1. Summary
Although anti-DNA antibodies have been decisively linked to the pathogenesis of lupus nephritis, the mechanisms have not been conclusively determined. Recently, we reported that anti-DNA antibodies may contribute to kidney damage by upregulation of proinflammatory genes in mesangial cells (MC), a process involving both Fc receptor dependent and independent pathways. In investigating the mechanism by which pathogenic anti-DNA antibodies modulate gene expression in MC, we found that the pathogenic anti-DNA antibody 1A3F bound to high mobility group binding protein 1 (HMGB1), an endogenous ligand for TLR2/4 and RAGE (receptor for advanced glycation end-products). Interestingly, HMGB1 treatment of MC induced a similar pattern of genes as stimulation with 1A3F. Furthermore, HMGB1 and 1A3F exhibited a synergistic proinflammatory effect in the kidney, where increased expression of HMGB1 was found in lupus patients but not in patients with other types of renal disease. TLR2/Fc and RAGE/Fc inhibited the proinflammatory effects of 1A3F on MC. Finally, we found enhanced susceptibility of lupus prone MRL-lpr/lpr (MRL/lpr) as compared to normal BALB/c derived MC to pathogenic anti-DNA antibody and LPS stimulation (in particular enhanced chemokine synthesis), in addition to significantly increased expression of TLR4. Our results suggest that gene upregulation in MC induced by nephritogenic anti-DNA antibodies is TLR2/4 and RAGE dependent. Finally, HMGB1 may act as a proinflammatory mediator in antibody induced kidney damage in systemic lupus erythematosus (SLE).
Keywords: Systemic Lupus Erythematosus, Autoantibodies, Inflammation
2. Introduction
One characteristic serologic feature of SLE is the presence of high titers of anti-nuclear antibodies, including antibodies to double stranded (ds) DNA. Elevated serum anti-dsDNA autoantibodies are often associated with kidney damage, or lupus nephritis [1,2]. Characterization of the antigenic specificity of anti-dsDNA antibodies indicates that reactivity with renal antigens is strongly predictive for their renal pathogenicity, or nephritogenicity [3]. Although immunoglobulin deposition in kidney glomeruli is a common feature in lupus nephritis, it still remains unclear how nephritogenic anti-dsDNA antibodies interact with kidney cells and result in injury.
Involvement of Fc receptors in antibody mediated nephritis has been intensively studied. In the murine nephrotoxic serum nephritis model as well as in NZB × NZW F1 mice which develop spontaneous lupus, signaling through Fc receptors is essential in activating infiltrating immune cells, while Fc-independent mechanisms participate in the interactions between the pathogenic antibodies and the resident kidney cells (mesangial cells and podocytes) [4–8]. In antibody induced nephritis, uncomplexed pathogenic anti-DNA or anti-glomerular basement membrane Abs alone, however, only produce moderate kidney damage. Fully penetrant disease often requires co-administration of adjuvants, indicating that activation of innate immunity mediators, especially toll-like receptors (TLR), may play an essential role in boosting the effect of pathogenic antibodies [5]. Recent studies investigating the pathogenesis of SLE have focused on the specific roles of TLR7 and TLR9, since these receptors recognize the same ligands (RNA or DNA related antigens) as lupus autoantibodies. B cells and dendritic cells can be activated by chromatin/ribonucleoprotein loaded antibodies, via co-stimulation of FcR and TLR9/7, respectively [9–11]. More recently, a duplicate TLR7 gene was determined to be responsible for the Yaa locus associated lupus manifestations in male BXSB mice [12,13]. The role of other TLRs in the pathogenesis of lupus and lupus nephritis has been less well investigated, and is a subject of great interest [14).
Besides microbial components, TLR ligands may arise endogenously from tissue injury and inflammation, apoptosis or necrosis of host cells [15]. A number of endogenous activation ligands have been identified for TLR2, TLR3, TLR4, TLR7 and TLR9 [9,16–18]. One such endogenous ligand for TLR2/4 is high mobility group binding protein 1 (HMGB1) [19,20]. HMGB1 belongs to a group of non-histone DNA binding proteins that can be passively released from the nucleus of necrotic or damaged cells [21]. In addition, activated monocytes/macrophages can actively secrete HMGB1 [22], which itself further induces the secretion of proinflammatory cytokines and chemokines. HMGB1 is a late mediator of endotoxin lethality via its receptors TLR2/4 or RAGE [23,24], while neutralizing HMGB1 ameliorates endotoxin induced acute inflammatory lung injury [25]. Increased expression of HMGB1 is found in target tissues, such as the skin lesions of lupus patients and the salivary glands of patients with Sjögren’s syndrome [26,27], while the translocation of HMGB1 to the cytoplasm and extracellular space was found to coincide with the peak of clinical disease in experimentally induced lesions of cutanteous lupus [28). Moreover, high titers of antibodies against HMGB proteins have been reported in patients with autoimmune diseases, including SLE [29–32]. These findings suggest a potential role of HMGB1 in the development of tissue damage in SLE.
Previously, we demonstrated that nephritogenic anti-DNA antibodies modulate gene expression in kidney MC [33]. In the current study, we further address the mechanism of gene regulation by pathogenic antibodies in MC. We investigate the role of HMGB1 and it’s receptors in antibody-MC interactions, and study the potential role of genetically determined susceptibility in this process.
