Abstract
Wnt signaling pathways are regulated both at the intracellular and extracellular levels. During embryogenesis, the in vivo effects of the secreted frizzled related protein (Sfrp) family of Wnt inhibitors are poorly understood. Here, we show that inactivation of Sfrp2 results in subtle limb defects in mice with mesomelic shortening and consistent shortening of all autopodal elements that is clinically manifested as brachydactyly. In addition, there is soft tissue syndactyly of the hindlimb. The brachydactyly is caused by decreased chondrocyte proliferation and delayed differentiation in distal limb chondrogenic elements. These data suggest that Sfrp2 can regulate both chondrogenesis and regression of interdigital mesenchyme in distal limb. Sfrp2 can also repress canonical Wnt signaling by Wnt1, Wnt9a, and Wnt4 in vitro. Sfrp2-/- and TOPGAL/Sfrp2-/- mice have a mild increase in beta-catenin and beta-galactosidase staining, respectively, in some phalangeal elements. This however does not exclude a potential concurrent effect on non-canonical Wnt signaling in the growth plate. In combination with what is known about BMP and Wnt signaling in human brachydactylies, our data establish a critical role for Sfrp2 in proper distal limb formation and suggest SFPR2 could be a novel candidate gene for human brachy-syndactyly defects.
Keywords: secreted-frizzled, Wnt, Sfrp2, brachydactyly, syndactyly
Introduction
The Wnt signaling pathway is extremely conserved throughout the animal kingdom. In vertebrates, with as many as nineteen distinct Wnt genes, ten Frizzled and a few additional receptors and co-receptors and multiple intracellular signal transduction pathways, the complexity of this signaling system has continued to emerge (www.stanford.edu/∼rnusse/wntwindow.html). In general, a number of Wnt proteins, usually referred to as canonical Wnts, including Wnt1, Wnt3a and Wnt8, preferentially transduce their signal by stabilizing beta-catenin inside the cell to regulate transcription by binding to members of the Tcf/Lef family of transcription factors; other Wnts, like Wnt4, Wnt5a and Wnt11, at least in lower vertebrates, seem to preferentially act through the Wnt/Ca2+ pathway or the planar-cell-polarity pathway and are referred to as non-canonical Wnts. Increasing evidence now suggests that Wnts ability to activate different signaling pathways is not dependent on the specific Wnt molecules but rather upon the kind of receptors on the cell surface they interact with (for review see (He, 2003; Mikels and Nusse, 2006b; Veeman et al., 2003)).
An additional layer of complexity is represented by a large group of secreted proteins whose function appears to inhibit and modulate Wnt binding to their receptors, Frizzled's (for review see (Kawano and Kypta, 2003)). This is achieved through at least two different mechanisms thus identifying two distinct classes of extracellular Wnt antagonists. Members of the Dickkopf (Dkk) family exert their inhibitory action via binding to the LDL-related proteins of Wnt co-receptors (Lrp5/Lrp6) (He et al., 2004), and are considered selective inhibitors of the beta-catenin canonical pathway. The second class of antagonistic molecules, i.e., members of the secreted frizzled-related proteins (Sfrps), Wnt inhibitory factor 1(Wif-1), and Cerberus, acts as extracellular decoy receptors by sequestering Wnts from binding to Frizzled receptors. Hence, this second group of molecules can interfere with both canonical and non-canonical Wnt signaling pathways and may be considered a more general class of inhibitors.
The family of secreted frizzled-related proteins contains five genes, Sfrp1 to Sfrp5, and three additional members, called Sizzled, Sizzled2, and Crescent. They share a cysteine-rich domain (CRD) that is homologous to the one found in the Frizzled receptor, and thus behave as secreted Wnt receptors. Initially, the Sfrps were also named secreted apoptosis related proteins (Sarps) by a group of investigators that described their apoptotic activity in vitro (Melkonyan et al., 1997). Following this initial description, a host of studies have implicated Sarps/Sfrps in the regulation of apoptosis of many tissues (Drake et al., 2003; He et al., 2005; Jones et al., 2000; Ko et al., 2002; Lee et al., 2006b; Lee et al., 2004b) and, as a consequence, further linked Wnt signaling to tumorigenic processes (He et al., 2005; Lee et al., 2004a; Ugolini et al., 2001; Zhou et al., 1998). However, their effect on apoptotic processes and cancer formation is more prominent in the adult organism. Accordingly, Sfrp1 null mice develop normally but, as adults, exhibit increased trabecular bone mineral density and volume due to inhibition of osteoblasts and osteocyte apoptosis (Bodine et al., 2005; Bodine et al., 2004). The mechanism by which Sfrps regulate cell death is still unclear.
During embryogenesis, the developmental function of the Sfrp's is less well understood. Their pattern of expression is restricted and has been detected in several tissues and organs during mouse and chick development (Hoang et al., 1998; Ladher et al., 2000; Leimeister et al., 1998). In several cases, it complemented that of Wnt signaling molecules and while this supported the hypothesis of their antagonistic role, it also raised the question of whether they might contribute to the morphogenetic gradient of Wnt growth factors (Kim et al., 2001; Lee et al., 2000). Others, instead, have suggested that, at low concentration, Sfrps might potentiate Wnt signaling rather than antagonizing it (Uren et al., 2000).
Skeletal development, especially during limb bud formation, is an important process where Wnts have been shown to participate in multiple essential roles, including mesenchymal cell condensation, synovial joint induction, and chondrocyte proliferation and differentiation (for review see (Church and Francis-West, 2002; Yang, 2003)). Currently, little is known about the contribution of the Sfrps during these processes. In particular, the function of one family member, Sfrp2, during embryogenesis and skeletal development has been elusive. Recent studies have shown that while homozygous Sfrp2 or Sfrp1 null mutant mice do not have embryonic defects, and hence, are functionally redundant, double Sfrp2/Sfrp1 mutant mice have defects in axis elongation and somite segmentation (Satoh et al., 2006).
In this study, we demonstrate that although Sfrp2 null mice are undistinguishable from their wild-type littermates at birth and exhibit normal patterning of mesenchymal condensations with normal joint formation, they show mild mesomelic shortening and brachydactyly due to decreased chondrocyte proliferation and delayed differentiation in distal limb skeletal elements, and hindlimb soft-tissue syndactyly due to lack of regression of interdigital mesenchyme.
Materials and Methods
RNA in situ hybridization
Mouse embryos and newborn pups were collected at the specified times. Limb buds were dissected, washed with cold PBS and fixed in 4% paraformaldehyde/PBS at 4° C for 12 hours. Paraffin embedding, sectioning and in situ hybridization were carried out as previously described (Morello et al., 2001). Anterior/posterior and/or dorsal/ventral, 5μm thick limb sections were used for hybridizations. Antisense and sense riboprobes were synthesized with T7, T3 or SP6 RNA polymerase in the presence of [α-35S]UTP (1,000 Ci/mmol; NEN). The Sfrp2-specific probe was generated from a mouse EST clone (IMAGE clone ID 536389; C. Leimeister). The following probes were described previously: type X Collagen (Elima et al., 1993) kindly provided by Dr. V. Lefebvre (Cleveland Clinic Foundation, Cleveland, OH), Indian Hedgehog (Bitgood and McMahon, 1995) and osteocalcin (Desbois et al., 1994) kindly provided by Dr. G. Karsenty (Baylor College of Medicine, Houston, TX). The mouse Sox9 probe corresponds to nucleotides 1129-1382 of GenBank accession No. NM_011448 (253bp NarI fragment). The mouse Gdf5 cDNA probe corresponds to nucleotides 1735-2266 of GenBank accession No. U08337 and was generated by RT-PCR amplification of NIH-3T3 cells RNA. Primers were: sense, 5′-TTCATCGACTCTGCCAAC-3′; antisense, 5′-CATACTCTTCTCTTCACCCC-3′.
Mice, constructs and northern blot
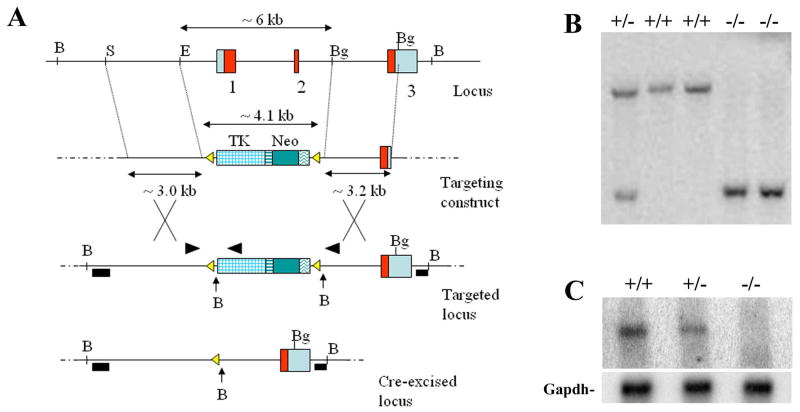
A mouse genomic SV129/Ev library cloned into the Xba I sites of Lambda FIX® II Vector from Stratagene (La Jolla, CA) was screened with a full-length Sfrp2 cDNA probe. Two clones containing the Sfrp2 gene, were isolated and utilized for the characterization of the gene locus. The targeting vector was designed to replace about 6kb of the Sfrp2 locus, including exons 1 and 2, with a cassette containing the neomycin resistance and the tymidine kinase (TK) selection markers flanked by loxP sites. After linearization, the vector was electroporated into AB2.2 embryonic stem (ES) cells. Homologous recombination was identified by Southern analysis using two external flanking probes and BamHI restriction digestion. Properly targeted ES cell clones were then electroporated with a Cre-recombinase expression plasmid; excision of the selection marker cassette was obtained in vitro, demonstrated by PCR (see Fig.2) and confirmed by Southern hybridization for the absence of the TK specific band. C57/Bl6 blastocysts were injected, chimeric mice generated and germ line transmission of the mutated allele was achieved. For the Northern blot, 10ug of total RNA extracted from E15.5 embryos were loaded per lane and the full-length Sfrp2 cDNA was used as probe. The mice utilized for all the experiments were F2 generation mice with a mixed genetic background, derived from the mating of F1 mice (50% Sv129Ev/50% C57Bl6). TOPGAL mice were obtained from Jackson laboratory (strain= Tg(Fos-lacZ)34EFu/J). They were mated with Sfrp2-/- mice (129Sv/Ev) and then TOPGAL/Sfrp2+/- were crossed to generate TOPGAL/Sfrp2-/- or TOPGAL/WT mice. All mice were housed in a specific pathogen free facility and under light, temperature- and humidity controlled conditions. The animal research complied with all relevant federal guidelines and institutional policies.
