fibrils were floated on droplets of 1% bovine serum albumin (BSA) to block non-specific binding, washed with distilled water, floated on droplets of 1 μg/mL streptavidin-colloidal gold conjugate (Invitrogen cat# A-32360), washed again, and stained with 1% uranyl acetate. This technique is similar to previously reported methods [19, 41, 42].
Circular dichroism spectroscopy
An AVIV 202 CD spectrometer (Aviv Biomedical, Lakewood, NJ, USA) was used with 0.1 cm path length quartz cells. To ensure that measurements were made on freshly assembled structures rather than on aged aggregates that could have formed during storage, immediately prior to CD experiments all peptides were dissolved in a small amount of neat TFA for 15 min, precipitated and washed extensively with diethyl ether, and dried for approximately 1 h in a vacuum desiccator. Peptides were then dissolved in degassed water, and their concentrations were adjusted to 1 mM using phenylalanine absorbance at 257 nm. Solutions of Q11 and mixtures containing 10% RGDS-Q11 or IKVAV-Q11 and 90% Q11 were diluted to a working concentration of 300 μM in CD-compatible buffer (4.3 mM KH2PO4, 1.4 mM Na2HPO4, 140 mM KF, pH 7.4) and analyzed immediately. Triplicate scans with a 1 nm step size were averaged at 25°C.
Oscillating rheometry
A Bohlin Gemini rheometer (Malvern Instruments Ltd., Malvern, UK) with an 8 mm parallel plate configuration was utilized. Gels were produced in situ on the bottom plate using a filter paper template with an 8 mm diameter hole. The template was pre-saturated with PBS, and peptide solutions were pipetted into it. The peptide solution was then gently overlaid with additional PBS and enclosed in a humidified chamber to minimize drying. After 40 min gelation, the PBS and template were removed, leaving a circular gel of 8 mm diameter. The top plate was lowered onto the gel until the top plate was completely and uniformly contacted, and frequency sweep measurements were performed from 0.01–10 Hz in a closed, humidified chamber at 0.1% strain. Three independent gels were tested at each formulation.
Endothelial cell culture
Primary human umbilical vein endothelial cells (HUVECs), Endothelial Growth Medium (EGM)-2, and subculture reagents were purchased from Lonza. HUVECs were maintained in EGM-2 containing 2% fetal bovine serum at 37°C/5% CO2 and subcultured according to the supplier’s protocols. To produce gels for culture experiments, 60μL of peptide solutions were pipetted into 24-well culture inserts (transparent PTFE membrane, 0.4 μm pore size, 1.13 cm2 area, Fisher cat# PICM-01250). Prior to adding medium and seeding cells, these solutions were incubated at 4°C overnight, which increased their viscosity so as to minimize surface perturbations upon PBS addition. Subsequent pipetting of PBS over these peptide solutions produced smooth gels with no surface features discernable by phase contrast microscopy. To adsorb fibronectin (FN), the gels were incubated under a solution of human plasma FN (Sigma cat# F2006, 100 μg/mL in EBM-2, Lonza cat# cc-3156) overnight.
Cell attachment, spreading, and growth
HUVECs were seeded at a density of 8,850 cells/cm2 on top of Q11 gels and on uncoated inserts. After one hour, the medium was removed, and unattached and loosely adherent cells were removed with serial PBS washes. Cells were fixed with 3.7% paraformaldehyde and permeabilized with 0.1% Triton X-100 for 30 min, respectively. Nuclei were stained with 10 ng/mL 4′,6-diamidino-2-phenylindole (DAPI), and f-actin was stained with phalloidin. Using epifluorescence microscopy, six non-overlapping images per gel were collected, and nuclei per gel were counted. Average projected areas were calculated by dividing the total phalloidin-stained area by the number of nuclei using ImageJ software. To measure cell growth, HUVECs were seeded on gels, cultured in serum-supplemented EGM-2 for 64 h, and analyzed with an MTS-based proliferation assay (Promega cat# G3582) according to the manufacturer’s protocols. Absorbance values were converted to the number of cells per unit area using a standard curve. Peptide cytotoxicity was measured using the same MTS assay after incubating confluent monolayers of HUVECs for 24 h in EGM-2 containing 0.01–1.0 mg/mL peptide. Higher peptide concentrations than these were not possible because gels were formed in the culture medium and disrupted the subsequent MTS assay. Controls included cultures without peptide, cultures that had been fixed with absolute ethanol, and wells without cells.
Peptide immunogenicity
C57BL/6 mice (6 weeks old, female, Taconic Farms, IN) were housed and fed in our institution’s Animal Facility. All Q11-based peptides were purified to >98% purity, and all peptide injections contained <0.06 EU/mL endotoxin, as measured by LAL chromogenic endpoint analysis (Lonza cat# 50-647U). Q11, RGDS-Q11, and 10% RGDS-Q11/90% Q11 were dissolved in sterile water. Peptide stocks were mixed 1:3 with sterile PBS to produce fibrillized peptides at concentrations of 2 mM. Mice were immunized subcutaneously with 100 μL of these solutions of fibrils and boosted after 4 weeks with an additional 50 μL of fibril solutions. Positive control mice received equivalent doses of an antigenic peptide from ovalbumin (OVA, residues 323–339, sequence ISQAVHAAHAEINEAGR, [43, 44], 91% purity) in complete Freund’s adjuvant (CFA), with boosting in incomplete Freund’s adjuvant (IFA). Negative control mice received injections of sterile PBS. After 5 weeks, the mice were sacrificed, their sera were collected, and anti-peptide IgG was detected in the sera using ELISA. To maintain identical peptide fibrillization, peptides were prepared for coating onto ELISA plates in the same way as they were for immunizations. Peptide samples were then diluted in PBS to 20 μg/mL and adsorbed onto high-binding ELISA plates overnight at 4°C. Washing steps were performed with 0.5% (v/v) Tween 20 in PBS (PBST), and plates were blocked with 1% BSA in PBST. Sera were serially diluted and applied to the plates. Bound mouse IgG was detected with peroxidase-conjugated goat anti-mouse IgG (H+L) (Jackson Immuno Research Laboratories cat# 115035003), and titers were determined by identifying the highest serum dilution having an absorbance greater than three standard deviations above the mean of the negative controls (PBS-injected mice). To ensure binding of the peptide to the plate, ELISA was additionally performed using streptavidin-coated plates (20μg/mL, coated overnight at 4°C) and biotinylated peptide fibrils. Biotinylated fibrils were created by co-assembling 1% biotinylated versions of the peptides into the samples (see Table 1). Mixed fibrils of Q11 and RGDS-Q11 contained biotinylated versions of both peptides, with 1% total biotinylated peptide. Serial dilutions of sera from negative control mice all had low background absorbances that were statistically identical to each other by ANOVA for dilutions as low as 1:10 for the adsorbed peptide ELISA and 1:100 for the bound peptide ELISA. Because of this, experimental groups were not analyzed in dilutions lower than this, and mice showing no positive titers were reported as having titers of 10−1 and 10−2, respectively. In all animal work, institutional guidelines for the care and use of laboratory animals were strictly followed under a protocol approved by our Institutional Animal Care and Use Committee.
