Abstract
Quantitative genetic approaches have been developed that allow researchers to determine which of two mechanisms, mutation accumulation (MA) or antagonistic pleiotropy (AP), best explain observed variation in patterns of senescence using classical quantitative genetic techniques. These include the creation of mutation accumulation lines, artificial selection experiments and the partitioning of genetic variances across age classes. This last strategy has received the lion's share of empirical attention. Models predict that inbreeding depression (ID), dominance variance and the variance among inbred line means will all increase with age under MA but not under those forms of AP that generate marginal overdominance. Here, we show that these measures are not, in fact, diagnostic of MA versus AP. In particular, the assumptions about the value of genetic parameters in existing AP models may be rather narrow, and often violated in reality. We argue that whenever ageing-related AP loci contribute to segregating genetic variation, polymorphism at these loci will be enhanced by genetic effects that will also cause ID and dominance variance to increase with age, effects also expected under the MA model of senescence. We suggest that the tests that seek to identify the relative contributions of AP and MA to the evolution of ageing by partitioning genetic variance components are likely to be too conservative to be of general value.
Keywords: ageing, quantitative genetics, inbreeding depression, dominance
1. Introduction
Senescence is generally thought to have evolved because the strength of selection for vitality declines with age (Medawar 1946, 1952; Hamilton 1966). Two non-exclusive genetic mechanisms have been proposed to explain the role that inheritance plays in this phenomenon. Mutation accumulation (MA; Medawar 1946, 1952) posits that de novo germline mutations arise that diminish survival or reproduction. While the effects of these mutations can occur at any age, the deleterious load at equilibrium will be greatest at late age because the strength of purifying selection diminishes with increased age. A second model, antagonistic pleiotropy (AP; Williams 1957), imagines that genes can contribute to the evolution of senescence if mutations arise that increase early-age vitality at the expense of late-age vitality. Hamilton (1966) and Charlesworth (1994, 2001) formalized much of this ageing theory by integrating it into demographic and population genetic frameworks.
Within the field of evolutionary demography, researchers have focused on assessing the relative degree to which each of these two mechanisms contributes to the evolution of senescence in both laboratory-based and natural populations. These efforts can be classified into two kinds of approaches: those that seek to understand the joint distribution of de novo mutational effects across ages (i.e. the mutational variance–covariance matrix or U-matrix; Pletcher et al. 1998; Mack et al. 2000; Yampolsky et al. 2000) and those that try to infer the joint distribution of age-specific allele effects that segregate in populations (the genotypic variance–covariance matrix or G-matrix). In the first instance, researchers have tested the underlying assumption of the MA model—namely, that there exist deleterious de novo mutations with effects confined to late ages—by maintaining populations over many generations under conditions intended to minimize selection. While mutation accumulation studies can be technically demanding and are feasible in only a few study systems, they are among the most direct means with which to evaluate the genetic mechanisms of ageing.
The other strategy is to characterize segregating genetic variance and covariance for age-specific traits at multiple ages either indirectly using artificial selection (e.g. Rose & Charlesworth 1981b; Luckinbill et al. 1984; Rose 1984; Partridge & Fowler 1992) or directly using quantitative trait locus (QTL) analyses (e.g. Nuzhdin et al. 1997; Leips & Mackay 2000; Leips et al. 2006; Wilson et al. 2006) or more traditional quantitative genetic methods. The last requires that extant patterns of standing genetic variation reflect past action of the two putative genetic mechanisms for ageing. The traditional quantitative genetic approach has been advocated, in particular, by Charlesworth & Hughes (hereafter referred to as CH; Hughes & Charlesworth 1994; Charlesworth & Hughes 1996), who extended existing population genetic models (e.g. Rose 1982; Charlesworth 1994) to illustrate how genetic variance components differ under the two mechanisms of senescence. CH suggested that, under MA, additive variance, dominance variance, homozygote variance and inbreeding depression (ID) should all increase with age. By contrast, working with the assumption that AP loci are overdominant (i.e. that they contribute to segregating genetic variation by heterozygote advantage), CH argue that, under AP, only additive variance is expected to increase with age (table 1).
Table 1.
Predictions of variance component change with age given the two models of ageing.
| variance component | description | quantitative genetic definitiona | MA | AP |
|---|---|---|---|---|
| VA | additive genetic variance (variance in main effects) | b | ↑ | ↑c |
| VH | homozygote variance (variance in inbred line means) | ↑ | no change | |
| VD | dominance variance (variance in dominance effects in heterozygotes) | ↑ | no change | |
| IDd | inbreeding depression (change in mean caused by inbreeding) | ↑ | no change | |
| δ | standardized inbreeding depression | e | ↑ | no change |
Following definitions from Falconer & Mackay (1996). We assume a biallelic locus i with allele frequencies pi1+pi2=1, where allele i2 is recessive.
The main effect is αi=ai+(pi2−pi1)di, a function of allele frequencies and additive and dominance effects (defined in the text).
VA was originally expected not to change with age under AP (Hughes & Charlesworth 1994); this was later amended to predict an increase with age under both models (Charlesworth & Hughes 1996).
Inbreeding depression depends upon genetic effects and allelic variation. For our purposes here, that qualifies it as a ‘variance component’. ID is proportional to the inbreeding coefficient F. ID is equal to B, or ‘inbreeding load’ in Charlesworth & Hughes (1996).
