Abstract
Danionella dracula is a new species of sexually dimorphic, miniature and highly developmentally truncated cyprinid fish. Compared with its close relative, the zebrafish Danio rerio, it lacks 44 bones or parts thereof and represents one of the most developmentally truncated vertebrates. Absence of the majority of bones appears to be due to developmental truncation via terminal deletion. In contrast to these larval-like features, D. dracula also shows several hyperossifications. Uniquely, among carp-like fishes, male D. dracula have a series of long, pointed odontoid processes on the jaws greatly resembling the jaw dentition of teleosts with true teeth. The anterior-most process in each jaw is extended as a canine-like fang projecting through the epithelium. True jaw teeth are absent from all 3700 species of cypriniforms and were lost at least in the Upper Eocene. It remains to be investigated, however, whether the conserved pathways to regulate tooth development in cypriniforms have been used in D. dracula to form and pattern the odontoid processes. This new species represents a remarkable example linking progenetic paedomorphosis via heterochronic change in developmental timing to the evolution of morphological novelties.
Keywords: Danionella, Cypriniformes, jaw teeth, miniaturization, developmental truncation, evolutionary novelty
1. Introduction
Miniaturization, the evolution of extremely small body size, has been linked to the appearance of morphological novelties (Hanken 1993; Hanken & Wake 1993). In vertebrates, this relationship is best documented for amphibians and fishes, the most intriguing recent example being the cypriniform genus Paedocypris (Kottelat et al. 2006; Britz & Conway 2009). Among cypriniforms (carp-like fishes), the most unusual morphological novelties are found in miniatures showing a high degree of developmental truncation (Rüber et al. 2007; Britz & Conway 2009). Here, we report the discovery of a new miniature species of cypriniform, Danionella dracula n. sp., which is closely related to the zebrafish (Danio rerio) and, even when adult, shows a remarkably larval-like skeleton. This developmental truncation is associated in this species with the evolution of astonishing novel morphological characters, all of them sexually dimorphic. The most extraordinary novelty of males of the new species is a series of odontoid processes in both the upper and lower jaws, some of which project through the epithelium as true teeth.
Jaw teeth are a defining character of gnathostomes and are almost universally present in various shapes and sizes among the 50 000 species of jawed vertebrates. Jaw teeth have been lost repeatedly during the evolution of gnathostomes, with turtles and birds being the most prominent examples. Among bony fishes, teeth are also absent from the jaws of all gonorynchiforms and cypriniforms, curimatid characiforms, and the majority of syngnathoids among teleosts. The zebrafish, a member of the cypriniforms, has become an important model organism for the study of the developmental genetic pathways that initiate tooth formation, and recent studies have demonstrated that essential parts of this pathway have been retained in the oral cavity during cypriniform evolution (Jackman & Stock 2006; Stock et al. 2006; Stock 2007). The newly discovered species D. dracula is morphologically closer to an oral dentition than any other cypriniform, but with the evolutionary acquisition of its odontoid processes, it has clearly followed an alternative route to the re-evolution of jaw teeth. The present paper describes this unique cypriniform fish, its unusual jaw anatomy and developmentally truncated skeleton, and highlights its potential for addressing more general questions about the evolutionary role of heterochrony to generate morphological diversity and the re-evolution of lost structures.
2. Material and methods
(a) Anatomical study
All sizes are of a standard length (SL; from tip of the lower jaw to the end of hypural complex in millimetres). Measurements were made at 10× magnification using a stereomicroscope equipped with an ocular micrometer. For the study of the skeleton, 10 males and 10 females were cleared and double stained for bone and cartilage (Taylor & Van Dyke 1985). For the histological study of the odontoid processes, a 16.7 mm male was embedded in paraplast, serially sectioned at 8 μm, and the sections were stained with Azan-Domagk. Specimens and sections were photographed with a Jenoptik ProgRes 12 C digital camera on a Zeiss Tessovar or a Zeiss Axiocam HRc on a Zeiss Discovery V20. One male was prepared for scanning electron microscopy; it was critical-point dried and sputter coated with palladium–gold and observed and photographed in a Philips Xl-30 FEG SEM. All specimens are deposited in the Natural History Museum (BMNH), London.
(b) Biological material, DNA isolation, PCR and DNA sequencing
To assess the phylogenetic position of D. dracula among rasborine cyprinids, analyses were performed based on 44 specimens of the Cyprinidae representing 39 species, one species of the Catostomidae, two species of the Cobitidae, two species of the Balitoridae, one species of the Gyrinocheilidae and one species of the Gonorynchiformes as a more distant out-group (see the electronic supplementary material for a list of specimens). The phylogenetic analyses were based on four mitochondrial genes (12S rRNA, 16S rRNA, cytochrome c oxidase subunit I (cox1) and cytochrome b (cytb)) and one nuclear gene (exon 3 of the recombinase-activating gene 1 (RAG1)).
The cytb nucleotide sequences published by Rüber et al. (2007) were complemented with cytb sequences newly determined for an additional seven Danionella specimens. The 12S rRNA, 16S rRNA, cox1 and RAG1 genes were PCR amplified and sequenced for 29 Rasborinae taxa, specifically for this study. In addition, several sequences were obtained from GenBank (see the electronic supplementary material for GenBank accession numbers).