Statistical analysis
Attachment, spreading, proliferation, cytotoxicity, and rheometry experiments were analyzed by one-way ANOVA with post-hoc comparisons. Experiments having residuals with equal variances (attachment, spreading, proliferation, and rheometry) were analyzed using Tukey’s HSD post-hoc comparisons, and experiments with normal but unequal variances (cytotoxicity) were analyzed using Tamhane’s T2 post hoc comparisons [45, 46].
RESULTS and DISCUSSION
Peptide synthesis, gelation, and bulk co-assembly
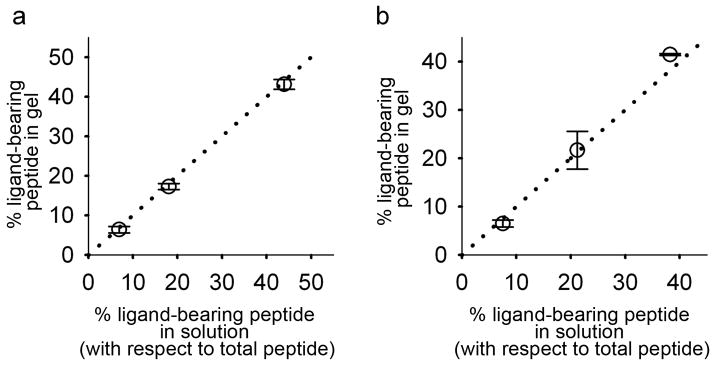
We encountered no significant synthetic difficulties for any of the peptides reported, and all Q11-based peptides were soluble in water at concentrations up to at least 40 mM. Self-supporting, translucent gels were formed, however, when aqueous solutions of 10–40 mM Q11 were overlaid with PBS. With careful pipetting, the underlying peptide layer and overlying PBS layer did not mix, and gelation was induced in the peptide layer by the diffusion of buffer constituents into it from the PBS. After gelation, the PBS layer could be removed and replaced with additional buffer or media. In addition, incubating Q11 solutions overnight at 4°C prior to the PBS overlay increased the viscosity of the Q11 solution slightly, resulting in an extremely smooth gel surface. These smooth gels allowed subsequent cell attachment, spreading, and proliferation experiments to be conducted without variability in gel topography. To investigate the co-assembly of multiple Q11-based peptides within these gels on a bulk scale, we measured the composition of peptide mixtures with HPLC before and after gelation (Figure 1). By comparing the peptide ratios of pre-gelled solutions with those that had been gelled and washed using PBS, it could be determined whether any of the co-assembled peptides leached out of the gels. It was found that RGDS-Q11 was quantitatively incorporated into Q11 matrices at every concentration tested, up to 44% RGDS-Q11/56% Q11. For IKVAV-Q11, no deviation was observed as high as 21% IKVAV-Q11/79% Q11, but at 38% the concentration of IKVAV-Q11 was 3.2% higher in gels than in pre-gelled solutions, indicating that at higher concentrations IKVAV-Q11 may disrupt the assembly of Q11. However, at lower levels of incorporation used in the subsequent experiments, 10 mol %, both RGDS-Q11 and IKVAV-Q11 were quantitatively incorporated. This feature enabled the production of gels with precisely defined incorporation of multiple peptides simply by mixing them in solution and inducing gelation.
Figure 1.
Quantitative bulk co-assembly of Q11 with RGDS-Q11 (a) and IKVAV-Q11 (b), as determined by comparing peptide ratios in gels with those of their corresponding pre-gelled peptide solutions using HPLC. The dotted line represents perfect matching between gels and solutions. Means ± SD with 9 gels represented per panel; 3 independent gels for each formulation.
Effect of ligand incorporation on fibrillization, secondary structure, and viscoelasticity of Q11 gels
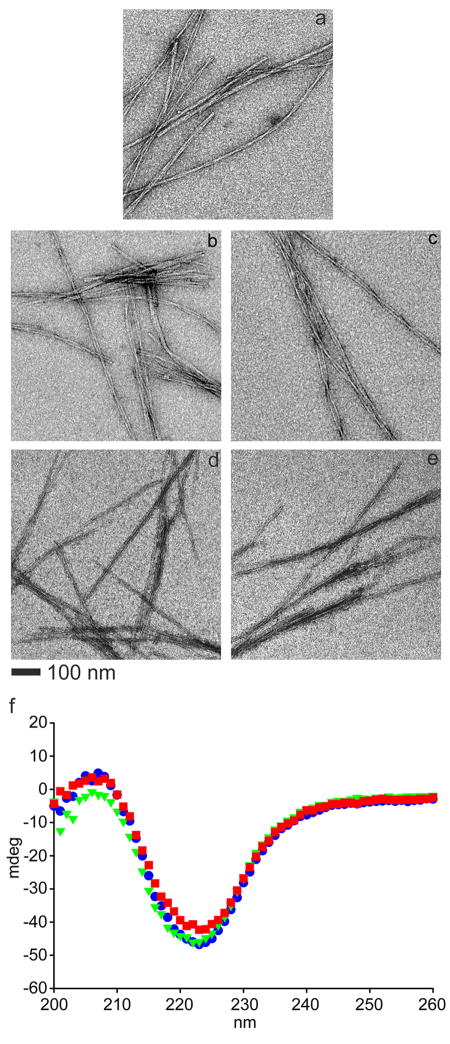
Cellular responses to ligand-presenting materials can be strongly affected by the material’s stiffness [24–26], so having independent control over both ligand incorporation and viscoelasticity is important for systematically optimizing such materials for driving a desired cell phenotype [9]. In order to determine whether the incorporation of ligand-bearing Q11 peptides altered the fibril morphology or stiffness of Q11-based gels, we employed TEM, circular dichroism, and oscillating rheometry. We focused on peptide mixtures containing 1:10 ratios of ligand-bearing peptides to Q11 based on previous investigations showing that this level of RGD incorporation significantly increased HUVEC proliferation in gels stiffened by native chemical ligation [13]. In addition, bulk analyses showed that this ratio was quantitatively incorporated, with no leaching or disruption of self-assembly. Scrambled versions of the ligand-bearing peptides (RDGS-Q11 and VAKVI-Q11) were also evaluated, as they were later employed as negative controls in subsequent cell culture experiments. Using TEM with negative staining, it was observed that none of these four peptides significantly altered the fibrillization of Q11 when combined in a 1:10 ratio with Q11 (Figure 2). Each peptide mixture containing 90% Q11 and 10% RGDS-Q11, IKVAV-Q11, RDGS-Q11 or VAKVI-Q11 formed fibrils having widths of approximately 10–20 nm and morphologies similar to 100% Q11 fibrils. There was no evidence of morphologically distinct subpopulations of fibrils that might indicate segregation of the peptides into mutually exclusive fibrils, so it was assumed that the mixed peptides formed co-assembled fibrils, a finding that was later verified using colloidal gold staining, described below. These fibril morphologies were also consistent with other previously reported β-sheet fibrillar peptides [47, 48]. Significant lateral aggregation was observed for all samples, although it is difficult to conclusively relate the observed interfibrillar interactions to those that may occur in the gels given that the samples were adsorbed and dried onto TEM grids.
