Abstract
The ‘independent action hypothesis’ (IAH) states that each pathogen individual has a non-zero probability of causing host death and that pathogen individuals act independently. IAH has not been rigorously tested. In this paper, we (i) develop a probabilistic framework for testing IAH and (ii) demonstrate that, in two out of the six virus–insect pathosystems tested, IAH is supported by the data. We first show that IAH inextricably links host survivorship to the number of infecting pathogen individuals, and develop a model to predict the frequency of single- and dual-genotype infections when a host is challenged with a mixture of two genotypes. Model predictions were tested using genetically marked, near-identical baculovirus genotypes, and insect larvae from three host species differing in susceptibility. Observations in early-instar larvae of two susceptible host species support IAH, but observations in late-instar larvae of susceptible host species and larvae of a less susceptible host species were not in agreement with IAH. Hence the model is experimentally supported only in pathosystems in which the host is highly susceptible. We provide, to our knowledge, the first qualitative experimental evidence that, in such pathosystems, the action of a single virion is sufficient to cause disease.
Keywords: virus, baculovirus, independent action, infection process, evolution, population genetics
1. Introduction
In the vast majority of pathosystems, it is unknown how many pathogen individuals cause infection of a host organism. Estimates of this number are necessary to predict the likelihood that hosts are infected by multiple genotypes, the magnitude of genetic drift (Chao 1990), the evolution of pathogen genotypes (Cooper et al. 2002) and the interaction between pathogen genotypes within diseased hosts. These interactions include competition (Turner & Chao 1999), complementation (Vignuzzi et al. 2006) and recombination (Gibbs et al. 2001). For viruses, there is no framework for predicting how many virions have caused infection. On the other hand, there is ample experimental evidence that the number of virions causing infection can be small, as shown by the data from two experimental approaches.
First, genetic drift can be very strong when low viral doses are used, suggesting that only a small number of virions cause infection. Ali et al. (2006) found that the number of genotypes present in an artificial plant virus population was strongly reduced during horizontal transmission by aphids, but no estimates of the number of virions causing infection were made. Smith & Crook (1988) detected single-genotype infections after administering a small virus dose, in a betabaculovirus–insect larvae pathosystem. It is not clear from Smith and Crook's analyses whether a single virion was causing infection, since the starting population used was poorly defined, available techniques for detecting genotypes were not sensitive enough and only a small sample size was analysed. Moreover, the experiments of Smith & Crook (1988) and Ali et al. (2006) lacked an interpretative framework that would allow estimation of the number of founders of viral infection in a host.
Second, results of dose–response experiments, characterized by values of the median infectious dose (ID50) or lethal dose (LD50), suggest that the number of virions causing infection is in some instances very small. For example, Bianchi et al. (2000) found an LD50 value of only 2.9 occlusion bodies (OBs), when challenging young Spodoptera exigua larvae with OBs of S. exigua multicapsid nucleopolyhedrovirus. (The number of OBs given per host was quantified, but note that one OB contains multiple virions.) Other authors have produced evidence for a low number of founders of disease using ID50 or LD50 values that are based on the biological activity of a virus (e.g. the number of plaque-forming units (PFU) in cell culture). Evidence produced in this way is not conclusive because dose values based on biological activity do not reflect the actual number of virions causing infection. For example, Spieker et al. (1996) found an LD50 of less than 0.01 PFU, yielding a conundrum if a PFU were interpreted as an individual virion, because at least one virion is needed to cause disease. The data suggest that the number may be small, but they cannot be interpreted in an absolute sense.
Although data from these two approaches suggest that only a small number of virions can cause infection, a quantitative understanding of this issue is lacking. This is a significant gap in our knowledge, because the number of infecting virions will determine what virus genotypes are ultimately present in infected hosts. The virus genotype distribution in infected hosts will affect virus population dynamics, population genetics and evolution. There is complementation between virus genotypes in some pathosystems, and this depends on complementary genetic content (López-Ferber et al. 2003; Vignuzzi et al. 2006). On the other hand, consider what could occur when two genetically identical virions challenge a host. The probability that each virion infects the host can be (i) bigger: there is cooperation between the two identical virions, (ii) smaller: there is antagonism between identical virions, or (iii) unchanged: there is no added benefit or cost to the number of identical virions present, as each acts on its own. In the latter case, the action of each virion is independent. Although independent action may be the commonly accepted null model, we are not aware of experimental data supporting it.
A rigorous test of a hypothesis should first be based on a clear formulation of the hypothesis, and second on the design of discriminating experiments that provide for a high probability of disagreement between the hypothesis and the data if the hypothesis is false (Hillborn & Mangel 1997; Turchin 2003). The independent action hypothesis (IAH) was originally formulated by Druett (1952) to provide a null model for the dose–response of mammals exposed to anthrax spores by inhalation. IAH states that each pathogen individual has a non-zero probability of causing infection and that conspecific pathogen individuals do not affect each other during this process (Druett 1952). IAH, as formulated by Druett and as tested here, concerns the entire sequence of pathogen-related events leading to infection: from initial host exposure to the pathogen until the full-blown, systemic infection by the pathogen, which results in host death. Thus, here we consider the entire chain of biological events leading to infection and reduce this complex process to a single probabilistic outcome: will a pathogen individual infect the host or not? Implied in IAH is the premise that each pathogen individual has an independent chance of infecting the host. In reality, the chance of a single virion causing disease may be small, but once IAH is supported by the data, the immediate corollary is that a single virion is sufficient to cause disease.
IAH offers an important initial null hypothesis of a pathogen's mode of action and perhaps has wide applicability. However, a rigorous experimental test of IAH has, to our knowledge, not been reported in any system. Here, we propose an interpretative framework and develop an experimental procedure for rigorously testing whether IAH pertains to a pathosystem. We provide strong qualitative evidence that in some systems there is indeed independent action. We consider the most straightforward embodiment of IAH: hosts are exposed to a fixed number of pathogen individuals (i.e. no variation in pathogen dose) and the probability that a pathogen individual infects the host is constant over hosts (i.e. no variation in host susceptibility). This is inherently a limited model, and if it is rejected, alternative hypotheses could take into account additional refinements. For example, the effects of relaxing the assumptions of a fixed dose or constant infection chance could be explored (Ridout et al. 1993; Ben-Ami et al. 2008). Alternatively, dependent action between conspecific individuals could be considered. Examples of dependent action are cooperation or antagonism, which could conceivably occur during any event in the infection process.