Figure 2.
(A) Schematic diagram showing the strategy used to obtain Sfrp2-/- mice. (B) Southern blot genotyping using the 5′ flanking probe and BamHI digestion. (C) Northern blot showing decreased or absent Sfrp2 mRNA transcript in heterozygous or homozygous null mice, respectively, normalized with Gapdh (bottom).
Staining of skeletal preparation
Skeletal preparations of newborns (P1), four day old (P4) and six month old mice were prepared and stained with Alcian Blue and Alizarin Red as previously described (McLeod, 1980). Sections from the TOPGAL transgenic mice were stained for B-galactosidase activity according to standard procedures.
Statistical analyses of bone length
Limb skeletal preparations were photographed with a Nikon 5.0 megapixel digital camera mounted atop of a Nikon SMZ1500 dissecting scope. All microscope and camera settings were identical for the capture of all images. Digital image files were uploaded to an Axiovision software (Carl Zeiss Vision, Munchen-Hallbergmoos, Germany) connected to a Zeiss Axioplan 2 scope and scale equivalency was achieved via transfer of stage micrometer calibration information. These procedures allowed for direct comparison of each image. Metacarpal lengths were measured, via Axiovision measurement tool, from the tip of the proximal epiphysis to the tip of the distal epiphysis. Digital lengths were calculated from the tip of the proximal epiphysis of the first phalanx to the distal tip of the third phalanx. These analyses were done at newborn, P4 and adult (6 month) stage. Data were collected on bone lengths in 11 distinct bone elements (4 metacarpals, 5 digits, ulna and radius) for each genotype (mutant and wt), for a total of 22 groups. Eight (n=8, at newborn and P4) and twelve (n=12, at 6 month) subject animals were measured in each group (genotype × bone combination). A two-way analysis of variance was conducted to determine the effect of the genotype while accounting for inherent differences between bones.
Proliferation, Tunel and Immunodetection assays
Pregnant female mice at E17.5 dpc were injected with bromodeoxyuridine (Brdu labeling reagent, 1ml/100grs of body weight, Zymed, S. San Francisco, CA) intraperitoneally and then sacrificed 2 hours later. Embryos were harvested, washed in cold PBS and processed for paraffin embedding. Brdu staining was carried out on paraffin sections of distal limbs using the Zymed BrdU staining kit (Zymed, S. San Francisco, CA) and following the manufacturer's recommendations. BrdU positive cells and total number of cells in selected fields were counted (n=3 mice). Serial sections from E17.5 embryos were also utilized for the detection of PCNA using a PCNA staining kit from Zymed (S. San Francisco, CA). Limb sections of E17.5 embryos were also utilized and stained with the Apoptag Fluorescein in situ Apoptosis Detection Kit (Chemicon, Temecula, CA), according to the kit instructions. Beta-catenin immunodetection was performed using a commercial antibody (BD Transduction Laboratories) that was revealed by a Vectastain ABC kit (Vector Laboratories, Burlingame, CA).
Cell Culture, Transfections and Reporter Gene assay
COS7 cells were cultured in complete Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and seeded in six-well plates with equal amounts of cells the day before the transfection. Cells were transfected at 70% confluency using Lipofectamine™ 2000 with PLUS™ reagent (Invitrogen, Carlsbad, CA) following the manufacturer's recommendations and fresh DMEM medium was added 5 hours after transfection. Co-transfections were performed in triplets with TOP/FOP-flash reporter plasmids, pIRES-Wnt1-GFP (supplied by Dr. Yingzi Yang, NIH, Bethesda, MD), pcDNA 3.1⊕-Sfrp2, pcDNA 3.1⊕-Wnt4-V5/ HisA, pcDNA 3.1⊕-Wnt5a-V5/ HisA, pcDNA 3.1⊕-Wnt5b-V5/ HisA, pcDNA 3.1⊕-Wnt9a-V5/HisA 0.4 μg each and 0.2 μg of β-galactosidase (Promega, Madison, WI) as indicated. The total amount of transfected DNA was kept constant at 1.4 μg per well by adding empty plasmid DNA (pcDNA3.1+ - Invitrogen). Transfected cells were harvested 48 hours after the transfection. Luciferase activity was measured by using D-Luciferin substrate (BD Biosciences, San Jose, CA). Readings on a Fluorstar OPTIMA (BMG Labtechnologies, Offenburg, Germany) were normalized for transfection efficiency measuring β-galactosidase activity by using Galacto-Light Plus ™ System (Applied Biosystems, Foster City, CA). The histogram shows average values with standard deviations from three independent transfections.
Results
Sfrp2 is persistently expressed during mouse limb development in a tissue and temporally-restricted fashion
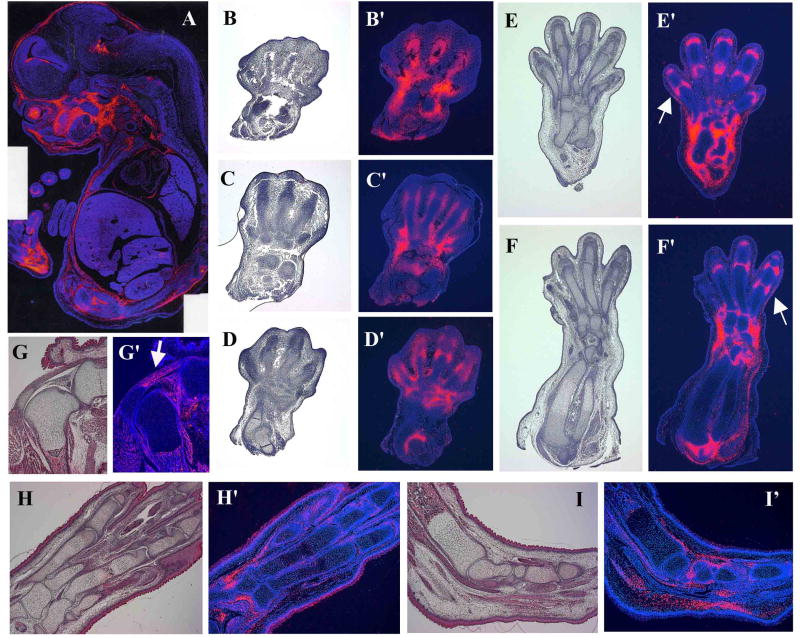
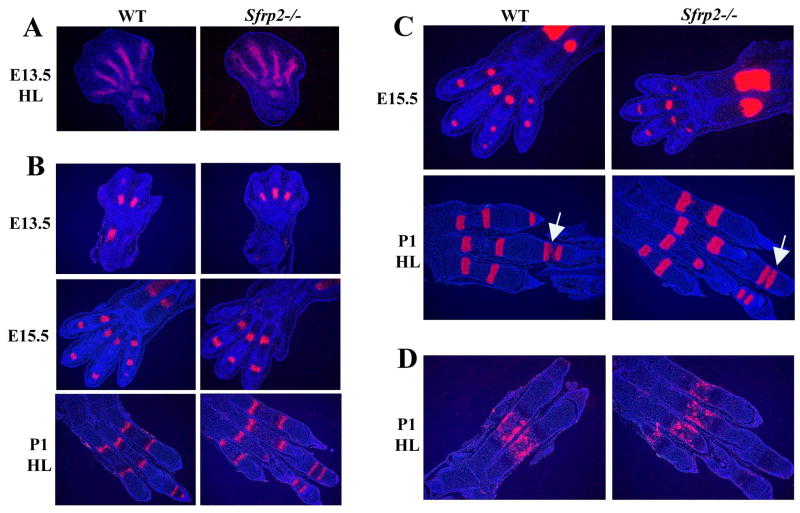
The Sfrp2 transcript is expressed in multiple tissues and organs during mouse embryonic development (Fig.1A), including the limb bud (Leimeister et al., 1998). In this tissue, its expression starts as early as E10.5 (Lescher et al., 1998) and persists until birth. We have analyzed the Sfrp2 expression pattern at different stages of mouse limb development, from E13.5 until birth, by RNA in situ hybridization. At E13.5, while distal limb patterning is still in progress, Sfrp2 shows a proximal-distal gradient of expression with the autopod expressing higher levels of transcript compared to the zeugopod and stylopod. In these last segments, Sfrp2 localizes in the already defined region of the elbow joint; in the autopod however, it is highly expressed in presumptive joints and in the mesenchymal tissue surrounding the forming joints in the carpal (Fig.1C′,D′) and tarsal region. More distally, its expression is detected in perichondrial cells that surround and line the chondrogenic condensations of the digital rays (Fig.1B′,C′). Its expression also begins to invade into the future phalangeal joint mesenchyme (Fig.1 D′). At E15.5 the expression in the perichondrium of the digits is lost and Sfrp2 now strongly localizes in the joint mesenchyme between the phalangeal elements and in the surrounding mesenchymal cells that will ultimately form ligaments and the synovial capsule (Fig.1E′,F′)(Dreyer et al., 2004). Interestingly, Sfrp2 expression seems to flank both distal and proximal epiphysis of metacarpals and phalangeal elements (Fig.1.E′,F′ arrows). The same is observed in the carpal/tarsal region. At E17.5 Sfrp2 is still expressed in the joints of the limb (Fig.1H′), although at much weaker levels. However, the tendons are now strongly positive, including the patellar tendon (Fig.1G′), the Achilles's tendon (Fig.1I′), and fibroblast of the plantar fascia (Fig.1I′). The synovial cells of the knee joint are also expressing Sfrp2. In the newborn mouse, residual expression of Sfrp2 is observed in the connective tissue between the metacarpal/metatarsal bones (data not shown). The expression detected in the hindlimb follows and parallels that observed in the forelimb. From this analysis and previously published data, we conclude that during early limb bud formation, Sfrp2 expression seems to mirror, in a proximal-distal fashion, the tissues that pattern the limb mesenchymal condensations specifically localizing to perichondrial, interdigital, and interzone mesenchyme. At later stages, its expression becomes predominant within tendons, synovial cells and ligaments.
