Abstract
Separation of duplicated centrosomes (spindle-pole bodies or SPBs in yeast) is a crucial step in the biogenesis of the mitotic spindle. In vertebrates, centrosome separation requires the BimC family kinesin Eg5 and the activities of Cdk1 and polo kinase; however, the roles of these kinases are not fully understood. In Saccharomyces cerevisiae, SPB separation also requires activated Cdk1 and the plus-end kinesins Cin8 (homologous to vertebrate Eg5) and Kip1. Here we report that polo kinase has a role in the separation of SPBs. We show that adequate accumulation of Cin8 and Kip1 requires inactivation of the anaphase-promoting complex-activator Cdh1 through sequential phosphorylation by Cdk1 and polo kinase. In this process, Cdk1 functions as a priming kinase in that Cdk1-mediated phosphorylation creates a binding site for polo kinase, which further phosphorylates Cdh1. Thus, Cdh1 inactivation through the synergistic action of Cdk1 and polo kinase provides a new model for inactivation of cell-cycle effectors.
Centrosomes function as microtubule organizing centres (MTOC) and are responsible for the bipolar nature of the mitotic spindle1. Misregulation of the centrosome cycle can produce an abnormal number of centrosomes and multi-polar spindles, and cause chromosome mis-segregation resulting in aneuploidy. Centrosomal abnormalities and aneuploidy are common in many types of cancers2.
In mammalian cells, centrosomes consist of a pair of centrioles and the surrounding dense fibrillar mass known as the pericentriolar material3. A cell inherits one centrosome from its progenitor but replicates it as it progresses through the cell cycle. The duplicated centrosomes remain tethered to each other by a linker until the time of mitotic entry, when the linkage between the sister centrosomes is severed and they move away from each other, eventually positioning themselves face-to-face, separated by an interdigitated array of microtubules. The structure and duplication of centrosomes have been studied in some detail; however, the process by which they separate is not well understood.
The spindle pole body (SPB), a multi-protein assembly, is the centrosome-equivalent in S. cerevisiae4. Yeast daughter cells inherit from the mother cells one SPB bearing a half-bridge (a structure that extends from one side of the SPB) and duplicate it as they traverse START (the point in late G1 that precedes DNA replication). The bridge between the duplicated SPBs is severed in late S phase, allowing SPBs to move away from each other and form a metaphase spindle. Breaking of the inter-SPB bridge requires the BimC family kinesins Cin8 and Kip1 (refs 5–7). Cells deficient in both Cin8 and Kip1 are unable to separate SPBs, and hence fail to assemble a spindle. Cells lacking Cdc28 (Cdk1)/Clb (mitotic kinase) activity also fail to separate duplicated SPBs, suggesting that in yeast, Cdk1 is also required for SPB separation8–10. A recent study in S. cerevisiae has shown that Cdh1, the activator of the E3 ubiquitin ligase APC (anaphase promoting complex), which targets Cin8 and Kip1 for proteolytic degradation, is a potent inhibitor of SPB separation. The Cdk1–cyclin B (Clb) complex was also shown to catalyse SPB separation by inactivating Cdh1 through phosphorylation, thereby allowing accumulation of Cin8 and Kip1 (ref. 9).
Here we show that inactivation of Cdh1 requires both Cdk1 and polo kinase. Phosphorylation of Cdh1 by Cdk1–Clb creates polo-box binding (PBB) sites that mediate recruitment of Cdc5 polo kinase and consequently, further phosphorylation of Cdh1. Hence, Cdk1 acts as a priming kinase for Cdc5 in the process of Cdh1 inactivation, which is necessary for Cin8/Kip1 accumulation and SPB separation.
RESULTS
Cdc5 polo kinase induces SPB separation in spindle-deficient cdc34 and cdc28 mutants
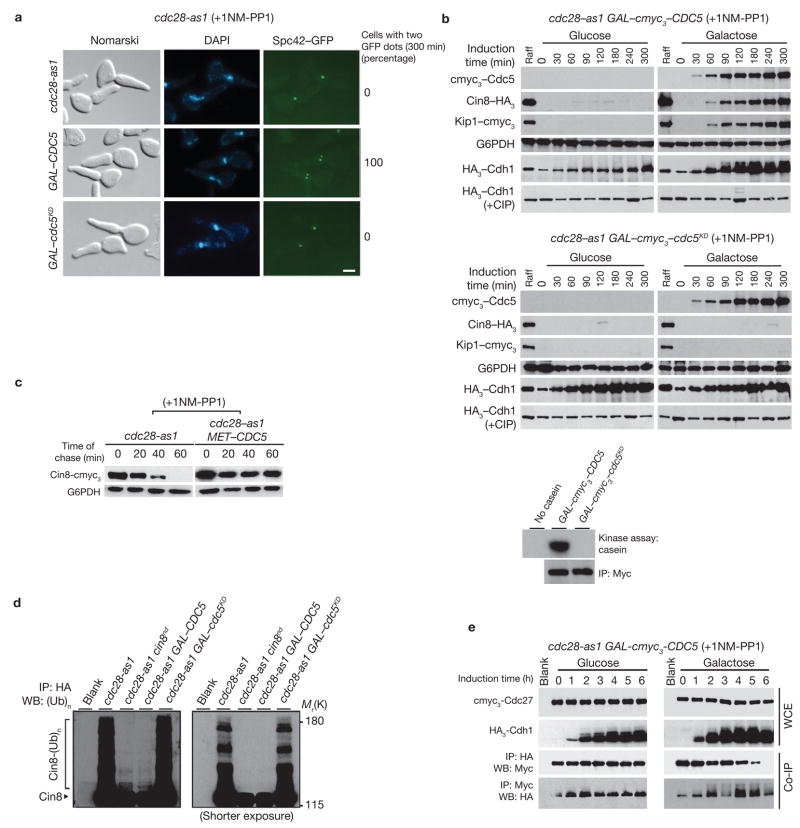
Previously9 we had characterized an allele of CDC28, cdc28-as1, carrying an F88G substitution in the ATP-binding pocket, which renders it sensitive to the ATP analogue 1NM-PP1 (ref. 11). In the presence of 1NM-PP1, cdc28-as1 cells are arrested in G2/M with an elongated bud and 2N DNA content, but are unable to separate SPBs. Our initial investigations had suggested that in addition to Cdc28, Cdc5 (yeast polo kinase) may also be important for SPB separation. To confirm this, Cdc5 or its kinase-dead version (Cdc5KD: N209A, ref. 12; Fig. 1a), was expressed from the GAL1 promoter in 1NM-PP1-treated cdc28-as1 cells carrying the GFP-tagged SPB component Spc42. Overexpression of Cdc5 allowed SPB separation (two Spc42–GFP spots) in all cells, whereas the Cdc5KD version did not, suggesting that even in the absence of mitotically active Cdc28, Cdc5 kinase can mediate SPB separation (Fig. 1a).
Figure 1.