In this paper, we first show that the number of pathogen individuals infecting a host can be predicted under IAH, which extends the usefulness of the theory beyond making predictions on dose–response relationships. The prediction for the number of individuals infecting a host follows from the proportion of hosts that has not been infected during a challenge experiment (i.e. host survival in our pathosystem). Given the association between host survival and the number of infecting pathogens, we devised a probabilistic framework for the expected frequency of dual-genotype infections, when hosts are challenged with a pathogen population containing two genotypes. This allows us to test the pertinence of IAH predictions. Note that although we use the term ‘genotypes’ to denote two sorts of distinguishable individuals of a particular pathogen, the approach we present is most applicable when said genotypes are in all other respects the same. In other words, the genotypes would ideally be identical except for a marker sequence.
Experimental data were obtained using virus–insect pathosystems: two genotypes of the alphabaculovirus Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) and three lepidopteran host species. This virus was chosen because of the relative ease with which marked virus genotypes can be generated (Luckow et al. 1993), and the availability of a variety of lepidopteran insect hosts and developmental stages with widely differing susceptibility to AcMNPV (Payne 1986; Possee et al. 1993). We previously developed baculovirus genotypes that are suitable for testing IAH (Zwart et al. 2008). These genotypes, from here on named A and B, are launched from bacterial artificial chromosomes encoding the entire AcMNPV genome (Luckow et al. 1993), and differ only in short (100 bp, less than 0.1% of the AcMNPV genome) non-coding marker sequences in a transcriptionally inactive region. These marker sequences allow for detecting genotype presence by means of a sensitive quantitative real-time PCR (qPCR)-based assay (Zwart et al. 2008).
It is important to carefully define what is understood by ‘infection’ for the pathosystems we have chosen. Here, we define infection as the process resulting in the production of viral transmission stages—OBs for baculoviruses—and virus-induced death of the host. We choose this definition because we wish to understand virus population genetics during horizontal transmission of a baculovirus, particularly the changes in genotype frequency due to genetic bottlenecks in infection. In our model pathosystems, OBs are responsible for horizontal transmission, and production of OBs in significant numbers occurs only in lethally infected hosts (e.g. Federici 1997). Note that we subsequently refer to virus-induced host death as simply ‘death’, since in our experimental system these are the only deaths we have to consider; the insect larvae used are free of other pathogens and attrition in our experimental set-up is negligible.
Alphabaculoviruses are transmitted as OBs, i.e. proteinaceous bodies containing many virions. The infectious unit is the virion (Federici 1997), and so we view virions as being ‘pathogen individuals’ in our modelling. The fact that MNPVs have multiple nucleocapsids per virion does not affect model predictions in our set-up. The OBs used in the challenge experiments were separately derived from bacmids; the virions they contain therefore have nucleocapsids of only a single genotype (Zwart et al. 2008). Despite the intricacies of the baculovirus transmission stage, the frequency of dual-genotype infections in this particular set-up can therefore be described with a general model. Here we demonstrate that the IAH model renders pertinent host survival-based estimates of the frequency of dual-genotype infections in two out of the six pathosystems investigated.
2. Material and methods
(a) The frequency of dual-genotype infection under independent action hypothesis
Under IAH, pathogen individuals act in a strictly independent manner (Druett 1952). The mean number of infecting pathogen individuals (λ) for a homogeneous population, or for any given genotype in a mixed pathogen population, is therefore
| (2.1) |
Here n is the number of pathogen individuals the host is challenged with (e.g. the number of pathogen individuals ingested by the host in our experimental system) and p the probability of infection for each pathogen individual. We assume here that infection entails pathogen-induced host death and the production of horizontal transmission stages (OBs) to a level at which they can be detected by a detection assay (qPCR in our set-up). Note that the mathematical model remains valid if we consider a series of successive events leading to infection, as long as during each event the pathogen individuals act independently of each other. All we would need to do is redefine the meaning of p to also include these additional probabilistic events in breaking the host defence barriers. The inclusion of these additional events would again result in host death and a measurable number of pathogen individuals contributing to the horizontal transmission stages generated in a host. Therefore, under IAH, as defined here, there is only a single probabilistic outcome that needs to be considered, host survival or death.
Typically, p is very small, requiring a large n to produce even low numbers of infecting pathogens. If the probability of infection for each pathogen individual is independent of that of the others, the number of infecting pathogen individuals per host has a Poisson distribution with a mean λ (Olkin et al. 1994). The probability of host survival after a pathogen challenge (S, i.e. proportion surviving) is then the zero term of the distribution of the number of infecting pathogens, that is
| (2.2) |
IAH thus inextricably links the number of infecting pathogen individuals to host survival. Importantly, this association is independent of the exact n and p values, which cannot always be experimentally determined.
It would be technically difficult, if not impossible, to empirically determine the number of infecting pathogen individuals in a host after a challenge. However, if the host is challenged with a pathogen population consisting of multiple genotypes, then the distribution of genotype frequencies in the horizontal transmission stages from individual dead hosts results from genetic drift and depends on the number of infecting pathogen individuals. Specifically, if mortality is high, then a large number of infecting pathogen individuals is expected. Consequently, there will be very few infections in which individuals of only a single pathogen genotype have contributed. At low mortality, on the other hand, the frequency of infections to which only a single genotype contributes is expected to be high, because, under IAH, the action of one pathogen individual is enough to cause infection. If, however, infection by multiple pathogen individuals is required to cause disease and IAH does not hold, then a heightened frequency of mixed-genotype infections, compared to the IAH prediction, is expected at any level of mortality.