3. Materials and Methods
3.1. Antibodies
1A3F is an IgG2a anti-dsDNA mAb derived from B6.Sle1 mice. The proinflammatory effects of 1A3F on MC are Fc dependent [33]. ZA8A3 is an IgG2a anti-nuclear negative mAb isolated from NZM 2410 mice [3]. Hybridoma cell lines were cultured in serum-free mAb medium and maintained in two-chamber flasks for mAb production (BD Bioscience, San Jose, CA). Supernatants from the hybridoma cultures were collected and the mAbs were purified using the Protein A based Montage mAb purification kit (Millipore, Billerica, MA).
A murine monoclonal anti-HMGB1 antibody (HAP46.5 (IgG1), Sigma, St. Louis, MO) was labeled with biotin using the EZ-link NHS-biotin reagent kit (Pierce, Rockford, IL).
3.2. Cells and Cell Culture
Primary MCs derived from 3–4 week old female MRL/lpr and BALB/c mice were isolated as previously described [33]. MC were maintained in culture in DMEM medium supplemented with amino acids, L-glutamine, sodium pyruvate and 20% FCS, at 37°C/5% CO2.
3.3. Reagents
Benzonase endonuclease was purchased from Novagen (Madison, WI). Recombinant human HMGB1 was purchased from Sigma. Results were confirmed with an additional HMGB1 preparation from ProteinOne (Bethesda, MD). HMGB1 is highly conserved, with more than 95% amino acid identity between human and rodent forms. Mouse recombinant RAGE/Fc and TLR2/Fc fusion proteins were purchased from R&D Systems (Minneapolis, MN).
All antibodies and reagents used for this study were tested for endotoxin contamination by quantitative chromogenic limulus amebocyte lysate assay (LAL) using the QCL-1000 kit from Biowhittaker (Walkersville, MD), and confirmed to contain less than 0.1 EU/ml.
3.4. Mice and injection protocol
Female 4–6 week old BALB/c mice were purchased from the Jackson Laboratory (Bar Harbor, ME). Mice were housed in the animal facility of the Albert Einstein College of Medicine. All studies were approved by the Albert Einstein College of Medicine Institute for Animal Studies. Mice were injected intravenously with a single injection of 300 µg purified antibody and/or intraperitoneally with 10 µg of HMGB1, with 4 mice in each group. Mice were sacrificed 24 hours after the injection.
3.5. Human kidney samples
Kidney needle biopsies (performed for clinical indications) were obtained from SLE or control renal disease patients, hospitalized at the Jacobi and Montefiore Medical Centers affiliated with Albert Einstein College of Medicine. Left-over tissue, following all necessary histopathological analyses, was used for this study, which was approved by the Committee on Clinical Investigations of the Albert Einstein College of Medicine. All lupus patients in the study fulfilled at least 4 of the 1982 revised American College of Rheumatology criteria for the diagnosis of SLE [34]. Normal kidney tissue (confirmed histologically) from six individuals was obtained from the National Disease Research Interchange tissue bank (Philadelphia, PA), and used as normal controls.
3.6. Real-time RT-PCR
Total RNA was extracted from treated cells using the RNeasy isolation kit (Qiagen, Valencia, CA), or from kidneys using Trizol (Invitrogen, Carlsbad, CA). PCR primers were designed using the PRIMER3 program (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi), based on published sequence data from the Ensembl database (http://www.ensembl.org/Mus_musculus/). At least one intron was included to avoid genomic DNA amplification. Amplicons ranged from 80 to 120 bp. Total RNA was reverse transcribed, and real-time PCR performed in triplicate by the SYBR green method and the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems, Warrington, UK), using the following conditions: 10 min at 95°C, and 40 cycles of 95°C for 10 s, 60°C for 20 s and 72°C for 30 s. The expression of each gene was normalized to 2 control genes.
3.7. PCR amplification
Mouse whole blood was used for isolating genomic DNA with a QIAGEN DNeasy blood & tissue kit (Qiagen, Valencia, CA). The murine complement receptor related protein (Crry) gene was used for PCR amplification. The primer sequences for Crry were:
forward: 5’CGCAGAATTCAATCTCTTTTCTTTGCCCAG-3’;
reverse: 5’TTGAGTTCAATGCACTGAGGAGG-3’.
PCR was performed using the following conditions: 94°C for 3 min, 35 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 45 s, and 72°C for 10 min. The size of amplicon is about 400 bp. PCR products, and in some cases, genomic DNA or antibody alone, were analyzed by running on agarose gels containing ethidium bromide.
3.8. Western blotting
HMGB1, mAb 1A3F and ZA8A3 were pre-treated with DNase I at 37°C for 30 min before Western blotting. Removal of DNA was confirmed by running the samples on an ethidium bromide stained agarose gel. HMGB1 was run in a 10–20% Tris-Glycine precast gel (Bio-Rad) in SDS Tris-Glycine buffer. Samples were transferred to a PVDF Immobilon-P membrane (Millipore) using a Criterion Trans-Blot Cell (Bio-Rad). The membrane was first blocked in 10% milk in PBS/Tween 20 for 3 h at room temperature (RT), and then incubated with antibody at 1 µg/ml for 1 h at RT. After repeated washes with PBS/Tween 20, horseradish peroxidase (HRP) conjugated secondary antibody goat anti-mouse IgG was applied at a 1:10000 dilution for 45 min at RT. The membrane was then thoroughly washed and developed with the ECL Western blotting detection kit, and exposed to Hyperfilm (GE Healthcare, Piscataway, NJ).