Ectopic expression of Cdc5 causes hyperphosphorylation of Cdh1 and SPB separation. (a) cdc28-as1 cells carrying GAL–CDC5 (US5255) or GAL–cdc5KD (kinase-dead; US5360) were synchronized by α-factor treatment in YEP+Raff and released into YEP+Raff+Gal containing 1NM-PP1 at 24 °C for 300 min. Numbers indicate percentage of cells with two Spc42–GFP dots at 300 min. The scale bar represents 2 μm. (b) Top panel: G1-synchronized cdc28-as1 cells carrying GAL–cmyc3–CDC5 (US5233) were released into YEP+Raff+Gal containing 1NM-PP1 for 6 h and cells were collected at the indicated times. Western blots show Cin8–HA3 and Kip1–cmyc3 levels, and immunoprecipitated HA3–Cdh1 (with and without alkaline phosphatase (CIP)). cdc28-as1 cells carrying GAL–cmyc3–cdc5KD (US5371) were treated as described for the top panel. Bottom panel: cmyc3–Cdc5 kinase was immunoprecipitated from cdc28-as1 cells carrying either GAL–cmyc3–CDC5 or GAL–myc3–cdc5KD and was used in an in vitro assay to determine kinase activity using casein as a substrate. (c) cdc28-as1 MET–CDC5 cells carrying GAL–CIN8–cmyc3 (US5310) were grown in Raff+Met medium containing α-factor for 2 h at 24 °C and then transferred to Raff+Gal+Met medium containing α-factor for 90 min to induce Cin8 expression. Cells were then released at 24 °C into methionine-deficient glucose medium containing 1NM-PP1 and cycloheximide (1 mg ml−1) to inhibit protein synthesis). The fate of Cin8 pulse was determined by western blotting. (d) G1-synchronized cdc28-as1, cdc28-as1 expressing non-degradable version of Cin8 (cin8nd) from endogenous promoter, cdc28-as1 GAL–CDC5 and cdc28-as1 GAL–cdc5KD (US5312, US5314, US5315, US5341) were released into medium containing 1NM-PP1 and the proteasome inhibitor MG132. All except cin8nd-carrying strains harboured endogenously-tagged CIN8–HA3. Ubiquitin conjugates of immunoprecipitated Cin8 were separated on a 6% SDS gel and detected by anti-ubiquitin antibodies. The blot on the right shows a shorter exposure. (e) G1-synchronized cdc28-as1 GAL–CDC5 cells carrying endogenously-tagged cmyc3–CDC27 and HA3–CDH1 (US5355) in raffinose medium were released into either glucose or galactose medium containing 1NM-PP1. Immunoprecipitates were analysed by western blotting. Full-length blots for b, c and e are presented in Supplementary Information, Fig. S5–1.
The accumulation of Cin8 and Kip1 within 60 min of Cdc5 expression (Fig. 1b) suggests that Cdc5 mediates SPB separation by stabilizing these kinesins. Cin8 and Kip1 accumulation was also accompanied by hyperphosphorylation of Cdh1 (Fig 1b). Cdh1 hyperphosphorylation and kinesin accumulation was not seen in 1NM-PP1-treated cdc28-as1 cells expressing Cdc5KD, suggesting that Cdc5 kinase activity is required for these responses (Fig. 1b, middle panel). These results indicate that Cin8 and Kip1 accumulation is probably due to Cdh1 inactivation caused by Cdc5-mediated phosphorylation. This is consistent with two additional observations: first, Cdc5 overexpression stabilized Cin8, although the Cin8 pulse dissipated rapidly in 1NM-PP1-treated cdc28-as1 (Fig. 1c) immunoprecipitated from 1NM-PP1-and second, although Cin8–HA3 treated cdc28-as1 was highly ubiquitinated, ectopic expression of Cdc5 abolished Cin8 ubiquitination (Fig. 1d). It has been shown previously that inactivation of Cdh1 by Cdc28-mediated phosphorylation decreases its binding to the APC complex13. We found that phosphorylation by Cdc5 also dampens, to some extent, the interaction between Cdh1 and the APC component Cdc27 (Fig. 1e).
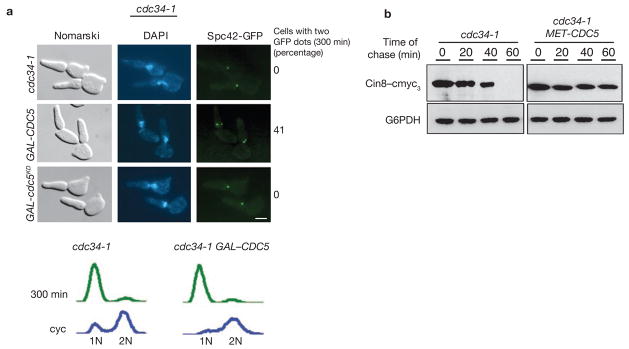
These results were further supported by ectopic expression of Cdc5 in cdc34-1 cells, which are unable to initiate S phase at restrictive temperature, arrest in late G1 (post-START) with duplicated but unseparated SPBs and are devoid of Cdc28–Clb kinase activity. Overexpression of Cdc5 in cdc34-1 cells leads to both stabilization of Cin8 and separation of SPBs (Fig. 2).
Figure 2.
Ectopic expression of Cdc5 causes spindle assembly in cdc34-1 cells. (a) cdc34-1 cells carrying GAL–CDC5 (US5276) or GAL–cdc5KD (US5308) were synchronized in G1 by α-factor treatment in YEP+Raff and released into YEP+Raff+Gal medium. The scale bar represents 2 μm. Upper Panel: Spc42–GFP signal and FACS profile of cells at 300 min. Numbers represent percentage of cells with distinctly visible Spc42–GFP dots representing two well-separated SPBs. (b) cdc34-1 MET–CDC5 cells carrying GAL–CIN8–cmyc3 (US5311) were grown in Raff+Met medium containing α-factor for 2 hrs at 24 °C and then transferred to Raff+Gal+Met medium containing α-factor for 90 min to induce Cin8 expression. Cells were then released at 37 °C into methionine-deficient glucose medium containing cycloheximide. The fate of Cin8 pulse was determined by western blotting.
PBB sites in Cdh1 are necessary for SPB separation
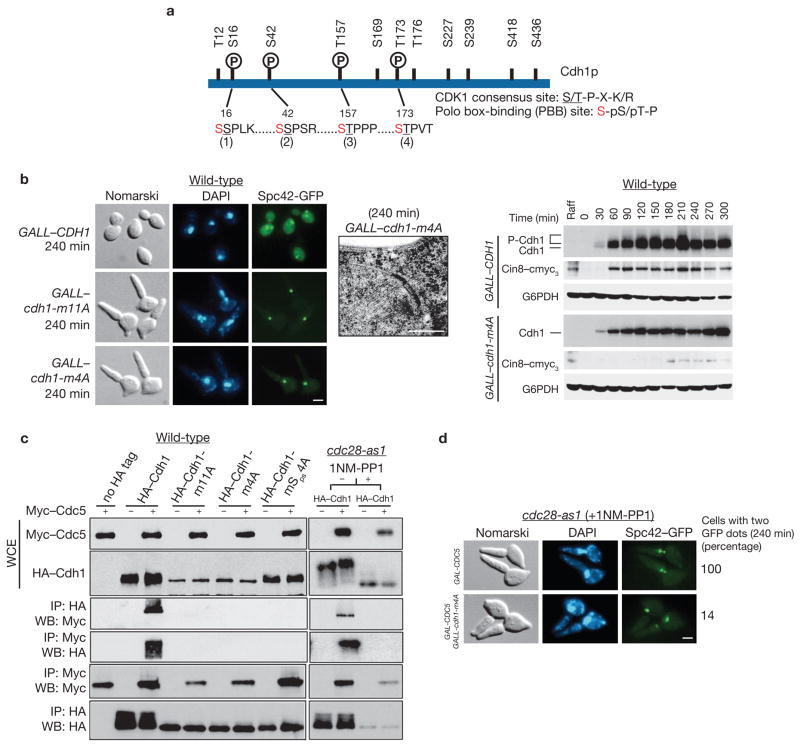
Cdh1 contains eleven Cdk1-phosphorylation sites (S/T X P K/R)13,14 (Fig. 3a). Wild-type cells expressing a mutant Cdh1 from the weak GAL10 promoter (GALL), in which all eleven Ser or Thr have been substituted by Ala (Cdh1–m11A), are not viable; these cells complete S phase but fail to separate SPBs (Fig. 3b). The sequence surrounding four of the eleven sites (Ser 16, Ser 42, Thr 157 and Thr 173), if phosphorylated, resembles the PBB sites (S-pS/pT-P, where ‘p’ denotes the phosphorylated form; Fig. 3a)15. To test whether these sites are important for SPB separation, we expressed in wild-type cells a GALL promoter-driven mutant form of Cdh1 (GALL–cdh1–m4A), in which Cdc28-phosphorylatable Ser and Thr residues (designated 1, 2, 3, 4 in Fig. 3a) were substituted by Ala. Cells expressing GALL–CDH1 separated their SPBs within 120 min when released synchronously from G1 arrest, accumulated Cin8 and showed Cdh1 hyperphosphorylation; however, GALL–cdh1–m4A cells were unable to separate SPBs, showed very weak hyperphosphorylation of Cdh1–m4A and failed to accumulate Cin8 (Fig. 3b). These results suggest that PBB sites are crucial for Cdh1 inactivation and hence for SPB separation (Supplementary Information, Fig. S1)
Figure 3.