Standardized inbreeding depression is ID divided by the outbred mean phenotype . δ is termed ‘ID’ under Charlesworth & Hughes (1996). CH expect both measures of ID to increase with age under MA but not AP. This presents somewhat of a quandary, however, when the same data generate opposite trends for ID and δ with age (e.g. Hughes et al. 2002). In these cases, the same data would seem to suggest different mechanisms of senescence depending upon how one scaled the data.
Estimates of these putatively diagnostic variance components are far easier to obtain than estimates of the U-matrix. Furthermore, this variance component approach can be applied to a far greater variety of biological systems, including humans. For these reasons, this variance partitioning strategy has become a popular way to determine the relative importance of MA versus AP in the evolution of senescence. These tests have been carried out not only in model systems in the laboratory (Mueller 1987; Hughes & Charlesworth 1994; Charlesworth & Hughes 1996; Promislow et al. 1996; Hughes et al. 2002; Rose et al. 2002; Snoke & Promislow 2003; Swindell & Bouzat 2006) but also across a broad taxonomic range of non-model systems, including (but not limited to) beetles (Fox et al. 2006), crustaceans (Yampolsky & Galimov 2005), birds (Charmantier et al. 2006; Brommer et al. 2007; Keller et al. 2008), ungulates (Wilson et al. 2007; Nussey et al. 2008) and snails (Escobar et al. 2008). These studies usually favour an MA interpretation based upon the predictions that arise from the models of CH. Rose et al. (2007), however, argue that these results are inconsistent and, in some cases, unreliable, owing to laboratory artefacts and inappropriate experimental designs. As a result, they question the use of the variance partitioning approach to determine evolutionary mechanisms.
Whether CH's model provides a suitable diagnostic test for these two theories of senescence also depends upon the generality of their genetic model for each mechanism. We do need clear and mutually exclusive predictions in order to determine the relative importance of MA and AP in the evolution of senescence. However, we suggest that the existing models may not provide definitive, testable predictions. While it is important to note that CH's model does hold under specific conditions, here we argue that it may not be general enough to account for a potentially important class of overdominant AP loci. These AP loci could generate patterns of genetic variance and ID that will appear as signatures of MA.
2. AP and genetic variation
We begin with a review of AP involving a single locus with two alleles. We then consider CH's model of AP, focusing in particular on key simplifying assumptions of their model. We will argue that these assumptions are unnecessary to generate balancing selection and variation for senescence by AP. However, these overly restrictive assumptions are required to ensure the particular patterns of genetic variation that CH believe are diagnostic of AP.
(a) AP and overdominance
Following standard quantitative genetic notation for genetic effects (Falconer & Mackay 1996; Lynch & Walsh 1998), imagine a single genetic locus i that has effects at two ages, x and y, where y>x. The locus has two alleles, 1 and 2, where allele 1 increases some fitness-related phenotype Z at age x when compared with allele 2. The phenotypic rankings of the two alleles are reversed at later age y. The genotypic values are given relative to age-specific means of the two homozygotes: the phenotypic difference between the best and worst homozygotes at age x is
| (2.1) |
where ai(x) is the allelic effect at age x. Likewise, the allelic effect at age y is given by
| (2.2) |
The value of heterozygotes may differ from the homozygote mean at either age; the age-specific difference at age j is the dominance deviation
| (2.3) |
The value di(j) quantifies the degree to which allele 1 is dominant to allele 2 (at locus i and age j).
Rose (1982) and Curtsinger et al. (1994) have shown that, under certain combinations of parameter values, AP can cause the fitness of the heterozygote to exceed the fitness of both homozygotes when averaged over all traits/ages/environments (marginal overdominance). This situation will cause alleles to equilibrate at intermediate frequencies, allowing single loci to generate large amounts of genetic variation. The exact amount and type of genetic variation that is expected to exist (additive, dominance, etc.) will depend upon the particular genetic effects that relate the genotype to the phenotype: ai(x), ai(y), di(x) and di(y). The likelihood that a stable polymorphism is maintained depends upon these genetic effects and upon the relevance of the trait/age/environment to fitness.
To further explore this, we use β to indicate the ratio of fitness differences between the best and worst homozygotes at locus i for traits x and y,
| (2.4) |
The polymorphism is protected by marginal overdominance if and only if the fitness of the heterozygote is greater than the fitness of both homozygotes. Following Rose (1982), we can define this necessary and sufficient condition in terms of genetic effects and selection differentials
| (2.5) |
The reversal in sign of allelic effects across ages will generate negative additive genetic covariance at that locus. If the locus is of major effect, or there are many loci that act in the same way, then this genetic covariance may be detected using standard quantitative genetic approaches (Lynch & Walsh 1998). If, however, allelic variation is maintained by mutation, then these loci will contribute weakly to the covariances and trade-offs might not be detected except, perhaps, by using artificial selection experiments of very long duration (e.g. Service et al. 1988).