Figure 1.
Sfrp2 expression pattern by mRNA in situ hybridization. Panel A shows Sfrp2 expression in a full embryo section at E15.5 (composite image). Panels B,C,D,E,F,G,H,I show hematoxylin and eosin stained serial sections of the corresponding panels B′,C′,D′,E′,F′,G′,H′,I′ which were hybridized with the Sfrp2 antisense probe. Sfrp2 specific signal was pseudo-colored in red. The hybridizations with the sense probe were negative and are not shown. (B′,C′,D′) E13.5 distal forelimb sections showing Sfrp2 expression in perichondrium surrounding the digits, in mesenchyme around the carpal bones and in forming joints. (E′,F′) At E15.5 Sfrp2 expression strongly localizes into joints as well as mesenchymal cells around them. Note expression flanking distal and proximal epiphysis of metacarpal and phalangeal elements (arrows). (G′,H′,I′) At E17.5 Sfrp2 expression is still present in joints, although weaker, and becomes more strongly localized in tendons and ligaments, including the achille's and patellar tendons (arrow in G′).
Homozygous Sfrp2 null mice are viable
In order to study the function of the Sfrp2 gene during mouse embryonic development we generated Sfrp2 null mice. For this purpose, a targeting construct designed to replace the first 2 exons of Sfrp2 with a floxed selection cassette (Fig.2A) was electroporated into mouse ES cells. Homologous recombination with the Sfrp2 locus was confirmed by Southern blot (data not shown) and positive ES cell clones were amplified and further electroporated with a Cre-recombinase expressing plasmid. Cre-excised clones were confirmed by both Southern analysis (Fig.2B) and PCR. Sfrp2 heterozygous mice in a mix 129Sv/C57black genetic background were obtained after germ line transmission of the targeted Sfrp2 allele from chimeric mice and were undistinguishable from their wild-type littermates. Homozygous Sfrp2 mutant mice were generated by intercross of heterozygous animals (Fig.2B) and were born at the expected Mendelian ratios. A Northern blot using total RNA extracted from whole embryos at E15.5 stage of development, showed decreased and absent specific Sfrp2 transcript in heterozygous and homozygous mutant mice, respectively, compared to wild-type mice (Fig.2C). At birth, the Sfrp2-/- mice were viable and showed no gross abnormalities; as adults, they were fertile and had a normal lifespan. Because Sfrp2 is expressed in several different tissues and organs during embryogenesis like kidney, eye, craniofacial mesenchyme, joints, and others, we performed a full histological analysis on the Sfrp2 null mice at 10 weeks of age. We found no morphological abnormalities and therefore decided to proceed with several functional tests, including complete blood counts, serum chemistries, urine analyses, electro-retinograms, X-ray analyses and full body PixiMus scans. The measurement of all these parameters yielded values that were similar to those obtained from wild-type littermate mice (data not shown).
Sfrp2 deficiency causes brachydactyly, mild mesomelic shortening and posterior soft-tissue syndactyly
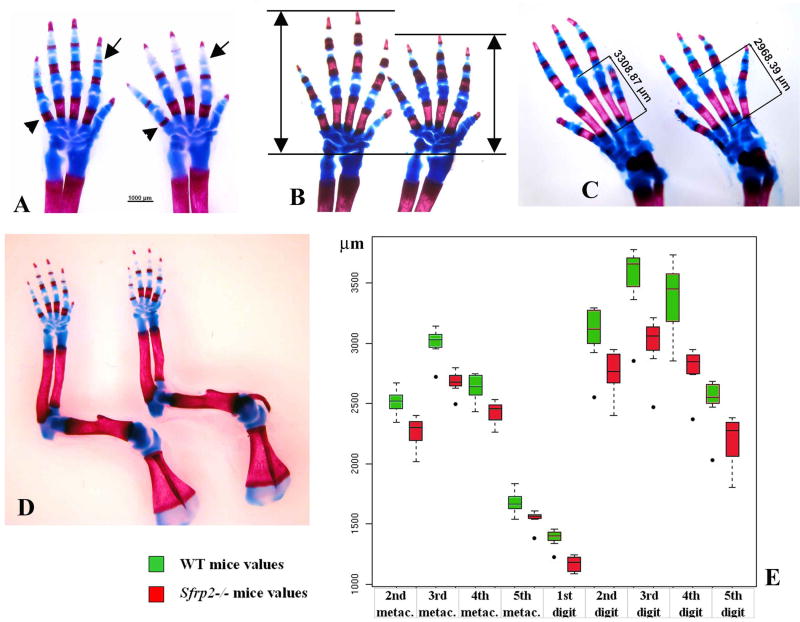
Although we found no obvious phenotype in the adult Sfrp2 null mice, we decided to study in detail the skeleton of the newborn mice because of the Sfrp2 dynamic expression pattern in this tissue. We stained newborn skeletal preparations with Alcian Blue that stains all cartilaginous elements, and Alizarin Red that stains calcified bone matrix. No bone patterning defects were seen in the Sfrp2 mutant mice compared to wild-type littermates, however the distal portion of the appendicular skeleton showed clear but subtle abnormalities. The metacarpal/metatarsal bones were shortened and the shortening also affected the phalangeal bones resulting in a decreased overall length of each digit (Fig.3A). This was particularly evident when observing the calcified diaphyses of these bones (Fig.3A arrowheads). Moreover, while the ossification centers of the second phalanges were fully formed in the wild-type newborns, these were consistently delayed and just emerging in the Sfrp2-/- newborns (Fig.3A arrows). We then analyzed similar skeletal preparations in 4 days old pups (P4) and noticed the same shortening that was observed at the newborn stage (Fig.3B). Both forelimbs and hindlimbs were equally affected (Fig.3C). Importantly, when other long bones of the forelimb were analyzed at P1 and P4, the length of the humerus in the mutant mice was similar to that of the wild-type counterparts; however, both ulna and radius showed a mild but consistent shortening in the Sfrp2-/- mice versus WT (see table1 and Fig.3D). Such shortening was similar, in percentage, to that observed in the metacarpals; however, the digits were the most severely affected. In order to quantify these limb defects, digital images were obtained from n=8 mice/genotype and analyzed with the Zeiss Axiovision software. The results showed a highly significant and consistent shortening of the bones of mutant animals (p < 10e-15), and this difference holds in each pair-wise comparison for the 11 bones (4 metacarpals, 5 digits, ulna and radius) in our study (Fig.3E). Importantly, the magnitude of decrease was not equal for all bones although the direction of change was always the same (mutant bones were always shorter on average).
Figure 3.
Brachydactyly observed in the Sfrp2-/- mice (in each panels A,B,C,D WT limbs are to the left and Sfrp2-/- to the right). (A) Skeletal preparation of distal limbs of newborn mice stained with Alcian blue (cartilage) and Alizarin red (calcified bone matrix). Note decreased length of all skeletal elements, most evident by looking at their calcified portions (arrowheads point to fifth metacarpal element). Also note consistent lack of ossification centers in the second phalangeal elements of Sfrp2-/- compared with WT littermate mice (arrows). (B) Forelimb skeletal preparations of four days old pups (P4). The overall length of autopod is consistently decreased in the mutant mice. (C) The same is observed in P4 hindlimb skeletal preparations (the length measurement of the third metatarsal is shown). (D) Newborn forelimb skeletal preparations showing mild mesomelic shortening in the mutant mice. (E) Statistical analysis of forelimb autopod bone lengths showing significant shortening of each skeletal elements in the Sfrp2-/- mice versus controls (n=8, p < 10e-15).
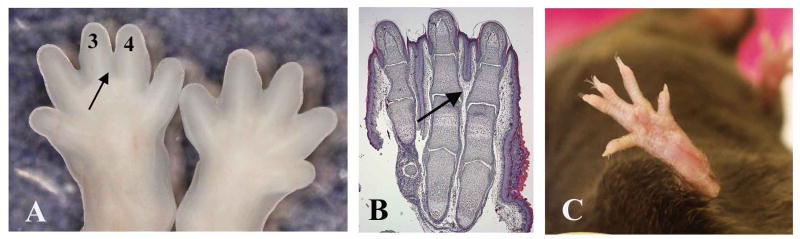
Distal limb skeletal preparations were also analyzed in adult male mice at 6 months of age (n=12) and the length of all skeletal elements (up to the first phalanx) was measured. Consistent with the previous data, adult Sfrp2-/- mice showed statistical significant shortening of distal skeletal elements compared to wild-type littermates (see Table1). This suggests that the loss of Sfrp2 causes a persistent effect on skeletal growth and homeostasis even in the postnatal period. No additional defects of the skeleton were noted, however, a hind limb, unilateral, soft-tissue syndactyly between the third and fourth digit (complete or incomplete) was observed. While this anomaly was 25% penetrant in the mixed genetic background, it presented as fully penetrant and often bilateral in the 129Sv pure background (Fig.4A-C).