Phosphorylation of Cdh1 by Cdc5 requires priming by Cdc28. (a) Schematic representation of the four Cdk phosphorylation sites constituting the PBB sites in Cdh1. Residues potentially phosphorylated by Cdc28 are underlined and designated 1, 2, 3 and 4. Ser residues (indicated in red) immediately preceding Cdc28 phosphorylation site are highly conserved in PBB sites. (b) G1-synchronized wild-type cells endogenously tagged with CIN8–cmyc3 carrying either GALL–CDH1, GALL–cdh1–m11A or GALL–cdh1–m4A (US4622, US4623, US4624) in YEP+Raff were released into YEP+Raff+Gal for 300 min and collected for Spc42–GFP visualization, immunoblotting and electron microscopy. The scale bar represents 2 μm (immunofluorescence) and 0.3 μm (electron microscopy). (c) Wild-type cells harbouring GAL–cmyc3–CDC5 cells and carrying either GALL–HA3–CDH1, GALL–HA3–cdh1–m11A, GALL–HA3–CDH1–m4A or GALL–HA3–cdh1–mSps4A (US4625, US4626, US4627, US4628) were first synchronized in G1 by α-factor treatment in raffinose medium and then released into either glucose or galactose medium for 180 min. In parallel, G1-synchronized cdc28-as1 cells carrying GALL–HA3–CDH1 and GAL–cmyc3–CDC5 (US5353) were released into either glucose or galactose medium with or without 1NM-PP1 for 240 min. Immunoprecipitates were analysed by western blotting. (d) G1-synchronized cdc28-as1 GAL–cmyc3–CDC5 cells with or without GALL–HA3–cdh1–m4A (US5354, US5354) in YEP+Raff were released into YEP+Raff+Gal medium. Numbers indicate percentage of cells with two well-separated Spc42–GFP dots at 240 min. The scale bar represents 2 μm. Full-length blots for b and c are shown in Supplementary Information, Fig. S5–1.
Next, we asked whether Cdc5 physically associates with Cdh1 and whether PBB sites indeed mediate this association. The PBB site contains two Ser residues, both important for Cdc5-binding15; the second Ser is phosphorylated by Cdc28 but requires the first Ser for binding to Cdc5. Cells carrying GAL–cmyc3–CDC5 and expressing GALL-driven HA3–Cdh1, HA3–Cdh1–m11A, HA3–Cdh1–m4A (Cdk1-phopsphorylatable Ser and Thr in the second position in each PBB site substituted with Ala) or HA3–Cdh1–mSps4A (Cdh1 with the first Ser of each PBB site substituted with Ala) were grown to exponential phase and cell extracts were used for reciprocal co-immunoprecipitation. Wild-type Cdh1 associated with Cdc5, whereas none of the Cdh1 mutants did so (Fig. 3c). This indicates that the putative PBB sites mediate interaction between Cdc5 and Cdh1 and that phosphorylation of these sites by Cdc28 is essential for this interaction (Fig. 3c). The latter conclusion is also consistent with the observation that 1NM-PP1 treatment of cdc28-as1 cells completely abolished the interaction between Cdc5 and Cdh1 (Fig. 3c, right panel). The importance of these PBB sites in Cdh1 for SPB separation is highlighted by our observation that the ability of ectopically expressed Cdc5 to induce SPB separation in 1NM-PP1-treated cdc28-as1 cells was severely compromised by co-expression of Cdh1–m4A (Fig. 3d).
Phosphorylation of Cdh1 by Cdc5 polo kinase requires priming by Cdc28 (Cdk1)
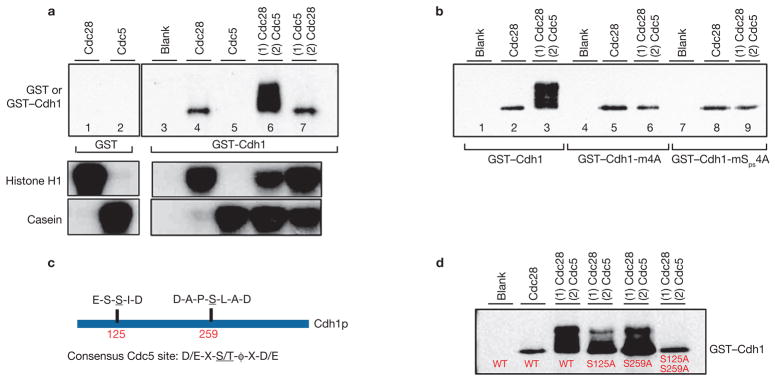
To investigate the relationship between Cdh1, Cdc28 and Cdc5 further, we asked whether Cdc5 is able to phosphorylate Cdh1 in vitro and whether phosphorylation of PBB sites by Cdc28 is a prerequisite for this. Cdc28–Clb2–HA3 and Cdc5–HA3 kinases were immunoprecipitated from nocodazole-treated cells and used in kinase reactions with bacterially expressed GST or GST–Cdh1 as substrates. Cdc28–Clb2 kinase, but not Cdc5 was clearly able to phosphorylate GST–Cdh1. Interestingly, when GST–Cdh1 was first phosphorylated by Cdc28–Clb2 and then subsequently exposed to Cdc5, significant hyperphosphorylation of GST–Cdh1 was seen (Fig. 4a). However, in a reciprocal experiment where GST–Cdh1 was first exposed to Cdc5 and subsequently used as a substrate for Cdc28–Clb, no hyperhosphorylation was observed; instead, phosphorylation corresponding to the GST–Cdh1 exposed to Cdc28–Clb2 alone was seen (Fig. 4a). Hyperphosphorylation by Cdc5 is dependent on PBB sites as GST–Cdh1–m4A and GST–Cdh1–mSps4A are not hyperphosphorylated by Cdc5 (Fig. 4b). These results suggest that Cdc28–Clb kinase phosphorylates Cdh1 at PBB sites and primes it for binding to Cdc5.
Figure 4.