Figure 2.
TEM of Q11-based peptide mixtures. Q11 (a), 10% RGDS-Q11/90% Q11 (b), 10% IKVAV-Q11/90% Q11 (c), 10% RDGS-Q11/90% Q11 (d), and 10% VAKVI-Q11/90% Q11 (e). Circular dichroism (f) of 100% Q11 (●, blue), 10% RGDS-Q11/90% Q11 (▼, green), and 10% IKVAV-Q11/90% Q11 (■, red). 300 μM peptide in 140 mM KF, 4.3 mM KH2PO4, 1.4 mM Na2HPO4, pH 7.4; 0.1 cm path length.
Using circular dichroism (CD) spectroscopy, we investigated whether the incorporation of 10% RGDS-Q11 or IKVAV-Q11 significantly changed Q11’s secondary structure. \Q11 displayed a positive band at 206 nm and a negative band at 224 nm (Figure 2f), consistent with previous reports for Q11 [49, 50] and the related peptide Ac-QQRFQWQFEQQ-Am [51], indicating a predominantly β-turn structure [47, 48]. This secondary structure is distinct from other self-assembling β-sheet peptides such as RADA16-I, which exhibits β-sheet structure [15]. The secondary structure of Q11 was maintained for mixtures of 10% RGDS-Q11 or IKVAV-Q11 with 90% Q11. In a further confirmation of the TEM results, these CD data indicated that the presence of the ligand-bearing peptides did not significantly alter the morphology of Q11 fibrils. Had the incorporation of the ligand-bearing peptides into Q11 fibrils significantly altered the β-sheet folding of Q11, one would expect to see more significant spectral changes. Additionally, had the ligand-bearing peptides assembled into distinct fibrils, one would expect to see additional spectral features added to the baseline of Q11’s spectrum. Note that the CD spectra have been plotted in terms of ellipticity (mdeg) rather than mean residue ellipticity owing to the fact that some samples were mixtures of different peptides.
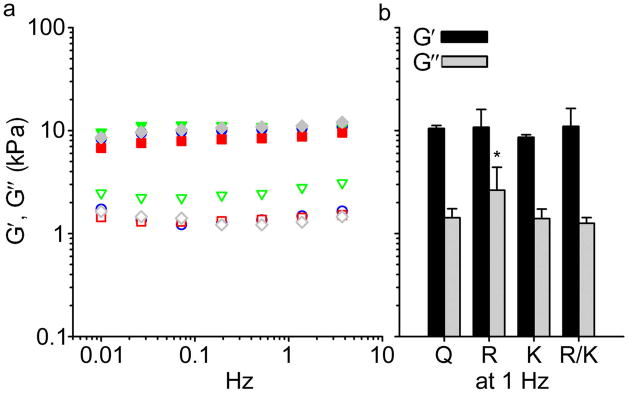
Having established that ligand incorporation did not alter fibril morphology, the viscoelasticity of Q11-based gels was subsequently investigated. In previous work, we found the stiffness of Q11 gels to be dependent on peptide concentration, with storage moduli ranging from 1–10 kPa for gels having peptide concentrations between 5–30 mM, respectively [13]. Such concentration-dependent stiffness has also been observed for other self-assembling peptide gels [17]. In the present work, gels containing 30 mM total peptide and 10% RGDS-Q11, 10% IKVAV-Q11, or 5% RGDS-Q11/5% IKVAV-Q11 were compared with unmodified Q11 gels. All four formulations demonstrated strain-insensitive behavior at oscillation frequencies between 0.01 Hz and 10 Hz, loss moduli (G″) that were about one order of magnitude smaller than storage moduli (G′), and no crossing of G′ and G″ at any frequency (Figure 3a). That is, each formulation exhibited the rheological characteristics of a cross-linked gel. Average storage moduli varied less than 2.5 kPa between groups, and evaluation of multiple gels did not reveal statistically significant differences between Q11 and ligand-bearing gels (Figure 3b). It is possible that statistical significance may be revealed with a larger sample size, but even if this were to be the case, the differences between groups are much smaller than those that have previously been found to affect cell behavior [25]. In the present experiments, the only statistically significant difference observed was for the loss modulus of the 10% RGDS-Q11/90% Q11 gels, which was slightly higher compared with the other gel formulations. However, small variances in G″ have not been previously associated with differential cell behavior. Because ligand incorporation had such a minimal impact on the gels’ viscoelasticity, this enables the simultaneous and independent adjustment of both ligand presentation and gel mechanics, for example by combining ligand-bearing peptides with covalent stiffening strategies [13] or by changing peptide concentration [17]. Having the ability to independently adjust both these aspects enables systematic adjustment of these matrices to achieve desired biological responses [9]. Moreover, RGDS and IKVAV are significantly different in terms of their chemical properties, RGDS being neutrally charged and hydrophilic, and IKVAV being positively charged and comparatively hydrophobic, suggesting that it would be possible to co-assemble a wide range of other peptide sequences within Q11 gels.
Figure 3.
Oscillating rheometry. Frequency sweep (a) and statistical comparison of multiple gels at 1 Hz (b). G′ (filled symbols) and G″ (open symbols) are shown for Q11 (●, ○, blue, labeled “Q” in (b)), 10% RGDS-Q11/90% Q11 (▼, ▽, green, “R”), 10% IKVAV-Q11/90% Q11 (■, □, red, “K”), and 5% RGDS-Q11/5% IKVAV-Q11/90% Q11 (◆, ◇, gray, “R/K”). All gels had 30 mM total peptide. *p < 0.05 by ANOVA with Tukey’s HSD post-hoc test. Means±SD; n=3 independent gels per group.