For studying RAGE expression, total cell lysates from MRL/lpr mesangial cells, B6 mouse lung and spleen were loaded, separated by SDS-PAGE, and transferred to a PVDF membrane as above. The membrane was blocked in 10% milk in PBS/Tween 20 for 3 h at RT, and then incubated with a rat anti-RAGE mAb (R&D Systems) at 5 µg/ml at 4°C overnight. After repeated washes with PBS/Tween 20, HRP conjugated goat anti-rat IgG was applied at a 1:8000 dilution for 45 min at RT. The membrane was then thoroughly washed and developed with the ECL Western blotting detection kit, and exposed to Hyperfilm.
3.9. Immunohistochemistry
Kidneys were fixed with formalin and embedded in paraffin. After deparaffinization, rehydration and antigen retrieval, four µm sections were first blocked with 2% BSA in PBS and avidin/biotin solution (Vector Laboratories, Burlingame, CA). Biotin labeled anti-HMGB1 was applied at 10 µg/ml at RT for 2 h, and developed with the Vectastain ABC kit (Vector Laboratories). Sections were then counterstained with Mayer’s hematoxylin (Sigma), dehydrated, and mounted with Permount (Fisher, Fairlawn, NJ) and coverslips.
3.10. ELISA
For detection of HMGB1 by ELISA, 96 well plates were coated with the HAP46.5 anti-HMGB1 mAb at 2 µg/ml overnight at 4°C. After blocking with 3% FCS, plates were incubated with samples at 37°C for 2 h. After repeated washes, plates were incubated with biotin labeled anti-HMGB1 antibody at 1 µg/ml for 1 h, followed by a 1:1000 dilution of streptavidin conjugated with alkaline phosphatase. Plates were developed with phosphatase substrate (Sigma) and read at OD 405 nm. CXCL1/KC ELISA was performed using a mouse KC Quantikine ELISA kit according to the manufacturer’s instructions (R&D Systems).
For determining mAb binding to HMGB1, 96 well plates were coated with recombinant HMGB1 at 5 µg/ml overnight at 4°C. For the dsDNA ELISA, plates were coated with 100 µg/ml of salmon sperm dsDNA at 37°C overnight. For mAb binding to RAGE and TLR2, plates were coated with RAGE/Fc or TLR2/Fc at 10 µg/ml overnight at 4°C. After blocking with 3% FCS, plates were incubated with the samples (mAbs) at 37°C for 2 h. After repeated washes, plates were incubated with a 1:1000 dilution of secondary alkaline phosphatase conjugated goat anti-mouse IgG antibody. Plates were developed with phosphatase substrate and read at OD 405 nm.
3.11. DNA microarray and data analysis
Total RNA was extracted from antibody treated cells using the RNeasy Isolation kit (Qiagen). Total RNA (70 ng) was then reverse transcribed to cDNA, labeled with biotin and fragmented using the NuGEN biotin system (NuGEN, San Carlos, CA) for hybridization on Affymetrix mouse genome genechips 430A 2.0 (Affymetrix, Santa Clara, CA), containing 22,600 probes representing over 14,000 well characterized genes. Genechip data was analyzed with Arrayassist 3.0 (Stratagene, La Jolla, CA). Briefly, raw data was normalized using the GC-RMA method and transformed into log2. Criteria for selecting differentially expressed genes were as follows: signal present in at least 6 out of 12 samples, fold change more than 2, and p-value < 0.05 (t-test).
3.12. Flow cytometry
Cells were detached from tissue culture plates with 2 mM EDTA, pelleted, and washed with medium. Cells were resuspended, and blocked with 1% BSA/PBS for 30 min. For detection of TLR2 and TLR4, cells were Fc-blocked with anti-mouse CD16/CD32 (FcãRIII/II; BD Pharmingen, San Diego, CA) at 40 µg/ml for 1 h at 4°C, followed by an anti-TLR2 or anti-TLR4 antibody (eBiosciences) at the appropriate concentration for 1 h at 4°C. Appropriate fluorochrome-conjugated secondary Ab was added for 30 min at 4°C. Cells were washed, resuspended in 1% paraformaldehyde and analyzed on a Beckon Dickinson Calibur flow cytometry instrument (BD Immunocytometry Systems, Mountain View, CA).
3.13. Statistical analysis
All results are expressed as mean ± standard deviation (SD). Data were first tested for normality. Student’s t-test (two-tailed) was then applied. The nonparametric Mann-Whitney U test was used for comparison of HMGB1 staining between lupus patients and the control group. A probability of <0.05 was assigned to reject the null hypothesis.