The expected frequency of mixed-genotype infections under IAH can be readily calculated. Assume that a host population is challenged with two genotypes, A and B, and that the number of pathogen individuals contributing to infection for each genotype (λA=pAnA and λB=pBnB) is Poisson distributed. The expected proportion of dual-genotype infected hosts, P(A∩B), divided by the expected probability of infection (one minus no infection, ), is the probability that both genotypes are present in an infected host, P′(A∩B)
| (2.3) |
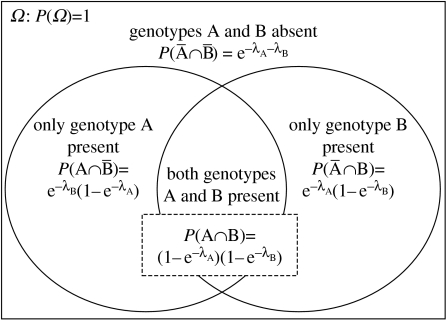
The justification for this equation is given in the electronic supplementary material. A Venn diagram (figure 1) illustrates the proportions of challenged hosts that contain no pathogen transmission stages, transmission stages of one pathogen genotype (A or B) or transmission stages of both pathogen genotypes, explaining in a non-mathematical guise the rationale of the above formula. Given (i) host survival, (ii) the frequencies of genotypes in the original pathogen population, and (iii) the relative infection probabilities of the pathogen genotypes, IAH predicts the expected frequency of dual-genotype infected hosts (figure 2). Comparing the experimentally observed frequency of dual-genotype infection with the model prediction allows for an unambiguous test of IAH.
Figure 1.
The IAH-based model for the frequency of dual-genotype infection; it is summarized in a Venn diagram, where Ω is the set of all possible outcomes. Note that for baculoviruses, we define infection as the production of viral transmission stages (OBs) and virus-induced death of the host.
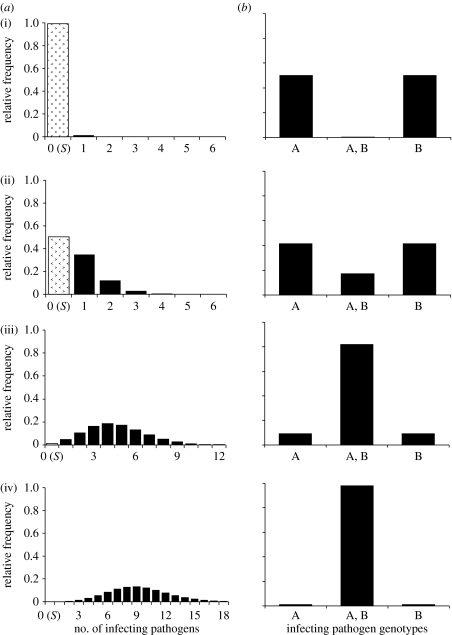
Figure 2.
Model predictions under IAH on the frequency distribution of (a) infecting pathogens and (b) the frequency of single- and dual-genotype infections in dead hosts. For all graphs, frequency is on the y-axis. The frequency that zero pathogens infect a host is also host survival (S) and denoted by the dotted bars in (a). A starting population with a 1 : 1 ratio of pathogen genotypes A and B and an equal infection chance (pA=pB) between these pathogen genotypes were assumed ((i) S=0.99, (ii) S=0.50, (iiii) S=0.01 and (iv) S=0.0001).
(b) Challenge experiments
All challenge experiments and qPCR analyses were basically performed as previously described (Zwart et al. 2008). Briefly, developmentally synchronous larvae were starved overnight and subsequently allowed to drink from small droplets of an OB suspension with a given concentration (minimally 24 larvae per dose). The larvae were then reared separately on a semi-synthetic diet, and upon death collected and individually stored. OBs were purified from cadavers of individual larvae and DNA was then extracted from these OBs. A separate qPCR reaction was then performed to determine the presence of genotypes A and B in the DNA sample.
We assume that there is no variation in the ingested dose. Therefore, OB concentration in the inoculum can be used as a proxy for dose. For determining the dose–response relationship for S. exigua third-instar larvae (L3), a different range of concentrations of a 1 : 1 OB mixture (A : B) were taken for different replicates (four replicates in total). The total number of replicates taken per concentration of OBs (OBs ml−1) was as follows: one replicate for 102; three replicates for 103; four replicates for 104; four replicates for 105; three replicates for 106; and one replicate for 107. Most replicates were taken in the intermediate OB concentration range (104–105), since this is the most variable and relevant range. At least 24 larvae were taken per concentration per replicate, and 24 larvae were also taken as non-virus controls. The best fitting p-value (1.12×10−5) was found by nonlinear regression (mortality=1−exp(−p·concentration), SPSS v. 15.0).
The infection probabilities for virions of genotypes A and B were likewise estimated by nonlinear regression on dose–response data obtained individually for each genotype in S. exigua L3 (Zwart et al. 2008), giving a ratio of the infection probabilities of the two genotypes (pA : pB=3.17). The exact ratio of the two genotypes in the OB mixture used for the experiments was also determined by qPCR (nA : nB=0.46; Zwart et al. 2008). The expected ratio λA : λB under the IAH model is therefore 1.46 : 1.
L3 and L5 larvae of three species were exposed to different doses of virus OBs. If there were less than the required number of cadavers (=24) for qPCR analysis, challenge experiments were repeated and the results of different replicates combined in analysis. For each instar (L3 or L5) of each species, 24 larvae were individually analysed by qPCR. In S. exigua and Trichoplusia ni, the fifth instar is the final larval instar, whereas for Mamestra brassicae it is the penultimate larval instar. An OB concentration resulting in the survival level S in the range 0.14–0.34 was used for S. exigua and T. ni, and S≈0.7 for M. brassicae. A lower survival rate could not be reached in M. brassicae L5.
