Abstract
Large sections of the 3′ untranslated region (UTR) of hepatitis C virus (HCV) were deleted from an infectious cDNA clone, and the RNA transcripts from seven deletion mutants were tested sequentially for infectivity in a chimpanzee. Mutants lacking all or part of the 3′ terminal conserved region or the poly(U–UC) region were unable to infect the chimpanzee, indicating that both regions are critical for infectivity in vivo. However, the third region, the variable region, was able to tolerate a deletion that destroyed the two putative stem–loop structures within this region. Mutant VR-24 containing a deletion of the proximal 24 nt of the variable region of the 3′ UTR was viable in the chimpanzee and seemed to replicate as well as the undeleted parent virus. The chimpanzee became viremic 1 week after inoculation with mutant VR-24, and the HCV genome titer increased over time during the early acute infection. Therefore, the poly(U–UC) region and the conserved region, but not the variable region, of the 3′ UTR seem to be critical for in vivo infectivity of HCV.
More than 170 million people worldwide are infected chronically with the hepatitis C virus (HCV; ref. 1), and HCV is the most common cause of chronic liver disease and hepatocellular carcinoma in many countries (2, 3). Furthermore, HCV-related liver failure frequently requires liver transplantation (2). In addition, HCV infection is associated with a number of extrahepatic diseases, most notably mixed cryoglobulinemia and glomerulonephritis (3). Thus, HCV infection is a major global public health problem, and there is an urgent need to understand the virus better and to develop vaccines and more effective treatment.
HCV has been classified as the sole member of the genus Hepacivirus within the Flaviviridae family (4). Like members of the genera Flavivirus and Pestivirus, HCV has a positive-sense single-stranded RNA genome that encodes a single long ORF bounded by 5′ and 3′ untranslated regions (UTR). The UTRs of HCV have highly conserved elements believed to be critical for RNA replication and packaging, as well as for polyprotein translation (5–8). Additionally, elements of the 5′ UTR function as an internal ribosome entry site for initiation of polyprotein translation (9). The 3′ UTR consists of a short sequence of variable length and composition (variable region), a polypyrimidine tract [poly(U–UC) region], and a highly conserved sequence of approximately 100 nt at the 3′ end (conserved region or 3′X; refs. 6–8 and 10). These three distinct regions were found among isolates of all six HCV genotypes (6–8, 10–12). The sequence of the conserved region has the potential to form three stem–loop structures, including a highly stable stem–loop of 46 nt at the 3′ end (6–8, 13). In many other positive-strand RNA viruses, including the related flaviviruses, conserved sequences or structures at or near the 3′ end represent cis elements critical for viral replication (reviewed in ref. 6). However, the importance of different conserved elements of the 3′ UTR of HCV has not been established experimentally.
The development of infectious cDNA clones of HCV (14–16) and the refinement of in vivo transfection techniques (16) have made it feasible to test the effect of various mutations on the ability of HCV to replicate in the only validated animal model, the chimpanzee. In the present study, we sequentially inoculated a chimpanzee with a series of mutants containing large deletions in the 3′ UTR and determined which mutations were lethal as a first step toward mapping critical regions of the 3′ UTR.
MATERIALS AND METHODS
Amplification and Cloning of the 3′ UTR Deletion Mutants of pCV-H77C.