4. Results
4.1. Pathogenic anti-DNA antibodies bind to HMGB1
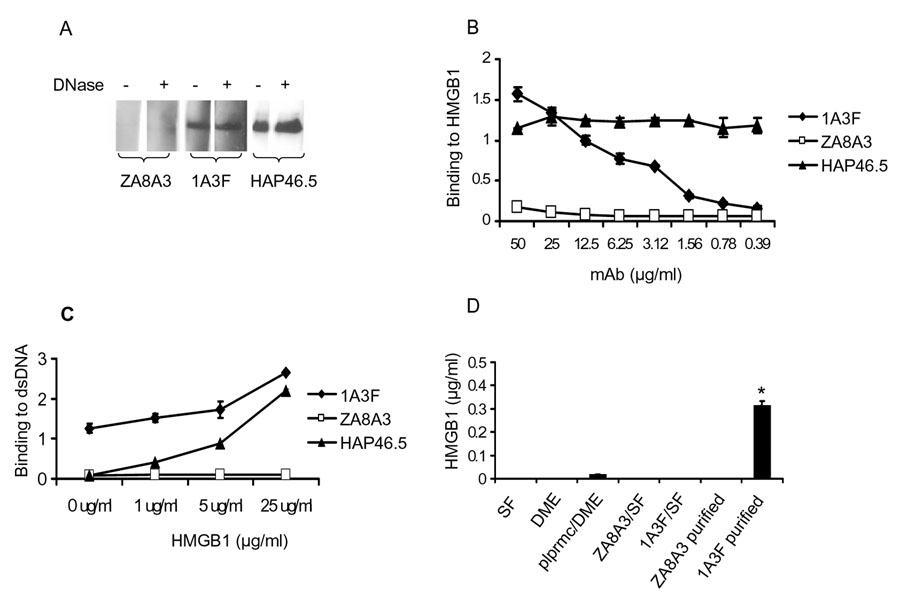
Previously, we and others have identified α-actinin as a cross-reactive antigen for pathogenic anti-DNA antibodies [35–40]. In more recent studies we found that mice immunized with α-actinin produce anti-chromatin antibodies. Further analysis revealed that this anti-chromatin response specifically targeted the chromatin associated protein HMGB1 [41]. As mentioned above, HMGB1 is an endogenous ligand for TLR2/4 and RAGE, and anti-HMGB antibodies have been reported in lupus patients. Therefore, we examined whether pathogenic anti-DNA antibodies mediate their proinflammatory effects via binding to HMGB1. We found that the pathogenic anti-DNA mAb 1A3F, but not the non-pathogenic isotype matched ZA8A3 mAb, binds strongly to HMGB1 (Figure 1A). Moreover, removal of DNA by DNase I treatment did not decrease the binding of 1A3F to HMGB1, indicating that the binding is not via a DNA bridge (Figure 1A). The binding of 1A3F to HMGB1 was dose dependent, as quantitatively measured by ELISA (Figure 1B). Furthermore, when preincubated with HMGB1, the binding reactivity of 1A3F to dsDNA was significantly enhanced (Figure 1C), suggesting that HMGB1 may act as a linker between this antibody and the DNA antigen.
Fig. 1.
Pathogenic anti-DNA antibodies bind to HMGB1. (A) HMGB1 was electrophoresed in a 10–20% Tris-Glycine gel, transferred to a PVDF membrane, and immunoblotted with the DNAse pretreated (+) or DNAse untreated (−) monoclonal antibodies indicated in the figure. HMGB1, ZA8A3, 1A3F and the HAP46.5 anti-HMGB1 mAb were pre-treated with DNase I at 37°C for 30 min before blotting. Removal of DNA was confirmed by running the samples on an ethidium bromide stained agarose gel. HMGB1 was blotted as a single band at ~31 kD. (B) Dose dependent binding of 1A3F to HMGB1 as shown by ELISA. Plates coated with HMGB1 (5 µg/ml) were incubated with serially diluted mAb starting at a concentration of 50 µg/ml. ZA8A3 is an anti-DNA negative, non-pathogenic control mAb. The HAP46.5 anti-HMGB1 mAb was used as a positive control. (C) The binding of 1A3F and HAP46.5 to dsDNA is enhanced by pre-incubating the antibody (5 µg/ml) with HMGB1 (1, 5, and 25 µg/ml) at 37°C for 1 h. (D) Measurement of HMGB1 content in the culture medium and purified antibodies by ELISA, as described in Materials and Methods. SF, serum-free mAb culture medium; DME, Dulbecco's modified Eagles medium; plprmc, primary MRL/lpr MCs; ZA8A3/1A3F SF, unpurified hybdridoma culture supernatant; ZA8A3/1A3F purified, affinity purified mAb (* p<0.01). Values on the y-axis in B to D are optical density units @405 nm, and the graphs depict the mean and SD.
Since HMGB1 can be released from damaged cells or secreted from activated lymphoid cells, it is possible that pathogenic antibodies can pick up free HMGB1 in cell culture medium (or in vivo). We then tested the HMGB1 content in our antibodies. Although HMGB1 was not detectable in the culture medium either from mAb hybridomas or MC, purified 1A3F, but not non-pathogenic ZA8A3, contains small amounts of HMGB1 (Figure 1D).