Total DNA extractions from muscle tissue of specimens fixed in 70–100 per cent ethanol were conducted using the Qiagen DNeasy Blood and Tissue kit following manufacturer's protocols. The complete cytb gene was PCR amplified with two versatile primers, Don-GluF and Don-ThrR (Rüber et al. 2004) and additional primers published by Rüber et al. (2007). For the remaining mitochondrial genes, approximately 380 bp of the 3′ end of the 12S rRNA was amplified with the primers L1091 and H1478 (Kocher et al. 1989), approximately 500 bp of the 5′ end of the 16S rRNA was amplified with the primers 16Sar and 16Sbr (Palumbi et al. 1991) and a 700 bp fragment of cox1 was amplified with the primers LCO1490 and HCO2198 (Folmer et al. 1994). Approximately 1500 bp of the nuclear RAG1 gene were amplified with the primers RAG1F1 and RAG1R1 (Lopez et al. 2004). PCR amplifications on a G-Storm GS1 PCR thermal cycler and sequencing reactions followed the procedures outlined in Rüber et al. (2007).
(c) Sequence alignment and phylogenetic analyses
Protein-coding nucleotide sequence datasets were aligned by eye. The 12S rRNA and 16S rRNA gene nucleotide sequence datasets were aligned following procedures given by Rüber & Zardoya (2005), resulting in an initial 12S rRNA alignment of 476 positions and an initial 16S rRNA alignment of 618 positions. The final DNA nucleotide sequence dataset used for all subsequent analyses consisted of a total of 3925 aligned positions (mtDNA: cytb 1131 bp, cox1 669 bp, 12S rRNA 284 bp, 16S rRNA 374 bp; nuclear DNA: RAG1 1467 bp). The Akaike information criterion implemented in Modeltest v. 3.7 (Posada & Crandall 1998) was used to determine the evolutionary model that best fits the dataset. The model selected was subsequently used for Bayesian inference (BI) and maximum-likelihood (ML) analyses. A BI of cyprinid phylogeny was performed with MrBayes v. 3.1.2 (Huelsenbeck & Ronquist 2001) by Metropolis coupled Markov chain Monte Carlo (MC3) sampling for 3 000 000 generations (two independent runs each with four simultaneous MC chains; chain temperature 0.2; sample frequency 300 under the GTR+I+Γ model as selected by Modeltest v. 3.7). The cytb dataset was run with seven data partitions (first, second and third codon positions for the combined mitochondrial protein-coding genes (cytb and cox1) and the nuclear RAG1, respectively, and the mitochondrial rRNA data), and model parameters were estimated independently for each of the respective data partitions using the unlink command in MrBayes v. 3.1.2. Tracer v. 1.4 (Rambaut & Drummond 2007) was used to plot the −log-likelihood scores against generation time to evaluate mixing, run convergence and the burn-in needed before reaching stationarity. We then used PAUP* v. 4.0b10 (Swofford 2002) to reconstruct the 50 per cent majority-rule consensus tree of the post burn-in trees (a burn-in of 150 000 generations was used, resulting in 10 000 post burn-in trees from the two independent runs). ML analyses were conducted with GARLI v. 0.94 (Zwickl 2006) under the GTR+I+Γ model (parameters not fixed) and using the default settings (best tree, −ln likelihood 57 456.92).
(d) Divergence time estimates
A time tree was constructed using penalized likelihood (PL), as implemented in r8s v. 1.70 (Sanderson 2003), based on the ML phylogram to date major cladogenetic events. The truncated Newton (TN) algorithm and the additive penalty function were used for the PL analyses. To find the optimal smoothing parameter (λ) for PL, cross validation was performed over a range of values of λ ranging from 100 to 102.8 in 15 steps (optimal λ=1.58). We used the oldest known fossil of the Cyprinidae (Patterson 1993), Parabarbus sp. from the Early Eocene (Ypresian, 49.0–54.8 Myr ago) as the calibration point to estimate divergence times between the Danionella species and applied a median age of the Ypresian (51.9 Myr ago) to calibrate the cyprinid tree. The most recent common ancestor (MRCA) of cyprinids and its sister group (stem group calibration) were used as the fixed ‘cyprinid root’. The 95% confidence intervals on time estimates were obtained through bootstrapping by (i) generating 100 bootstrapped datasets in Phylip v. 3.6 (Felsenstein 1989), (ii) estimating the ML tree for each of the bootstrapped date sets in PAUP* under the topological constraint of the original ML tree and under the model parameters obtained with Modeltest, and (iii) converting the 100 phylograms into chronograms under PL in r8s as described above.
In accordance with article 8.6 of the International Code of Zoological Nomenclature, copies of the PDF file of this work have been deposited in the following five publicly accessible libraries: (i) Raffles Museum of Biodiversity Research, Singapore, (ii) BMNH, London, (iii) Swedish Museum of Natural History, Stockholm, (iv) California Academy of Sciences, San Francisco, and (v) National Museum of Natural History, Smithsonian Institution, Washington DC.