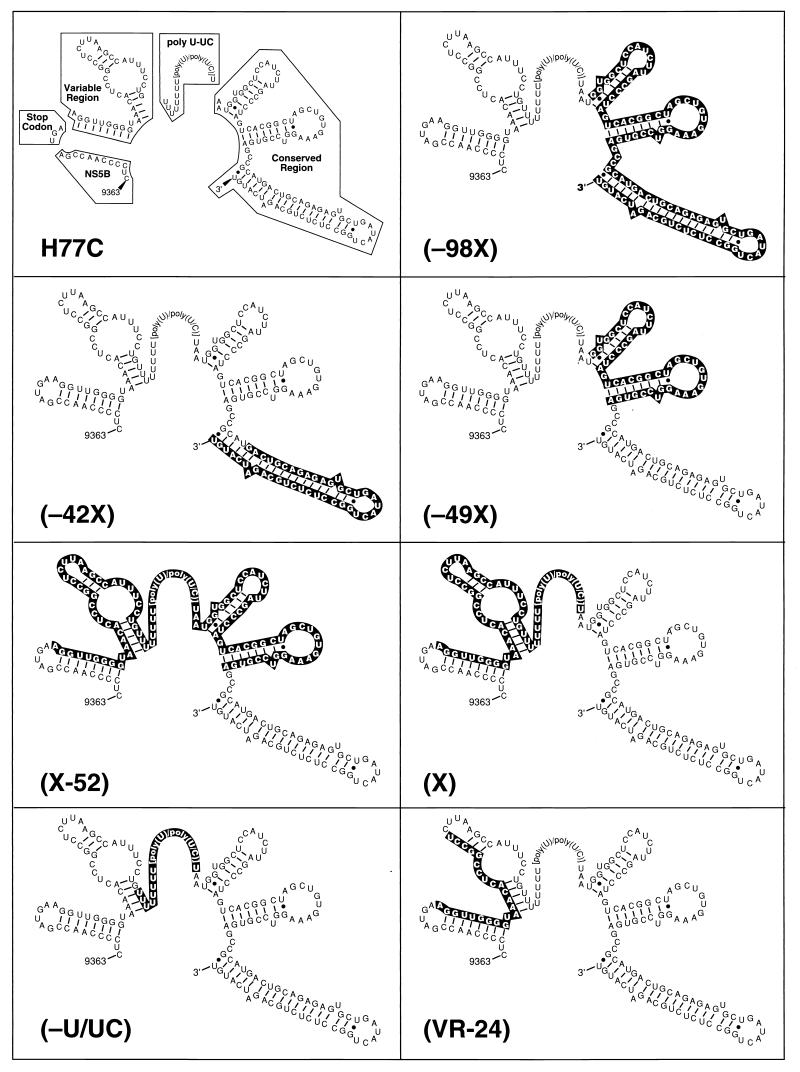
We generated mutated 3′ UTRs of the infectious cDNA clone of HCV genotype 1a (H77C; ref. 15) by using standard PCR procedures and/or synthesized oligonucleotides. The mutated 3′ UTR was subcloned into a TA vector (Invitrogen), substituted for the entire 3′ UTR of H77C (15) contained in a pGEM vector (pH77-3′UTR), or substituted for the corresponding sequence of H77C. Multiple clones were screened by sequence analysis. Subcloned fragments with the correct sequence were then cloned into the original infectious clone, H77C. The seven mutants (Fig. 1) were constructed with the oligonucleotides listed in Table 1 as follows. For −98X, the mutated 3′ UTR was amplified from H77C by PCR with primers H9386F and H9501R, purified, and cloned into H77C by using AflII/XbaI sites. For −42X, hybridized cDNA of two complementary oligonucleotides (H9517S-ODN and H9557R-ODN) were cloned into pH77-3′UTR by using NheI/XbaI sites and then cloned into H77C by using AflII/XbaI sites. For X-52, the mutated 3′ UTR was generated by an extension PCR procedure. The 5′ fragment from H77C was amplified with primers H9120S and H9377R, purified, extended at the 3′ end by PCR with antisense primer H(X-52)R, and then cloned into H77C by using NdeI/XbaI sites. For X, a 5′ fragment was PCR amplified from H77C with primers H9120S and H9377R, purified, extended at the 3′ end by PCR with antisense primer HXR, and finally cloned into H77C by using NdeI/XbaI sites. For −49X, a 5′ fragment was PCR amplified from H77C with primers H9386F and H9513R, purified, extended by PCR with antisense primer H-49XR, and cloned into H77C by using AflII/XbaI sites. For −U/UC, hybridized oligonucleotides [(−U/UC)S-ODN and (−U/UC)R-ODN] were cloned into pH77-3′UTR by using AflII/NheI sites and transferred into H77C by using AflII/XbaI sites. For VR-24, the mutated 3′ UTR was amplified from H77C with primers H9120S-II and H9377R-II, purified, and cloned into H77C by using NdeI/AflII sites.