(b) The CH model of AP and senescence
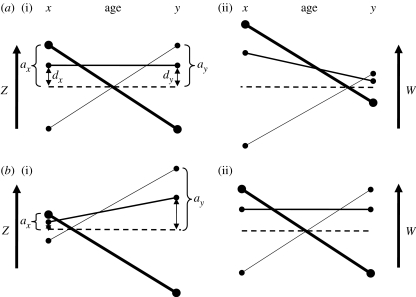
CH applied the overdominance model to antagonism between traits expressed at different ages. There are many possible relationships between genetic effects that can cause overdominance but CH consider only a specific form of this model, where ai(x)=−ai(y), such that the effects of the two homozygotes on age-specific phenotypes are constrained to be equal and opposite over both ages. The effects of dominance on phenotypes are held positive and constant across ages, di(x)=di(y)>0 (figure 1a(i)). CH chose these particular effect values as they provide the simplest model of AP (Hughes & Charlesworth 1994; Charlesworth & Hughes 1996). While we recognize the need to simplify models to make them tractable, we will argue that if we relax these simplifying assumptions we obtain results that are possibly more evolutionarily relevant than the CH model. Our alternative model of AP has very different quantitative genetic consequences.
Figure 1.
AP models for marginal overdominance. (a) Age-specific genetic effects on (i) phenotype and (ii) fitness under the Charlesworth–Hughes (CH) model of AP. (b) Age-specific genetic effects on (i) phenotype and (ii) fitness under a more stable model of AP. The thick solid lines indicate the genetic value of homozygote 11, the thin solid lines are the values of the alternative homozygote 22, and the intermediately solid lines indicate the value of the heterozygotes. The dashed lines are average of homozygote values. CH assume equal additive effects on phenotypes across ages (indicated by the vertical lengths of brackets on a(i)). They also assume equal dominance effects (double-headed arrows). This requires that ID and dominance variance will not change with age (equation (2.1) in the text). (a(ii)) The additive and dominance effects on the fitness scale decline with age because early-age traits (x) are more relevant to fitness than late-age traits (y). All else being equal, (b(ii)) the stability of these polymorphisms increases with increased symmetry of homozygote effects on the scale of fitness. (b(i)) With ageing, maximum stability is reached when additive and dominance effects on the phenotypic scale are greatest at late age. This causes additive genetic variance, dominance variance and ID to increase with age.
CH reasonably argue from Hamilton (1966) that the strength of selection declines with age (i.e. βi<1). Weighting these genetic effects by fitness, they calculate allele frequencies conditioned upon a stable interior equilibrium. They then predict the magnitude of several genetic variance components, as well as the potential for ID, using equilibrium allele frequencies and genetic effects (table 1). Doing the same for a model of MA, they are able to contrast quantitative genetic predictions for MA versus AP models. They find that the dominance variance and ID will increase with age for MA but not for AP. Recall that their model assumes that the additive and dominance effects on phenotype are constant across ages and that a stable polymorphism is achieved under AP. When we weight these genotypic values by fitness, however, the genetic effects must be reduced at late age (figure 1a(ii)).
(c) A more stable model of senescence caused by overdominant AP
Curtsinger et al. (1994) showed that AP is most likely to generate marginal overdominance if the allelic effects are equal and opposite across traits when they are inversely weighted by fitness; in our notation and applied to age, this is when ai(y)=(ai(x)/βi) (figure 1b(ii)). Viewed from the perspective of fitness, this is the simplest relevant model of AP as well as the most stable. This model requires that the additive effects on phenotype increase with age (figure 1b(i)).
In reality, we have no idea how common these loci are compared with the loci modelled by CH. However, we can imagine that if there were variation among AP loci in the change of the magnitude of allelic effects across ages, then selection would more effectively remove genetic variation generated by loci with symmetrical phenotypic effects (i.e. CH-type loci) than with symmetric effects on fitness (our AP model). As a result, loci with increased allelic effects with age will have an outsized effect on patterns of genetic variation. This is essentially a ‘selective sieve’ argument similar to those that have previously been used to explain why we should expect negative genetic correlations across life-history traits (Falconer & Mackay 1996) and why we should expect that dominance and epistatic variance should be greatest in fitness traits (Mukai et al. 1972; Crnokrak & Roff 1995; Roff & Emerson 2006). Below, we explore the consequence of these kinds of AP loci on the putatively diagnostic patterns of genetic variation that have been described by CH.
(d) Implications of these AP models for genetic variance components
CH originally suggested that increases in additive genetic variance with age indicate MA loci (Charlesworth 1990; Hughes & Charlesworth 1994), because this model predicts more equitable allele frequency (and thus greater allelic variation) at late age due to higher mutational load. Because allele frequencies should not change with age under AP, it was believed that these loci would not cause additive genetic variance to change with age. In subsequent work, Charlesworth & Hughes (1996) show that, under AP, the later-acting trait will have much higher additive genetic variance at late age, too. The reason for this has to do with how dominance effects contribute towards additive genetic variation. Under random mating, additive genetic variance is the variation among main effects, which in the two allele models considered by CH is simply the variance in allele frequencies (heterozygosity) multiplied by the square of the main effects (table 1). The main effect increases with the difference between the frequency of the recessive and dominant alleles. For traits acting early, the dominant allele will be more common than the recessive allele, making the main effect α less than the additive effect a. For traits acting at late age, the reverse is true (the common allele is recessive) and the main effect will exceed the additive effect. Recall that under CH's model, the additive effects are constant over all ages, meaning that main effects must increase with age. Because the additive genetic variance is proportional to the square of the main effects, CH's model predicts an increase in additive genetic variance with age under AP. In this way, they show that additive genetic variance is not diagnostic of MA versus AP.