Visualization of ligand presentation on Q11 fibrils
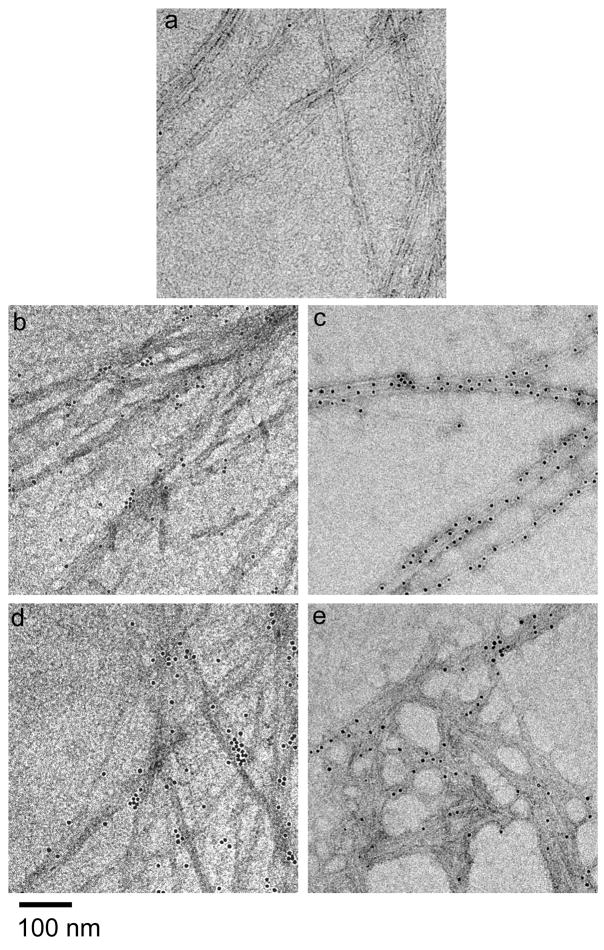
Using N-terminally biotinylated peptides, avidin-conjugated colloidal gold, and TEM, the display of the ligands by the peptide fibrils was visualized. Fibrils of biotin-RGDS-Q11, biotin-IKVAV-Q11, and mixtures of the ligand-bearing peptides with Q11 all stained specifically and strongly with avidin-gold compared to Q11 (Figure 4). This indicated that the N-termini of these peptides, and presumably their N-terminal ligands, were located on the surface of the fibrils. In the TEM images, some amount of resolution was sacrificed during blocking and gold labeling (see methods), but the fibrils were clearly visible. The gold labeling was distributed along the surface of the fibrils for each of the biotinylated groups, and for mixtures of peptides there was not significant evidence of segregation into distinct fibrils. This indicated that the peptides co-assembled into fibrils containing both peptides. Although it was clear that the RGDS and IKVAV ligands were present on the fibril surface, it was not clear whether all of them were present or whether there existed some degree of heterogeneity in fibril structure. However, the morphologic similarities between the ligand-bearing co-assemblies and pure Q11 (Figure 2), as well as the gold labeling, suggested that most of the ligand sequences were not buried within the peptide fibrils. Such burial might be expected to induce greater morphological changes than those observed. Subsequent cell culture experiments, described below, further supported that these ligands were functionally available.
Figure 4.
Ligands were displayed on the surface of Q11 fibrils, as evidenced by the labeling of biotinylated ligand-bearing peptides with streptavidin-colloidal gold. Q11 fibrils showed minimal background gold staining (a), whereas 10% biotin-RGDS-Q11/90% Q11 (b), 100% biotin-RGDS-Q11 (c), 10% biotin-IKVAV-Q11/90% Q11 (d) and 100% biotin-IKVAV-Q11/90% Q11 (e) bound significant levels of avidin-gold.
Modulation of cell attachment, spreading, and growth in HUVEC cultures
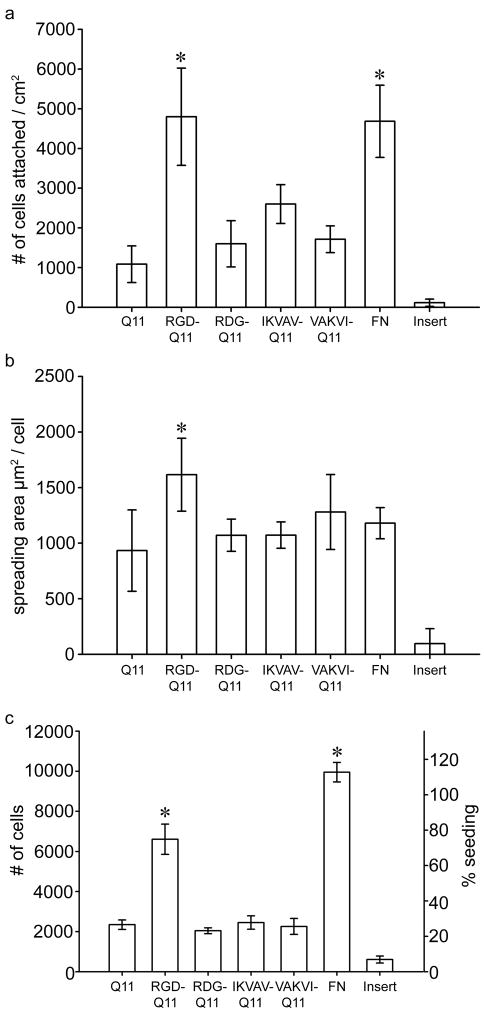
The behavior of HUVECs on Q11 gels was modulated by the inclusion of RGDS-Q11 and IKVAV-Q11 in the matrix. One hour after seeding, the attachment of HUVECs to 30mM peptide gels containing 10% RGDS-Q11 was fivefold greater than on Q11 gels. In addition, cell attachment was as extensive on RGDS-bearing gels as on fibronectin-adsorbed Q11 gels, a highly adhesive surface (Figure 5a). HUVECs did not attach, spread, or proliferate to any degree on unmodified culture inserts. The response to RGDS-Q11 was specific, as the scrambled RDGS-Q11 peptide abolished this effect. Collectively, these cell attachment data in conjunction with the TEM and CD data indicated that the RGDS ligand was presented by the peptide fibrils and available for specific cell binding, even in serum-containing medium. The inclusion of 10% IKVAV-Q11 also increased HUVEC attachment, but to a lesser degree. This increased attachment was statistically significant by t-test compared with Q11 and the scrambled VAKVI-Q11 (p= 0.002 and 0.01, respectively), but it barely failed to reach statistical significance by ANOVA when compared within the experiment as a whole (p= 0.06 with Tukey’s HSD post-hoc test). These results are interpreted to mean that IKVAV-Q11 had a detectable but small effect on HUVEC attachment. This small effect of IKVAV on HUVEC adhesion was not altogether unexpected, because in previous work it was found that surface-bound IKVAV only slightly increased endothelial cell attachment, while at the same time inducing a more spindle-shaped, migratory morphology [36–38]. The similar response induced by IKVAV-Q11 on HUVECs, along with the sequence specificity indicated by the scrambled peptide control, suggested that the IKVAV sequence was also displayed by the Q11 fibrils. To observe a more dramatic effect of this peptide on different cell types, it may be interesting in future work to investigate Q11-based matrices in cultures of primary neurons or neuronal cell lines, as IKVAV has been shown to modulate neuronal cell behavior in a variety of different biomaterials contexts [40, 52–54].
Figure 5.
HUVEC behavior on Q11 gels. Attachment (a) and spreading (b) at 1 h (n=4 independent gels; means±SD). Cell numbers (c) at 64 h (n=5, means±SD). *p < 0.05 by ANOVA with Tukey’s HSD post-hoc test.