Figure 4.
Syndactyly observed in the Sfrp2-/- mice. (A) E15.5 distal hind limbs showing unilateral syndactyly between the third and fourth digit. (B) Newborn hind limb section stained with hematoxylin and eosin. The arrow points to failed inter-digital mesenchyme regression with absence of cleavage between third and fourth digit. (C) Incomplete hindlimb syndactyly in an adult Sfrp2 null mouse.
Our observations suggest an essential role for Sfrp2 in the control of chondrocytes function during endochondral bone formation of distal limb skeletal elements and show a proximal-distal gradient of effect, with normal stylopod and the autopod more affected than the zeugopod. The syndactyly instead indicates that Sfrp2 contributes to the regression of interdigital mesenchyme in the hindlimb.
Lack of Sfrp2 causes decreased chondrocyte proliferation and delayed differentiation in distal limb skeletal elements
The observed decrease in appendicular distal bone length could be the result of an altered mesenchymal condensation or chondrocyte differentiation, an increase in the apoptotic rate of chondrocytes, a decrease in their proliferation rate or a combination of these events. In order to evaluate these possibilities we first analyzed the expression of Sox9, an early marker of mesenchymal condensation, in the digital rays of mouse limb autopods at E13.5. No differences were noted when comparing Sfrp2-/- vs WT littermate limb sections (Fig.5A). We then analyzed the expression of two chondrocyte differentiation markers, Indian hedgehog (Ihh) and type X Collagen (Col10a1), at different time points during development by RNA in situ hybridization. Ihh is a marker of pre-hypertrophic chondrocytes while type X Collagen is specific for hypertrophic chondrocytes. While both of these markers showed a comparable pattern of expression at all stages of differentiation that were analyzed (Fig.5B,C), type X collagen hybridization at the newborn stage showed an increased distance between proximal and distal growth plate of the first phalangeal element of wild-type versus the Sfrp2 mutant littermates (Fig.5C, arrows). The phalangeal delay in chondrocyte hypertrophy compared to wild-type is already detectable at E17.5 (Fig.6A). At newborn stage the expression level of Osteocalcin, a marker of mature osteoblasts differentiation, was unchanged between the two genotypes (Fig.5D).
Figure 5.
mRNA in situ hybridization analysis of mesenchymal, cartilage and bone differentiation markers at E13.5, E15.5 and newborn (P1) stage (HL= hindlimb). (A) Sox9 and (B) Indian hedgehog antisense probes show comparable pattern of expression between Sfrp2-/- and WT controls. (C) Type X collagen is normally expressed, however the probe detected a delay in chondrocyte differentiation of Sfrp2-/- mice demonstrated by the smaller distance between the hypertrophic zones in the metaphyses of the first phalanx at P1 (arrows). (D) The antisense probe specific for Osteocalcin shows no overt difference at P1 between the two genotypes.
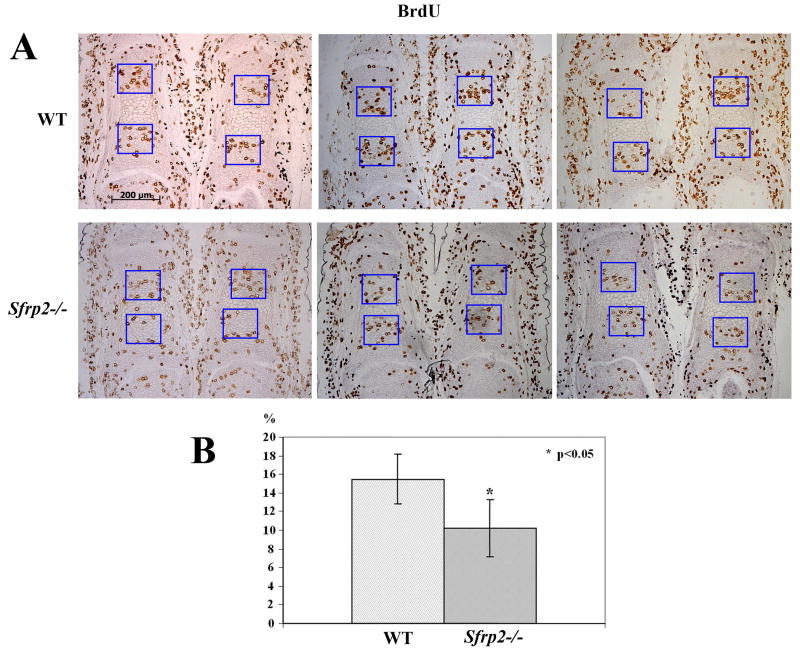
Figure 6.
Chondrocyte proliferation defect in Sfrp2-/- distal limb elements. (A) BrdU labeling shows decreased number of BrdU positive proliferating chondrocytes in E17.5 first phalanx sections of Sfrp2-/- versus controls. Also note the consistent hypertrophy delay in mutant phalanges. (B) Percentage of chondrocytes positively labeled with BrdU staining. The difference between WT and Sfrp2-/- mice (n=3) was statistically significant (p<0.05).
Next, we performed a Tunel assay to assess the pool of apoptotic chondrocytes in the distal limb of the Sfrp2 mutant mice. We decided to assay embryo sections at E17.5 days of development, a stage where chondrocytes have already reached hypertrophy in the autopod region but vascular invasion of metacarpal bones is just beginning. In all sections that we analyzed an equivalent number of apoptotic cells were observed in Sfrp2 null mice compared to wild-type littermates (data not shown). The majority of the apoptotic cells were localized in the zone of chondrocyte hypertrophy.
Lastly, we analyzed the chondrocyte rate of proliferation by injecting pregnant mice with bromodeoxyuridine (BrdU) at 17.5 days of gestation and harvesting the embryos for paraffin-embedded tissue sectioning and BrdU staining. We counted the number of BrdU positive cells versus the total number of cells present inside equivalent square fields arbitrarily drawn on growth plate sections within the zone of proliferating chondrocytes. We observed a consistently decreased number of BrdU positive cells in the first phalangeal elements of the Sfrp2 mutant mice compared to the same bones of the littermate controls (Fig.6A). Cells were counted in at least two sections from n=3 mice/genotype and the difference was statistically significant (p<0.05) (Fig.6B). We then observed similar findings on the metacarpal bones and in the first phalangeal elements at P1; although these data didn't reach statistical significance, the trend was clear (data not shown). To independently confirm the decreased rate of chondrocyte proliferation, we also stained E17.5 distal limb sections for proliferating cell nuclear antigen (PCNA), a cell mitotic marker. PCNA staining confirmed our BrdU studies, showing fewer PCNA positive cells in Sfrp2 mutant compared to wt littermate control phalangeal sections (data not shown).
From these experiments we conclude that chondrocyte apoptosis is unaffected in Sfrp2 mutant mice compared to wild-type littermates; however, chondrocytes from the distal portion of the Sfrp2 null mouse limbs have both a proliferation defect and a differentiation delay compared to chondrocytes in the same location of WT mice.
Sfrp2 regulation of nuclear beta-catenin levels in vitro and in vivo
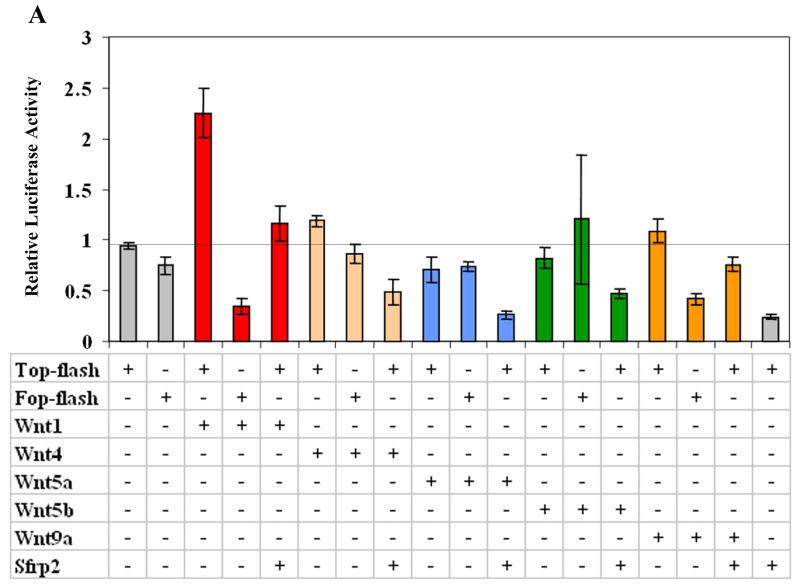
Because the control of cell proliferation by Wnts is usually positively regulated by beta-catenin, we decided to test whether Sfrp2 is able to inhibit canonical Wnt signaling as suggested by others (Lee et al., 2000; Topol et al., 2003). To do this we used COS7 cells and the in vitro LEF/TCF reporter (TOP/FOP-FLASH) assay where increase in transcription of a luciferase gene is stimulated by intranuclear accumulation of beta-catenin. We tested Wnt1, Wnt4, Wnt9a (previously known as Wnt14), Wnt5a, and Wnt5b. Both Wnt4 and Wnt9a were recently shown to act through the canonical pathway (Guo et al., 2004) and have an expression pattern overlapping that of Sfrp2 during joint development, while Wnt5a and Wnt5b are strongly expressed in the growth plate. Wnt1 as well as Wnt4 and Wnt9a, although to a lesser extent, up-regulated TOP-FLASH activity and hence can act via canonical signaling in our experimental system; independent co-transfection of Sfrp2 with all of these Wnts led to a significant decrease of the luciferase activity (Fig.7A). Both Wnt5a (as expected) and Wnt5b did not appear to signal through the canonical Wnt pathway in our transfection assays and, if anything, were mild inhibitors of it (Fig.7A).