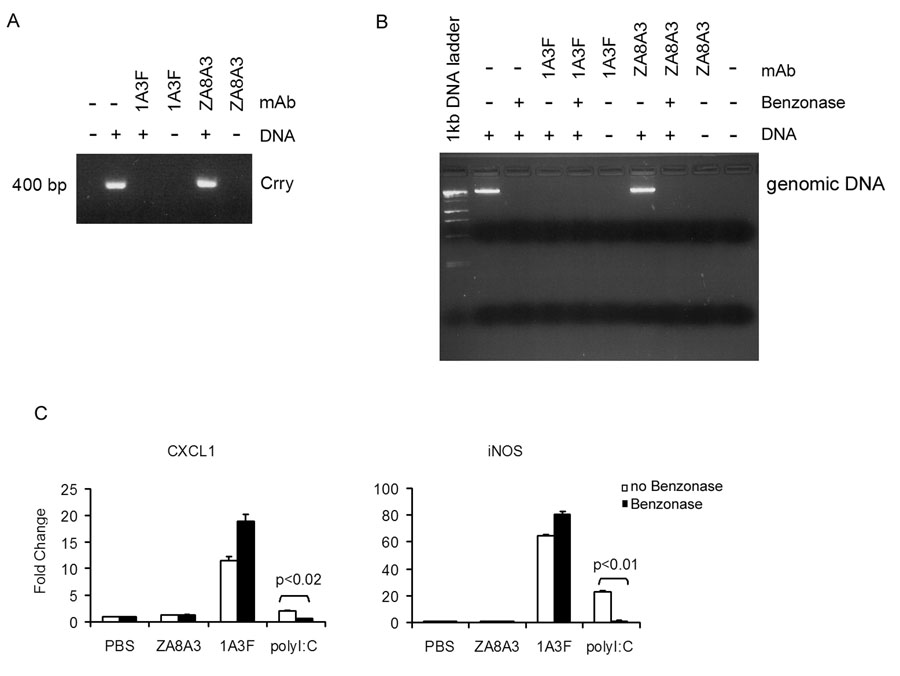
Next, we determined if there is a possible role for bound DNA in the stimulatory effect of 1A3F on MC. It has been reported that anti-DNA Abs can protect DNA bound in the binding site from DNase digestion [42]. To test if there is DNA contained in the 1A3F binding site, we used two approaches: PCR amplification and direct staining for DNA with ethidium bromide. First, a PCR amplification was performed with the purified antibodies as a template, using primers for a randomly selected gene, that encoding complement receptor related protein (Crry). The PCR reaction amplified a band ~400 bp from mouse whole blood genomic DNA (Figure 2A); however, no Crry product was amplified using the antibody 1A3F or ZA8A3. Moreover, when genomic DNA was preincubated with 1A3F, but not the control antibody ZA8A3, there was no amplification as well, suggesting either that the PCR reaction was blocked in the presence of 1A3F or that DNA was degraded by the antibody (Figure 2A). We then examined whether benzonase, an endonuclease that degrades all forms of nucleic acids (single and double stranded RNA and DNA), can degrade DNA in the presence of 1A3F. Using direct staining for DNA with ethidium bromide, we found that DNA preincubated with 1A3F was still degraded by benzonase (Figure 2B). Moreover, DNA incubated with 1A3F alone (without benzonase) was also degraded, indicating that 1A3F contains DNase activity, a property previously observed in some other anti-DNA antibodies [43]. Finally, we found that the stimulatory effect of 1A3F in MC did not decrease following benzonase treatment (Figure 2C). Taken together, these studies failed to demonstrate DNA contained in the purified antibody preparations. Moreover, the gene induction effect by the 1A3F pathogenic antibody is not likely to be mediated by DNA.
Fig. 2.
The 1A3F antibody preparation does not contain DNA. (A) PCR amplification of potential DNA contained in anti-DNA mAb 1A3F. Genomic DNA (0.25 µg) from mouse whole blood, either alone or preincubated with 1A3F or the control mAb ZA8A3 (10 µg/ml) at 37°C for 30 min, was subjected to PCR amplification using primers for the murine gene for complement receptor related protein Crry. The PCR products were analyzed on a 2% agarose gel. (B) 1A3F does not protect DNA from digestion by benzonase. Mouse genomic DNA (0.5 µg) preincubated with or without 1A3F or control mAb was further treated with benzonase (12.5 U) at 37°C for 2 h. The reaction mixtures were analyzed on agarose gels stained with ethidium bromide. (C) 1A3F, ZA8A3 and poly I:C (as a positive control) were pre-treated with benzonase endonuclease for 24 h to degrade all forms of nucleic acids before incubation with MC. Gene expression was then measured by real-time PCR in MCs incubated with antibodies for 6 h. Data displayed here shows one representative experiment of at least 3 independent repeats with similar results. Relative mean fold changes and SD are shown.