3. Results
(a) Danionella dracula sp. nov.
(i) Material
3.1.1.1 Holotype
BMNH 2008.1.1.1, male, 16.3 mm SL; Myanmar: Kachin state: stream at Sha Du Zup between Mogaung and Tanai, collected 15 April 2007, Zaw Zaw Htun.
3.1.1.2 Paratypes
BMNH 2008.1.1.2–99, 98 specimens, 32 males and 66 females, 11.2–15.4 mm SL; same data as holotype. BMNH 2008.1.1.100–119, 20 specimens cleared and double stained, 10 males and 10 females, 13.1–16.7 mm SL; same locality data as holotype.
(ii) Etymology
The species name dracula alludes to the long tooth-like fangs in the jaws in males of the new species and was inspired by Count Dracula in Bram Stoker's novel. A noun in apposition.
(iii) Distribution
The species is known only from a small stream near Sha Du Zup in northern Myanmar (Burma).
(iv) Diagnosis
Danionella dracula is distinguished from its congeners, Danionella translucida and Danionella mirifica, by: the presence of a single bone in the upper jaw (versus two); the presence of a single row of 6–13 odontoid processes in males on the dorsal surface of the dentary and on the ventral surface of the upper jaw, the anterior-most processes of which are large and canine-like (versus absence of processes); the lack of the maxillary–mandibular cartilage (versus its presence); the possession of 7+8, 8+7 or 8+8 principal caudal fin rays (versus 9+9), 3–4 dorsal procurrent rays (versus 5–6) and 2–3 ventral procurrent rays (versus 4–6).
(v) Description
Known maximum size of 16.7 mm SL. Body elongate, body depth contained 6–7.5 times in standard length. Abdominal region almost circular in cross section, body behind anal fin laterally compressed. Caudal peduncle long, narrow, 1.7–2 times in body depth and laterally compressed. Head and eye large, mouth supraterminal. Upper and lower jaws massive in large males, resulting in longer preorbital distance. Nostrils well developed. Lateral line canals and pores on head and body absent. Dorsal fin short-based, situated opposite posterior half of long-based anal fin. Caudal fin furcate with remnants of larval-fin-fold in front of its dorsal and ventral margins. Remnant of pre-anal larval-fin-fold present in females only. Anus and genital papilla of males located distinctly anterior to anal fin between pelvic fins; in females at typical position in front of anal fin. Window (pseudotypanum) present in body musculature rendering pigmented surface of lateral side of anterior gas bladder chamber visible. Body muscles greatly thinned out at lateral side of posterior gas bladder chamber. Scales absent.
Morphometric information based on 10 males (including holotype) and 10 females is presented in table 1 in the electronic supplementary material.
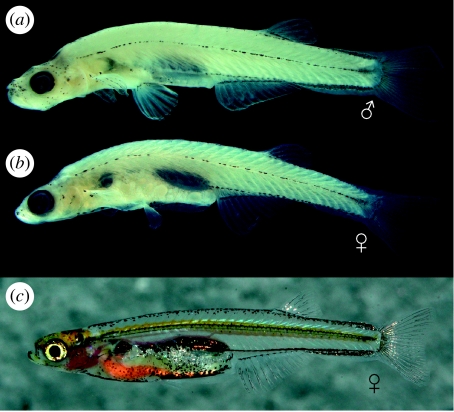
Pigmentation in alcohol specimens is restricted to five rows of melanophores (figure 1): a broad, mid-dorsal row from head to caudal-fin base; a mid-lateral row along horizontal septum from shoulder girdle to hypural plate; a ventral row from in front of and slightly above anal-fin base along ventral larval-fin-fold to the end of hypural plate; a row along anal-fin base; an abdominal, mid-ventral row from ventral tip of cleithrum to anus; and melanophores capping dorsal and dorsolateral surface of gas bladder chambers and their connecting duct. In life, body is colourless and largely translucent (figure 1c), except for melanophore patterns described above and a thin yellow line running along body at level of neural tube.
Figure 1.
Lateral view of D. dracula. (a) Holotype (BMNH 2008.1.1.1), male, 16.3 mm. (b) Paratype (BMNH 2008.2-99), female, 14.0 mm. (c) Live female.