Figure 1.
(On the opposite page.) The 3′ UTR deletion mutants of an infectious cDNA clone of HCV (pCV-H77C; ref. 15; figure adapted from Kolykhalov et al., ref. 6). The deleted sequences are shown in black bars. Sequences are shown starting at position 9,363 in the NS5B. The 3′ UTR of H77C has 225 nt, consisting of the ORF stop codon, a short sequence of 40 nt (variable region), a poly(U–UC) region of 81 nt, and a 3′ terminal sequence of 101 nt (conserved region). Sequences in the 3′ end of NS5B and in the variable region of the 3′ UTR could potentially form two stem–loop structures, and sequences of the conserved region of the 3′ UTR could potentially form three stem–loop structures.
Table 1.
Oligonucleotides used for constructing the deletion mutants of the 3′ UTR of an HCV infectious clone (pCV-H77C)
| Designation | Sequence (5′ → 3′)* |
|---|---|
| H9386F | GTCCAAGCTTACGCGTAAACACTCCGGCCTCTTAAGCCATTTCCTG |
| H9501R | CGTCGAATTCTAGATTAAAGAAGGGAAAAAGAAAGGAAAAAAAGAAGGAAAGG |
| H9517S-ODN | GCCCTAGTCACGGCTAGCTGTGAAAGGTCCGTGAGCCGCATCTAGAGACG |
| H9557R-ODN | CGTCTCTAGATGCGGCTCACGGACCTTTCACAGCTAGCCGTGACTAGGGC |
| H9120S | GTCCGCGCTAGGCTTCTGTCCAGAGGAGGCAG |
| H9377R | TCATCGGTTGGGGAGGAGGTAGATG |
| HXR | CGTCTCTAGACATGATCTGCAGAGAGGCCAGTATCAGCACTCTCTGCAGTCATGCGGCTCACGGACCTTTCACAGCTAGCCGTGACTAGGGCTAAGATGGAGCCACCATTTCATCGGTTGGGGAGGAGGTAGATG |
| H(X-52)R | CGTCTCTAGACATGATCTGCAGAGAGGCCAGTATCAGCACTCTCTGCAGTCATGCGGCTCATCGGTTGGGGAGGAGGTAGATG |
| H9513R | ATGCGGCATTAAAGAAGGGAAAAAGAAAGGAAAAAAAGAAGGAAAGG |
| H-49XR | CGTCTCTAGACATGATCTGCAGAGAGGCCAGTATCAGCACTCTCTGCAGTCATGCGGCATTAAAGAAGGGAAAAAGAAAGG |
| (-U/UC)S-ODN | ACTCCGGCCTCTTAAGCCATTTCCTGAATGGTGGCTCCATCTTAGCCCTAGTCACGGCTAGCTGTGAAAGGT |
| (-U/UC)R-ODN | ACCTTTCACAGCTAGCCGTGACTAGGGCTAAGATGGAGCCACCATTCAGGAAATGGCTTAAGAGGCCGGAGT |
| H9120S-II | GTCCGCGCTAGGCTTCTGTCCAGAGGAG |
| H9377R-II | TTTTTTCGTCTTAAGTCATCGGTTGGGGAGGAGGTAGATG |
HCV-specific sequences are shown in plain text. Non-HCV-specific sequences are shown in bold face. Cleavage sites used for cDNA cloning are underlined.
Large-Scale Preparation of the 3′ UTR Deletion Mutants of pCV-H77C, Followed by Complete Sequence Analysis.