Phosphorylation of Cdh1 by Cdc28 and Cdc5. (a) Cdc28–Clb2–HA3 and Cdc5–HA3 immunoprecipitated from nocodazole-treated wild-type (WT) cells (US1165, US3259) were used in kinase reactions with GST or GST–Cdh1 as substrates. For the two-step kinase reaction (lanes 6 and 7; see Methods), ‘1’ denotes the first kinase used and ‘2’ denotes the second kinase used after removal of the first kinase. To demonstrate substrate specificities, Cdc28–Clb2 and Cdc5 kinase activities were assayed using histone H1 and casein as substrates. (b) Phosphorylation of GST–Cdh1, GST–Cdh1–m4A and GST–Cdh1–mSps4A by immunoprecipitated Cdc28–Clb2–HA3 and Cdc5–HA3. Designations ‘1’ and ‘2’ in lanes 3, 6 and 9 represent sequential exposure to the two kinases, as described in a. (c) Schematic representation of putative Cdc5 phosphorylation sites in Cdh1. (d) The blot shows sequential phosphorylation of GST–Cdh1, GST–Cdh1–S125A, GST–Cdh1–S259A and GST–Cdh1–S125A S259A by Cdc28–Clb2–HA3 and Cdc5–HA3 kinases.
The Cdh1 sequence was examined to determine which residues in Cdh1 are phosphorylated by Cdc5. This revealed two short motifs resembling the consensus Cdc5 phosphorylation site D/E-X-S/T-Φ-X-D/E, where ‘Φ’ denotes a hydrophobic residue16 (Fig. 4c). To test whether these sites are indeed phosphorylated by Cdc5, bacterially expressed mutant versions of GST–Cdh1, in which Ser 125 and Ser 259 had been changed (singly or in combination) to Ala, were used as substrates in in vitro kinase assays using Cdc28–Clb2 and Cdc5 (Fig. 4a). GST–Cdh1 was hyperphosphorylated strongly and single mutants GST–Cdh1–S125A or GST–Cdh1–S259A phosphorylated to some extent, whereas the double-mutant GST–Cdh1–S125A S259A failed to be hyperphosphorylated, suggesting that both sites are phosphorylated by Cdc5 (Fig. 4d). However, Ser 125 and Ser 259 residues did not influence Cdh1 binding to Cdc5 as Cdh1 and Cdh1–S125A S259A bound equally well to Cdc5 (Supplementary Information, Fig. S2). Taken together, these results implicate Cdc5, together with Cdc28–Clb, in SPB separation due to its capacity to phosphorylate and thus inactivate Cdh1.
Cdc5 deficiency delays spindle assembly
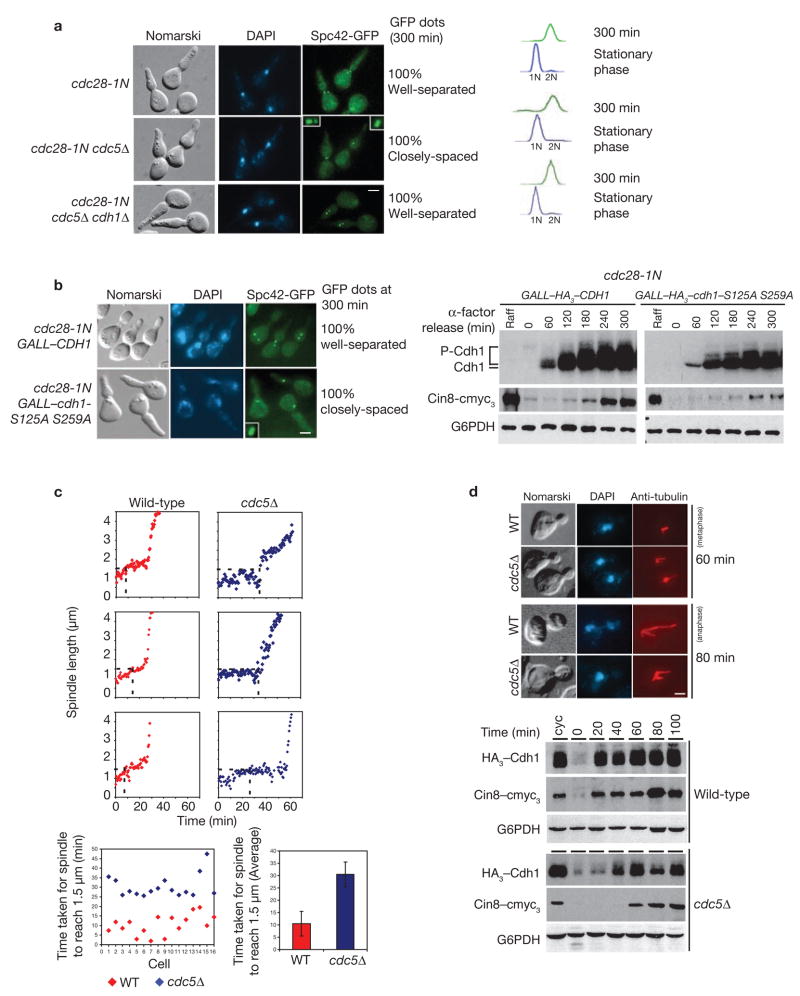
To determine whether Cdc5 function is essential for SPB separation, we compared cdc28-1N and cdc28-1N cdc5Δ double mutants (kept alive by GAL–CDC5) carrying SPC42–GFP for their ability to separate SPBs at the restrictive temperature. We used the cdc28-1N strain for this experiment because, as with 1NM-PP1-treated cdc28-as1, it is unable to initiate mitosis and arrests at the same stage of the cell cycle with 2N DNA content and an undivided nucleus; but, in contrast to cdc28-as1, it efficiently assembles a short spindle17. As expected, cdc28-1N cells separated SPBs and arrested uniformly in G2/M by 180 min after release from G1 arrest (Fig. 5a). However, in most of the cdc28-1N cdc5Δ cells, GFP signals revealed SPBs as a bilobed entities, indicating that they had separated but remained close to each other (Fig. 5a). Hence, in the absence of Cdc5, cdc28-1N cells are able to break the inter-SPB bridge but fail to move them apart to assemble a characteristic short spindle. Interestingly, cdc2-8-1N cdc5Δ cells can be readily cured of this defect by deletion of CDH1 (Fig. 5a, bottom panel), indicating that Cdc5 has an important role in efficient separation of SPBs, probably by inactivation of Cdh1.
Figure 5.
Absence of Cdc5 delays assembly of short spindle. (a) Stationary-phase cells of cdc28–1N, cdc28–1N cdc5Δ GAL–CDC5 and cdc28–1N cdc5Δ GAL–CDC5 cdh1Δ strains (US4362, US5259, US5364) were obtained by growth on YEP+Raff+Gal medium at 24 °C for 3 days and were subsequently transferred to YEP+Glu medium at 37 °C. Spc42–GFP signals and FACS in cells at 300 min are shown. (b) cdc28–1N carrying either GALL–CDH1 or GALL–cdh1–S125A S259A (US5369, US5372) were synchronized in G1 by α-factor treatment in Raffinose medium and then released into Raff+Gal medium at 37 °C. Samples were collected to analyse the pattern of Spc42–GFP fluorescence and for western blotting. Insets in a and b show a magnified view of Spc42–GFP dots. (c) Spindle length distribution in wild-type and cdc5Δ cells expressing Spc42–GFP (US3786, US5282) was determined by live-cell imaging (3 examples are shown from a total of 16 cells imaged documented in Supplementary Information, Fig. S3). The distance between the Spc42–GFP dots is plotted versus time. The vertical dotted lines represent the time at which the spindle first attains a stable length of 1.5 μm before a phase of rapid increase sets in. The bottom panels show the distribution of time taken to reach a spindle length of 1.5 μm in various cells. (d) Wild-type and cdc5Δ GAL–CDC5 cells carrying HA3–Cdh1 and Cin8–cmyc3 (US5322, US4970) were synchronized in G1 by α-factor treatment in YEP+Raff+Gal medium and then released into YEP+Glu medium. Western blots and tubulin immunofluorescence staining of samples collected at various times are shown. Scale bars represent 2 μm (a, b, d). The spindle length distribution is shown in Supplementary Information, Fig. S4a. Full-length blots for b and d are documented in Supplementary Information, Fig. S5–2.