(vi) Osteology
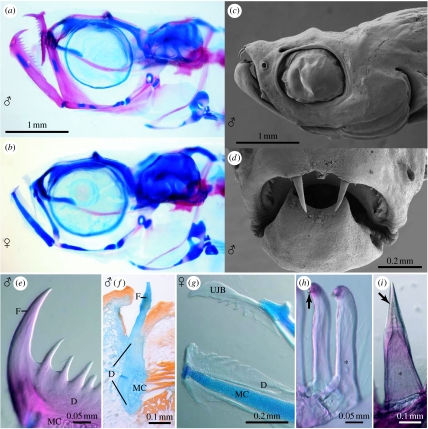
The cleared and double-stained specimens revealed that the skull, hyopalatine arch, gill arches and endoskeletal shoulder girdle are mostly cartilaginous with perichondral ossifications giving the skeleton an overall larval appearance. The following bones and cartilages are absent in D. dracula: kinethmoid; preethmoid; vomer; dermethmoid; nasal; parietal; intercalar; extrascapular; infraorbitals 2–5; angular; coronomeckelian; ectopterygoid; metapterygoid and pars metapterygoidea; urohyal; basibranchial 4 cartilage; hypobranchials 1–3 and hypobranchial cartilages 1–3; sclerotic bones; post-temporal; postcleithrum; mesocoracoid; pectoral radials 3–4 and pectoral radial cartilages 3–4; pectoral distal radial cartilages; pelvic radials 1–3 and pelvic radial cartilages 1–3; supraneurals 2 and 5–9 and supraneural cartilages 5–9; middle radials in dorsal and anal fins; epineurals; epipleurals; uroneural 2; and scales. The upper jaw consists only of one bilaterally paired bone (figure 2a,b), representing either the premaxilla or the maxilla only, or a fusion of these two bones. Each of the jaws in males bears a longitudinal series of 6–13 processes that gradually decrease in size posteriorly and greatly resemble the jaw dentition of other teleosts. The most anterior of these odontoid processes is fang-like (figure 2a,e). Females have only poorly ossified jaw bones with rudimentary fangs in both jaws and only few, short serrations (figure 2g). Scanning electron microscopy of the head demonstrates further that the upper fangs in males project approximately 0.3 mm beyond the epithelium and are curved ventrally (figure 2c,d). By contrast, the lower fangs have only their distal tips exposed. Our histological study of the processes confirms that the fangs pierce the epidermal covering (figure 2f) and demonstrates that the odontoid processes are extensions of the dentary and upper jaw bone with no demarcation between the processes and these bones. It further shows that the processes lack an enameloid cusp and a pulp cavity (figure 2f), in contrast to the four pharyngeal teeth on the heavily ossified ceratobranchial 5 in D. dracula (figure 2h) or the jaw teeth of a characiform otophysan (figure 2i). They are therefore not true teeth. In addition to the jaws, the entopterygoid of the hyopalatine arch and the basipterygium of the pelvic girdle exhibit a sexual dimorphism, being grossly enlarged and hyperossified in males. The axial skeleton includes a total of 36–37 vertebrae, consisting of 15–16 abdominal and 20–22 caudal vertebrae. Ribs are present on vertebrae 5–12; dorsal-fin rays ii, 4–5, i; anal-fin rays ii, 9–11, i; principal caudal-fin rays 7+8, 8+7 or 8+8 plus 3–4 dorsal and 2–3 ventral procurrent rays; pectoral fin rays i, 4, ii and pelvic fin rays i, 3, ii in females and i, 4, i in males. There are three brachiostegal rays.
Figure 2.
Danionella dracula. (a) Male, paratype, 16.2 mm (BMNH 2008.1.1.100-119), cleared and double stained, head skeleton, lateral view. (b) Female, paratype, 14.7 mm (BMNH 2008.1.1.100-119), cleared and double stained, head skeleton, lateral view. (c) Male, paratype, 14.8 mm (BMNH 2008.1.1.2-99), scanning electron micrograph, lateral view. (d) Same as (c), but close-up of mouth in anterior view. (e) Same as (a), but close-up of odontoid processes at anterior tip of lower jaw. (f) Male, 16.7 mm, histological section through canine-like odontoid process of the lower jaw, illustrating lack of pulp cavity and enameloid cusp. (g) Female, paratype, 15.4 mm (BMNH 2008.1.1.100-119), cleared and double stained, close-up of jaws with serrate edges on jaw bones and rudimentary canine-like odontoid processes, lateral view. (h) Same as (b), but close-up of pharyngeal jaw teeth with pulp cavity (marked with asterisk) and enameloid cusp (marked with arrow). (i) Hoplias malabaricus (BMNH 2005.7.5.778-863), 44 mm, cleared and double stained, close-up of individual premaxillary tooth with pulp cavity (marked with asterisk) and enameloid cusp (marked with arrow) for comparison with (e). D, dentary; F, fang-like odontoid process; MC, Meckel's cartilage; UJB, upper jaw bone.
(vii) Molecular phylogenetics and divergence time estimates
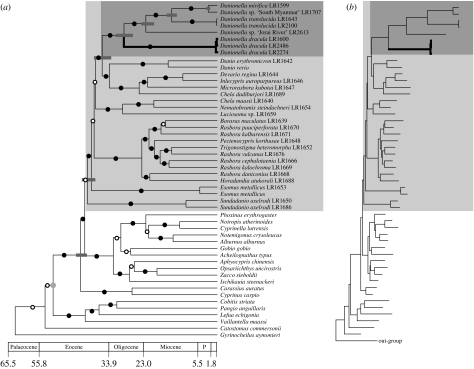
Our molecular phylogenetic analyses to determine the phylogenetic position of D. dracula demonstrate that Danionella is a derived member of the Rasborinae clade A (sensu Rüber et al. 2007), supporting previous morphological (Roberts 1986; Fang 2003) and molecular phylogenetic (Rüber et al. 2007; Conway et al. 2008) hypotheses. Danionella dracula was recovered as the sister group of the remaining Danionella species. A relaxed molecular clock approach revealed that Danionella split from its sister group at approximately 36.4 (95% confidence interval 34.6–38.7) Myr ago. The time window for the evolution of the strikingly tooth-like processes unique to D. dracula was determined as 29.5 (95% confidence interval 27.4–31.9) Myr since it split from the MRCA of Danionella (figure 3).