Each mutant was transformed into DH5α competent cells (GIBCO/BRL) and selected on LB agar plates containing 100 μg/ml ampicillin (Sigma). Several colonies were cultured in LB liquid containing ampicillin at 30°C for ≈18–20 h. After small-scale preparation (Wizard Plus Minipreps DNA Purification Systems, Promega), a plasmid preparation with the expected digestion pattern was retransformed to select a single clone, and large-scale preparation of plasmid DNA was performed with a Qiagen plasmid Maxi kit (Chatsworth, CA) as described (15). The complete HCV sequence of the final preparation was determined. Both strands of DNA were sequenced with the PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems) by using AmpliTaq DNA polymerase, FS (Perkin–Elmer) and about 60 specific sense and antisense primers. Each mutant was stable; the digestion pattern was as expected after retransformation, and the complete sequence was the expected one.
Intrahepatic Transfection of a Chimpanzee with Transcribed RNA.
In each of two transcription reactions, 10 μg of plasmid DNA, made linear with XbaI (Promega), was transcribed in a 100-μl reaction volume with T7 RNA polymerase (Promega) at 37°C for 2 h as described (15), and then 5 μl of the final reaction mixture was analyzed by agarose gel electrophoresis and ethidium bromide staining (15). Each transcription mixture was diluted with 400 μl of ice-cold PBS without calcium or magnesium and then immediately frozen on dry ice and stored at −80°C. Within 24 h, both transcription mixtures were injected into the same chimpanzee by percutaneous intrahepatic injection guided by ultrasound. Each inoculum was injected into several separate areas of the liver. If the chimpanzee did not become infected, the same transfection was repeated once. After two negative results, the next mutant was inoculated into the same chimpanzee by following the same protocol. Injections were performed at weeks 0 and 3 with mutant −98X; at weeks 8 and 10 with −42X; at weeks 16 and 18 with X-52; at weeks 22 and 27 with X; at weeks 33 and 35 with −49X; at weeks 38 and 42 with −U/UC; and finally at week 46 with mutant VR-24. The chimpanzee was maintained in an approved facility under conditions that met or exceeded all requirements for animal use.
Serum samples were collected weekly from the chimpanzee and monitored for liver enzyme levels by standard procedures. Anti-HCV antibodies were monitored by the second-generation ELISA (Abbott), and HCV RNA was monitored by a sensitive reverse transcription–nested PCR assay with AmpliTaq Gold DNA polymerase (11, 17). Samples were scored as negative for HCV RNA if two independent tests on 100 μl of serum were negative. The genome equivalent titer of HCV in positive samples was determined by reverse transcription–nested PCR on 10-fold serial dilutions of the extracted RNA (11). The 3′ region of the VR-24 mutant genomes (nucleotides 9,311–9,517 of H77C) were amplified from the infected chimpanzee at weeks 2 and 5 post inoculation (p.i.) by reverse transcription–nested PCR as described (15) and analyzed by direct sequencing and TA vector cloning of PCR products.
RESULTS
We deleted selected regions of the 3′ UTR of pCV-H77C (Fig. 1). Infectivity of RNA transcripts from seven mutants was evaluated by consecutive intrahepatic transfection of a chimpanzee. The entire genome of each mutant was sequenced before transcription to ensure that the only changes from the wild type were the engineered ones in the 3′ UTR. Mutants were considered nonviable if viremia, as measured by viral RNA in the serum, was not detected within 2 weeks of the repeat transfection. Because an HCV infection would compromise the chimpanzee for further transfections, the order of the mutants was chosen such that the mutants we believed had the least chance of replicating were inoculated first.