Figure 7.
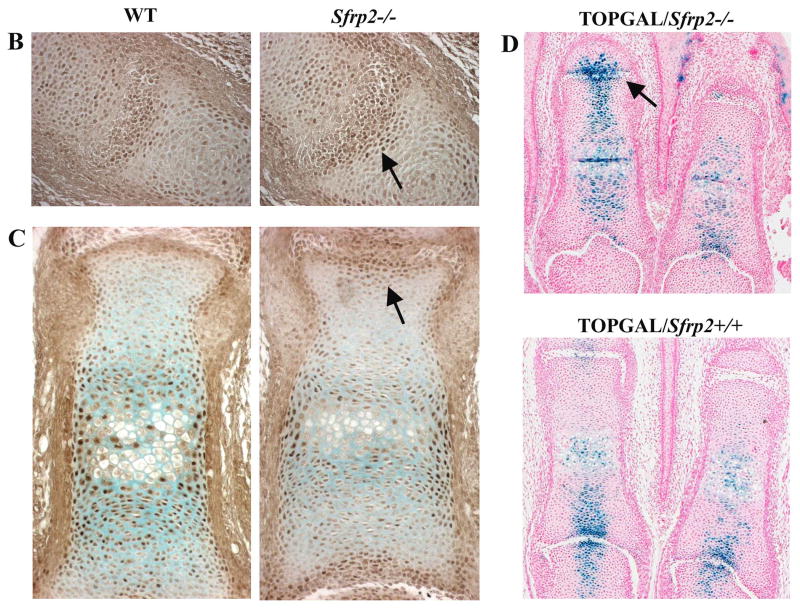
Sfrp2 modulation of beta-catenin in vitro and in vivo. (A) Transfection assay using the Top/Fop-flash luciferase reporter system. Wnt1and to a lesser extent, Wnt4 and Wnt9a activated the Top-flash reporter gene, demonstrating that they can signal via the canonical pathway and co-transfection with Sfrp2 inhibited such activation. Wnt5a and Wnt5b had no effect on the reporter gene, suggesting they are not signaling through the canonical pathway in our experimental system. (B) Sections of interphalangeal joint at E15.5 stained with a beta-catenin specific antibody. Note subtle increased accumulation of beta-catenin in the epiphyseal region (arrow) of the Sfrp2-/- mouse (right panel). (C) Sections of first phalanx at E17.5 stained with the same antibody. Notice the increase beta-catenin localization in the epiphyseal area (arrow) in the mutant mouse versus the WT. (D) Sections of first phalangeal elements of TOPGAL/Sfrp2 trangenic mice, showing increased β-galactosidase staining in the articular chondrocytes of some interphalangeal joints of the Sfrp2 null mice (arrow).
According to these results and based on the Sfrp2 expression pattern, specific regions of the Sfrp2 null mice are expected to have increased canonical Wnt activity. Western blot analysis on E11.5 and E15.5 autopod lysates failed to reveal significant difference in levels of stabilized beta-catenin between the mutant and wt control embryos. However, immunodetection of beta-catenin on limb sections at E15.5 and E17.5 suggested an increase in beta-catenin nuclear accumulation in interphalangeal joints of Sfrp2 null mice versus WT littermate controls (Fig. 7B,C arrows). To further evaluate Wnt signaling in Sfrp2 null mice, we crossed them with the TOPGAL transgenic mice. These mice express β-galactosidase in the presence of the lymphoid enhancer binding factor 1/transcription factor 3 (LEF/TCF) mediated signaling pathway and activated β-catenin (DasGupta and Fuchs, 1999) and are a reporter strain for canonical Wnt signaling. Although the number of β-galactosidase positive cells varied among sections from different transgenic embryos, articular chondrocytes of some interphalangeal joints at E17.5 expressed the reporter gene in higher intensity and number in TOPGAL/Sfrp2-/- vs TOPGAL/Sfrp2+/+ mice (Fig.7D arrow).
These experiments confirm that Sfrp2 can inhibit canonical Wnt signaling in vitro and in vivo and suggest that a mild alteration of beta-catenin localization in distal limb chondrocytes likely contributes to the observed phenotype.
Discussion
Loss of function and mis-expression studies overexpressing Wnt during chondrogenesis have unraveled a complex and dynamic Wnt signaling network regulating joint and endochondral bone formation in the limb. However, the embryonic role of extracellular antagonists of the Wnt signaling, with the exception of Dickkopf1 (Mukhopadhyay et al., 2001), is still poorly defined. Members of the secreted frizzled related family of proteins have a widespread and specific expression pattern throughout development, including during limb development (Leimeister et al., 1998; Lescher et al., 1998). Moreover, their putative antagonistic function of both canonical and non-canonical Wnt activity makes them potential regulators of embryonic morphogenesis. We generated Sfrp2 null mice but while others were unable to reveal the phenotype, we detected a shortening of the distal bony elements of the limbs due to decrease in chondrocyte proliferation and differentiation together with a hindlimb soft-tissue syndactyly. These data suggest that Sfrp2 is not alone required for regulating mesenchymal condensation, patterning or joint morphogenesis. Instead, Sfrp2 may fine tune Wnt activity in restricted areas of the embryo, including growth plate and interdigital mesenchyme.
Sfrp2 role during limb skeletal formation
During long bone formation Wnts have been shown to regulate mesenchymal condensation and proliferation, synovial joint induction and chondrocyte proliferation and maturation (Yates et al., 2005). The antagonistic function of Sfrp2 raises the question of potential Wnt targets specificity during the longitudinal growth of distal limb bone elements in the Sfrp2-/- mice. To address this we need to consider the Wnt molecules that are co-expressed with Sfrp2 during this process. While beta-catenin becomes downregulated in condensing pre-chondrogenic mesenchymal cells (Ryu et al., 2002), it remains highly expressed in mesenchymal cells surrounding the anlagen condensations, induced perhaps by Wnt9a, Wnt4 and Wnt16. These genes are expressed in an overlapping and sometimes complementary fashion in the mesenchyme surrounding chondrogenic condensations, in zones of future joint formation and then in cells that will form ligaments, tendons, and synovial capsules (Guo et al., 2004). This expression pattern is very similar to that of Sfrp2, as seen in Figure 1. Hence, loss of Sfrp2 antagonism on Wnt signaling would predict further accumulation of beta-catenin in the digital perichondrium, and thereby limit the number of cells that are recruited into mesenchymal prechondrogenic condensations. This would decrease the normal pool of proliferating chondrocytes and contribute to the shortening of digits. In addition, it has been recently suggested that Wnt signaling regulates cell fate commitment of mesenchymal precursor cells: increased beta-catenin levels would favor a commitment toward the osteoblastic lineage while decreased beta-catenin levels would enhance the chondrocytic lineage formation (Day et al., 2005; Hill et al., 2005).
Wnt5a is also expressed in the limb mesenchyme and assumes a proximal-distal gradient of expression during limb outgrowth (Dealy et al., 1993). After mesenchymal condensations appear, its expression becomes excluded from such condensations and restricted to presumptive perichondrial cells and mesenchymal cells next to them, especially along the digits (Chimal-Monroy et al., 2002; Hartmann and Tabin, 2000; Yamaguchi et al., 1999); this is overlapping to Sfrp2 expression. Additionally, in the mouse growth plate, Wnt5a was described at the boundary between proliferating and pre-hypertrophic chondrocytes, contiguous to Wnt5b expression, where its signaling is needed for coordinating both proliferation and differentiation of growth plate chondrocytes and ensure longitudinal long bone growth (Yang et al., 2003). When over-expressed in non-hypertrophic chondrocytes of transgenic mice (under the regulation of the type II collagen promoter (Col2a1)), Wnt5a caused shortening of limb skeletal elements with thicker cartilage and delayed chondrocyte hypertrophy and ossification (Yang et al., 2003). Interestingly, Wnt5a can activate or inhibit canonical beta-catenin signaling depending on which receptor it interacts with (Mikels and Nusse, 2006a; Topol et al., 2003). In this context, Sfrp2 may modulate Wnt5a activity so that Sfrp2 null mice would have altered Wnt5a signaling in chondrocytes and an associated decreased proliferation and delayed differentiation. This is consistent with the shortening and delayed ossification that we observed.