Figure 3.
Reconstructed phylogeny of the Cyprinidae using ML based on an alignment of 3925 positions of both mitochondrial and nuclear DNA nucleotide sequence data using Chanos chanos (Gonorynchiformes) as out-group. (a) Time tree obtained under a relaxed molecular clock using PL and a fossil calibration point (stem group of cyprinids; the node used for calibration is indicated with a grey circle). The 95% confidence intervals for selected nodes are shown with grey bars. The Rasborinae clade A and the genus Danionella are highlighted with a grey and a dark grey box, respectively. Branches with a posterior probability of 0.95 or above are shown with a black circle and those less than 0.95 with a white circle. (b) Phylogram highlighting the long branches of the Danionella species.
4. Discussion
(a) Miniaturization, developmental truncation and evolutionary novelty
The evolution of very small adult body size, miniaturization, is widespread among animals and has been cited as a prominent evolutionary process leading to phyletic diversification (Hanken & Wake 1993). Some of the most impressive cases of miniaturization occur in teleost fishes (Weitzman & Vari 1988; Hanken & Wake 1993), which also include the smallest vertebrates maturing at sizes below 10 mm (Winterbottom & Emery 1981; Watson & Walker 2004; Pietsch 2005; Kottelat et al. 2006). Teleosts provide convincing examples for the claim that the evolution of extremely small size is frequently associated not only with reduction and structural simplification, but also with hyperossification, the evolution of morphological novelty and with increased morphological variability (Hanken & Wake 1993). Of the 2400 species of Cyprinidae, 36 are considered miniatures including the smallest vertebrate species Paedocypris progenetica (Kottelat et al. 2006), and cyprinids are thus a model group of teleosts for the study of evolution of miniaturization. A recent analysis of the phylogenetic relationships of the cyprinid subfamily Rasborinae (Rüber et al. 2007) has suggested that miniaturization evolved several times independently in that group, which includes more than 63 per cent of all cyprinid miniatures (23 out of 36). At least in cypriniforms, miniaturization events have resulted in tiny organisms of two different types (Rüber et al. 2007; Britz & Conway 2009): the ‘proportioned dwarfs’ (tiny, but otherwise more or less identical copies of their larger ancestors) and the ‘developmentally truncated miniatures’ (tiny and resembling early developmental stages of their larger ancestors). For cypriniforms, Rüber et al. (2007) and Britz & Conway (2009) identified an unexpected correlation between developmentally truncated miniatures and the evolution of morphological novelties. This finding is supported and exemplified by D. dracula.
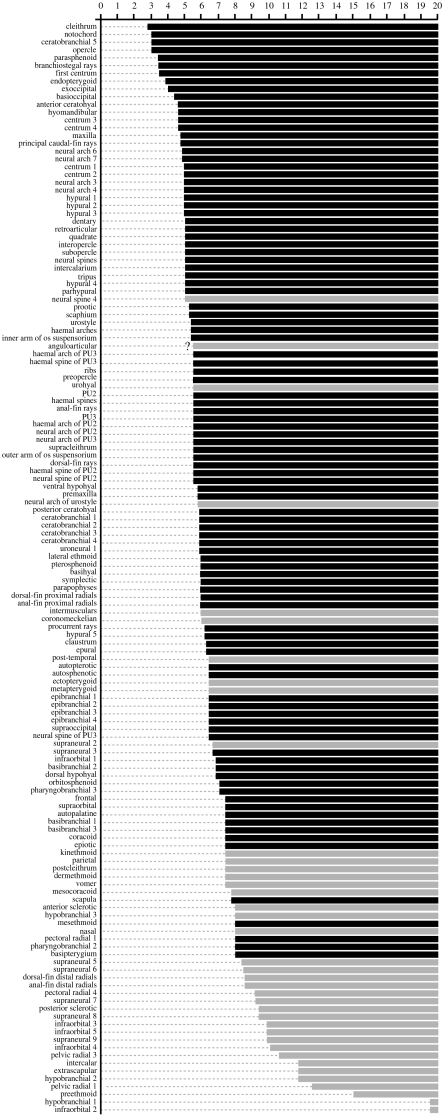
Forty-four bones or parts thereof fail to develop in D. dracula, compared with its close relative the zebrafish D. rerio, which is roughly twice its length. To evaluate the degree of developmental truncation, we compared the missing bones in D. dracula with the sequence of ossification of the closely related zebrafish for which a comprehensive dataset is available (Cubbage & Mabee 1996; Bird & Mabee 2003). Our analysis shows that with few exceptions the missing bones in D. dracula are those that ossify late in the ontogenetic trajectory of the zebrafish (figure 4). Their lack in D. dracula thus suggests a relatively simple case of developmental truncation, i.e. loss of terminal stages in ontogeny. There are another nine bones or bone parts, however, which are missing in D. dracula and are not readily explained by simple truncation. Of these nine, two characters (absence of neural spine 4, intermusculars) are uniquely shared among Asian miniature cypriniforms with Paedocypris and another two (absence of post-temporal and ectopterygoid) with Paedocypris and Sundadanio. Along with additional characters, these losses have been interpreted as evidence for a close relationship of the three genera, with Danionella as the sister group of Paedocypris (see discussion in Britz & Conway 2009). Absence of the urohyal and the metapterygoid is a derived character of all Danionella species and absence of the coronomeckelian bone seems to be an autapomorphy of D. dracula.