To determine whether the inability of cdc28-1N cdc5Δ to efficiently separate SPBs is linked to the state of Cdc5-phosphorylation sites in Cdh1 (Ser 125 and Ser 259), CEN plasmids carrying either GALL–CDH1 or GALL–cdh1–S125A S259A were introduced into cdc28-1N cells. Cells were synchronized in G1 by a-factor treatment and then released into the galactose medium at 37 °C. As expected, expression of Cdh1 in cdc28-1N cells (with active Cdc5) did not interfere with SPB separation; however, expression of Cdh1–S125A S259A (not phosphorylated by Cdc5) compromised the efficiency of separation (Fig. 5b). This is consistent with the lower level of Cdh1–S125A S259A phosphorylation and diminished level of Cin8 in cdc28-1N cells carrying GALL–cdh1–S125A S259A, compared with those carrying GALL–CDH1 (Fig. 5b), even though both Cdh1 and Cdh1–S125A S259A bind Cdc5 equally well (Supplementary Information, Fig. S2). Hence, a lower level of Cin8 in the absence of phosphorylation by Cdc5 may be sufficient to break the inter-SPB link, but Cdc5 function is necessary to boost its abundance for proper segregation of SPBs and assembly of a characteristic spindle in cdc28-1N cells.
Although the cdc28-1N mutant is proficient in SPB separation, it is a defective kinase and may exacerbate the effect of Cdc5 deficiency on SPB separation. Therefore, we analysed SPB separation and short-spindle formation (approximately 1.5 μm) by live-cell imaging in wild-type and cdc5Δ (kept alive by GAL–CDC5) cells harbouring Spc42–GFP. We took 1.5 μm as our ‘target length’ because it is generally considered to be the characteristic length of a short spindle18, after which the second phase of rapid elongation begins (anaphase). Cells were subjected to time-lapse microscopy in glucose medium at 24 °C. Compared with wild-type, cdc5Δ cells were consistently delayed in attaining a spindle length of 1.5 μm (Fig. 5c; Supplementary Information, Figs S3, S4a); however, they eventually formed anaphase spindles as seen in wild-type cells (‘0 min’ in these experiments is the time-point preceding that at which SPBs can be seen as two distinct entities for the first time). On average, the wild-type strain took approximately 10 min to assemble a short spindle of 1.5 μm length, whereas, cdc5Δ cells attained this length at approximately 25–30 min. The delay in short-spindle formation in cdc5Δ cells was consistent with their sluggishness in phosphorylating Cdh1 (Fig. 5d). This delay seems to be caused by delayed accumulation of Cin8 (Fig. 5d) as it can be alleviated by expression of proteolytic-resistant Cin8 (Supplementary Information, Fig. S4b).
Cdc5 becomes essential for SPB separation in absence of Acm1
Despite the finding that Cdc5 induces SPB separation by inactivating Cdh1 (Figs 1–3), its deficiency only delays spindle assembly but does not entirely prevent it. As Cdc5-deficient cells can accumulate Cin8 to some extent, it is possible that Cdh1 is partially inhibited by some other mechanism. Recently, it was shown that Cdh1 forms a complex with three other proteins, Acm1, Bmh1 and Bmh2, during S phase19,20, and that the Acm1–Bmh1–Bmh2 complex acts as a negative regulator of APCCdh1. Acm1 is a non-essential protein whose deficiency does not significantly affect cell growth, SPB separation or spindle formation (Fig. 6a, left panel). To determine whether Acm1 is the negative regulator that allows low-level accumulation of Cin8 and therefore sluggish spindle assembly in cdc5Δ cells, we analysed acm1Δ, cdc5Δ and cdc5Δ acm1Δ cells (kept alive by GAL–CDC5) expressing Spc42–GFP, released from G1 arrest into glucose medium at 24 °C. As expected, acm1Δ cells separated SPB efficiently, whereas cdc5Δ cells, as observed before, were sluggish in doing so (Fig. 6a). However, cdc5Δ acm1Δ cells contained only a single Spc42–GFP spot, indicating their failure to separate SPBs. Analysis by electron microscopy showed that these cells contain duplicated SPBs still attached by a bridge (Fig. 6a). Consistent with this, Cin8 levels were very low in cdc5Δ acm1Δ cells, compared with cdc5Δ cells (Fig. 6a, right panel). It should be noted that although both cdc5Δ and acm1Δ strains show normal buds, the buds in cdc5Δ acm1Δ mutant are hyperpolarized. This may indicate that the double mutant is deficient in Cdc28–Clb kinase activity, perhaps due to their failure to inactivate Cdh1, which can also target mitotic Clbs for degradation. Indeed, deletion of Cdh1 in cdc5Δ acm1Δ cells not only restored their ability to assemble a spindle but also suppressed hyperpolarization of the buds (Fig. 6b). These results suggest that Acm1, together with Cdc28–Clb and Cdc5 polo kinase, participates in the inactivation of Cdh1 during S phase and that in the absence of Acm1, Cdc5 becomes essential for SPB separation.
Figure 6.
Functional redundancy between Cdc5 and APCCdh1-inhibitor Acm1 in SPB separation and involvement of Cdc5 in nuclear export of Cdh1. (a) acm1Δ, cdc5Δ and cdc5Δ acm1Δ cells carrying GAL–CDC5 (US5300, US5301, US5302) with endogenously tagged CIN8–cmyc3 were synchronized in G1 in YEP+Raff+Gal and then released into YEP+Glu medium at 24 °C. Samples were collected for Spc42–GFP visualization, electron microscopy and western blotting. The scale bar represents 2 μm. Numbers indicate percentage of cells with two Spc42–GFP dots at 80 min for acm1Δ and cdc5Δ cells, and at 160 min for cdc5Δ acm1Δ cells (as doubling time in these cells is twice as long). EM micrograph shows the state of SPBs at 240 min. Scale bar for EM image, 0.2 μm. Insets show a magnified view of Spc45–GFP dots. (b) Stationary phase cells of cdc5Δ acm1Δ and cdc5Δ acm1Δ cdh1Δ strains harbouring GAL–CDC5 (US5302, US5376) were obtained by growth on YEP+Raff+Gal medium at 24 °C for three days and were subsequently inoculated into YEP+Glu at 24 °C. Cells at 240 min are shown. (c) G1-synchronized cdc28-as1 GAL–CDC5 (US5373) and msn5Δ cdc28-as1 GAL–CDC5 (US5374) cells were released into YEP+Raff+Gal containing 1NM-PP1 for 300 min. Samples were collected for Cdh1–GFP visualization and immunofluorescence staining. Scale bars represent 2 μm (a, b, c). Numbers represent percentage of cells with nuclear Cdh1–GFP and spindles at 300 min. Full-length blots for section a are presented in Supplementary Information, Fig. S5–2.