Interestingly, a member of the Bmp family of secreted proteins encoded by the mouse locus brachypodism, Growth differentiation factor 5 (Gdf5) shows an expression pattern similar to that of Sfrp2. Gdf5 regulates skeletal elements size by increasing proliferation within the epiphyseal cartilage and loss of function mutations cause severe reductions in the length of appendicular bones, especially the digits (Storm and Kingsley, 1999). This role seems to parallel that of Sfrp2. Thus, evidence points to the existence of at least two signaling pathways, Bmp and Wnt that show proximal-distal limb gradients of action and together contribute to the elongation of long bones. Moreover, functional and phenotypic data suggest that Wnt and Bmp signaling can interact. While Gdf5 signals primarily through the serine/threonine kinase receptor BmpR1b (Baur et al., 2000), Wnt5a can bind to the receptor tyrosine kinase Ror2 (Oishi et al., 2003) and ultimately, BmpRIb and Ror2 form a ligand independent heteromeric complex to modulate Gdf5 signaling (Sammar et al., 2004). This raises the question of whether there is competitive versus synergistic action between Bmp and Wnt pathways. While Gdf5 was normally expressed (see Supplementary Data Figure 1), we currently don't know if its downstream signaling pathway was affected in the Sfrp2-/- mice. Considering that the human orthologs of these factors, CDMP1 (GDF5), BMPR1B and ROR2 have all been implicated in autosomal dominant non-syndromic brachydactylies, it is not unreasonable to hypothesize that they may genetically interact. Additionally, the frizzled domain of Sfrps appears to regulate the activity of certain matrix metalloproteinases responsible for the degradation of Chordin, a known Bmp inhibitor (Lee et al., 2006a), suggesting a novel and more direct interaction between Bmp and Wnt pathways in the extracellular matrix.
Other Wnt inhibitors have been shown to play a role during limb skeletal formation. Sfrp3 (Frzb-1) is normally expressed in mesenchymal and chondrocytic cells and its over-expression in the chick limb causes a shortening of skeletal elements, joint fusion and delay in chondrocyte differentiation (Enomoto-Iwamoto et al., 2002). Similarly, it was shown that Sfrp1 is a negative regulator of Wnt signaling for the appropriate timing of chondrocytes differentiation in the growth plate (Gaur et al., 2006). This suggests that Sfrp3 and Sfrp1 may exert opposite effects to Sfrp2 in the growth plate and such a coordinated action involving different Sfrps has already being hypothesized elsewhere (Yoshino et al., 2001). Dickkopf1 (Dkk1) is a canonical Wnt signaling inhibitor expressed in the developing limb. Dkk1 null mice show distal limb defects including fusion of digits and the appearance of ectopic pre- and post-axial digital elements (Mukhopadhyay et al., 2001). These data indicate that Dkk1, similarly to the Sfrp2, regulates both cell death and cell proliferation (Mukhopadhyay et al., 2001), and further stress the importance of fine Wnt signaling modulation for proper development of distal limb elements. Moreover, the coordinated expression of Dkk1 and Bmp2 suggests again a putative Bmp-Wnt genetic interaction during the process of digit formation. This was already demonstrated during the genesis of the limb apical ectodermal ridge (Soshnikova et al., 2003).
Finally, Wnt signaling and β-catenin stabilization are important for the beginning of synovial joint induction (Guo et al., 2004). Sfrp2 expression throughout joint morphogenesis raises the question of its potential role during this process. We analyzed joint sections at different embryonic stages, in the newborn and the adult skeleton but never observed joint fusions or abnormalities of the articular surfaces (data not shown). In situ hybridization with a probe specific for Gdf5, a known early marker of joint formation, showed a comparable expression pattern between Sfrp2 mutant and control mice at E13.5 and E15.5 (see Supplementary Data Figure1). From these observations, we conclude that Sfrp2 is dispensable from joint formation, and perhaps functional redundancy involving other putative Wnt-inhibitors compensates for its loss during this process. Alternatively, joint morphogenesis may be less sensitive to Wnt signaling dysregulation compared to growth plate maturation.
Syndactyly in the Sfrp2-/- mice
Syndactylies are characterized by fusion of soft-tissue between the fingers and/or toes with or without bony fusion and are generally caused by an apoptotic defect. The Sfrp2-/- mice consistently show a posterior syndactyly, always between the third and fourth digit, which can be complete or incomplete. It is 25% or 100% penetrant in the mixed C57bl/129Sv and 129Sv pure genetic background, respectively and this may be due to the presence of different modifier alleles. This phenotype is consistent with a failed regression of interdigital mesenchyme between these two digits, likely due to defective programmed cell death although we were not able to demonstrate this. This is in accordance with the initial in vitro isolation and description of the Sfrps as secreted apoptosis related proteins (SARPs) (Melkonyan et al., 1997). As mentioned previously, Dkk1 is also involved in modulating apoptosis in distal limb and this function was shown to be modulated by Bmp signaling (Grotewold and Ruther, 2002). Bmp signaling is in fact considered necessary for apoptosis in the mouse limb (Guha et al., 2002). We have analyzed expression levels of Bmp2, Bmp4 and Bmp7 by RNA in situ hybridization on E13.5 and E15.5 hind limb autopod sections collected from 129Sv embryos and observed no overt differences between Sfrp2 mutant and littermate control mice (see Supplementary Data Figure 2). This may suggest that perhaps other genes regulating interdigital tissue apoptosis could be involved and a Bmp-independent apoptotic pathway may be associated with the syndactyly in the Sfrp2-/- mice.
SFRP2 and human brachydactyly
Non-syndromic brachydactylies are dominantly inherited genetic traits in which selected skeletal elements of the hand and foot are shortened or sometime missing; in addition, but more rarely, there can be fusion of elements causing either symphalangism or soft-tissue syndactyly. They were classified by Bell in 1951 (Temtamy and McKusick, 1978) into several recognizable distinct types. Mutations in a few genes, including Indian hedgehog (IHH), Bmp-receptor 1b, ROR2, CDMP1 (the human ortholog of Gdf5) and HOXD13, have been described in several probands affected with brachydactyly type A1, type A2, type B, type C, and type D/E respectively (Gao et al., 2001; Johnson et al., 2003; Lehmann et al., 2003; Oldridge et al., 2000; Polinkovsky et al., 1997). These data in combination with the Sfrp2-/- mouse phenotype suggest that both dysregulation of Bmp and Wnt signaling can cause brachydactyly and lends support to the genetic interaction of these signaling pathways. Mutations in CDMP1 (GDF5) can be associated with a clinical spectrum that includes acromesomelic dysplasia or shortening of both autopodal and zeugopodal elements. This again parallels what is seen in the Sfrp2 mutant mice albeit of greater severity. The human phenotype suggests that BMP and WNT signaling may be antagonistic at the level of digital chondrogenesis. While loss of function of CDMP1 causes brachydactyly C, gain of function of ROR2, a putative Wnt receptor, likely causes dominantly inherited brachydactyly B (van Bokhoven et al., 2000). This observation is consistent with the brachydactyly observed with the loss of a putative Wnt inhibitor, Sfrp2. This hypothesis would be further supported with the finding of human mutations in SFRP2 in a brachydactyly/syndactyly patient. The effects on multiple phalangeal elements in the Sfrp2-/- mice point to human brachydactyly type E as a potential candidate disorder for screening.
Supplementary Material
(A,B,C,D) mRNA in situ hybridization using a Gdf5 antisense probe (specific signal was pseudo colored in yellow) at E13.5 and E15.5 showing a comparable level of expression between Sfrp2-/- and WT control mice. (E,F) In situ hybridization using the Sfrp2 antisense probe confirmed absence of Sfrp2 expression in the Sfrp2 null mice.
mRNA in situ hybridization using antisense probes for Bmp2, Bmp4 and Bmp7 on hind limb autopod sections obtained from E13.5 129Sv embryo pups. No significant differences were noted between Sfrp2-/- and control mice.
Acknowledgments
We are grateful to Kevin Preuss for helping setting up the transfection assay and Monique Land for editorial assistance. We also thank Dr. C. Leimeister, Wuerzburg, Germany, Dr. V. Lefebvre, Cleveland, OH, Dr. G. Karsenty, Houston, TX and Dr. Yingzi Yang, NHGRI, Bethesda for sharing reagents with us. This work was supported by the NIH HD22657 (BL), ES11253 (BL).