(c) Binomial test for comparing experimental data to model predictions
In order to test whether the qPCR data conformed to IAH, the frequency of dual-genotype infections observed in experiments was compared with that predicted by the model, using an exact one-tailed binomial test (SPSS v. 15.0). The observed host survival was used to calculate λ (equation (2.2)), which was partitioned into λA and λB in the ratio 1.46 : 1 (see above). The expected frequency of dual-genotype infection in dead hosts, P′(A∩B), is predicted by equation (2.3). This is the probability of success in the binomial test. The number of host cadavers with both genotypes present (as determined by qPCR) was the number of successes observed, whereas the total number of host cadavers analysed was the number of observations. A significance level of 0.05 was used.
(d) Testing for the production of transmission stages in surviving larvae
We operationally defined infection in the experimental system as virus-induced host death and production of transmission stages (OBs). The 1 : 1 correspondence between virus-induced death and appreciable production of OBs, which is well supported in the literature, was verified in S. exigua. L3 and L5 larvae were challenged with OBs as described above. Larvae were collected and stored at −20°C: infected larvae at death, and ‘surviving’ larvae when they began transitioning to the pre-pupa stage. In our experiments, we have never seen larvae die after transition into the pre-pupal stage, so these larvae would probably all have survived into pupation. We randomly selected four non-virus controls, 12 pre-pupa survivors and four viral deaths, and performed OB purification and DNA extraction as described above. We screened ‘purified OB samples’ for the presence of OBs by phase-contrast light microscopy.
Genotype presence was determined by qPCR on DNA extracted from these larval samples, as described above. For a positive qPCR control, 1010 copies of the pGEM-luc plasmid (Promega), containing the firefly luciferase sequence, were added to all samples during DNA extraction (at the start of proteinase K treatment; see Zwart et al. 2008). The luciferase sequence in this plasmid was detected by qPCR, using the forward primer 5′-TGTTGGGCGCGTTATTTATC-3′, reverse primer 5′-AGGCTGCGAAATGTTCATACT-3′ and the same qPCR conditions (i.e. Zwart et al. 2008).
3. Results
(a) Dose–response relationship upon challenge of S. exigua L3 larvae with AcMNPV
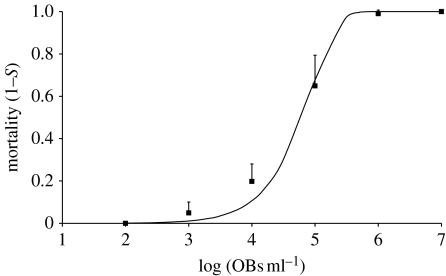
We first considered whether the dose–response relationship observed is compatible with IAH, as IAH should lead to a distinct dose–response curve (Druett 1952). A 1 : 1 mix of AcMNPV genotypes A and B was used to challenge S. exigua third-instar (L3) larvae, a permissive lepidopteran host species (Bianchi et al. 2000), by droplet feeding (Hughes & Wood 1981; Zwart et al. 2008). The observed dose–response relationship does not only depend on whether IAH holds, but also on the between-host variation in the ingested dose (Ridout et al. 1993) and variation in susceptibility (Ridout et al. 1993; Ben-Ami et al. 2008). Discriminating between departure from IAH and the effects of variation in host susceptibility is therefore problematic (e.g. Ben-Ami et al. 2008). The dose–response data in S. exigua L3 (figure 3) are, however, compatible with IAH considering the good fit to the data, which would further improve if between-host and between-cohort variations in dose and susceptibility are accounted for in the model (Ridout et al. 1993; Ben-Ami et al. 2008).
Figure 3.
Dose–response data for AcMNPV in S. exigua L3 larvae. On the x-axis is the occlusion body (OB) concentration per ml (log), and on the y-axis the mortality. Bioassay mortality data (squares), with the standard deviation, and the fitted dose–response relationship under IAH (curve) are displayed. See §2 for the number of replicates per concentration. Note that, for simplicity, OB concentration was taken as dose, rather than as an estimate of the number OBs or virions ingested.
(b) The frequency of dual-genotype infection in S. exigua L3 larvae
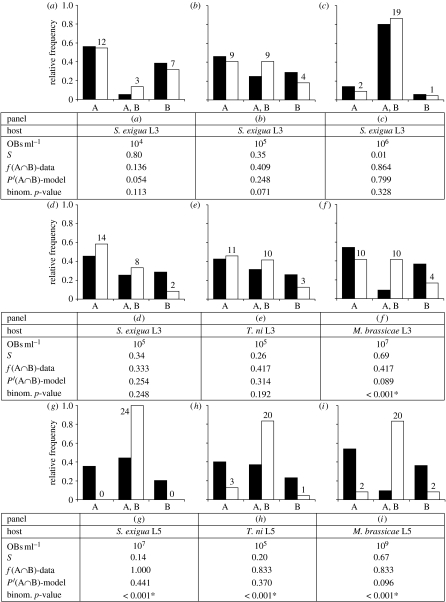
The frequency of dual-genotype infection was then determined for three different doses in S. exigua L3 (figure 4a–c). The frequencies of dual-genotype infection in cadavers were not significantly different from model predictions of P′(A∩B). We therefore conclude that IAH is supported for the challenge of S. exigua L3 larvae with AcMNPV, both by the experimental results on genotype occurrence in cadavers, and by the dose–response data. This is the first report, to our knowledge, of a thorough experimental test of IAH in any pathosystem.
Figure 4.
Model predictions and experimental data for the frequency of dual-genotype infections. The presence of genotypes A and B was determined at different doses in (a–c) S. exigua L3 (22 larvae per dose), and in different host species in (d–f) L3 and (g–i) L5 larvae (24 larvae per treatment). Model predictions are depicted with black bars. Experimental data are depicted with white bars, and the number of observations noted above each column. OBs ml−1 is the concentration of OBs used in the challenge experiments. S is survival and f(A∩B) is the observed frequency of dual-genotype infection in cadavers. P′(A∩B) is the predicted frequency of dual-genotype infection, given host mortality, under IAH. Binom. p-value is the observed significance of a binomial test comparing f(A∩B) with P′(A∩B). An asterisk marks significant departure from IAH. (a–c) It is demonstrated that IAH is not rejected in the third larval instar of S. exigua, irrespective of dose or mortality. (d–f) It is shown that IAH is not rejected in L3 of S. exigua and T. ni, but is rejected in L3 of semi-permissive M. brassicae. (g–i) Significant departure from IAH in challenge experiments with fifth larval instars in all three species is shown.