Mutants in which the 3′ terminal 98 nt (−98X), the 3′ terminal 42 nt (−42X), or the proximal 49 nt (−49X) of the conserved region of the 3′ UTR was deleted failed to replicate in the chimpanzee, indicating that the conserved region was critical for infectivity in vivo. Moreover, mutants containing only the 3′ terminal 49 nt (X-52) or 101 nt (X) of the 3′ UTR were also not viable in vivo, indicating that the conserved region by itself was not sufficient and that either the poly(U–UC) region or the variable region also was required. Deletion of the poly(U–UC) region (−U/UC) destroyed infectivity, whereas removal of the proximal 24 nt of the variable region (VR-24) of the 3′ UTR did not. We had not detected HCV RNA, anti-HCV, or alanine aminotransferase elevation in the chimpanzee during the 46 weeks of the experiment before inoculation of mutant VR-24. In contrast, the chimpanzee became viremic 1 week after inoculation with mutant VR-24 and had an HCV genome titer of 102 genome equivalents per ml, which increased to 104 genome equivalents per ml during weeks 2–6 p.i. Thus, the conserved region and the poly(U–UC) region of the 3′ UTR, but not the proposed secondary structure of the variable region, which was disrupted in VR-24, seem to be critical for infectivity.
Viral genomes recovered from the chimpanzee 2 and 5 weeks after inoculation of mutant VR-24 were sequenced from the 3′ end of NS5B through the 3′ UTR of HCV. The consensus sequence of the variable region of the 3′ UTR was identical to that of mutant VR-24 at weeks 2 and 5 p.i. Moreover, all 9 and 16 clones obtained at weeks 2 and 5 p.i., respectively, from the infected chimpanzee had the engineered deletion of 24 nt within the variable region, indicating that the infection was caused by the VR-24 mutant and that the deletion was maintained. Interestingly, a G to A mutation was observed at position 9,376 within the ORF stop codon (UGA and UAA). Direct sequence analysis of PCR amplicons showed significant quasispecies at this position, and 6 of 9 clones generated from amplicons by TA cloning had the G to A mutation at week 2 p.i. At week 5 p.i., A was dominant at this position in the direct sequence, and 15 of 16 clones analyzed had this mutation. No other mutations were detected within the HCV sequence analyzed from the infected chimpanzee.
DISCUSSION
Deletion of large segments of any region of the 3′ UTR, except for the variable region, destroyed the ability of an infectious cDNA clone of HCV to replicate in a chimpanzee. Although these experiments showed that all but one of the designated “regions” plays a critical role in replication in vivo, they did not provide information about the relevant functions. Because HCV does not replicate sufficiently well in cell culture to allow biochemical analysis, the identification of functions depends on indirect analysis using artificial systems combined with deductions based on comparisons with related viruses that are more amenable to study.
Although the chimpanzee model is somewhat limited in its ability to define functions, it can provide important information about the biological relevance of results obtained by in vitro studies. In this instance, results from the chimpanzee study disagree with conclusions from in vitro studies that suggested that transcripts missing the 3′ terminal conserved region were able to replicate in cell culture (18, 19). In fact, three different mutants deleted for all or part of the conserved region were unable to infect a chimpanzee. Mutant −98X, which lacks the entire conserved region, was the first mutant injected. Thus, even if it replicated inefficiently in the chimpanzee, it should have been detected eventually, because the chimpanzee was studied by reverse transcription–PCR for 46 weeks before the infectious mutant was tested. Additionally, the HCV genome sequences recovered at weeks 48 and 51 were exclusively those of the VR-24 mutant. Therefore, it seems that not only was mutant −98X unable to mount a robust infection, it was unable even to sustain itself in vivo, suggesting that the conserved region of the 3′ UTR was indispensable for virus multiplication in the chimpanzee.