Contract grant sponsor: NIH;
Contract grant number: HD22657 (BL) and ES11253 (BL)
References
- Baur ST, Mai JJ, Dymecki SM. Combinatorial signaling through BMP receptor IB and GDF5: shaping of the distal mouse limb and the genetics of distal limb diversity. Development. 2000;127(3):605–619. doi: 10.1242/dev.127.3.605. [DOI] [PubMed] [Google Scholar]
- Bitgood MJ, McMahon AP. Hedgehog and Bmp genes are coexpressed at many diverse sites of cell-cell interaction in the mouse embryo. Dev Biol. 1995;172(1):126–138. doi: 10.1006/dbio.1995.0010. [DOI] [PubMed] [Google Scholar]
- Bodine PV, Billiard J, Moran RA, Ponce-de-Leon H, McLarney S, Mangine A, Scrimo MJ, Bhat RA, Stauffer B, Green J, Stein GS, Lian JB, Komm BS. The Wnt antagonist secreted frizzled-related protein-1 controls osteoblast and osteocyte apoptosis. J Cell Biochem. 2005;96(6):1212–1230. doi: 10.1002/jcb.20599. [DOI] [PubMed] [Google Scholar]
- Bodine PV, Zhao W, Kharode YP, Bex FJ, Lambert AJ, Goad MB, Gaur T, Stein GS, Lian JB, Komm BS. The Wnt antagonist secreted frizzled-related protein-1 is a negative regulator of trabecular bone formation in adult mice. Mol Endocrinol. 2004;18(5):1222–1237. doi: 10.1210/me.2003-0498. [DOI] [PubMed] [Google Scholar]
- Chimal-Monroy J, Montero JA, Ganan Y, Macias D, Garcia-Porrero JA, Hurle JM. Comparative analysis of the expression and regulation of Wnt5a, Fz4, and Frzb1 during digit formation and in micromass cultures. Dev Dyn. 2002;224(3):314–320. doi: 10.1002/dvdy.10110. [DOI] [PubMed] [Google Scholar]
- Church VL, Francis-West P. Wnt signalling during limb development. Int J Dev Biol. 2002;46(7):927–936. [PubMed] [Google Scholar]
- DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126(20):4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8(5):739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Dealy CN, Roth A, Ferrari D, Brown AM, Kosher RA. Wnt-5a and Wnt-7a are expressed in the developing chick limb bud in a manner suggesting roles in pattern formation along the proximodistal and dorsoventral axes. Mech Dev. 1993;43(23):175–186. doi: 10.1016/0925-4773(93)90034-u. [DOI] [PubMed] [Google Scholar]
- Desbois C, Hogue DA, Karsenty G. The mouse osteocalcin gene cluster contains three genes with two separate spatial and temporal patterns of expression. J Biol Chem. 1994;269(2):1183–1190. [PubMed] [Google Scholar]
- Drake JM, Friis RR, Dharmarajan AM. The role of sFRP4, a secreted frizzled-related protein, in ovulation. Apoptosis. 2003;8(4):389–397. doi: 10.1023/a:1024181203729. [DOI] [PubMed] [Google Scholar]
- Dreyer SD, Naruse T, Morello R, Zabel B, Winterpacht A, Johnson RL, Lee B, Oberg KC. Lmx1b expression during joint and tendon formation: localization and evaluation of potential downstream targets. Gene Expr Patterns. 2004;4(4):397–405. doi: 10.1016/j.modgep.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Elima K, Eerola I, Rosati R, Metsaranta M, Garofalo S, Perala M, De Crombrugghe B, Vuorio E. The mouse collagen X gene: complete nucleotide sequence, exon structure and expression pattern. Biochem J. 1993;289(Pt 1):247–253. doi: 10.1042/bj2890247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto-Iwamoto M, Kitagaki J, Koyama E, Tamamura Y, Wu C, Kanatani N, Koike T, Okada H, Komori T, Yoneda T, Church V, Francis-West PH, Kurisu K, Nohno T, Pacifici M, Iwamoto M. The Wnt antagonist Frzb-1 regulates chondrocyte maturation and long bone development during limb skeletogenesis. Dev Biol. 2002;251(1):142–156. doi: 10.1006/dbio.2002.0802. [DOI] [PubMed] [Google Scholar]
- Gao B, Guo J, She C, Shu A, Yang M, Tan Z, Yang X, Guo S, Feng G, He L. Mutations in IHH, encoding Indian hedgehog, cause brachydactyly type A-1. Nat Genet. 2001;28(4):386–388. doi: 10.1038/ng577. [DOI] [PubMed] [Google Scholar]
- Gaur T, Rich L, Lengner CJ, Hussain S, Trevant B, Ayers D, Stein JL, Bodine PV, Komm BS, Stein GS, Lian JB. Secreted frizzled related protein 1 regulates Wnt signaling for BMP2 induced chondrocyte differentiation. Journal of cellular physiology. 2006;208(1):87–96. doi: 10.1002/jcp.20637. [DOI] [PubMed] [Google Scholar]
- Grotewold L, Ruther U. The Wnt antagonist Dickkopf-1 is regulated by Bmp signaling and c-Jun and modulates programmed cell death. Embo J. 2002;21(5):966–975. doi: 10.1093/emboj/21.5.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha U, Gomes WA, Kobayashi T, Pestell RG, Kessler JA. In vivo evidence that BMP signaling is necessary for apoptosis in the mouse limb. Dev Biol. 2002;249(1):108–120. doi: 10.1006/dbio.2002.0752. [DOI] [PubMed] [Google Scholar]
- Guo X, Day TF, Jiang X, Garrett-Beal L, Topol L, Yang Y. Wnt/beta-catenin signaling is sufficient and necessary for synovial joint formation. Genes Dev. 2004;18(19):2404–2417. doi: 10.1101/gad.1230704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann C, Tabin CJ. Dual roles of Wnt signaling during chondrogenesis in the chicken limb. Development. 2000;127(14):3141–3159. doi: 10.1242/dev.127.14.3141. [DOI] [PubMed] [Google Scholar]
- He B, Lee AY, Dadfarmay S, You L, Xu Z, Reguart N, Mazieres J, Mikami I, McCormick F, Jablons DM. Secreted frizzled-related protein 4 is silenced by hypermethylation and induces apoptosis in beta-catenin-deficient human mesothelioma cells. Cancer Res. 2005;65(3):743–748. [PubMed] [Google Scholar]
- He X. A Wnt-Wnt situation. Dev Cell. 2003;4(6):791–797. doi: 10.1016/s1534-5807(03)00165-5. [DOI] [PubMed] [Google Scholar]
- He X, Semenov M, Tamai K, Zeng X. LDL receptor-related proteins 5 and 6 in Wnt/beta-catenin signaling: arrows point the way. Development. 2004;131(8):1663–1677. doi: 10.1242/dev.01117. [DOI] [PubMed] [Google Scholar]
- Hill TP, Spater D, Taketo MM, Birchmeier W, Hartmann C. Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev Cell. 2005;8(5):727–738. doi: 10.1016/j.devcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Hoang BH, Thomas JT, Abdul-Karim FW, Correia KM, Conlon RA, Luyten FP, Ballock RT. Expression pattern of two Frizzled-related genes, Frzb-1 and Sfrp-1, during mouse embryogenesis suggests a role for modulating action of Wnt family members. Dev Dyn. 1998;212(3):364–372. doi: 10.1002/(SICI)1097-0177(199807)212:3<364::AID-AJA4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Johnson D, Kan SH, Oldridge M, Trembath RC, Roche P, Esnouf RM, Giele H, Wilkie AO. Missense mutations in the homeodomain of HOXD13 are associated with brachydactyly types D and E. Am J Hum Genet. 2003;72(4):984–997. doi: 10.1086/374721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SE, Jomary C, Grist J, Stewart HJ, Neal MJ. Modulated expression of secreted frizzled-related proteins in human retinal degeneration. Neuroreport. 2000;11(18):3963–3967. doi: 10.1097/00001756-200012180-00012. [DOI] [PubMed] [Google Scholar]
- Kawano Y, Kypta R. Secreted antagonists of the Wnt signalling pathway. J Cell Sci. 2003;116(Pt 13):2627–2634. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- Kim AS, Lowenstein DH, Pleasure SJ. Wnt receptors and Wnt inhibitors are expressed in gradients in the developing telencephalon. Mech Dev. 2001;103(12):167–172. doi: 10.1016/s0925-4773(01)00342-2. [DOI] [PubMed] [Google Scholar]
- Ko J, Ryu KS, Lee YH, Na DS, Kim YS, Oh YM, Kim IS, Kim JW. Human secreted frizzled-related protein is down-regulated and induces apoptosis in human cervical cancer. Exp Cell Res. 2002;280(2):280–287. doi: 10.1006/excr.2002.5649. [DOI] [PubMed] [Google Scholar]
- Ladher RK, Church VL, Allen S, Robson L, Abdelfattah A, Brown NA, Hattersley G, Rosen V, Luyten FP, Dale L, Francis-West PH. Cloning and expression of the Wnt antagonists Sfrp-2 and Frzb during chick development. Dev Biol. 2000;218(2):183–198. doi: 10.1006/dbio.1999.9586. [DOI] [PubMed] [Google Scholar]
- Lee AY, He B, You L, Dadfarmay S, Xu Z, Mazieres J, Mikami I, McCormick F, Jablons DM. Expression of the secreted frizzled-related protein gene family is downregulated in human mesothelioma. Oncogene. 2004a;23(39):6672–6676. doi: 10.1038/sj.onc.1207881. [DOI] [PubMed] [Google Scholar]
- Lee CS, Buttitta LA, May NR, Kispert A, Fan CM. SHH-N upregulates Sfrp2 to mediate its competitive interaction with WNT1 and WNT4 in the somitic mesoderm. Development. 2000;127(1):109–118. doi: 10.1242/dev.127.1.109. [DOI] [PubMed] [Google Scholar]
- Lee HX, Ambrosio AL, Reversade B, De Robertis EM. Embryonic dorsal-ventral signaling: secreted frizzled-related proteins as inhibitors of tolloid proteinases. Cell. 2006a;124(1):147–159. doi: 10.1016/j.cell.2005.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JL, Chang CJ, Chueh LL, Lin CT. Secreted frizzled related protein 2 (sFRP2) decreases susceptibility to UV-induced apoptosis in primary culture of canine mammary gland tumors by NF-kappaB activation or JNK suppression. Breast cancer research and treatment. 2006b;100(1):49–58. doi: 10.1007/s10549-006-9233-9. [DOI] [PubMed] [Google Scholar]