Figure 4.
Diagram to illustrate the relationship between sequence of ossification of D. rerio compiled from Cubbage & Mabee (1996) and Bird & Mabee (2003) and losses and reductions in the skeleton of adult D. dracula. Numbers along the top refer to sizes in millimetres at which ossifications appear in the zebrafish. Names of bones on the left side of the diagram are arranged in sequence of ossification in the zebrafish from top (earliest) to bottom (latest). First appearance of ossification in the zebrafish is shown as horizontal bars at given lengths. Black bars represent ossifications present and grey bars ossifications absent in D. dracula. No information was provided by above authors for pectoral radials 2 and 3, pelvic radial 2, middle radials of dorsal and anal fin and uroneural 2, which are all absent in D. dracula. Question mark at bar of the anguloarticular indicates lack of separate information for angular and articular in Cubbage & Mabee (1996).
The large number of undeveloped skeletal structures highlights the larval-like neurocranial and branchial skeleton of D. dracula and makes it even more developmentally truncated than Paedocypris, which lacks 41 bones or bone parts when compared with the zebrafish (Britz & Conway 2009). This theme of loss and reduction in D. dracula contrasts with pronounced hyperossification restricted to its sexually dimorphic skeletal structures, the jaws, hyopalatine arch and pelvic fin skeleton. The most remarkable morphological novelties of all are the canine-like fangs on the jaws that project far beyond the epithelium and the other odontoid processes in series with them. The biological function of these jaw modifications, which can be easily mistaken for true teeth, is at present unknown.
With its highly developmentally truncated skeleton, D. dracula is a clear case of progenetic paedomorphosis through heterochrony, as defined by Gould (1977). Heterochrony, the change in developmental timing of characters, has been considered an important evolutionary mechanism for the creation of morphological diversity (e.g. de Beer 1958; Gould 1977; Raff & Wray 1989; Smith 2003), but the two different types of paedomorphosis, progenesis (paedomorphosis produced by an acceleration of maturation) and neoteny (paedomorphosis produced by a retardation of somatic development), have been assigned quite different evolutionary roles. For example, De Beer (1958, p. 64) argued that progenesis is ‘of no significance in progressive evolution’, while Gould (1977, p. 345) considered it to have ‘great macro-evolutionary potential’, but, as he admitted, only in ‘exceedingly rare’ cases. Danionella dracula is certainly of considerable interest in this context, as it combines a predominantly larval/juvenile skeleton with the highly progressive development of its jaws, hyopalatine arch and pelvic girdle skeleton. It is conceivable that developmental truncation in this species has facilitated the evolution of these novel structures by freeing large parts of the skeleton from developmental constraints, dissociating developmentally linked pathways and creating a greater potential for more dramatic changes. This potential, one would have to conclude, remained unused by the other two species of Danionella, D. translucida and D. mirifica. Both are also developmentally truncated to a similar degree as D. dracula, but do not show the same skeletal modifications. Danionella dracula may thus serve as a convincing example for the evolution of spectacular morphological novelty not only via miniaturization but also by a heterochronic shift towards extreme progenetic paedomorphosis.
(b) Danionella dracula and the re-evolution of teeth
None of the 3700 described cypriniform species have jaw teeth, although several predatory species have evolved a dorsally directed pointed process on each dentary at their symphysis (Howes 1978, 1979, 1980). Superficially, these symphysial processes resemble the canines of the lower jaw in D. dracula. The processes, however, are covered by cornified skin and fit into a groove in the upper jaw, which lacks any projections. Our study clearly demonstrates that the fangs and processes in D. dracula are not true teeth. Their condition is, however, still more similar to that of toothed jaws than in any other cypriniform. The fangs pierce the skin as true teeth and the remaining odontoid processes are arranged in an anteroposterior series resembling the jaw dentition of typical teleosts. Jackman & Stock's (2006) recent claim that the evolutionary reacquisition of teeth in cypriniforms might be relatively easy is challenged by the jaw structure of D. dracula. In line with Dollo's law (Dollo 1883; Gould 1970), which states that complex structures do not re-evolve with the same complexity once they have been lost, this species has formed tooth-like structures as extensions from the body of the dentary and upper jaw bone. However, as Jackman & Stock (2006) and Stock et al. (2006), however, have demonstrated in the zebrafish, the developmental genetic pathways to regulate tooth formation have been conserved in the oral cavity during the evolution of cypriniforms. It remains to be tested whether these conserved pathways may have been deployed to initiate and pattern the development of its odontoid processes. Because the new species belongs to the same clade as the vertebrate model organism zebrafish, it offers an interesting opportunity to unravel the genetic mechanisms that have led to its spectacular tooth-like jaw modifications and its highly developmentally truncated skeleton.