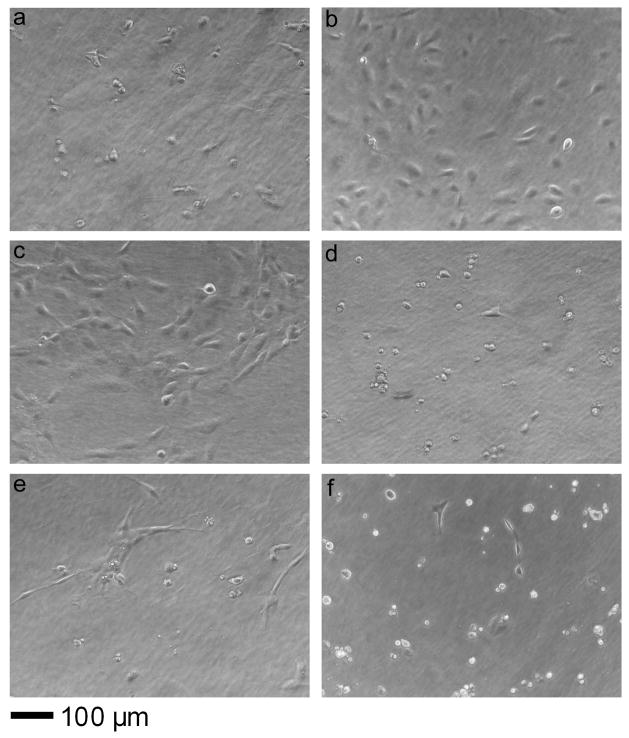
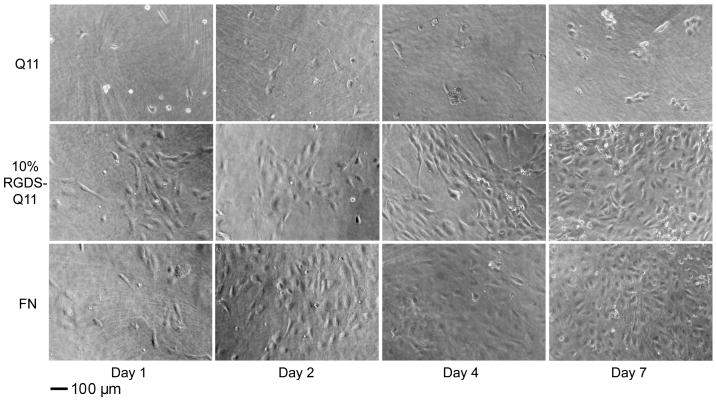
Cell spreading at short time points was less affected than overall attachment by the inclusion of ligands (Figure 5b). At one hour, only 10% RGDS-Q11 gels showed a significant increase in spreading over unmodified Q11, and this difference was relatively small. At later time points, however, much larger differences in cell spreading and morphology were observed by phase contrast microscopy. After three days, FN-coated and 10% RGDS-Q11 gels showed significantly more cell coverage in comparison to unmodified Q11, the scrambled RDGS-Q11 control surface, or IKVAV-containing gels (Figure 6). On FN-coated and RGDS-Q11-containing matrices, HUVECs exhibited morphologies approaching a normal cobblestone appearance, whereas the IKVAV-Q11-containing surfaces promoted a more spindle-shaped, elongated morphology with far fewer cells. With an MTS-based proliferation assay, cell growth was quantified at this time point (3 days, Figure 5c), showing that 10% RGDS-Q11 gels supported increased cell growth at a rate between Q11 and fibronectin-coated Q11. Similarly to cell attachment, the scrambled RDGS-Q11 did not produce this effect. Proliferation was not measured directly, so it was not determined whether the higher cell numbers after 3 days were a result of improved early adhesion or an increase in proliferation, but it is likely that both mechanisms contributed. Looking more closely at the RGDS-containing gels, additional cultures were followed for up to seven days, after which both RGDS-containing and fibronectin-adsorbed gels exhibited confluent monolayers with normal cobblestone morphology (Figure 7). This was in contrast to unfunctionalized Q11 gels, which showed only sparse clusters containing a few cells each. Taken together, the cell attachment, spreading, and growth data indicated that RGDS-Q11 assembled in such a way that the RGDS ligand was specifically available on the surface of the fibrils for modulating cell behavior. The effect of IKVAV was more subtle, as proliferation was not increased, but similar to previous observations [36], HUVECs on IKVAV-containing gels exhibited a more spindle-shaped morphology, contrasting with the cobblestone appearance on FN or RGDS-containing gels. It has been found previously that IKVAV may promote a more migratory phenotype of endothelial cells [36, 55], which is consistent with our findings. This aspect could be further clarified with more focused cell migration or invasion assays. Additionally, previous work has indicated that IKVAV promotes active collagen IV metalloproteases, which could liberate additional soluble signals from the ECM [36]. With a defined system such as the one presented here, this mechanism could be more systematically explored.
Figure 6.
Phase contrast images of HUVEC cultures at 64 h on Q11 (a), fibronectin-adsorbed Q11 (b), 10% RGDS-Q11/90% Q11 (c), 10% RDGS-Q11/90% Q11 (d), 10% IKVAV-Q11/90% Q11 (e), and 10% VAKVI-Q11/90% Q11 (f).
Figure 7.
Phase contrast images of HUVEC cultures on Q11, 10% RGDS-Q11/90% Q11, and fibronectin-adsorbed Q11 (FN) at days 1,2,4, and 7.
Non-cytotoxicity and non-immunogenicity of Q11-based peptides
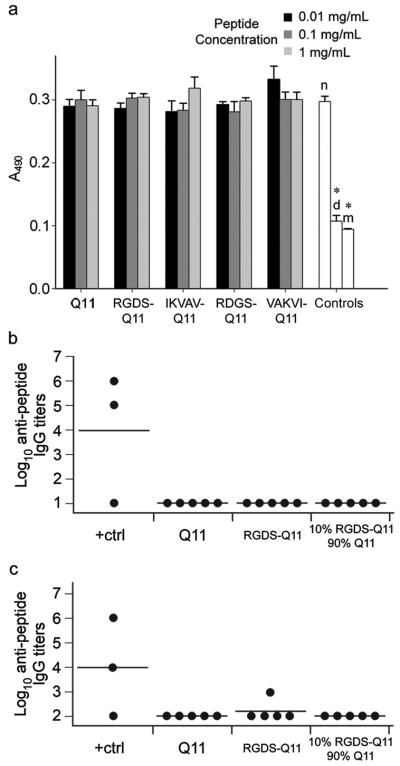
Glutamine-rich β-sheet-forming peptides that are significantly longer than Q11 can be cytotoxic, especially when the polyglutamine stretches are longer than 37 residues [56]. Understanding the cytotoxicity of Q11 and its derivatives is necessary for guiding the future development of Q11-based peptides as biomaterials, and also for determining that the variable cell attachment, spreading, and growth of HUVECs observed in the present study did not arise from differences in cytotoxicity. In confluent cultures of HUVECs, none of the Q11-based peptides were found to be cytotoxic at any concentration tested (Figure 8a), eliminating this possibility and clearing the path for employing these peptides in vitro.
Figure 8.