- Lee JL, Lin CT, Chueh LL, Chang CJ. Autocrine/paracrine secreted Frizzled-related protein 2 induces cellular resistance to apoptosis: a possible mechanism of mammary tumorigenesis. J Biol Chem. 2004b;279(15):14602–14609. doi: 10.1074/jbc.M309008200. [DOI] [PubMed] [Google Scholar]
- Lehmann K, Seemann P, Stricker S, Sammar M, Meyer B, Suring K, Majewski F, Tinschert S, Grzeschik KH, Muller D, Knaus P, Nurnberg P, Mundlos S. Mutations in bone morphogenetic protein receptor 1B cause brachydactyly type A2. Proc Natl Acad Sci U S A. 2003;100(21):12277–12282. doi: 10.1073/pnas.2133476100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leimeister C, Bach A, Gessler M. Developmental expression patterns of mouse sFRP genes encoding members of the secreted frizzled related protein family. Mech Dev. 1998;75(12):29–42. doi: 10.1016/s0925-4773(98)00072-0. [DOI] [PubMed] [Google Scholar]
- Lescher B, Haenig B, Kispert A. sFRP-2 is a target of the Wnt-4 signaling pathway in the developing metanephric kidney. Dev Dyn. 1998;213(4):440–451. doi: 10.1002/(SICI)1097-0177(199812)213:4<440::AID-AJA9>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- McLeod MJ. Differential staining of cartilage and bone in whole mouse fetuses by alcian blue and alizarin red S. Teratology. 1980;22(3):299–301. doi: 10.1002/tera.1420220306. [DOI] [PubMed] [Google Scholar]
- Melkonyan HS, Chang WC, Shapiro JP, Mahadevappa M, Fitzpatrick PA, Kiefer MC, Tomei LD, Umansky SR. SARPs: a family of secreted apoptosis-related proteins. Proc Natl Acad Sci U S A. 1997;94(25):13636–13641. doi: 10.1073/pnas.94.25.13636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikels AJ, Nusse R. Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PLoS biology. 2006a;4(4):e115. doi: 10.1371/journal.pbio.0040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikels AJ, Nusse R. Wnts as ligands: processing, secretion and reception. Oncogene. 2006b;25(57):7461–7468. doi: 10.1038/sj.onc.1210053. [DOI] [PubMed] [Google Scholar]
- Morello R, Zhou G, Dreyer SD, Harvey SJ, Ninomiya Y, Thorner PS, Miner JH, Cole W, Winterpacht A, Zabel B, Oberg KC, Lee B. Regulation of glomerular basement membrane collagen expression by LMX1B contributes to renal disease in nail patella syndrome. Nat Genet. 2001;27(2):205–208. doi: 10.1038/84853. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay M, Shtrom S, Rodriguez-Esteban C, Chen L, Tsukui T, Gomer L, Dorward DW, Glinka A, Grinberg A, Huang SP, Niehrs C, Belmonte JC, Westphal H. Dickkopf1 is required for embryonic head induction and limb morphogenesis in the mouse. Dev Cell. 2001;1(3):423–434. doi: 10.1016/s1534-5807(01)00041-7. [DOI] [PubMed] [Google Scholar]
- Oishi I, Suzuki H, Onishi N, Takada R, Kani S, Ohkawara B, Koshida I, Suzuki K, Yamada G, Schwabe GC, Mundlos S, Shibuya H, Takada S, Minami Y. The receptor tyrosine kinase Ror2 is involved in non-canonical Wnt5a/JNK signalling pathway. Genes Cells. 2003;8(7):645–654. doi: 10.1046/j.1365-2443.2003.00662.x. [DOI] [PubMed] [Google Scholar]
- Oldridge M, Fortuna AM, Maringa M, Propping P, Mansour S, Pollitt C, DeChiara TM, Kimble RB, Valenzuela DM, Yancopoulos GD, Wilkie AO. Dominant mutations in ROR2, encoding an orphan receptor tyrosine kinase, cause brachydactyly type B. Nat Genet. 2000;24(3):275–278. doi: 10.1038/73495. [DOI] [PubMed] [Google Scholar]
- Polinkovsky A, Robin NH, Thomas JT, Irons M, Lynn A, Goodman FR, Reardon W, Kant SG, Brunner HG, van der Burgt I, Chitayat D, McGaughran J, Donnai D, Luyten FP, Warman ML. Mutations in CDMP1 cause autosomal dominant brachydactyly type C. Nat Genet. 1997;17(1):18–19. doi: 10.1038/ng0997-18. [DOI] [PubMed] [Google Scholar]
- Ryu JH, Kim SJ, Kim SH, Oh CD, Hwang SG, Chun CH, Oh SH, Seong JK, Huh TL, Chun JS. Regulation of the chondrocyte phenotype by beta-catenin. Development. 2002;129(23):5541–5550. doi: 10.1242/dev.129.23.5541. [DOI] [PubMed] [Google Scholar]
- Sammar M, Stricker S, Schwabe GC, Sieber C, Hartung A, Hanke M, Oishi I, Pohl J, Minami Y, Sebald W, Mundlos S, Knaus P. Modulation of GDF5/BRI-b signalling through interaction with the tyrosine kinase receptor Ror2. Genes Cells. 2004;9(12):1227–1238. doi: 10.1111/j.1365-2443.2004.00799.x. [DOI] [PubMed] [Google Scholar]
- Satoh W, Gotoh T, Tsunematsu Y, Aizawa S, Shimono A. Sfrp1 and Sfrp2 regulate anteroposterior axis elongation and somite segmentation during mouse embryogenesis. Development. 2006;133(6):989–999. doi: 10.1242/dev.02274. [DOI] [PubMed] [Google Scholar]
- Soshnikova N, Zechner D, Huelsken J, Mishina Y, Behringer RR, Taketo MM, Crenshaw EB, 3rd, Birchmeier W. Genetic interaction between Wnt/beta-catenin and BMP receptor signaling during formation of the AER and the dorsal-ventral axis in the limb. Genes Dev. 2003;17(16):1963–1968. doi: 10.1101/gad.263003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm EE, Kingsley DM. GDF5 coordinates bone and joint formation during digit development. Dev Biol. 1999;209(1):11–27. doi: 10.1006/dbio.1999.9241. [DOI] [PubMed] [Google Scholar]
- Temtamy SA, McKusick VA. The genetics of hand malformations. Birth Defects Orig Artic Ser. 1978;14(3):i–xviii. 1–619. [PubMed] [Google Scholar]
- Topol L, Jiang X, Choi H, Garrett-Beal L, Carolan PJ, Yang Y. Wnt-5a inhibits the canonical Wnt pathway by promoting GSK-3-independent beta-catenin degradation. J Cell Biol. 2003;162(5):899–908. doi: 10.1083/jcb.200303158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugolini F, Charafe-Jauffret E, Bardou VJ, Geneix J, Adelaide J, Labat-Moleur F, Penault-Llorca F, Longy M, Jacquemier J, Birnbaum D, Pebusque MJ. WNT pathway and mammary carcinogenesis: loss of expression of candidate tumor suppressor gene SFRP1 in most invasive carcinomas except of the medullary type. Oncogene. 2001;20(41):5810–5817. doi: 10.1038/sj.onc.1204706. [DOI] [PubMed] [Google Scholar]
- Uren A, Reichsman F, Anest V, Taylor WG, Muraiso K, Bottaro DP, Cumberledge S, Rubin JS. Secreted frizzled-related protein-1 binds directly to Wingless and is a biphasic modulator of Wnt signaling. J Biol Chem. 2000;275(6):4374–4382. doi: 10.1074/jbc.275.6.4374. [DOI] [PubMed] [Google Scholar]
- van Bokhoven H, Celli J, Kayserili H, van Beusekom E, Balci S, Brussel W, Skovby F, Kerr B, Percin EF, Akarsu N, Brunner HG. Mutation of the gene encoding the ROR2 tyrosine kinase causes autosomal recessive Robinow syndrome. Nat Genet. 2000;25(4):423–426. doi: 10.1038/78113. [DOI] [PubMed] [Google Scholar]
- Veeman MT, Axelrod JD, Moon RT. A second canon. Functions and mechanisms of beta-catenin-independent Wnt signaling. Dev Cell. 2003;5(3):367–377. doi: 10.1016/s1534-5807(03)00266-1. [DOI] [PubMed] [Google Scholar]
- Yamaguchi TP, Bradley A, McMahon AP, Jones S. A Wnt5a pathway underlies outgrowth of multiple structures in the vertebrate embryo. Development. 1999;126(6):1211–1223. doi: 10.1242/dev.126.6.1211. [DOI] [PubMed] [Google Scholar]
- Yang Y. Wnts and wing: Wnt signaling in vertebrate limb development and musculoskeletal morphogenesis. Birth Defects Res C Embryo Today. 2003;69(4):305–317. doi: 10.1002/bdrc.10026. [DOI] [PubMed] [Google Scholar]
- Yang Y, Topol L, Lee H, Wu J. Wnt5a and Wnt5b exhibit distinct activities in coordinating chondrocyte proliferation and differentiation. Development. 2003;130(5):1003–1015. doi: 10.1242/dev.00324. [DOI] [PubMed] [Google Scholar]
- Yates KE, Shortkroff S, Reish RG. Wnt influence on chondrocyte differentiation and cartilage function. DNA Cell Biol. 2005;24(7):446–457. doi: 10.1089/dna.2005.24.446. [DOI] [PubMed] [Google Scholar]
- Yoshino K, Rubin JS, Higinbotham KG, Uren A, Anest V, Plisov SY, Perantoni AO. Secreted Frizzled-related proteins can regulate metanephric development. Mech Dev. 2001;102(12):45–55. doi: 10.1016/s0925-4773(01)00282-9. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Wang J, Han X, Zhou J, Linder S. Up-regulation of human secreted frizzled homolog in apoptosis and its down-regulation in breast tumors. Int J Cancer. 1998;78(1):95–99. doi: 10.1002/(sici)1097-0215(19980925)78:1<95::aid-ijc15>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A,B,C,D) mRNA in situ hybridization using a Gdf5 antisense probe (specific signal was pseudo colored in yellow) at E13.5 and E15.5 showing a comparable level of expression between Sfrp2-/- and WT control mice. (E,F) In situ hybridization using the Sfrp2 antisense probe confirmed absence of Sfrp2 expression in the Sfrp2 null mice.
mRNA in situ hybridization using antisense probes for Bmp2, Bmp4 and Bmp7 on hind limb autopod sections obtained from E13.5 129Sv embryo pups. No significant differences were noted between Sfrp2-/- and control mice.