Acknowledgments
We thank U. Tin Win, Hein Aquarium, Yangon, for helping to collect the type series and NSF's Cypriniform Tree of Life project (DEB 0431326) for financial support to K.W.C. The collecting trip to Myanmar was funded from the collection enhancement grant scheme of BMNH, London. Erwin Schraml, Munich, provided the photo of the live D. dracula. We are grateful to W. Maier, Tübingen, for access to his histology laboratory and M. Meinert for preparing the histological sections.
Supplementary Material
Table 1. Summary of specimens, ID=identification number (personal collection of first author), and GenBank accession numbers of the species used.
Table 2. Selected morphometric information on 10 males and 10 females of Danionella dracula including the holotype.
References
- Bird N.C., Mabee P.M. Developmental morphology of the axial skeleton of the zebrafish, Danio rerio (Ostariophysi: Cyprinidae) Dev. Dyn. 2003;228:337–357. doi: 10.1002/dvdy.10387. doi:10.1002/dvdy.10387 [DOI] [PubMed] [Google Scholar]
- Britz, R. & Conway, K. W. 2009 Osteology of Paedocypris, a miniature and highly developmentally truncated fish (Teleostei: Ostariophysi: Cyprinidae). J. Morphol. (doi:10.1002/jmor.10698) [DOI] [PubMed]
- Conway K.W., Chen W.-J., Mayden R.L. The ‘celestial pearl danio’ is a miniature Danio (s.s) (Ostariophysi: Cyprinidae): evidence from morphology and molecules. Zootaxa. 2008;1686:1–28. [Google Scholar]
- Cubbage C.C., Mabee P.M. Development of the cranium and paired fins in the zebrafish Danio rerio (Ostariophysi: Cyprinidae) J. Morphol. 1996;229:121–160. doi: 10.1002/(SICI)1097-4687(199608)229:2<121::AID-JMOR1>3.0.CO;2-4. doi:10.1002/(SICI)1097-4687(199608)229:2<121::AID-JMOR1>3.0.CO;2-4 [DOI] [PubMed] [Google Scholar]
- de Beer G.R. Clarendon Press; Oxford, UK: 1958. Embryos and ancestors. [Google Scholar]
- Dollo L. Les lois de l'évolution. Bull. Soc. Belge Géol. Paleont Hydrol. 1883;7:164–166. [Google Scholar]
- Fang F. Phylogenetic analysis of the Asian cyprinid genus Danio. Copeia. 2003;2003:714–728. doi:10.1643/IA03-131.1 [Google Scholar]
- Felsenstein J. Phylip—phylogeny inference package (v. 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- Gould S.J. Dollo on Dollo's law: irreversibility and the status of evolutionary laws. J. Hist. Biol. 1970;3:189–212. doi: 10.1007/BF00137351. doi:10.1007/BF00137351 [DOI] [PubMed] [Google Scholar]
- Gould S.J. Belknap; Cambridge, MA: 1977. Ontogeny and phylogeny. [Google Scholar]
- Hanken J. Adaptation of bone growth to miniaturization of body size. In: Hall B.K., editor. Bone, bone growth. vol. 7. CRC Press; Boca Raton, FL: 1993. pp. 79–104. [Google Scholar]
- Hanken J., Wake D.B. Miniaturization of body size: organismal consequences and evolutionary significance. Annu. Rev. Ecol. Syst. 1993;24:501–519. doi:10.1146/annurev.es.24.110193.002441 [Google Scholar]
- Howes G.J. The anatomy and relationships of the cyprinid fish Luciobrama macrocephalus (Lacepède) Bull. Br. Mus. Nat. Hist. (Zool.) 1978;34:1–64. [Google Scholar]
- Howes G.J. Notes on the anatomy of Macrochirichthys macrochirus (Velenciennes), 1844, with comments on the Cultrinae (Pisces, Cyprinidae) Bull. Br. Mus. Nat. Hist. (Zool.) 1979;36:147–200. [Google Scholar]
- Howes G.J. The anatomy, phylogeny and classification of the bariliine cyprinid fishes. Bull. Br. Mus. Nat. Hist. (Zool.) 1980;37:129–198. [Google Scholar]
- Huelsenbeck J.P., Ronquist F.R. MrBayes: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. doi:10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Jackman W.R., Stock D.W. Transgenic analysis of Dlx regulation in fish tooth development reveals evolutionary retention of enhancer function despite organ loss. Proc. Natl Acad. Sci. USA. 2006;103:19390–19395. doi: 10.1073/pnas.0609575103. doi:10.1073/pnas.0609575103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher T.D., Thomas W.K., Meyer A., Edwards S.V., Pääbo S., Villablanca F.X., Wilson A.C. Dynamics of mitochondrial DNA: evolution in animals—amplification and sequencing with conserved primers. Proc. Natl Acad. Sci. USA. 1989;86:6196–6200. doi: 10.1073/pnas.86.16.6196. doi:10.1073/pnas.86.16.6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottelat M., Britz R., Tan H.H., Witte K.E. Paedocypris, a new genus of southeast Asian cyprinid fish with a remarkable sexual dimorphism, comprises the world's smallest vertebrate. Proc. R. Soc. B. 2006;273:895–899. doi: 10.1098/rspb.2005.3419. doi:10.1098/rspb.2005.3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez J.A., Chen W.J., Orti G. Esociform phylogeny. Copeia. 2004;2004:449–464. doi:10.1643/CG-03-087R1 [Google Scholar]