It is known that Cdh1 is localized to the nucleus during G1, but its Msn5-dependent export to the cytoplasm during S phase is important for the stability of its nuclear substrates21. Movement of Cdh1 out of the nucleus is dependent on its phosphorylation by Cdc28–Clb as Cdh1–m11A expressed in wild-type cells, or Cdh1 expressed in 1NM-PP1-treated cdc28-as1 cells, remains exclusively nuclear and in both cases cells fail to assemble a spindle (data not shown). As Cdh1 is also phosphorylated by Cdc5, we asked whether nuclear export is an indispensible aspect of Cdc5-mediated inactivation of Cdh1 or whether Cdc5 is able to inactivate Cdh1 even if it is retained in the nucleus. To test this, G1-synchronized cdc28-as1 and cdc28–as1 msn5Δ cells expressing GAL promoter-driven CDC5 were released into galactose medium containing 1NM-PP1, and Cdh1 localization and the state of the spindle was determined. Ectopic expression of Cdc5 in 1NM-PP1-treated cdc28-as1 cells caused Cdh1 to localize in the cytoplasm (Fig. 6c, upper left panel), suggesting that Cdc5 can induce Cdh1 export despite a marked deficiency in Cdc28–Clb activity. As Cdh1 export is dependent on Msn5, GFP–Cdh1 remained in the nucleus in msn5Δ cdc28-as1 cells expressing GAL–CDC5 (Fig. 6c, lower left panel). Despite the presence of Cdh1 in the nucleus, these cells (100%) were able to assemble a spindle (Fig. 6c, lower right panel). From these results we conclude that although Cdc5 participates in the nuclear export of Cdh1, it is capable of inactivating Cdh1 even if Cdh1 is retained in the nucleus.
Cdc5 stability is dependent on Cdc28–Clb activity
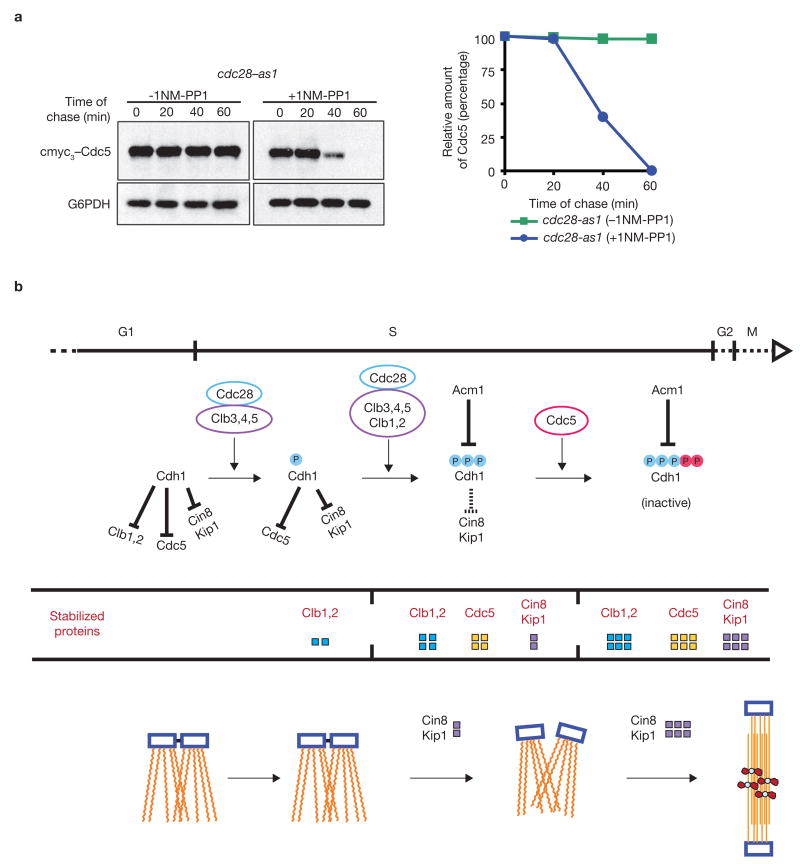
It is clear from our observations that Cdc5 has an important role in the inactivation of Cdh1. However, if Cdc5 itself is targeted for destruction by Cdh1 (ref. 22), then what prevents proteolytic degradation of Cdc5? The finding that overexpression of Cdc5 in 1-NM-PP1 treated cdc28-as1 mutants leads to Cin8 accumulation (Fig. 1) may indicate that Cdc5 is normally unstable in this mutant. Indeed, Cdc5 levels are very low in cdc28-as1 cells (data not shown). To determine whether Cdc28 mitotic kinase is responsible for preventing Cdc5 degradation, we monitored the fate of a Cdc5 pulse (expressed from GAL–CDC5 construct) in cdc28-as1 mutants in the presence or absence of 1NM-PP1. The Cdc5 pulse was stable in untreated cdc28-as1 cells, whereas treatment with 1NM-PP1 caused a marked decline in the level of Cdc5 (Fig. 7a), suggesting that its stability is dependent on Cdc28–Clb kinase. Hence the Cdc28–Clb kinase complex is responsible for both stabilizing Cdc5 and catalysing its binding to Cdh1 by priming the PBB sites.
Figure 7.
Cdc5 is unstable in 1NM-PP1 treated cdc28-as1 cells. (a) cdc28-as1 cells (US3723) were synchronized in YEP+Raff medium with α-factor treatment and galactose was added to induce Cdc5 for 30 min. Subsequently, cells were released into cycloheximide-containing YEP+Glu medium, with or without 1NM-PP1, and the fate of Cdc5 pulse was determined by western blotting. The abundance of Cdc5 was quantified using a densitometer. (b) A scheme for the regulation of SPB separation involving Cdc28, Cdc5, Cdh1, Acm1 and microtubule binding proteins Cin8 and Kip1. Low level accumulation of Cin8 and Kip1 is sufficient to sever the inter-SPB bridge. Further separation of SPBs for the assembly of a characteristic short spindle requires greater accumulation of these kinesin motors, mediated by Acm1 and Cdc5 polo kinase. Full-length blots are presented in Supplementary Information, Fig. S5–2.
DISCUSSION
Cells entering G1 after mitosis inherit an active APCCdh1 complex from the previous cycle, which targets a number of proteins, including Clb1, Clb2, Cin8, Kip1 and Cdc5, for proteolytic destruction (ref. 23). For successful entry into mitosis in the subsequent division cycle, cells need to re-accumulate these proteins. Therefore, cells must ensure that Cdh1 is inactivated as they progress towards M phase. It has been shown previously that the Cdc28–Clb3, –Clb4 and –Clb5 are collectively responsible for accumulation of the mitotic Clb1 and Clb2 by inactivating Cdh1 (ref. 24). However, cdc28-as1 cells treated with 1NM-PP1 show a substantial amount of Clb2, but continue to degrade Cin8 and Kip1 in a Cdh1-dependent manner, and are unable to separate the SPBs. This suggests that Cdh1 is not fully inactivated in these cells. On the basis of our results, we propose a regulatory scheme for Cdh1 inactivation leading to SPB separation during a normal cell cycle (Fig. 7b): inactivation of Cdh1 (fully active during G1) is initiated in early S phase by Cdc28–Clb3, –Clb4 and –Clb5, which leads to accumulation of Clb1, Clb2 and Cdc5 polo kinase. Collective phosphorylation of Cdh1 by Cdc28–Clb kinase complexes then creates PBB sites which allow binding of Cdc5 to Cdh1. Cdc5 in turn phosphorylates Cdh1 at Ser 125 and Ser 258, rendering it completely inactive and allowing accumulation of microtubule-associated proteins, such as Cin8, and subsequently, separation of SPBs. Thus Cdc28/Cdk1 acts as a priming kinase for the recruitment of Cdc5 polo kinase to Cdh1.