(c) The frequency of dual-genotype infection in L3 or L5 larvae of three host species
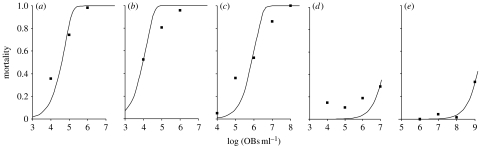
To test the generality of the IAH model for AcMNPV, we determined whether IAH can also predict the outcome of AcMNPV challenge in a later instar of the same host, and in other host species. We used L3 and L5 larvae of S. exigua, T. ni (another permissive host) and M. brassicae (a semi-permissive host). Here, we found that host compatibility and larval instar modulate the applicability of the IAH model. L3 larvae of S. exigua and T. ni showed a frequency of dual-genotype infection which was not significantly different from the IAH model predictions (figure 4d,e). However, the frequency of dual-genotype infection was significantly greater in L3 larvae of the semi-permissive M. brassicae (figure 4f) and in all of the challenge experiments with L5 larvae (figure 4g–i). For the sake of later discussion, we also provide dose–response data for all five pathosystems, those other than S. exigua L3 (figure 5).
Figure 5.
Dose–response relationships in semi-permissive larvae and L5. On the x-axes are the concentrations (log) of OBs in the inoculum, and on the y-axes the mortality for single replicates of dose–response bioassay data of (a) T. ni L3, (b) T. ni L5, (c) S. exigua L5, (d) M. brassicae L3 and (e) M. brassicae L5. Experimental data are denoted by squares and the IAH model for dose–response is given by the curve. This is done only for comparative purposes, using nonlinear regression (see §2). Note that only (a) T. ni L3 dual-genotype infection frequency data support the IAH model. The data suggest that in other instances, dose–response curves are shallower. Whether the T. ni L5 or M. brassicae L5 data are really shallower cannot be ascertained from these data alone.
(d) Production of transmission stages in surviving larvae
In both S. exigua L3 and L5 larvae, OBs were observed only in samples from larvae that died following virus challenge (table 1). Virus genotypes A, B or both could likewise be detected in only DNA samples from larvae that died of baculovirus infection during the challenge experiment (table 1).
Table 1.
Occlusion body and genotype presence in survivors of virus challenge. (For S. exigua L3 and L5 larvae, the presence of OBs and virus genotypes was determined in dead and surviving larvae following virus challenge, and non-virus controls. Status indicates whether the data are from fatalities, survivors or non-virus controls. The total larval samples analysed are the total number of larvae analysed with both light microscopy (OBs present) and qPCR (genotypes present). DNA samples were spiked with a plasmid containing the luciferase sequence during the extraction procedure, as a positive qPCR control. All samples were positive for the luciferase control. Host mortality was 0.33 for L3 and 0.58 for L5.)
| genotypes present | |||||||
|---|---|---|---|---|---|---|---|
| instar at challenge | OBs ml−1 | status | total larval samples analysed | OBs present | A only | A and B | B only |
| L3 | 0 | non-virus control | 4 | 0 | 0 | 0 | 0 |
| L3 | 105 | survivors | 12 | 0 | 0 | 0 | 0 |
| L3 | 105 | fatalities | 4 | 4 | 1 | 3 | 0 |
| L5 | 107 | survivors | 12 | 0 | 0 | 0 | 0 |
| L5 | 107 | fatalities | 4 | 4 | 0 | 4 | 0 |
4. Discussion
Based upon the idea of independent action of virions, and using simplifying assumptions on the variability of ingested dose and susceptibility to virus in our test systems, we have conducted the first explicit test, to our knowledge, of the pertinence of IAH in virus–host interactions. We find that in the two most permissive systems that we tested—early instar larvae of susceptible species—the frequency of dual-genotype infection conformed to the predictions of our simple model, confirming IAH. The demonstration that virus genotype frequencies in S. exigua L3 at three different doses, differing by two orders of magnitude, correspond to predictions based on IAH renders it highly plausible that in this system virions act independently. The frequency of dual-genotype infection is determined by the number of infecting viruses. Therefore, especially at low challenge doses and a high survival rate, the plausible number of founders for virus infection of a host is 1. The confirmation of model predictions is, to the best of our knowledge, the first good experimental evidence that the action of a single virion is sufficient to cause infection. This does not mean that every virion that manages to bypass initial host defences will contribute to infection. Rather, virions appear to be operating independently in the sequence of events leading to a full-blown infection of host larvae. On the other hand, with early instar larvae of the least susceptible insect species, and with late-instar larvae (which are generally more resistant than early instar larvae; Engelhard & Volkman 1995) of any of the three tested species, we find that the frequency of dual-genotype infections is higher than that predicted by IAH. Thus, in these cases, the data are not congruent with the IAH-based model. This does not necessarily mean that IAH is not applicable in these instances. It does mean that the combination of IAH with simplifying assumptions on ingested virus dose and variability in susceptibility to virus infection leads to incorrect and, very low predictions of the frequency of mixed-genotype infections.
If an insect larvae is challenged with baculovirus OBs, there are three possible outcomes: (i) although the insect is exposed to the virus (i.e. OBs are ingested), no viral invasion takes place, (ii) the virus does invade the insect, but there is little to no OB production, and the insect does not die (so-called ‘sublethal infection’; Goulson & Cory 1995), or (iii) the virus invades the insect, there is prolific OB formation and the host dies (our definition of infection). We consistently observed a higher frequency of dual-genotype infection than that predicted by the IAH model, even when the discrepancy between the data and model prediction was not significant (figure 4). This higher frequency of dual-genotype infection could be owing to the qPCR assay detecting virus genotypes that had not invaded the larvae (e.g. OB remnants in the midgut), or sublethally infected the host. There would then be an incongruity between our measure of infection (host death) and our measure of infection by a particular genotype (genotype presence in host transmission stages, as determined by qPCR).