However, Charlesworth & Hughes (1996) point out that the variation in homozygous effects VH should have diagnostic value because these effects more directly indicate the relationship between additive effects and allele frequency (table 1). Under MA, relaxed selection at late age increases heterozygosity, thereby causing an increase in VH. Under AP, however, allele frequencies do not change with age and dominance does not matter (as it does with VA). Because their model assumes that additive effects do not change with age under AP, they show that VH should not be affected by age. Under our model of AP, however, the loci that generate the VH will tend to have greater additive effects at late age than at early age. This will cause VH to increase with age with AP, just as it would with MA.
Owing to the perceived limitations of additive genetic variation tests, and possibly to the relative difficulty of deriving the completely inbred lines necessary for estimating VH, researchers have turned to CH's predictions regarding age-related changes in non-additive genetic variance. However, these predictions too may be problematic. Consider the age-related changes in ID and dominance variance. Both should increase under MA but not under AP, according to CH's model. In general, ID and dominance variance will increase with heterozygosity and with the magnitude of the dominance deviations (table 1). For simplicity, CH chose age-independent dominance effects, di(x)=di(y)>0, in their AP model. However, this ensured that neither ID nor VD could change with age (recall that allele frequencies cannot change with age, as traits at both ages are determined by the same locus). While this choice follows from a desire for parsimony (a single value is easier to model than two), what do we know about the pleiotropic effects of dominance when additive values reverse?
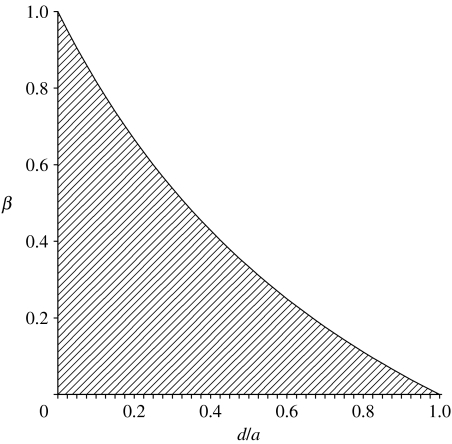
We could argue that, in this case, trait dominance will be higher at late age than at early age because, for some parameter space, an increase in dominance at late age is necessary in order to maintain polymorphism by AP. There is no parameter space where the reverse, a decrease in dominance, is necessary. Let us simplify equation (2.5) to illustrate this by setting the magnitude of additive effects on phenotype equal at both ages (as with CH). Dominance d at young age x becomes d+Δd at later age y. Rearranging equation (2.5), we find that if the heterozygote is the most fit genotype, then the necessary condition for a change in dominance with age is
| (2.6) |
Dominance increases in age when β<(a−d)/(a+d), where d is the dominance deviation at early age. In figure 2, we illustrate the critical selection ratio below which an increase in dominance with age is necessary in order for AP to maintain a protected polymorphism. If initial dominance is low relative to additive effects and if selection is much less intense at late age compared with early age, then dominance must increase with age to maintain polymorphism. When initial dominance is high, then an increase in dominance is less critical. In any case, if selection diminishes with age, then an age-related increase in dominance will always increase opportunities for more loci to contribute to genetic variation by marginal overdominance. The increased dominance with age will lead to age-related increases in ID and dominance variance (table 1)—exactly what CH show us we should expect if MA caused ageing.
Figure 2.
AP conditions that require increased dominance with age. The shaded region indicates that dominance must increase with age in order to generate variation by balancing selection. The horizontal axis is the amount of dominance at early age divided by the additive effect (e.g. 0 corresponds to additivity and 1 to complete dominance). The vertical axis is the fitness relevance of the phenotype at late age divided by that at early age. This assumes that additive effects are constant across age. Large differences in ages (low β) will require dominance to increase unless early-age dominance is very high.
Let us return to our earlier logic regarding stability and argue that symmetry on the fitness scale is the more natural way to model balancing selection by AP (i.e. ai(y)=(ai(x)/βi)). After all, this model is more likely to generate marginal overdominance because the homozygote effects on fitness are equal and opposite at the two ages. What is most likely to happen to the dominance deviations in this case? If we constrain the ratio of dominance to additive effects on phenotype to remain constant with age, then dominance deviations should increase (figure 1b(i)), causing ID and dominance variance to increase as well. For single traits, theory argues that as allelic values diverge for loci affecting fitness, heterozygote values should tend towards the better homozygote (Wright 1934; Kacser & Burns 1981). Applying this logic to each trait or age independently in the AP model, we expect that if additive effects increase with age, then dominance effects will too, or di(y)>di(x)>0.
Dominance effects on pleiotropy are unlikely to be so plastic, however. Keightley & Kacser (1987) have argued that physiological constraints will cause the direction of dominance to change when the fitness ranking of homozygotes changes across traits or environments. If so, then we can expect that di(y)>0>di(x) or di(y)<0<di(x). This has two consequences relevant to our discussion. First, polymorphisms are not likely to become stable (Curtsinger et al. 1994) and, even with AP, genetic variance will depend upon mutational input. Second, if an AP-facilitated polymorphism was somehow made stable despite unfavourable dominance effects, then any ID caused by the AP locus would be positive at one age and negative at the other. More realistically, MA might generate some positive ID at all ages so that, when taken together, AP and MA effects could lead to very different patterns of ID at early and late ages, despite contributions from AP. Moorad (2005) has argued that this model of dominance constraint could be tested by comparing the additive and dominance genetic correlations across life-history traits. If AP were capable of causing marginal overdominance, then dominance correlations should tend to be greater than additive genetic correlations. The physiological constraint argument of Keightley & Kacser (1987) against AP predicts that the two correlations should be identical. This is probably not a feasible test for ageing-related AP, as breeding designs that can resolve these non-additive genetic correlations may require thousands of families (Lynch & Walsh 1998, pp. 635–636).