This is consistent with the findings of a previous study15, in which pSer/pThr was originally identified as a PBB site using a proteomic approach, and suggested a mechanism by which polo kinase is recruited to specific sites in response to Cdk1 phosphorylation. Such a synergistic scheme involving Cdk and polo kinase has a number of parallels in other systems25. It should be noted that, although our results strongly suggest a role for Cdc5 polo kinase in Cdh1 inactivation, resulting in SPB separation, the possibility that Cdc5 also contributes to SPB separation by phosphorylating specific SPB-associated protein(s) necessary for the breaking of inter-SPB bridge cannot be ruled out. Such a model would be consistent with the observation that Cdc5 is localized to the SPBs during late S phase and early mitosis.
Inactivation of polo kinase is known to result in monopolar spindles in other systems, suggesting its involvement in centrosome separation26, but its exact role has not been elucidated. Cdc5 polo kinase has never been implicated in SPB separation in S. cerevisiae prior to this study because cdc5 mutants or cdc5Δ cells do assemble a spindle, albeit inefficiently, and go on to arrest in telophase22. Our results suggest that Cdc5 is non-essential for SPB separation in wild-type cells because Acm1 (Fig. 6) partially inhibits Cdh1 and promotes sluggish accumulation of Cin8. The failure of cdc5Δ acm1Δ double mutants to separate SPBs is consistent with this notion. Hence Cdc28–Clb, Cdc5 polo kinase and Acm1–Bmh1–Bmh2 cooperatively inactivate Cdh1 to allow accumulation of Cin8 and Kip1, resulting in timely separation of SPBs. It is unclear why cells would use three concerted ways to inactivate Cdh1; perhaps this elaborate mechanism has some relevance to substrate selection or the timing of their proteolytic destruction. Nevertheless, our findings provide a new model for the inactivation of important mitotic regulators by the synergistic action of Cdk1 and polo kinase. It remains to be seen whether the role of polo kinase in centrosome separation in mammalian cells also involves modulation of the proteolytic machinery.
METHODS
Yeast strains and plasmids
Strains used in this study are congenic to the wild-type strain W303. Standard molecular genetic techniques such as gene transplacement, gene disruption, tetrad dissection, PCR-based site-directed mutagenesis and tagging of endogenous genes were used to construct plasmids and strains with various genotypes. Southern blot analysis or PCR-based genotyping was used to confirm gene disruptions and transplacements.
Growth conditions and cell synchronization
Cells were grown in yeast extract peptone (YEP) supplemented with 2% glucose or 4% raffinose and 2% galactose. Methionine (final concentration 24 μg ml−1) was added in experiments requiring repression of the MET3 promoter. cdc28-as1 cells were treated with the Cdc28 kinase inhibitor 1NM-PP1 (500 nM, Cellular Genomics) for arrest with no spindle. Cells treated with the proteasome inhibitor MG132 carried an erg6 disruption to facilitate permeability of the inhibitor. For synchronization at G1, exponential-phase cells were grown at 24 °C in medium containing α-factor for bar1Δ cells (1 μg μl−1) or for BAR1 cells (5 μg μl−1) for 2.5–3 h. Cells were then filtered, washed and resuspended in fresh medium.
Immunoblotting and immunoprecipitation
Preparation of whole-cell extracts and protein precipitation by TCA for immunoblotting, and preparation of cell extracts for immunoprecipitations were carried out as described previously27. Immunodetection of HA or c-myc epitope-tagged fusion proteins was carried out using anti-mouse HA (1:1000) and anti-rabbit-Myc (1:1000) antibodies (Santa Cruz Biotechnology). Anti-HA antibody or anti-Myc agarose-conjugated beads (Santa Cruz Biotechnology) were used for co-immunoprecipitation. The enhanced chemiluminescence kit from Santa Cruz Biotechnology was used for all western blot analyses according to manufacturer’s instructions.
Detection of ubiquitin conjugates in vivo
G1-synchronized cells were released into medium containing 50 μM MG132. Cells were collected and lysed in lysis buffer containing 10 mM N-ethylmaleimide to inhibit deubiquitination. Pre-cleared protein extract (2 mg) was immunoprecipitated with mouse anti-HA agarose-conjugated beads (Santa Cruz Biotechnology) and proteins were separated on a 6% gel. After immunoblotting, the membrane was autoclaved at 121 °C for 10 min to expose buried polyubiquitin epitopes. Ubiquitin conjugates were detected by rabbit anti-ubiquitin antibody (1: 100; Sigma).
Kinase assays
Protein extracts were prepared as described above. Cdc28–Clb2–HA3 and Cdc5–HA3 were immunoprecipitated from 150 μg extracts with anti-HA agarose-conjugated beads (Santa Cruz) at 4 °C. The beads were then washed three times with RIPA buffer (1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 50 mM Tris-Hcl pH 7.2) followed by two washes with 25 mM MOPS. Kinase assay buffer (4 μl) containing inhibitors (10 mM Hepes pH 7.4, 750 mM KCl, 50 mM MgCl2, 5 mM EGTA, 5 mM DTT, 1 mM PMSF, 20 μg ml−1 leupeptin, 40 μg ml−1 aprotinin, complete protease inhibitor cocktail, 15 mM PNPP, 50 mM NaF, 1 mM sodium orthovanadate and 50 mM β-glycerophosphate) was added to the beads. Purified GST-fusion protein (substrate, 7.5–20 μg), 1 mM ATP (4 μl), 250 mM MOPS (1 μl, pH 7.2) and 1 μCi μl−1 γ-32ATP (1.1 μl) were then added give a final volume of 20 μl. Histone H1 (2 μg, Roche) and dephosphorylated casein (20 μg, Sigma) were used as substrate controls in assays of kinase activities of Cdc28–Clb2 and Cdc5, respectively. Kinase reactions were incubated at room temperature for 30 min and subsequently terminated by addition of SDS-sample buffer and boiling for 5 min. GST–fusion proteins were resolved on 8% gels, whereas histone H1 and casein were resolved on 12% gels for autoradiography.
For the two-step kinase assay, after the initial 30-min kinase reaction with the ‘first’ kinase bound to agarose-conjugated beads, the beads were pelleted by centrifugation at 550g for 4 min. The supernatant, which consisted of the kinase assay cocktail with the GST–fusion protein was immediately transferred to anti-HA agarose-conjugated beads bound to the ‘second’ kinase. This reaction was incubated for a further 15 min before SDS-sample buffer was added.
Flow cytometry, immunofluorescence and time-lapse imaging
Analysis of DNA content by flow cytometry, visualization of spindles by tubulin immunofluorescence and visualization of fluorescence-tagged cells were performed as described previously27. For time-lapse imaging, cells were placed between a coverslip and a thin layer of 2% agarose containing 2% glucose in low immunofluorescence yeast nitrogen base with complete drop-out medium supplemented with adenine. Microscopy was performed in an enclosed chamber maintained at 31.5 °C with a Zeiss Axio inverted microscope equipped with a Plan-Apochromat 100X (1.4 numerical aperture) objective and a Yokogawa CSU22 spinning disk confocal system with dual line argon krypton ion laser (wavelength 488 nm/568 nm). Stacks of images were taken at 30-s intervals (7 planes spaced 0.5 μm apart) with a Cascade:512B camera (Roper Scientific), acquired and analysed with Metamorph software (Universal Imaging). Spindle lengths were measured as pole-to-pole distances using the caliper function in Metamorph software. It should be noted that some mutants such as cdc28–1N cdc5Δ lost viability rapidly with the repeated exposures to light required for time-lapse microscopy, and therefore could not be subjected to live-cell imaging for detailed analysis.