- Palumbi S., Martin A., Romano S., McMillan W., Stice L., Grabowski G. University of Hawaii Press; Honolulu, HI: 1991. The simple fools guide to PCR. [Google Scholar]
- Patterson C. Teleostei. In: Benton M.J., editor. The fossil record 2. Chapman & Hall; London, UK: 1993. pp. 621–656. [Google Scholar]
- Pietsch T.W. Dimorphism, parasitism, and sex revisited: modes of reproduction among deep-sea ceratioid anglerfishes (Teleostei: Lophiiformes) Ichthyol. Res. 2005;52:207–236. doi:10.1007/s10228-005-0286-2 [Google Scholar]
- Posada D., Crandall K. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. doi:10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Raff R.A., Wray G.A. Heterochrony: developmental mechanisms and evolutionary results. J. Evol. Biol. 1989;2:409–434. doi:10.1046/j.1420-9101.1989.2060409.x [Google Scholar]
- Rambaut, A. & Drummond, A. J. 2007 Tracer v1.4. See http://beast.bio.ed.ac.uk/Tracer
- Roberts T. Danionella translucida, a new genus and species of cyprinid fish from Burma, one of the smallest living vertebrates. Environ. Biol. Fishes. 1986;16:231–241. doi:10.1007/BF00842977 [Google Scholar]
- Rüber L., Zardoya R. Rapid cladogenesis in marine fishes revisited. Evolution. 2005;59:1119–1127. doi:10.1554/04-394 [PubMed] [Google Scholar]
- Rüber L., Britz R., Tan H.H., Ng P.K.L., Zardoya R. Evolution of mouthbrooding and life-history correlates in the fighting fish genus Betta. Evolution. 2004;58:799–813. doi: 10.1111/j.0014-3820.2004.tb00413.x. doi:10.1554/03-364 [DOI] [PubMed] [Google Scholar]
- Rüber L., Kottelat M., Tan H.H., Ng P.K.L., Britz R. Evolution of miniaturization and the phylogenetic position of Paedocypris, comprising the world's smallest vertebrate. BMC Evol. Biol. 2007;7:38. doi: 10.1186/1471-2148-7-38. doi:10.1186/1471-2148-7-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson M.J. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. doi:10.1093/bioinformatics/19.2.301 [DOI] [PubMed] [Google Scholar]
- Smith K.K. Time's arrow: heterochrony and the evolution of development. Int. J. Dev. Biol. 2003;47:613–621. [PubMed] [Google Scholar]
- Stock D.W. Zebrafish dentition in comparative context. J. Exp. Zool. B Mol. Dev. Evol. 2007;308:1–27. doi: 10.1002/jez.b.21187. [DOI] [PubMed] [Google Scholar]
- Stock D.W., Jackman W.R., Trapani J. Developmental genetic mechanisms of evolutionary tooth loss in cypriniform fishes. Development. 2006;133:3127–3137. doi: 10.1242/dev.02459. doi:10.1242/dev.02459 [DOI] [PubMed] [Google Scholar]
- Swofford D.L. Sinauer; Sunderland, MA: 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods), v. 4.0b10. [Google Scholar]
- Taylor W.R., Van Dyke G.C. Revised procedures for staining and clearing small fishes and other vertebrates for bone and cartilage study. Cybium. 1985;9:107–119. [Google Scholar]
- Watson W., Walker H.J. The world's smallest vertebrate, Schindleria brevipinguis, a new paedomorphic species in the family Schindleriidae (Perciformes: Gobioidei) Rec. Aust. Mus. 2004;56:139–142. [Google Scholar]
- Weitzman S.H., Vari R.P. Miniaturization in South American freshwater fishes; an overview and discussion. Proc. Biol. Soc. Wash. 1988;101:444–465. [Google Scholar]
- Winterbottom R., Emery A.R. A new genus and two new species of gobiid fishes (Perciformes) from the Chagos Archipelago, central Indian Ocean. Environ. Biol. Fishes. 1981;6:139–149. doi:10.1007/BF00002777 [Google Scholar]
- Zwickl, D. J. 2006 GARLI—genetic algorithm for rapid likelihood inference. See http://www.bio.utexas.edu/faculty/antisense/garli/Garli.html
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1. Summary of specimens, ID=identification number (personal collection of first author), and GenBank accession numbers of the species used.
Table 2. Selected morphometric information on 10 males and 10 females of Danionella dracula including the holotype.