We have ruled out this possibility by demonstrating that there is little if any transmission stage production in the surviving hosts, and that virus genotypes could not be detected in surviving hosts (table 1). Others have already demonstrated that sublethal baculovirus infections are low-level infections (Burden et al. 2002; see also Hughes et al. 1997), corroborating our findings. In most viral pathosystems, systemic viraemia is the hallmark of productive infection, rather than host death. The rate of ‘infection’, and therefore ‘survival’, will then follow from the rate at which any viral genotype is systemically detected, as determined by PCR for example.
The higher frequency of dual-genotype infection that we consistently observed may suggest that other factors affect the frequency of dual-genotype infection. An IAH-based model that includes these factors—e.g. variation in ingested dose and differences in host susceptibility—might be able to describe the experimental results when the model formulated here fails to do so. Therefore, further models need to be formulated and tested before IAH itself, and not just the model we have formulated here, can be categorically rejected for late-instar larvae and semi-permissive species.
In all cases in which the IAH model was rejected, shallow dose–response curves were observed, suggesting variability in host susceptibility (figure 5). Shallow dose–response curves have been reported for AcMNPV infection of late-instar larvae (Bianchi et al. 2000, 2002) and semi-permissive larvae (Hernandez-Crespo et al. 2001). Our data suggest that there may be an agreement between the two methodologies for determining whether IAH holds: considering dose–response data as originally proposed (Druett 1952) and determining the presence of pathogen genotypes in virus transmission stages as proposed here. A combination of the two methods is therefore probably the most powerful way to test the applicability of IAH in a particular system.
The failure of the IAH model to predict the frequency of dual-genotype infection in four out of six pathosystems investigated raises the issue of what the main difference between these systems is. If there is really dependent action in these instances—which remains to be further tested—larval resistance mechanisms to baculoviruses could be the reason for this. A number of resistance mechanisms have been described, and they tend to increase with the progress in development, both within and between instars (Engelhard & Volkman 1995; Washburn et al. 1998; Hoover et al. 2002). This age-dependent resistance may help explain our findings in L5.
But what exactly is the reason that the IAH model fails in these instances? The entire disease process can be divided up into at least two steps to help identify potential reasons: (i) initial invasion of the host and (ii) breaking systemic host resistance mechanisms, resulting in host death and prolific OB production. If resistance mechanisms act on initial invasion, this does not necessarily lead to the failure of IAH; it only becomes harder for a virion to get into the host (host mortality at a given dose is lower, but the response still conforms to IAH). On the other hand, if a resistance mechanism acts on the breaking of the resistance of the host, this may lead to dependent action. For example, if a minimum or threshold number of virions must invade the host in order to successfully infect it, the frequency of dual-genotype infected hosts at a given level of host mortality would be increased, compared with an IAH-based model. Such a threshold could arise if there were a host resistance mechanism that could be overcome by pathogen swamping. However, such a threshold would cause a steeper dose–response curve (Zeise et al. 1987), which is contradictory to our data. In our experiments, rejection of IAH was accompanied by a shallower dose–response than that expected based on equation (2.2) under IAH (figure 5). Therefore, the observed combination of a high frequency of dual-genotype infections and a shallow dose–response curve cannot be attained by this simple extension of the IAH model. Other models should therefore be considered and tested.
There is more than one way to formulate a testable null hypothesis for IAH. In this paper, we focus on a null hypothesis that is derived from elementary assumptions on the action of individual virions, resulting in a Poisson model (equation (2.3)). A strength of this approach is the intrinsic linkage between dose–response and dual-genotype infection in a single model. A null hypothesis could, alternatively, be formulated by simply considering the proportion of challenged larvae infected with genotype A, and those infected with genotype B, and constructing a null hypothesis solely on the basis of this information. In the case of independence, one would expect that the proportion of larvae with both viruses, P(A∩B), is simply the product P(A)P(B). There are differences between this null hypothesis and the one elaborated in the paper in the use of prior information, and the reasoning that is used in the construction of the null hypothesis. The null hypothesis elaborated in this paper uses prior information on the relative concentrations of the two viruses in the inoculum and their infectiveness, while it uses the Poisson theory for calculating the expected founder numbers (λA and λB) of types A and B as an intermediate step in the calculation of the expected frequencies of infection with virus types A and B. The alternative formulation of a null hypothesis neither uses prior information on virus potency in the inoculum nor involves an estimation of founder number as an intermediate step in the calculation of the probability of dual infection. As this latter test does not involve the linkage between independent action and dose–response in the calculation of the expected frequency of dual-genotype infection, it has less power to reject the null hypothesis of independence. An advantage of this formulation of the null hypothesis is that it can be carried out when there is no prior information on infectivity and frequencies of the virus genotypes in the inoculum. In the case of our data, the lesser power of the second formulation of null hypothesis was shown by the overall lower p-values, especially with L5 larvae, and lack of rejection of the null hypothesis of IAH for T. ni L5 (p=0.065).
A recent report has shown that complementation between RNA virus genotypes is important for pathogenesis (Vignuzzi et al. 2006). Our findings show that the infection process of a DNA virus does not require complementation between conspecific individuals. The two findings are of course not contradictory; complementation is thought to occur between genotypes, whereas IAH, as tested here, concerns the interactions between conspecific individuals with virtually identical genomes. For example, IAH may very well apply to a clonal RNA virus population, although this conjecture may be hypothetical given the speed with which diversity is generated in RNA virus populations (e.g. Cuevas et al. 2005).
The experimental technique of challenging hosts with a mixture of pathogen genotypes and subsequently determining genotype frequencies is available in many pathosystems (Lenhoff et al. 1998; Marks et al. 2005; Carrasco et al. 2007), and techniques for tracking genotypes of human viruses are also readily available (Giannini et al. 1999). The approach used here for testing independent action could therefore be widely used.