Cytotoxicity (a) and immunogenicity (b, c) of Q11-based peptides. In (a), controls included no peptide (“n”), dead cells fixed with ethanol (“d”), and medium only (no cells, “m”). *p < 0.05 compared to no peptide control by ANOVA with Tamhane’s T2 post-hoc test (n=6, means±SD). In (b) and (c), IgG titers against the injected peptide fibrils were measured by ELISA on plates coated with adsorbed peptide (b) or with peptide bound via streptavidin-biotin (c). Controls: PBS injection (negative) and OVA peptide in CFA (positive). See methods for determination of titers. Lines represent average titers for each group, and each data point represents an individual mouse.
To understand the applicability of Q11-based materials for vivo applications, it is necessary to characterize their immunogenicity. Aggregation, polymerization, or assembly can increase peptide and protein immunogenicity, often unpredictably [27]. For ligand-bearing self-assembled β-sheet fibrillar biomaterials, previous reports have demonstrated minimal inflammatory responses in vivo [2, 30, 31], but an investigation of IgG production against such scaffolds has not previously been reported. To evaluate immunogenicity, we immunized and boosted C57BL/6 mice with Q11, RGDS-Q11, or mixtures of the two peptides, and we measured the production of anti-peptide IgG in the mice’s sera using ELISA. IKVAV-Q11 was not evaluated owing to its more subtle activity in the cell culture experiments. Despite the fact that the Q11-based peptides formed fibrils of non-native amino acid sequences, very low immunogenicity was found (Figure 8b, c). To ensure that these low readings were not a result of poor peptide binding to the ELISA plates, IgG levels were measured both on plates coated with adsorbed peptide (Figure 8b) and on plates where the peptides were bound via streptavidin-biotin (Figure 8c). These assays were sensitive to dilutions as low as 1:10 for the adsorbed peptide and 1:100 for the bound peptide (see methods). Only one mouse produced an IgG titer above background for RGDS-Q11, detectable only with the bound peptide ELISA (Figure 8c). No mice in the Q11 or 10% RGDS-Q11/90% Q11 groups showed positive responses, in contrast with the positive control mice, two of which showed an intense response with high titers of anti-OVA IgG, both by the adsorbed peptide ELISA and the bound peptide ELISA. One positive control mouse resulted in a failed immunization, illustrating the immunological variability of wild-type animals. These results indicated that RGDS-Q11, Q11, and mixtures of the two were minimally immunogenic, an important finding given that the RGDS sequence has in some cases enhanced the immunogenicity of other peptides [57, 58], and multivalent repetition of epitopes can lead to strong B cell responses [28, 59]. Owing to the minimal levels of anti-peptide IgG found in the experiments described here, neither of these two processes appeared to be significantly engaged for the Q11-based peptides tested. However, the single mouse from the RGDS-Q11 group showing a small but measurable anti-peptide IgG titer indicated that there is a potential for self-assembled, ligand-bearing, fibrillar peptide materials to interact with the immune system to some degree. Interestingly, even though one mouse from the RGDS-Q11 group showed a positive response, no positive responses were found in mixtures of the RGDS-Q11 peptide with Q11. A larger study will need to be conducted to determine the extent to which the density of RGDS on the surface of the fibrils affects immunogenicity. Further, it is not known what impact, if any, low levels of anti-peptide IgG production may have on the various biomedical applications envisioned for these materials, including scaffolds for regenerative medicine or coatings for implants. It is possible that some degree of anti-peptide IgG production may be tolerated in such settings, especially if the materials’ lifetime is limited by its degradation. Currently, these aspects are not well understood and will need to be clarified through future work as these materials progress towards clinical applications. Nevertheless, the findings reported here for RGDS-Q11 and Q11/RGDS-Q11 mixtures are to our knowledge the first direct evidence of ligand-bearing β-sheet fibrils demonstrating low immunogenicity. Coupled with their non-cytotoxicity, modular construction, and ability to display different ligands without significantly altering viscoelasticity, this builds a strong rationale for further developing these peptides and related peptides for in vivo applications, including scaffolds for regenerative medicine and bioactive coatings for biomaterials.
CONCLUSIONS
We report here a co-assembling set of peptides based on the sequence of Q11 that form integrated hydrogels, where the display of multiple ligands may be adjusted simply by mixing different peptides in solution and inducing gelation. RGDS-Q11 and IKVAV-Q11 were quantitatively incorporated into background gels of Q11 in a wide range of peptide ratios, and ligand incorporation had no significant impact on fibril morphology or secondary structure at levels capable of influencing HUVEC behavior. Gel viscoelasticity was minimally changed upon ligand inclusion, allowing ligand incorporation to be adjusted independently of gel mechanics. Both RGDS-Q11 and IKVAV-Q11 were presented on the surface of co-assemblies with Q11, and these ligands modulated HUVEC behavior in vitro. RGDS-Q11 significantly affected HUVEC attachment, spreading, and proliferation, while IKVAV-Q11 had a small effect on cell attachment and a subtle influence on cell morphology. Scrambled peptides for both RGDS and IKVAV abolished these effects, indicating that these responses were specific, even in serum-containing medium. Finally, none of the peptides investigated were cytotoxic, and Q11 and RGDS-Q11 were minimally immunogenic in mice. We anticipate that modular strategies such as the approach reported here will become increasingly important in designing biomaterials that are both complexly bioactive and tunable.