4.2. Pathogenic anti-DNA antibodies synergize with HMGB1 in the induction of kidney proinflammatory responses
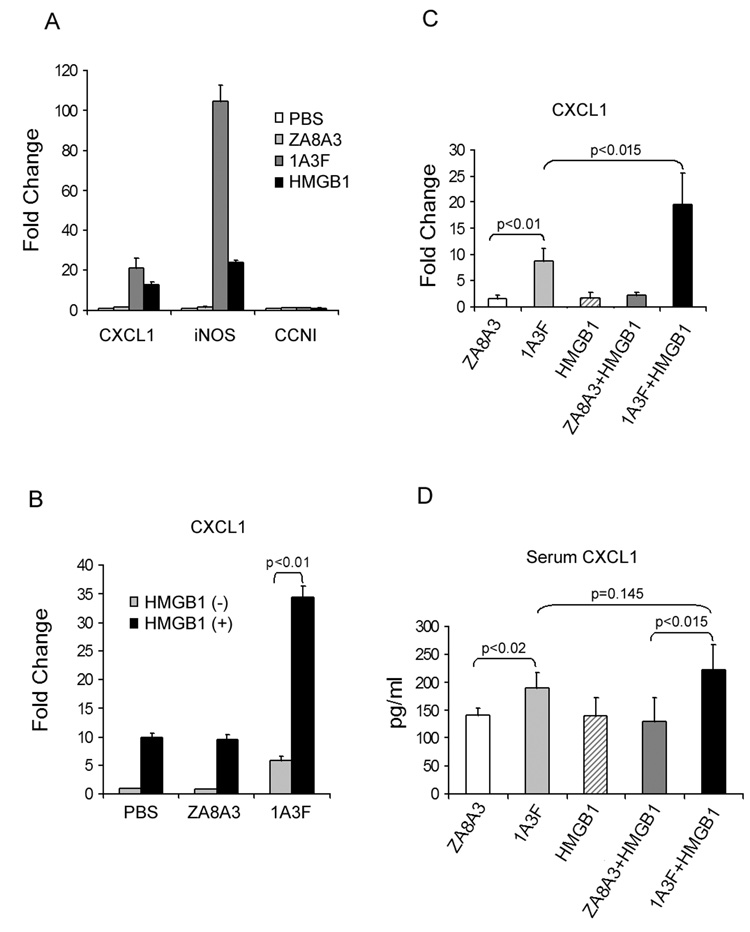
Given that pathogenic anti-DNA antibodies bind to HMGB1 and stimulate a proinflammatory response in MC and that this effect did not appear DNA dependent, we theorized that the stimulatory effect by 1A3F might be mediated via its binding to HMGB1. Indeed, HMGB1 treatment also induced upregulation of many genes in MC, including CXCL1 and iNOS, which were also activated by the pathogenic antibody 1A3F (Figure 3A). Furthermore, there was a synergistic effect when cells were treated with 1A3F together with HMGB1, as demonstrated by the upregulation of CXCL1 (Figure 3B).
Fig. 3.
Pathogenic anti-DNA antibody synergizes with HMGB1 in the induction of proinflammatory responses in kidney cells. MRL/lpr MCs were incubated with HMGB1 (10 µg/ml) or mAb (50 µg/ml), alone (A) or together (B), for 6 h before gene expression was measured by real-time PCR. The gene for Cyclin I (CCNI) was used in A as a control. C, BALB/c mice were injected with 10 µg of HMGB1 intraperitoneally and/or 300 µg of mAb intravenously, 24 h before the kidneys were removed (4 mice per group). Gene induction in the treated cells (A, B) or kidneys (C) was analyzed by real-time PCR. Mean fold changes and SD are shown. D, Serum CXCL1 was measured by ELISA in mice injected with antibody, HMGB1, or both. Values shown represent the mean and SD.
The synergy between pathogenic antibody and HMGB1 was then confirmed in vivo (Figure 3C and 3D). BALB/c mice injected with 1A3F and HMGB1, either alone or in combination, were examined for kidney chemokine expression by real-time PCR. Although mice receiving HMGB1 alone did not show significant gene upregulation in the kidneys, an increase in CXCL1 (p<0.015) was found when HMGB1 was co-administered with the pathogenic antibody 1A3F, but not with the control antibody ZA8A3 (Figure 3C). Moreover, CXCL1 was elevated in the serum from mice injected with 1A3F alone (190±27 pg/ml vs 140±13 pg/ml in mice injected with ZA8A3, p<0.02) or together with HMGB1 (222±45 pg/ml vs 129±44 pg/ml in mice receiving ZA8A3+HMGB1, p<0.015). Serum CXCL1 levels were higher in mice receiving the combined injection of 1A3F+HMGB1 as compared with 1A3F alone, although the difference did not reach statistical significance (222±45 pg/ml vs. 190±27 pg/ml, p=0.145) (Figure 3D). No significant increase in proteinuria in injected mice was detected, presumably since the mice received a low dose of antibody and were sacrificed after a relatively short time (24 h). The degree of glomerular immunoglobulin deposition was similar in mice receiving 1A3F alone, or together with HMGB1 (data not shown).