Knowing whether or not IAH holds is of great importance to understanding the population genetics of a particular pathogen. If pathogens are sparsely distributed in the environment, and if their relationship with a host is characterized by independent action, then this pathogen is likely to occur singly in hosts, resulting in vastly different evolutionary pressures on the pathogen then when it is usually competing with other pathogens (species or genotypes) in the same host. When disease initiated by single individuals is common, a sustainable exploitation strategy of the host could be selected for, aiming at a high reproductive output of the virus, and long survival of the host. When disease caused by multiple pathogen individuals is common, a pre-emptive and more wasteful exploitation strategy of the host, aiming at maximization of resource capture in the host in competition with others, would be more advantageous from the perspective of maximizing pathogen fitness. Considering the possibility that host disease initiated by a single or small number of individuals can occur on a regular basis, and its many ramifications, may therefore be highly relevant. Knowing whether IAH applies to a particular pathosystem may be indispensable to understanding pathogen evolution. Testing IAH is technically and conceptually possible, but testing IAH should go beyond testing the model that we have formulated here. Data contradictory to IAH in the current framework may perhaps be reconciled with IAH under more elaborate frameworks. Such frameworks may include the effects of variability among hosts and process details of the interaction between pathogens and their hosts.
Acknowledgments
We thank Frans Van Aggelen, Andre Gidding and Leo Koopman for providing S. exigua and M. brassicae eggs. We thank Mart de Jong, Cajo ter Braak, Kelli Hoover, Jaap Molenaar, Paul Struik and Gorben Pijlman for their comments on the manuscript. We also thank four anonymous referees for their detailed and constructive criticism. M.P.Z. was supported by a grant from the C.T. de Wit Graduate School for ‘Production Ecology and Resource Conservation’. J.S.C. would like to thank NSERC for funding.
Supplementary Material
References
- Ali A., Li H.Y., Schneider W.L., Sherman D.J., Gray S., Smith D., Roossinck M.J. Analysis of genetic bottlenecks during horizontal transmission of cucumber mosaic virus. J. Virol. 2006;80:8345–8350. doi: 10.1128/JVI.00568-06. doi:10.1128/JVI.00568-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ami F., Regoes R.R., Ebert D. A quantitative test of the relationship between parasite dose and infection probability across different host–parasite combinations. Proc. R. Soc. B. 2008;275:853–859. doi: 10.1098/rspb.2007.1544. doi:10.1098/rspb.2007.1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi F., Snoeijing I., Van der Werf W., Mans R.M.W., Smits P.H., Vlak J.M. Biological activity of SeMNPV, AcMNPV, and three AcMNPV deletion mutants against Spodoptera exigua larvae (Lepidoptera: Noctuidae) J. Invertebr. Pathol. 2000;75:28–35. doi: 10.1006/jipa.1999.4907. doi:10.1006/jipa.1999.4907 [DOI] [PubMed] [Google Scholar]
- Bianchi F., Vlak J.M., Rabbinge R., Van der Werf W. Biological control of beet armyworm, Spodoptera exigua, with baculoviruses in greenhouses: development of a comprehensive process-based model. Biol. Control. 2002;23:35–46. doi:10.1006/bcon.2001.0989 [Google Scholar]
- Burden J.P., Griffiths C.M., Cory J.S., Smith P., Sait S.M. Vertical transmission of sublethal granulovirus infection in the Indian meal moth, Plodia interpunctella. Mol. Ecol. 2002;11:547–555. doi: 10.1046/j.0962-1083.2001.01439.x. doi:10.1046/j.0962-1083.2001.01439.x [DOI] [PubMed] [Google Scholar]
- Carrasco P., Daros J.A., Agudelo-Romero P., Elena S.F. A real-time RT-PCR assay for quantifying the fitness of tobacco etch virus in competition experiments. J. Virol. Methods. 2007;139:181–188. doi: 10.1016/j.jviromet.2006.09.020. doi:10.1016/j.jviromet.2006.09.020 [DOI] [PubMed] [Google Scholar]
- Chao L. Fitness of RNA virus decreased by Muller's ratchet. Nature. 1990;348:454–455. doi: 10.1038/348454a0. doi:10.1038/348454a0 [DOI] [PubMed] [Google Scholar]
- Cooper V.S., Reiskind M.H., Miller J.A., Shelton K.A., Walther B.A., Elkinton J.S., Ewald P.W. Timing of transmission and the evolution of virulence of an insect virus. Proc. R. Soc. Lond. B. 2002;269:1161–1165. doi: 10.1098/rspb.2002.1976. doi:10.1098/rspb.2002.1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas J.M., Moya A., Sanjuan R. Following the very initial growth of biological RNA viral clones. J. Gen. Virol. 2005;86:435–443. doi: 10.1099/vir.0.80359-0. doi:10.1099/vir.0.80359-0 [DOI] [PubMed] [Google Scholar]
- Druett H.A. Bacterial invasion. Nature. 1952;170:288. doi: 10.1038/170288a0. doi:10.1038/170288a0 [DOI] [PubMed] [Google Scholar]
- Engelhard E.K., Volkman L.E. Developmental resistance in 4th-instar Trichoplusia ni orally inoculated with Autographa californica M nuclear polyhedrosis virus. Virology. 1995;209:384–389. doi: 10.1006/viro.1995.1270. doi:10.1006/viro.1995.1270 [DOI] [PubMed] [Google Scholar]
- Federici B.A. Baculovirus pathogenesis. In: Miller L.K., editor. The baculoviruses. Plenum Press; New York, NY: 1997. pp. 33–59. [Google Scholar]
- Giannini C., Giannelli F., Monti M., Careccia G., Marrocchi M.E., Laffi G., Gentilini P., Zignego A.L. Prevalence of mixed infection by different hepatitis C virus genotypes in patients with hepatitis C virus-related chronic liver disease. J. Lab. Clin. Med. 1999;134:68–73. doi: 10.1016/s0022-2143(99)90055-0. doi:10.1016/S0022-2143(99)90055-0 [DOI] [PubMed] [Google Scholar]
- Gibbs M.J., Armstrong J.S., Gibbs A.J. Recombination in the hemagglutinin gene of the 1918 ‘Spanish flu’. Science. 2001;293:1842–1845. doi: 10.1126/science.1061662. doi:10.1126/science.1061662 [DOI] [PubMed] [Google Scholar]
- Goulson D., Cory J.S. Sublethal effects of baculovirus in the cabbage moth, Mamestra brassicae. Biol. Control. 1995;5:361–367. doi:10.1006/bcon.1995.1042 [Google Scholar]
- Hernandez-Crespo P., Sait S.M., Hails R.S., Cory J.S. Behavior of a recombinant baculovirus in lepidopteran hosts with different susceptibilities. Appl. Environ. Microbiol. 2001;67:1140–1146. doi: 10.1128/AEM.67.3.1140-1146.2001. doi:10.1128/AEM.67.3.1140-1146.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillborn, R. & Mangel, M. 1997 The ecological detective; confronting models with data Monographs in Population Biology, vol. 28, p. 315. Princeton, NJ: Princeton University Press.