Acknowledgments
This research was supported in part by The American Heart Association (grant no. 0665218B), the National Science Foundation (grant no. CHE-0802286), and the National Institutes of Health (NIBIB, grant no. EB007335). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Biomedical Imaging and BioEngineering or the National Institutes of Health. We thank Karl Matlin and Jose Moyano for helpful discussions. TEM and CD were performed at the University of Chicago Electron Microscopy Facility and Biophysics Core Facility, respectively. We thank Dr. Dale Schaefer for access to the rheometer.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kisiday J, Jin M, Kurz B, Hung H, Semino C, Zhang S, et al. Self-assembling peptide hydrogel fosters chondrocyte extracellular matrix production and cell division: Implications for cartilage tissue repair. Proc Natl Acad Sci U S A. 2002;99:9996–10001. doi: 10.1073/pnas.142309999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, et al. Selective differentiation of neural progenitor cells by high-epitope density nanofibers. Science. 2004;303:1352–1355. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 3.Ramachandran S, Yu YB. Peptide-based viscoelastic matrices for drug delivery and tissue repair. BioDrugs. 2006;20:263–269. doi: 10.2165/00063030-200620050-00001. [DOI] [PubMed] [Google Scholar]
- 4.Law B, Weissleder R, Tung CH. Peptide-based biomaterials for protease-enhanced drug delivery. Biomacromolecules. 2006;7:1261–1265. doi: 10.1021/bm050920f. [DOI] [PubMed] [Google Scholar]
- 5.Segers VFM, Lee RT. Local delivery of proteins and the use of self-assembling peptides. Drug Discov Today. 2007;12:561–568. doi: 10.1016/j.drudis.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Hsieh PCH, MacGillivray C, Gannon J, Cruz FU, Lee RT. Local controlled intramyocardial delivery of platelet-derived growth factor improves postinfarction ventricular function without pulmonary toxicity. Circulation. 2006;114:637–644. doi: 10.1161/CIRCULATIONAHA.106.639831. [DOI] [PubMed] [Google Scholar]
- 7.Zhang S, Holmes TC, DiPersio CM, Hynes RO, Su X, Rich A. Self-complementary oligopeptide matrices support mammalian cell attachment. Biomaterials. 1995;16:1385–1393. doi: 10.1016/0142-9612(95)96874-y. [DOI] [PubMed] [Google Scholar]
- 8.Horii A, Wang X, Gelain F, Zhang S. Biological designer self-assembling peptide nanofiber scaffolds significantly enhance osteoblast proliferation, differentiation and 3-D migration. PLoS ONE. 2007;2:e190. doi: 10.1371/journal.pone.0000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collier JH. Modular self-assembling biomaterials for directing cellular responses. Soft Matter. 2008;4:2310–2315. doi: 10.1039/b805563g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 11.Haines-Butterick L, Rajagopal K, Branco M, Salick D, Rughani R, Pilarz M, et al. Controlling hydrogelation kinetics by peptide design for three-dimensional encapsulation and injectable delivery of cells. Proc Natl Acad Sci U S A. 2007;104:7791–7796. doi: 10.1073/pnas.0701980104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kretsinger JK, Haines LA, Ozbas B, Pochan DJ, Schneider JP. Cytocompatibility of self-assembled β-hairpin peptide hydrogel surfaces. Biomaterials. 2005;26:5177–5186. doi: 10.1016/j.biomaterials.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 13.Jung JP, Jones JL, Cronier SA, Collier JH. Modulating the mechanical properties of self-assembled peptide hydrogels via native chemical ligation. Biomaterials. 2008;29:2143–2151. doi: 10.1016/j.biomaterials.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gelain F, Bottai D, Vescovi A, Zhang S. Designer self-assembling peptide nanofiber scaffolds for adult mouse neural stem cell 3-dimensional cultures. PLoS ONE. 2006;1:e119. doi: 10.1371/journal.pone.0000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genove E, Shen C, Zhang S, Semino CE. The effect of functionalized self-assembling peptide scaffolds on human aortic endothelial cell function. Biomaterials. 2005;26(16):3341–3351. doi: 10.1016/j.biomaterials.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 16.Holmes TC, de Lacalle S, Su X, Liu G, Rich A, Zhang S. Extensive neurite outgrowth and active synapse formation on self-assembling peptide scaffolds. Proc Natl Acad Sci U S A. 2000;97:6728–6733. doi: 10.1073/pnas.97.12.6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sieminski A, Was A, Kim G, Gong H, Kamm R. The stiffness of three-dimensional ionic self-assembling peptide gels affects the extent of capillary-like network formation. Cell Biochem Biophys. 2007;49:73–83. doi: 10.1007/s12013-007-0046-1. [DOI] [PubMed] [Google Scholar]
- 18.Kasai S, Ohga Y, Mochizuki M, Nishi N, Kadoya Y, Nomizu M. Multifunctional peptide fibrils for biomedical materials. Peptide Sci. 2004;76:27–33. doi: 10.1002/bip.10565. [DOI] [PubMed] [Google Scholar]
- 19.Gras SL, Tickler AK, Squires AM, dDevlin GL, Horton MA, Dobson CM, et al. Functionalised amyloid fibrils for roles in cell adhesion. Biomaterials. 2008;29:1553–1562. doi: 10.1016/j.biomaterials.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 20.Hartgerink JD, Beniash E, Stupp SI. Self-assembly and mineralization of peptide-amphiphile nanofibers. Science. 2001;294:1684–1688. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 21.Storrie H, Guler MO, Abu-Amara SN, Volberg T, Rao M, Geiger B, et al. Supramolecular crafting of cell adhesion. Biomaterials. 2007;28:4608–4618. doi: 10.1016/j.biomaterials.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 22.Guler MO, Hsu L, Soukasene S, Harrington DA, Hulvat JF, Stupp SI. Presentation of RGDS epitopes on self-assembled nanofibers of branched peptide amphiphiles. Biomacromolecules. 2006;7:1855–1863. doi: 10.1021/bm060161g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chau Y, Luo Y, Cheung AC, Nagai Y, Zhang S, Kobler JB, et al. Incorporation of a matrix metalloproteinase-sensitive substrate into self-assembling peptides - a model for biofunctional scaffolds. Biomaterials. 2008;29:1713–1719. doi: 10.1016/j.biomaterials.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 24.Discher DE, Janmey P, Wang Y-l. Tissue Cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 25.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 26.Pelham RJ, Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci U S A. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maas C, Hermeling S, Bouma B, Jiskoot W, Gebbink MFBG. A role for protein misfolding in immunogenicity of biopharmaceuticals. J Biol Chem. 2007;282:2229–2236. doi: 10.1074/jbc.M605984200. [DOI] [PubMed] [Google Scholar]