4.3. Activation of MC by HMGB1 and 1A3F engages TLR2, RAGE, and MyD88
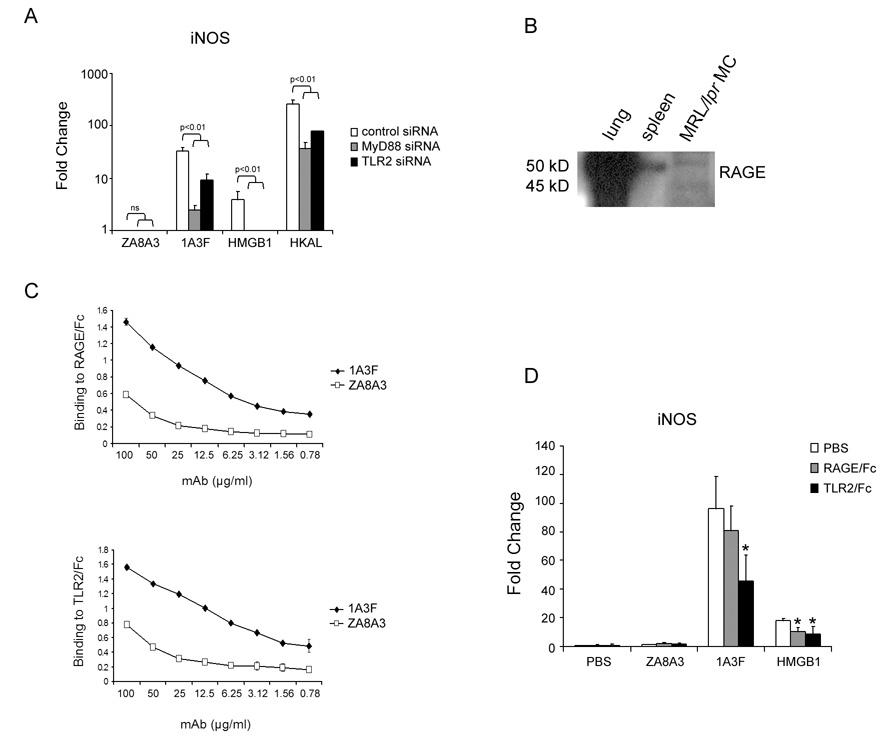
Studies by Park, Kokkola, and colleagues indicate that HMGB1 interacts with multiple receptors including TLR2, TLR4 and RAGE [20,24]. Therefore, if pathogenic antibodies act to upregulate gene expression in MC through HMGB1, blockade of any of these HMGB1 receptors or their downstream signaling adaptors should prevent activation of MC. To test this hypothesis, we first adopted a siRNA technique to knock down the expression of TLR2 as well as MyD88, a crucial signaling adaptor in TLR signaling, prior to antibody stimulation. Confirming successful siRNA transfection was our demonstration that downregulation of TLR2 and MyD88 significantly reduced the level of inflammatory gene responses in MC induced by heat killed Acholeplasma laidlawii (HKAL), a ligand for TLR2 [44] (Figure 4A). We found that in TLR2 or MyD88 siRNA transfected MCs, upregulation of iNOS induced by 1A3F or HMGB1 was significantly reduced (Figure 4A). Unfortunately, we were not successful in knocking down TLR4 using a siRNA approach, while TLR4/Fc was not available to us.
Fig. 4.
TLR2, RAGE, and MyD88 are involved in MC stimulation by HMGB1 and pathogenic anti-DNA antibody. (A) Transfection of MRL/lpr MCs with siRNA targeting at TLR2, MyD88, or a control sequence. Forty-eight hours after transfection, cells were treated with 50 µg/ml of antibody (1A3F and ZA8A3), 10 µg/ml of HMGB1, or 107 cells/ml of HKAL (TLR2 ligand) for 6 h. Transcript levels of iNOS induced in the treated cells were measured by real-time PCR. Data displayed here shows one representative experiment of at least 3 independent repeats with similar results. Mean and SD are shown. (B) MRL/lpr MC express the HMGB1 receptor RAGE. Lysates from C57Bl/6 mouse lung and spleen tissues and MRL/lpr mesangial cells were separated by SDS-PAGE and blotted with rat anti-mouse RAGE antibody. RAGE expression was detected as a band ~50 kD. In lung tissue and MCs, an additional band at ~45 kD was detected. (C) Binding of 1A3F to RAGE and TLR2, as demonstrated by ELISA. Plates were coated with RAGE/Fc or TLR2/Fc (10 µg/ml). Serial dilutions of antibody beginning from 50 µg/ml were applied, and antibody binding detected with goat anti-mouse IgG labeled with alkaline phosphatase. Data displayed here shows one representative experiment of 3 independent repeats with similar results. Values on the y-axis in this Figure are optical density units @405 nm, and the graphs depict the mean and SD. (D) Pathogenic anti-DNA mAb induced gene expression in MCs can be inhibited by blocking antibody binding to RAGE and TLR2. MCs were preincubated with culture medium containing PBS (control), RAGE/Fc or TLR2/Fc at 10 µg/ml for 1 h. PBS, 1A3F (100 µg/ml), or HMGB1 (10 µg/ml) was then added to the culture for 6 h. Real-time PCR was performed to analyze the expression of iNOS in treated MCs. Data displayed here shows one representative experiment of 3 independent repeats with similar results. Mean and SD are shown (* p<0.05 vs. PBS treatment).
Next, the role of RAGE in anti-DNA antibody mediated MC stimulation was further investigated. First, we found expression of RAGE in MCs, which showed two bands at approximately 45 kD and 50 kD (corresponding to the non-glycosylated and glycosylated forms of the protein). The control B6 mouse lung tissue strongly expressed RAGE, while spleen only demonstrated a weak band at 50 kD (Figure 4B). Second, 1A3F, but not the non-pathogenic ZA8A3, binds to both RAGE/Fc and TLR2/Fc as measured by ELISA (Figure 4C). This is consistent with the 1A3F mAb interacting with RAGE and TLR2 via its bound HMGB1. Finally, pre-incubation of MCs with the recombinant fusion proteins RAGE/Fc or TLR2/Fc substantially inhibited gene induction by 1A3F (Figure 4D). These studies strongly support a role for HMGB1 signaling via TLR2, RAGE, and MyD88 in MC stimulation by pathogenic anti-DNA antibodies.