3. Discussion
CH (Hughes & Charlesworth 1994; Charlesworth & Hughes 1996) have presented alternative predictions that are meant to allow investigators to discriminate between two proposed pathways for the evolution of senescence. They have argued that two quantitative genetic parameters—namely, age-specific ID and dominance variance—have diagnostic properties that allow us to discriminate between MA and AP. Here, we point to two potential challenges facing studies that attempt to test these predictions. First, AP could have played a dominant role in the evolution of senescence without leaving traces in extant patterns of genetic variation. An allele that has an antagonistic effect as described by Williams (1957)—that is, it increases vitality at early age at the expense of late-age vitality—will have one of two fates. It may be advantageous or deleterious with respect to lifetime fitness at all frequencies, in which case its frequency will approach fixation or loss. Any genetic variation will be maintained by mutational pressure. Alternatively, the new allele and its counterpart allele can generate marginal overdominance by a form of balancing selection that is stable when allele frequencies are intermediate. The likelihood that variation at an AP locus is generated this way is dependent upon dominance parameters (Curtsinger et al. 1994). However, the pattern of dominance pleiotropy required for this to happen has been dismissed elsewhere as unlikely (Keightley & Kacser 1987), suggesting that allelic variation with antagonistically pleiotropic effects across early and late ages is most likely maintained by recurrent mutation. While most AP loci are unlikely to generate marginal overdominance, it is possible that a few loci can and that these may generate important amounts of genetic variation for early- and late-age fitness traits. It is important to note that, at best, CH's tests will detect signatures of only the set of AP loci that generates marginal overdominance.
Quantitative genetic measures, such as ID and dominance variance, depend upon both allele frequencies and genetic effects. Under AP models of marginal overdominance, allele frequencies do not change with age, meaning that changes in genetic variance components are caused only by changes in the relationships between genotypes and phenotypes. Charlesworth & Hughes (1996) modelled a simple case of no change in allelic effects with age. They found that non-additive genetic measures are age-independent and, thus, qualitatively different from what we should expect from MA models. We have argued that this parsimony does not take into account the variation in age-related changes of allelic effects. AP loci that have increased phenotypic effects with age will contribute more to genetic variation than those with unchanged or reduced effects with age. When dominance effects are considered, this more inclusive perspective of overdominance AP reveals that ID and dominance variance may increase with age. Other variance components, such as additive genetic variation (Charlesworth & Hughes 1996) and the variation among inbred lines (in this paper), have been shown to give similar qualitative results with both MA and AP loci. In this light, we are led to conclude that none of the diagnostic tests proposed by CH allow us to distinguish AP from MA effects on ageing, even when the two mechanisms of ageing correspond exactly to the balancing selection/purifying selection dichotomy as envisioned by CH. For this reason, we may more accurately interpret age-related increases in additive genetic variance, inbred line mean variance, dominance variance and ID as evidence for some evolutionary mechanism for senescence.
While genetic variance partitioning approaches may be of limited value for understanding the evolution of ageing, they can still be useful. First of all, studies of genetic variance components for vital rates are a key piece of information if we are to predict the short-term evolutionary trajectories of a population under selection. For example, if reproduction is deliberately delayed to some late age (as in the classic experiment of Rose 1984, will life expectancy increase, and, if so, how quickly?
Perhaps more informative, however, are studies of the variance–covariance matrix for novel mutations, U. With accurate measures of U, we can better understand some of the underlying genetic constraints acting on senescence. If we find deleterious mutations for which age-specific effects are limited to late age and which have no beneficial pleiotropic effects, we could accept the existence of age-specific mutations that contribute to senescence, as posited by Medawar (1946, 1952). Similarly, finding age-specific mutations with beneficial effects early and deleterious effects late (or the reverse) would provide evidence for the type of mutations needed to generate senescence under Williams's (1957) model. What is less clear (and perhaps less testable) is whether such antagonistically pleiotropic mutations are likely to ever account for the enormous genetic variation for ageing that we observe in natural populations.
Mutation accumulation experiments can also reveal important age-related patterns of pleiotropy that do not fit into the basic MA/AP paradigm. For example, existing studies show us that most mutations are pleiotropic across ages but have the same direction of effect with respect to fitness (e.g. Pletcher et al. 1998, 1999; Mack et al. 2000; Yampolsky et al. 2000; Gong et al. 2006). They also report that the mean and variance of mutational effects tend to decrease with age. Both findings have precipitated new theoretical insights into how senescence evolves (e.g. Charlesworth 2001; Moorad & Promislow 2008). However, it should be noted that the distributions of mutational effects estimated by experiments may not well reflect the mutation input in real populations if mutations exhibit different properties in mutation accumulation lines. This issue may be particularly relevant to studies of ageing if deleterious mutations of large effect that would be rapidly purged in real populations persist in the experimental populations and have a profoundly different effect on the correlation of vitality traits across ages.