- Hoover K., Grove M.J., Su S.Z. Systemic component to intrastadial developmental resistance in Lymantria dispar to its baculovirus. Biol. Control. 2002;25:92–98. doi:10.1016/S1049-9644(02)00041-5 [Google Scholar]
- Hughes P.R., Wood H.A. A synchronous peroral technique for the bioassay of insect viruses. J. Invertebr. Pathol. 1981;37:154–159. doi:10.1016/0022-2011(81)90069-0 [Google Scholar]
- Hughes D.S., Possee R.D., King L.A. Evidence for the presence of a low-level, persistent baculovirus infection of Mamestra brassicae insects. J. Gen. Virol. 1997;78:1801–1805. doi: 10.1099/0022-1317-78-7-1801. [DOI] [PubMed] [Google Scholar]
- Lenhoff R.J., Luscombe C.A., Summers J. Competition in vivo between a cytopathic variant and a wild-type duck hepatitis B virus. Virology. 1998;251:85–95. doi: 10.1006/viro.1998.9394. doi:10.1006/viro.1998.9394 [DOI] [PubMed] [Google Scholar]
- López-Ferber M., Simón O., Williams T., Caballero P. Defective or effective? Mutualistic interactions between virus genotypes. Proc. R. Soc. Lond. B. 2003;270:2249–2255. doi: 10.1098/rspb.2003.2498. doi:10.1098/rspb.2003.2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckow V.A., Lee S.C., Barry G.F., Olins P.O. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol. 1993;67:4566–4579. doi: 10.1128/jvi.67.8.4566-4579.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H., Van Duijse J.J.A., Zuidema D., Van Hulten M.C.W., Vlak J.M. Fitness and virulence of an ancestral white spot syndrome virus isolate from shrimp. Virus Res. 2005;110:9–20. doi: 10.1016/j.virusres.2005.01.002. doi:10.1016/j.virusres.2005.01.002 [DOI] [PubMed] [Google Scholar]
- Olkin I., Gleser L.J., Derman C. Macmillan; New York, NY: 1994. Probability models and applications. [Google Scholar]
- Payne C.C. Insect pathogenic viruses as pest control agents. Fortschr. Zool. 1986;32:183–200. [Google Scholar]
- Possee R.D., Hirst M., Jones L.D., Bishop D.H.L., Cayley P.J. Field tests of genetically engineered baculoviruses. In: Beadle D.J., Bishop D.H.L., Copping L.G., Dixon G.K., Holloman D.W., editors. Opportunities for molecular biology in crop protection. The British Crop Protection Council; Farnham, UK: 1993. pp. 23–33. [Google Scholar]
- Ridout M.S., Fenlon J.S., Hughes P.R. A generalized one-hit model for bioassays of insect viruses. Biometrics. 1993;49:1136–1141. doi:10.2307/2532255 [Google Scholar]
- Smith I.R.L., Crook N.E. In vivo isolation of baculovirus genotypes. Virology. 1988;166:240–244. doi: 10.1016/0042-6822(88)90165-1. doi:10.1016/0042-6822(88)90165-1 [DOI] [PubMed] [Google Scholar]
- Spieker J.O., Yuill T.M., Burgess E.C. Virulence of six strains of duck plague virus in eight waterfowl species. J. Wildl. Dis. 1996;32:453–460. doi: 10.7589/0090-3558-32.3.453. [DOI] [PubMed] [Google Scholar]
- Turchin P. Complex population dynamics: a theoretical/empirical synthesis. Monographs in Population Biology. vol. 35. Princeton University Press; Princeton, NJ: 2003. p. 456. [Google Scholar]
- Turner P.E., Chao L. Prisoner's dilemma in an RNA virus. Nature. 1999;398:441–443. doi: 10.1038/18913. doi:10.1038/18913 [DOI] [PubMed] [Google Scholar]
- Vignuzzi M., Stone J.K., Arnold J.J., Cameron C.E., Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–348. doi: 10.1038/nature04388. doi:10.1038/nature04388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn J.O., Kirkpatrick B.A., Haas-Stapleton E., Volkman L.E. Evidence that the stilbene-derived optical brightener M2R enhances Autographa californica M nucleopolyhedrovirus infection of Trichoplusia ni and Heliothis virescens by preventing sloughing of infected midgut epithelial cells. Biol. Control. 1998;11:58–69. doi:10.1006/bcon.1997.0572 [Google Scholar]
- Zeise L., Wilson R., Crouch E.A.C. Dose–response relationships for carcinogens: a review. Environ. Health Perspect. 1987;73:259–308. doi: 10.1289/ehp.8773259. doi:10.2307/3430618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart M.P., Van Oers M.M., Cory J.S., Van Lent J.W.M., Van der Werf W., Vlak J.M. Development of a quantitative real-time PCR to determine genotype frequencies for studies in baculovirus population biology. J. Virol. Methods. 2008;148:146–154. doi: 10.1016/j.jviromet.2007.10.022. doi:10.1016/j.jviromet.2007.10.022 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.