Although it is still possible that the conserved region of the 3′ UTR is not required for RNA replication in cell culture, most data in the literature argue against this conclusion. Conserved terminal genome sequences or structures of RNA viruses typically have a critical role for RNA replication and/or packaging (4). Although the 3′ terminal 80–100 nt of different flaviviruses are heterogeneous in sequence, they all form putative stem–loop structures and have several conserved sequence elements upstream (4). Mutagenesis analysis of infectious cDNA clones of different flaviviruses showed that such sequences in the conserved region of the 3′ UTR were critical for virus replication; mutations or deletions resulted in significant biological effects on viral growth or virulence (20–25). The 3′ terminal sequence of 98 nt is highly conserved among the different variants of HCV (6, 8, 10, 12), and, similar to other viruses of the Flaviviridae family, a membrane-associated replicase complex is thought to initiate replication at the 3′ end of the HCV and to synthesize a complementary negative-strand RNA (4). The NS5B protein is believed to be the RNA-dependent RNA polymerase ultimately responsible for HCV RNA replication (26, 27). However, several research groups recently found that cellular proteins, including polypyrimidine tract-binding protein, interact specifically with elements within the conserved region of the 3′ UTR (28–31). These findings suggested that cellular proteins may act in concert with viral proteins at the 3′ end of the HCV genome, thus imposing additional sequence restraints on the conserved region of the 3′ UTR.
In contrast to the universal occurrence of conserved 3′ terminal sequences among positive-strand RNA viruses, the inclusion of a polypyrimidine tract within the 3′ UTR sequence is unique to HCV and GBV-B (6, 8, 32); therefore, there is no precedent for speculation as to its function. Because the mutant lacking the poly(U–UC) region was not viable in a chimpanzee, this region is critical for infectivity of HCV. The polypyrimidine tract-binding protein can interact with the poly(U–UC) sequences in addition to sequences within the conserved region (31). The poly(U–UC) region was found in all isolates of HCV to date, including the prototype (HCV-1; J.B., unpublished data), even though HCV-1 was previously reported to terminate with a poly(A) tail (33). The length of the poly(U–UC) tract seems to be highly variable (6, 8, 10, 12, 15, 16); however, it should be noted that PCR artifacts might contribute to this heterogeneity (15). Kolykhalov et al. (14) reported that HCV RNA transcripts from an infectious cDNA clone with a poly(U–UC) region of 133 nt seemed to have a replicative advantage in chimpanzees over those with a poly(U–UC) region of 75 nt, suggesting that the length of the poly(U–UC) region might influence viral replication.
Although the variable region of the 3′ UTR of HCV is very heterogeneous in length and composition among different isolates (34, 35), it is highly conserved among clones obtained from the same viral strain (6, 15, 16). It has been suggested that the proximal sequence of this region was important for HCV replication (34). However, the deletion mutant lacking the proximal 60% of the variable region was viable in the chimpanzee. Previously, we found that a chimeric infectious cDNA clone of genotype 1b, which contained several mutations within the distal 40% of the variable region, was viable in vivo (16), suggesting further that the sequence of this region may not be critical. Studies with other flaviviruses showed that variable sequences typically found between the ORF and the conserved sequences of the 3′ UTR of flaviviruses were not critical for viral replication (20, 22, 24). Our finding that an intact variable region is not critical for HCV replication in a chimpanzee is consistent with these conclusions. In the present study, the initial course of the infection with mutant VR-24 in a chimpanzee was similar to the course observed previously with the wild-type clone in two chimpanzees (ref. 15; J.B., unpublished data). More follow up is warranted to determine whether the in vivo phenotype of the 3′ UTR deletion mutant differs in any way from that of the wild-type virus.
Mutagenesis studies of HCV can be performed in chimpanzees, the only animal model for HCV. In vivo studies are the most definitive for evaluating infectivity and virulence, but they do not provide information about the relevant functions. Thus, the development of an efficient in vitro propagation system for HCV is required to perform more detailed analysis. The in vivo study provided information about whether predicted structures of the HCV 3′ UTR were biologically relevant. For example, deletion of the 3′ terminal stem–loop structure was deleterious, suggesting that this highly stable stem–loop structure was needed. In contrast, a mutant in which two predicted stem–loop structures were eliminated from the variable region of the 3′ UTR was viable in the chimpanzee, indicating that, if these secondary structures exist, they are not critical. Future in vivo studies could provide important information on other biologically relevant structures, as well as relevant protein sequences, in the HCV genome. Because the poly(U–UC) region and the conserved region of the HCV 3′ UTR were critical for infectivity, sequences within these regions and/or viral and host factors that interact with such sequences could represent targets for therapeutic agents against HCV.