In the light of the issues that we have raised here, where does this leave the empirical biologist interested in discriminating between the evolutionary models of senescence? There are still several assays that can provide definitive evidence for both MA and AP. QTL analyses can identify individual loci that appear to have antagonistic pleiotropic effects on age-specific fitness (Nuzhdin et al. 1997; Leips et al. 2006; Wilson et al. 2006). Tests of AP that look for trade-offs between early- and late-age fitness traits by detecting negative genetic correlations (e.g. Rose & Charlesworth 1981a; Tatar et al. 1996) can offer evidence for a role of the more inclusive AP model in senescence that does not depend upon overdominance. These tests should be viewed as conservative because trade-offs need not necessarily generate detectable patterns in genetic correlations (Dickerson 1955; Charlesworth 1990; Houle 1991). Finally, artificial selection experiments have demonstrated clear trade-offs between early- and late-age fitness traits, suggesting that at least some standing genetic variation for lifespan is due to genes with antagonistically pleiotropic effects (Rose 1984; Zwaan et al. 1995; Partridge et al. 1999). What about measurements of genetic variation for senescence? There is no doubt that such studies can help us determine the extent to which natural variation in rates of senescence is due to genetic versus environmental factors. Such studies are intrinsically valuable. But in the light of the results presented here, we are led to conclude that such studies cannot allow us to distinguish whether MA, AP or some other evolutionary force has contributed to such genetic variation.
Acknowledgments
This research was funded by National Science Foundation grant 0717234. We wish to thank David Hall, Troy Day, Kelly Dyer and two anonymous reviewers for their helpful discussion and commentary.
References
- Brommer J.E., Wilson A.J., Gustafsson L. Exploring the genetics of aging in a wild passerine bird. Am. Nat. 2007;170:643–650. doi: 10.1086/521241. doi:10.1086/521241 [DOI] [PubMed] [Google Scholar]
- Charlesworth B. Optimization models, quantitative genetics, and mutation. Evolution. 1990;44:520–538. doi: 10.1111/j.1558-5646.1990.tb05936.x. doi:10.2307/2409433 [DOI] [PubMed] [Google Scholar]
- Charlesworth B. Cambridge University Press; Cambridge, UK: 1994. Evolution in age-structured populations. [Google Scholar]
- Charlesworth B. Patterns of age-specific means and genetic variances of mortality rates predicted by the mutation-accumulation theory of ageing. J. Theor. Biol. 2001;210:47–65. doi: 10.1006/jtbi.2001.2296. doi:10.1006/jtbi.2001.2296 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Hughes K.A. Age-specific inbreeding depression and components of genetic variance in relation to the evolution of senescence. Proc. Natl Acad. Sci. USA. 1996;93:6140–6145. doi: 10.1073/pnas.93.12.6140. doi:10.1073/pnas.93.12.6140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charmantier A., Perrins C., McCleery R.H., Sheldon B.C. Age-dependent genetic variance in a life-history trait in the mute swan. Proc. R. Soc. B. 2006;373:225–232. doi: 10.1098/rspb.2005.3294. doi:10.1098/rspb.2005.3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crnokrak P., Roff D.A. Dominance variance—associations with selection and fitness. Heredity. 1995;75:530–540. doi:10.1038/hdy.1995.169 [Google Scholar]
- Curtsinger J.W., Service P.M., Prout T. Antagonistic pleiotropy, reversal of dominance, and genetic-polymorphism. Am. Nat. 1994;144:210–228. doi:10.1086/285671 [Google Scholar]
- Dickerson G.E. Genetic slippage in response to selection for multiple objectives. Cold Spring Harb. Symp. Quant. Biol. 1955;20:213–224. doi: 10.1101/sqb.1955.020.01.020. [DOI] [PubMed] [Google Scholar]
- Escobar J.S., Jarne P., Charmantier A., David P. Outbreeding alleviates senescence in hermaphroditic snails as expected from the mutation-accumulation theory. Curr. Biol. 2008;18:906–910. doi: 10.1016/j.cub.2008.04.070. doi:10.1016/j.cub.2008.04.070 [DOI] [PubMed] [Google Scholar]
- Falconer D.S., Mackay T.F.C. Addison-Wesley Longman Limited; Essex, UK: 1996. Introduction to quantitative genetics. [Google Scholar]
- Fox C.W., Scheibly K.L., Wallin W.G., Hitchcock L.J., Stillwell C., Smith B.P. The genetic architecture of life span and mortality rates: gender and species differences in inbreeding load of two seed-feeding beetles. Genetics. 2006;174:763–773. doi: 10.1534/genetics.106.060392. doi:10.1534/genetics.106.060392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y., Thompson J.N., Jr, Woodruff R.C. Effect of deleterious mutations on life span in Drosophila melanogaster. J. Gerontol. A Biol. Sci. Med. Sci. 2006;61:1246–1252. doi: 10.1093/gerona/61.12.1246. [DOI] [PubMed] [Google Scholar]