Electron microscopy
Cells for electron microscopy were prepared for thin sectioning by high-pressure freezing and freeze substitution as described previously28. Serial thin sections were viewed on a Philips CM10 electron microscope and images were captured with Gatan digital camera and viewed with Digital Micrograph software.
Supplementary Material
Acknowledgments
We are grateful to Janet Meehl and Michele Jones for their invaluable help with some of the experiments. We wish to thank David Morgan, Wolfgang Zachariae, Simonetta Piatti, Matthias Peter, Christopher Hardy and Kyung Lee for plasmids and strains, Mark Hall for valuable advice and Chee Peng Ng for help with electron microscopy. The work in MW laboratory was supported by NIH grant GM51312. U.S. is an adjunct faculty member of the Department of Pharmacology, National University of Singapore. The U.S. lab is supported by the Biomedical Research Council of A*STAR (Agency for Science, Technology and Research), Singapore.
Footnotes
Note: Supplementary Information is available on the Nature Cell Biology website.
AUTHOR CONTRIBUTIONS
K.C. and L.H.H. performed all experiments; T.H.G. and M.W. carried out the electron microscopy analysis; U.S. and K.C. planned the project and analysed the data.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Zimmerman W, Sparks CA, Doxsey SJ. Amorphous no longer: the centrosome comes into focus. Curr Opin Cell Biol. 1999;11:122–128. doi: 10.1016/s0955-0674(99)80015-5. [DOI] [PubMed] [Google Scholar]
- 2.Nigg EA. Origins and consequences of centrosome aberrations in human cancers. Int J Cancer. 2006;119:2717–2723. doi: 10.1002/ijc.22245. [DOI] [PubMed] [Google Scholar]
- 3.Sterns T. Centrosome duplication: a centriolar pas de deux. Cell. 2001;105:417–420. doi: 10.1016/s0092-8674(01)00366-x. [DOI] [PubMed] [Google Scholar]
- 4.Jaspersen SL, Winey M. The budding yeast spindle pole body: structure, duplication and function. Annu Rev Cell Dev Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- 5.Blangy A, et al. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83:1159–1169. doi: 10.1016/0092-8674(95)90142-6. [DOI] [PubMed] [Google Scholar]
- 6.Lane HA, Nigg EA. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of centrosomes. J Cell Biol. 1996;135:1701–1713. doi: 10.1083/jcb.135.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoyt MA, He L, Loo KK, Saunders WS. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J Cell Biol. 1992;118:109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim HH, Goh PY, Surana U. Spindle pole body separation in Saccharomyces cerevisiae requires dephosphorylation of the tyrosine 19 residue of Cdc28. Mol Cell Biol. 1996;16:6385–6397. doi: 10.1128/mcb.16.11.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crasta K, Huang P, Morgan G, Winey M, Surana U. Cdk1 regulates centrosome separation by restraining proteolysis of microtubule-associated proteins. EMBO J. 2006;25:2551–2563. doi: 10.1038/sj.emboj.7601136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitch I, et al. Characterization of four B-type cyclin genes of the budding yeast Saccharomyces cerevisiae. Mol Biol Cell. 1992;3:805–818. doi: 10.1091/mbc.3.7.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bishop AC, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 12.Bartholomew CR, et al. Cdc5 interacts with the Wee1 kinase in budding yeast. Mol Cell Biol. 2001;21:4949–4959. doi: 10.1128/MCB.21.15.4949-4959.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zachariae W, Schwab M, Nasmyth K, Seufert W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 1998;282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- 14.Hall MC, Warren EN, Borchers CH. Multi-kinase phosphorylation of the APC/C activator Cdh1 revealed by mass spectrometry. Cell Cycle. 2004;3:1278–1284. doi: 10.4161/cc.3.10.1153. [DOI] [PubMed] [Google Scholar]
- 15.Elia AEH, Cantley LC, Yaffe MB. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- 16.Nakajima H, Toyoshima-Morimoto F, Taniguchi E, Nishida E. Identification of a consensus motif for Plk (polo-like kinase) phosphorylation reveals Myt1 as a Plk1 substrate. J Biol Chem. 2003;278:25277–25280. doi: 10.1074/jbc.C300126200. [DOI] [PubMed] [Google Scholar]
- 17.Surana U, et al. The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell. 1991;65:145–161. doi: 10.1016/0092-8674(91)90416-v. [DOI] [PubMed] [Google Scholar]
- 18.Yeh E, et al. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Sacchromyces cerevisiae. J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez JS, Jeong D-E Choi E, Billings BM, Hall MC. Acm1 is a negative regulator of the Cdh1-dependent anaphase-promoting complex/cyclosome in budding yeast. Mol Cell Biol. 2006;26:9162–9176. doi: 10.1128/MCB.00603-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dial JM, Petrotchenko EV, Borchers CH. Inhibition of APCCdh1 activity by Cdh1/Acm1/Bmh1 ternary complex formation. J Biol Chem. 2007;282:5237–5248. doi: 10.1074/jbc.M606589200. [DOI] [PubMed] [Google Scholar]
- 21.Jaquenoud M, van Drogen F, Peter M. Cell cycle-dependent nuclear export of Cdh1p may contribute to the inactivation of APC/CCdh1. EMBO J. 2002;21:6515–6526. doi: 10.1093/emboj/cdf634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charles JF, et al. The Polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr Biol. 1998;8:497–507. doi: 10.1016/s0960-9822(98)70201-5. [DOI] [PubMed] [Google Scholar]
- 23.Castro A, Bernis C, Vigneron S, Labbe JC, Lorca T. The anaphase-promoting complex: a key factor in the regulation of cell cycle. Oncogene. 2005;24:314–325. doi: 10.1038/sj.onc.1207973. [DOI] [PubMed] [Google Scholar]
- 24.Yeong FM, Lim HH, Wang Y, Surana U. Early expressed Clb proteins allow accumulation of mitotic cyclin by inactivating proteolytic machinery during S phase. Mol Cell Biol. 2001;21:5071–5081. doi: 10.1128/MCB.21.15.5071-5081.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Vugt MA, Medema RH. Getting in and out of mitosis with Polo-like kinase-1. Oncogene. 2005;24:2844–2859. doi: 10.1038/sj.onc.1208617. [DOI] [PubMed] [Google Scholar]
- 26.Dai W, Wang Q, Traganos F. Polo-like kinases and centrosome regulation. Oncogene. 2002;21:6195–6200. doi: 10.1038/sj.onc.1205710. [DOI] [PubMed] [Google Scholar]
- 27.Yeong FM, Lim HH, Padmashree CG, Surana U. Exit from mitosis in budding yeast: biphasic inactivation of the Cdk1–cyclin B2 mitotic kinase and the role of Cdc20. Mol Cell. 2000;5:501–511. doi: 10.1016/s1097-2765(00)80444-x. [DOI] [PubMed] [Google Scholar]
- 28.Winey M, et al. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.