Acknowledgments
We thank J. Elser and L. Rasmussen (Science Applications International Corporation, Frederick, MD) for assistance in sequence analysis, M. Shapiro (Bioqual) for providing animal care, R. Engle (Georgetown University, Rockville, MD) for performing the second-generation ELISA, and C. L. Apgar for technical assistance. This study was supported in part by National Institutes of Health Contracts N01-AI-52705, N01-AI-45180, and N01-CO-56000.
ABBREVIATIONS
- HCV
hepatitis C virus
- p.i.
post inoculation
- UTR
untranslated region
References
- 1.World Health Organization. Wkly Epidemiol Rec. 1997;72:65–72. [Google Scholar]
- 2.Hoofnagle J H. Hepatology. 1997;26:15S–20S. doi: 10.1002/hep.510260703. [DOI] [PubMed] [Google Scholar]
- 3.Houghton M. In: Fields Virology. 3rd. Ed. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott–Raven; 1996. pp. 1035–1058. [Google Scholar]
- 4.Rice C M. In: Fields Virology. 3rd. Ed. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott–Raven; 1996. pp. 931–959. [Google Scholar]
- 5.Bukh J, Purcell R H, Miller R H. Proc Natl Acad Sci USA. 1992;89:4942–4946. doi: 10.1073/pnas.89.11.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kolykhalov A A, Feinstone S M, Rice C M. J Virol. 1996;70:3363–3371. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka T, Kato N, Cho M-J, Shimotohno K. Biochem Biophys Res Commun. 1995;215:744–749. doi: 10.1006/bbrc.1995.2526. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka T, Kato N, Cho M-J, Sugiyama K, Shimotohno K. J Virol. 1996;70:3307–3312. doi: 10.1128/jvi.70.5.3307-3312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemon S M, Honda M. Semin Virol. 1997;8:274–288. [Google Scholar]
- 10.Yamada N, Tanihara K, Takada A, Yorihuzi T, Tsutsumi M, Shimomura H, Tsuji T, Date T. Virology. 1996;223:255–261. doi: 10.1006/viro.1996.0476. [DOI] [PubMed] [Google Scholar]
- 11.Bukh J, Apgar C L, Engle R, Govindarajan S, Hegerich P A, Tellier R, Wong D C, Elkins R, Kew M C. J Infect Dis. 1998;178:1193–1197. doi: 10.1086/515683. [DOI] [PubMed] [Google Scholar]
- 12.Tokita H, Okamoto H, Iizuka H, Kishimoto J, Tsuda F, Miyakawa Y, Mayumi M. J Gen Virol. 1998;79:1847–1857. doi: 10.1099/0022-1317-79-8-1847. [DOI] [PubMed] [Google Scholar]
- 13.Blight K J, Rice C M. J Virol. 1997;71:7345–7352. doi: 10.1128/jvi.71.10.7345-7352.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolykhalov A A, Agapov E V, Blight K J, Mihalik K, Feinstone S M, Rice C M. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 15.Yanagi M, Purcell R H, Emerson S U, Bukh J. Proc Natl Acad Sci USA. 1997;94:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yanagi M, St. Claire M, Shapiro M, Emerson S U, Purcell R H, Bukh J. Virology. 1998;244:161–172. doi: 10.1006/viro.1998.9092. [DOI] [PubMed] [Google Scholar]
- 17.Yanagi M, Bukh J, Emerson S U, Purcell R H. J Infect Dis. 1996;174:1324–1327. doi: 10.1093/infdis/174.6.1324. [DOI] [PubMed] [Google Scholar]
- 18.Dash S, Halim A-B, Tsuji H, Hiramatsu N, Gerber M A. Am J Pathol. 1997;151:363–373. [PMC free article] [PubMed] [Google Scholar]