- Hamilton W.D. Moulding of senescence by natural selection. J. Theor. Biol. 1966;12:12–45. doi: 10.1016/0022-5193(66)90184-6. doi:10.1016/0022-5193(66)90184-6 [DOI] [PubMed] [Google Scholar]
- Houle D. Genetic covariance of fitness correlates—what genetic correlations are made of and why it matters. Evolution. 1991;45:630–648. doi: 10.1111/j.1558-5646.1991.tb04334.x. doi:10.2307/2409916 [DOI] [PubMed] [Google Scholar]
- Hughes K.A., Charlesworth B. A genetic analysis of senescence in Drosophila. Nature. 1994;367:64–66. doi: 10.1038/367064a0. doi:10.1038/367064a0 [DOI] [PubMed] [Google Scholar]
- Hughes K.A., Alipaz J.A., Drnevich J.M., Reynolds R.M. A test of evolutionary theories of aging. Proc. Natl Acad. Sci. USA. 2002;99:14 286–14 291. doi: 10.1073/pnas.222326199. doi:10.1073/pnas.222326199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacser H., Burns J.A. The molecular basis of dominance. Genetics. 1981;97:639–666. doi: 10.1093/genetics/97.3-4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P.D., Kacser H. Dominance, pleiotropy and metabolic structure. Genetics. 1987;117:319–329. doi: 10.1093/genetics/117.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller L.F., Reid J.M., Arcese P. Testing evolutionary models of senescence in a natural population: age and inbreeding effects on fitness components in song sparrows. Proc. R. Soc. B. 2008;275:597–604. doi: 10.1098/rspb.2007.0961. doi:10.1098/rspb.2007.0961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leips J., Mackay T.F. Quantitative trait loci for life span in Drosophila melanogaster: interactions with genetic background and larval density. Genetics. 2000;155:1773–1788. doi: 10.1093/genetics/155.4.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leips J., Gilligan P., Mackay T.F. Quantitative trait loci with age-specific effects on fecundity in Drosophila melanogaster. Genetics. 2006;172:1595–1605. doi: 10.1534/genetics.105.048520. doi:10.1534/genetics.105.048520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckinbill L.S., Arking R., Clare M.J., Cirocco W.C., Buck S.A. Selection for delayed senescence in Drosophila melanogaster. Evolution. 1984;38:996–1003. doi: 10.1111/j.1558-5646.1984.tb00369.x. doi:10.2307/2408433 [DOI] [PubMed] [Google Scholar]
- Lynch M., Walsh J.B. Sinauer Associates; Sunderland, MA: 1998. Genetics and analysis of quantitative traits. [Google Scholar]
- Mack P.D., Lester V.K., Promislow D.E.L. Age-specific effects of novel mutations in Drosophila melanogaster—II. Fecundity and male mating ability. Genetica. 2000;110:31–41. doi: 10.1023/a:1017538505627. doi:10.1023/A:1017538505627 [DOI] [PubMed] [Google Scholar]
- Medawar P.B. Old age and natural death. Modern Q. 1946;1:30–56. [Google Scholar]
- Medawar P.B. H. K. Lewis; London, UK: 1952. An unsolved problem of biology. [Google Scholar]
- Moorad, J. A. 2005 A quantitative investigation of dominance plasticity for a developmental threshold trait in a laboratory population of Tribolium castaneum PhD dissertation, Department of Biology, Indiana University, Bloomington, IN.
- Moorad J.A., Promislow D.E.L. A theory of age-dependent mutation and senescence. Genetics. 2008;179:2061–2073. doi: 10.1534/genetics.108.088526. doi:10.1534/genetics.108.088526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller L.D. Evolution of accelerated senescence in laboratory populations of Drosophila. Proc. Natl Acad. Sci. USA. 1987;84:1974–1977. doi: 10.1073/pnas.84.7.1974. doi:10.1073/pnas.84.7.1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai T., Chigusa S.I., Mettler L.E., Crow J.F. Mutation rate and dominance of genes affecting viability in Drosophila melanogaster. Genetics. 1972;72:335–355. doi: 10.1093/genetics/72.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussey D.H., Wilson A.J., Morris A., Pemberton J., Clutton-Brock T., Kruuk L.E. Testing for genetic trade-offs between early- and late-life reproduction in a wild red deer population. Proc. R. Soc. B. 2008;275:745–750. doi: 10.1098/rspb.2007.0986. doi:10.1098/rspb.2007.0986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzhdin S.V., Pasyukova E.G., Dilda C.L., Zeng Z.B., Mackay T.F. Sex-specific quantitative trait loci affecting longevity in Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 1997;94:9734–9739. doi: 10.1073/pnas.94.18.9734. doi:10.1073/pnas.94.18.9734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L., Fowler K. Direct and correlated responses to selection on age at reproduction in Drosophila melanogaster. Evolution. 1992;46:76–91. doi: 10.1111/j.1558-5646.1992.tb01986.x. doi:10.2307/2409806 [DOI] [PubMed] [Google Scholar]