- 28.Fehr T, Bachmann MF, Bucher E, Kalinke U, Padova FE, Lang AB, et al. Role of repetitive antigen patterns for induction of antibodies against antibodies. J Exp Med. 1997;185:1785–1792. doi: 10.1084/jem.185.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenberg AS. Effects of protein aggregates: An immunologic perspective. AAPS Journal. 2006;8:E501–E507. doi: 10.1208/aapsj080359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis ME, Hsieh PCH, Takahashi T, Song Q, Zhang SG, Kamm RD, et al. Local myocardial insulin-like growth factor 1 (IGF-1) delivery with biotinylated peptide nanofibers improves cell therapy for myocardial infarction. Proc Natl Acad Sci U S A. 2006;103:8155–8160. doi: 10.1073/pnas.0602877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kapadia MR, Chow LW, Tsihlis ND, Ahanchi SS, Eng JW, Murar J, et al. Nitric oxide and nanotechnology: a novel approach to inhibit neointimal hyperplasia. J Vasc Surg. 2008;47:173–182. doi: 10.1016/j.jvs.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruoslahti E, Pierschbacher MD. New perspectives in cell adhesion: RGD and integrins. Science. 1987;238:491–497. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 33.Hersel U, Dahmen C, Kessler H. RGD modified polymers: biomaterials for stimulated cell adhesion and beyond. Biomaterials. 2003;24:4385–4415. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- 34.Adams DN, Kao EYC, Hypolite CL, Distefano MD, Hu WS, Letourneau PC. Growth cones turn and migrate up an immobilized gradient of the laminin IKVAV peptide. J Neurobiol. 2005;62:134–147. doi: 10.1002/neu.20075. [DOI] [PubMed] [Google Scholar]
- 35.Tysseling-Mattiace VM, Sahni V, Niece KL, Birch D, Czeisler C, Fehlings MG, et al. Self-assembling nanofibers inhibit glial scar formation and promote axon elongation after spinal cord injury. J Neurosci. 2008;28:3814–3823. doi: 10.1523/JNEUROSCI.0143-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grant DS, Kinsella JL, Fridman R, Auerbach R, Piasecki BA, Yamada Y, et al. Interaction of endothelial cells with a laminin A chain peptide (SIKVAV) in vitro and induction of angiogenic behavior in vivo. J Cell Physiol. 1992;153:614–625. doi: 10.1002/jcp.1041530324. [DOI] [PubMed] [Google Scholar]
- 37.Berat R, Remy-Zolghadry M, Gounou C, Manigand C, Tan S, Salto C, et al. Peptide-presenting two-dimensional protein matrix on supported lipid bilayers: An efficient platform for cell adhesion. Biointerphases. 2007;2:165–172. doi: 10.1116/1.2821954. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura M, Mie M, Mihara H, Nakamura M, Kobatake E. Construction of multi-functional extracellular matrix proteins that promote tube formation of endothelial cells. Biomaterials. 2008;29:2977–2986. doi: 10.1016/j.biomaterials.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 39.Freitas VM, Vilas-Boas VF, Pimenta DC, Loureiro V, Juliano MA, Carvalho MR, et al. SIKVAV, a laminin alpha1-derived peptide, interacts with integrins and increases protease activity of a human salivary gland adenoid cystic carcinoma cell line through the ERK ½ signaling pathway. Am J Pathol. 2007;171:124–138. doi: 10.2353/ajpath.2007.051264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kleinman HK, Weeks BS, Cannon FB, Sweeney TM, Sephel GC, Clement B, et al. Identification of a 110-kDa nonintegrin cell surface laminin-binding protein which recognizes an a chain neurite-promoting peptide. Arch Biochem Biophys. 1991;290:320–325. doi: 10.1016/0003-9861(91)90547-v. [DOI] [PubMed] [Google Scholar]
- 41.Melchor JP, Van Nostrand WE. Fibrillar Amyloid beta-protein mediates the pathologic accumulation of its secreted precursor in human cerebrovascular smooth muscle cells. J Biol Chem. 2000;275:9782–9791. doi: 10.1074/jbc.275.13.9782. [DOI] [PubMed] [Google Scholar]
- 42.Matsumura S, Uemura S, Mihara H. Construction of biotinylated peptide nanotubes for arranging proteins. Molecular BioSystems. 2005;1:146–148. doi: 10.1039/b504516a. [DOI] [PubMed] [Google Scholar]
- 43.Renz H, Bradley K, Larsen GL, McCall C, Gelfand EW. Comparison of the allergenicity of ovalbumin and ovalbumin peptide 323–339. Differential expansion of V beta-expressing T cell populations. J Immunol. 1993;151:7206–7213. [PubMed] [Google Scholar]
- 44.Seroogy CM, Soares L, Ranheim EA, Su L, Holness C, Bloom D, et al. The gene related to anergy in lymphocytes, an E3 ubiquitin ligase, is necessary for anergy induction in CD4 T cells. J Immunol. 2004;173:79–85. doi: 10.4049/jimmunol.173.1.79. [DOI] [PubMed] [Google Scholar]
- 45.Tamhane AC. Multiple comparisons in model I: one-way ANOVA with unequal variances. Communications in Statistics. 1977;A6:15–32. [Google Scholar]
- 46.Tamhane AC. Multiple comparisons. In: Ghosh S, Rao CR, editors. Handbook of Statistics, Design and Analysis of Experiments. Amsterdam: Elsevier Science; 1996. pp. 587–630. [Google Scholar]
- 47.Brahms S, Brahms J. Determination of protein secondary structure in solution by vacuum ultraviolet circular dichroism. J Molec Biol. 1980;138:149–178. doi: 10.1016/0022-2836(80)90282-x. [DOI] [PubMed] [Google Scholar]
- 48.Bush CA, Sarkar SK, Kopple KD. Circular dichroism of β-turns in peptides and proteins. Biochemistry. 1978;17:4951–4954. doi: 10.1021/bi00616a015. [DOI] [PubMed] [Google Scholar]
- 49.Collier JH, Messersmith PB. Enzymatic modification of self-assembled peptide structures with tissue transglutaminase. Bioconjugate Chem. 2003;14:748–755. doi: 10.1021/bc034017t. [DOI] [PubMed] [Google Scholar]
- 50.Collier JH, Messersmith PB. Self-assembling polymer-peptide conjugates: Nanostructural tailoring. Adv Mater. 2004;16:907–910. [Google Scholar]
- 51.Aggeli A, Bell M, Boden N, Keen JN, McLeish TCB, Nyrkova I, et al. Engineering of peptide beta-sheet nanotapes. J Mater Chem. 1997;7:1135–1145. [Google Scholar]
- 52.Richard BL, Nomizu M, Yamada Y, Kleinman HK. Identification of synthetic peptides derived from laminin α1 and α2 chains with cell type specificity for neurite outgrowth. Exp Cell Res. 1996;228:98–105. doi: 10.1006/excr.1996.0304. [DOI] [PubMed] [Google Scholar]
- 53.Nomizu M, Weeks BS, Weston CA, Kim WH, Kleinman HK, Yamada Y. Structure-activity study of a laminin α1 chain active peptide segment Ile-Lys-Val-Ala-Val (IKVAV) FEBS Lett. 1995;365:227–231. doi: 10.1016/0014-5793(95)00475-o. [DOI] [PubMed] [Google Scholar]
- 54.Kato K, Utani A, Suzuki N, Mochizuki M, Yamada M, Nishi N, et al. Identification of neurite outgrowth promoting sites on the laminin alpha 3 chain G domain. Biochemistry. 2002;41:10747–10753. doi: 10.1021/bi020180k. [DOI] [PubMed] [Google Scholar]
- 55.Schnaper HW, Kleinman HK, Grant DS. Role of laminin in endothelial cell recognition and differentiation. Kidney Intl. 1993;43:20–25. doi: 10.1038/ki.1993.5. [DOI] [PubMed] [Google Scholar]
- 56.Chen S, Berthelier V, Yang W, Wetzel R. Polyglutamine aggregation behavior in vitro supports a recruitment mechanism of cytotoxicity. J Molec Biol. 2001;311:173–182. doi: 10.1006/jmbi.2001.4850. [DOI] [PubMed] [Google Scholar]
- 57.Yano A, Onozuka A, Asahi-Ozaki Y, Imai S, Hanada N, Miwa Y, et al. An ingenious design for peptide vaccines. Vaccine. 2005;23:2322–2326. doi: 10.1016/j.vaccine.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 58.Yano A, Onozuka A, Matin K, Imai S, Hanada N, Nisizawa T. RGD motif enhances immunogenicity and adjuvanicity of peptide antigens following intranasal immunization. Vaccine. 2003;22:237–243. doi: 10.1016/s0264-410x(03)00561-9. [DOI] [PubMed] [Google Scholar]
- 59.Bachmann MF, Rohrer UH, Kundig TM, Burki K, Hengartner H, Zinkernagel RM. The influence of antigen organization on B cell responsiveness. Science. 1993;262:1448–1451. doi: 10.1126/science.8248784. [DOI] [PubMed] [Google Scholar]