- 19.Yoo B J, Selby M J, Choe J, Suh B S, Choi S H, Joh J S, Nuovo G J, Lee H-S, Houghton M, Han J H. J Virol. 1995;69:32–38. doi: 10.1128/jvi.69.1.32-38.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khromykh A A, Westaway E G. J Virol. 1997;71:1497–1505. doi: 10.1128/jvi.71.2.1497-1505.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai C-J, Bray M, Men R, Cahour A, Chen W, Kawano H, Tadano M, Hiramatsu K, Tokimatsu I, Pletnev A, et al. Clin Diagn Virol. 1998;10:173–179. doi: 10.1016/s0928-0197(98)00035-x. [DOI] [PubMed] [Google Scholar]
- 22.Mandl C W, Holzmann H, Meixner T, Rauscher S, Stadler P F, Allison S L, Heinz F X. J Virol. 1998;72:2132–2140. doi: 10.1128/jvi.72.3.2132-2140.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandl C W, Aberle J H, Aberle S W, Holzmann H, Allison S L, Heinz F X. Nat Med. 1998;4:1438–1440. doi: 10.1038/4031. [DOI] [PubMed] [Google Scholar]
- 24.Men R, Bray M, Clark D, Chanock R M, Lai C-J. J Virol. 1996;70:3930–3937. doi: 10.1128/jvi.70.6.3930-3937.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeng L, Falgout B, Markoff L. J Virol. 1998;72:7510–7522. doi: 10.1128/jvi.72.9.7510-7522.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Behrens S-E, Tomei L, De Francesco R. EMBO J. 1996;15:12–22. [PMC free article] [PubMed] [Google Scholar]
- 27.Lohmann V, Korner F, Herian U, Bartenschlager R. J Virol. 1997;71:8416–8428. doi: 10.1128/jvi.71.11.8416-8428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inoue Y, Miyazaki M, Ohashi R, Tsuji T, Fukaya K, Kouchi H, Uemura T, Mihara K, Namba M. Biochem Biophys Res Commun. 1998;245:198–203. doi: 10.1006/bbrc.1998.8315. [DOI] [PubMed] [Google Scholar]
- 29.Ito T, Lai M M C. J Virol. 1997;71:8698–8706. doi: 10.1128/jvi.71.11.8698-8706.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito T, Tahara S M, Lai M M C. J Virol. 1998;72:8789–8796. doi: 10.1128/jvi.72.11.8789-8796.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsuchihara K, Tanaka T, Hijikata M, Kuge S, Toyoda H, Nomoto A, Yamamoto N, Shimotohno K. J Virol. 1997;71:6720–6726. doi: 10.1128/jvi.71.9.6720-6726.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simons J N, Pilot-Matias T J, Leary T P, Dawson G J, Desai S M, Schlauder G G, Muerhoff A S, Erker J C, Buijk S L, Chalmers M L, et al. Proc Natl Acad Sci USA. 1995;92:3401–3405. doi: 10.1073/pnas.92.8.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han J H, Shyamala V, Richman K H, Brauer M J, Irvine B, Urdea M S, Tekamp-Olson P, Kuo G, Choo Q-L, Houghton M. Proc Natl Acad Sci USA. 1991;88:1711–1715. doi: 10.1073/pnas.88.5.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han J H, Houghton M. Nucleic Acids Res. 1992;20:3520. doi: 10.1093/nar/20.13.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tokita H, Okamoto H, Iizuka H, Kishimoto J, Tsuda F, Lesmana L A, Miyakawa Y, Mayumi M. J Gen Virol. 1996;77:293–301. doi: 10.1099/0022-1317-77-2-293. [DOI] [PubMed] [Google Scholar]