- Partridge L., Prowse N., Pignatelli P. Another set of responses and correlated responses to selection on age at reproduction in Drosophila melanogaster. Proc. R. Soc. B. 1999;266:255–261. doi: 10.1098/rspb.1999.0630. doi:10.1098/rspb.1999.0630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletcher S.D., Houle D., Curtsinger J.W. Age-specific properties of spontaneous mutations affecting mortality in Drosophila melanogaster. Genetics. 1998;148:287–303. doi: 10.1093/genetics/148.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletcher S.D., Houle D., Curtsinger J.W. The evolution of age-specific mortality rates in Drosophila melanogaster: genetic divergence among unselected lines. Genetics. 1999;153:813–823. doi: 10.1093/genetics/153.2.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promislow D.E.L., Tatar M., Khazaeli A., Curtsinger J.W. Age-specific patterns of genetic variance in Drosophila melanogaster. I. Mortality. Genetics. 1996;143:839–848. doi: 10.1093/genetics/143.2.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roff D.A., Emerson K. Epistasis and dominance: evidence for differential effects in life-history versus morphological traits. Evolution. 2006;60:1981–1990. doi:10.1554/06-165.1 [PubMed] [Google Scholar]
- Rose M.R. Antagonistic pleiotropy, dominance, and genetic variation. Heredity. 1982;48:63–78. doi:10.1038/hdy.1982.7 [Google Scholar]
- Rose M.R. Laboratory evolution of postponed senescence in Drosophila melanogaster. Evolution. 1984;38:1004–1010. doi: 10.1111/j.1558-5646.1984.tb00370.x. doi:10.2307/2408434 [DOI] [PubMed] [Google Scholar]
- Rose M.R., Charlesworth B. Genetics of life-history in Drosophila melanogaster. I. Sib analysis of adult females. Genetics. 1981a;97:172–186. doi: 10.1093/genetics/97.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M.R., Charlesworth B. Genetics of life history in Drosophila melanogaster. II. Exploratory selection experiments. Genetics. 1981b;97:187–196. doi: 10.1093/genetics/97.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M.R., Drapeau M.D., Yazdi P.G., Shah K.H., Moise D.B., Thakar R.R., Rauser C.L., Mueller L.D. Evolution of late-life mortality in Drosophila melanogaster. Evolution. 2002;56:1982–1991. doi: 10.1111/j.0014-3820.2002.tb00124.x. doi:10.1554/0014-3820(2002)056[1982:EOLLMI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Rose M.R., Rauser C.L., Benford G., Matos M., Mueller L.D. Hamilton's forces of natural selection after forty years. Evolution. 2007;61:1265–1276. doi: 10.1111/j.1558-5646.2007.00120.x. doi:10.1111/j.1558-5646.2007.00120.x [DOI] [PubMed] [Google Scholar]
- Service P.M., Hutchinson E.W., Rose M.R. Multiple genetic mechanisms for the evolution of senescence in Drosophila melanogaster. Evolution. 1988;42:708–716. doi: 10.1111/j.1558-5646.1988.tb02489.x. doi:10.2307/2408862 [DOI] [PubMed] [Google Scholar]
- Snoke M.S., Promislow D.E. Quantitative genetic tests of recent senescence theory: age-specific mortality and male fertility in Drosophila melanogaster. Heredity. 2003;91:546–556. doi: 10.1038/sj.hdy.6800353. doi:10.1038/sj.hdy.6800353 [DOI] [PubMed] [Google Scholar]
- Swindell W.R., Bouzat J.L. Inbreeding depression and male survivorship in Drosophila: implications for senescence theory. Genetics. 2006;172:317–327. doi: 10.1534/genetics.105.045740. doi:10.1534/genetics.105.045740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M., Promislow D.E., Khazaeli A.A., Curtsinger J.W. Age-specific patterns of genetic variance in Drosophila melanogaster. II. Fecundity and its genetic covariance with age-specific mortality. Genetics. 1996;143:849–858. doi: 10.1093/genetics/143.2.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G.C. Pleiotropy, natural selection, and the evolution of senescence. Evolution. 1957;11:398–411. doi:10.2307/2406060 [Google Scholar]
- Wilson R.H., Morgan T.J., Mackay T.F.C. High-resolution mapping of quantitative trait loci affecting increased life span in Drosophila melanogaster. Genetics. 2006;173:1455–1463. doi: 10.1534/genetics.105.055111. doi:10.1534/genetics.105.055111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A.J., Nussey D.H., Pemberton J.M., Pilkington J.G., Morris A., Pelletier F., Clutton-Brock T.H., Kruuk L.E.B. Evidence for a genetic basis of aging in two wild vertebrate populations. Curr. Biol. 2007;17:2136–2142. doi: 10.1016/j.cub.2007.11.043. doi:10.1016/j.cub.2007.11.043 [DOI] [PubMed] [Google Scholar]
- Wright S. Physiological and evolutionary theories of dominance. Am. Nat. 1934;68:25–53. [Google Scholar]
- Yampolsky L.Y., Galimov Y.R. Evolutionary genetics of aging in Daphnia. Zh. Obshch. Biol. 2005;66:416–424. [PubMed] [Google Scholar]
- Yampolsky L.Y., Pearse L.E., Promislow D.E.L. Age-specific effects of novel mutations in Drosophila melanogaster—I. Mortality. Genetica. 2000;110:11–29. doi: 10.1023/a:1017582625191. doi:10.1023/A:1017582625191 [DOI] [PubMed] [Google Scholar]
- Zwaan B., Bijlsma R., Hoekstra R.E. Direct selection on life span in Drosophila melanogaster. Evolution. 1995;49:649–659. doi: 10.1111/j.1558-5646.1995.tb02301.x. doi:10.2307/2410318 [DOI] [PubMed] [Google Scholar]