We demonstrated above that 1A3F binds to RAGE-Fc and TLR2-Fc. To confirm that this binding was specific, we further investigated the antigenic specificities of 1A3F using 70 common self and foreign antigens in a multiplex proteome assay [45]. While in this assay as well we confirmed that 1A3F binds to nuclear antigens including chromatin and dsDNA, no significant binding was demonstrated to multiple other antigens, including cardiolipin, collagen II, collagen IV, elastin, and vimentin. However, 1A3F did bind to the non-HMGB1 associated, non-nuclear antigen heparan sulfate (data not shown). Therefore, we believe that the binding of 1A3F to RAGE and TLR2 is specific; furthermore, clearly at least some of the reactivity of 1A3F with autoantigens is not mediated by HMGB1 binding.
4.4. HMGB1 is overexpressed in lupus kidneys
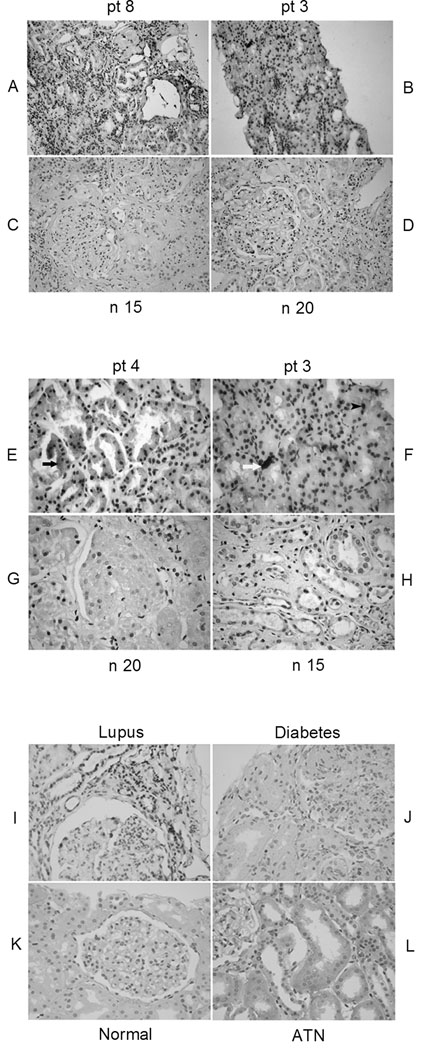
To ascertain whether HMGB1 is released or accumulated at the sites of organ damage, we performed immunohistological staining for HMGB1 in the kidney tissues from lupus patients. (It was not possible to study murine lupus kidneys due to unacceptably high background staining with the available anti-mouse HMGB1 antibody). In renal biopsy sections from patients with active lupus glomerulonephritis (WHO classes III, IV, and V) increased expression of HMGB1 was prominent (Figure 5 A, B, E, F), in contrast to the minimal staining observed in normal individuals (Figure 5 C, D, G, H). Staining of HMGB1 in lupus kidneys was observed in glomerular cells, tubular cells, as well as infiltrates of mononuclear cells. Increased HMGB1 staining was seen both in the nucleus, as well as in the cytoplasm. In some areas, there was also extra-cellular expression of HMGB1 (Figure 5). Eight of 12 lupus patients showed overexpression of HMGB1, while only one of six samples was weakly positive in normal individuals (p<0.01). Kidneys from patients with diabetic nephropathy and acute tubular necrosis demonstrated only very low to absent expression of HMGB1 (Figure 5 I–L), indicating that overexpression of HMGB1 in kidney is not a universal finding or a non-specific marker of kidney disease. It is important to note that while no staining was observed by immunohistochemistry in normal kidney tissue using the HAP46.5 anti-HMGB1 mAb, the same antibody stained strongly mesangial cells or kidney tissue lysates by Western blot (data not shown). Therefore, this particular HMGB1 antibody does detect the protein in normal kidney, but the level of expression is not sufficient to be detected by immunohistochemistry on intact normal tissue (at least relatively to lupus kidneys with active nephritis). Finally, HMGB1 was not detected in serum from active and inactive lupus patients, or in normal controls (data not shown).
Fig. 5.
Increased expression of HMGB1 in kidneys of lupus patients. Representative immunohistochemical staining of HMGB1 in the kidney from three lupus patients with WHO class III (patient 4), IV (patient 8), and V (patient 3) nephritis (A, B, E, F) and normal controls (C, D, G, H). Intrinsic renal cells and infiltrates of mononuclear cells from lupus patients exhibited significantly increased cytoplasmic (black arrow) (E), nuclear (black arrow head) (F) and extracellular (white arrow) (F) expression of HMGB1. pt, patient; n, normal control. (Magnification, A-D, 10x; E-L, 40×). (I,J,K,L) Immunohistochemistry analysis of HMGB1 expression in non-SLE diseased kidneys compared to lupus kidneys with nephritis. Kidney paraffin sections from normals, patients with lupus nephritis (Class IV), diabetic nephropathy, and acute tubular necrosis (2 subjects in each group) were stained for the expression of HMGB1 using biotin labeled anti-HMGB1 antibody. HMGB1 staining appears as a dark brown color. Sections were counter stained with hematoxylin in blue. The kidneys from patients with lupus nephritis exhibited strong staining for HMGB1; kidneys with acute tubular necrosis (ATN) showed only minimal HMGB1 expression in the cytoplasm of tubular cells. Both diabetic and normal kidneys are negative for extranuclear HMGB1 staining. Shown here are representative images from each disease category.
4.5. Genetic background affects the renal response to pathogenic antibody
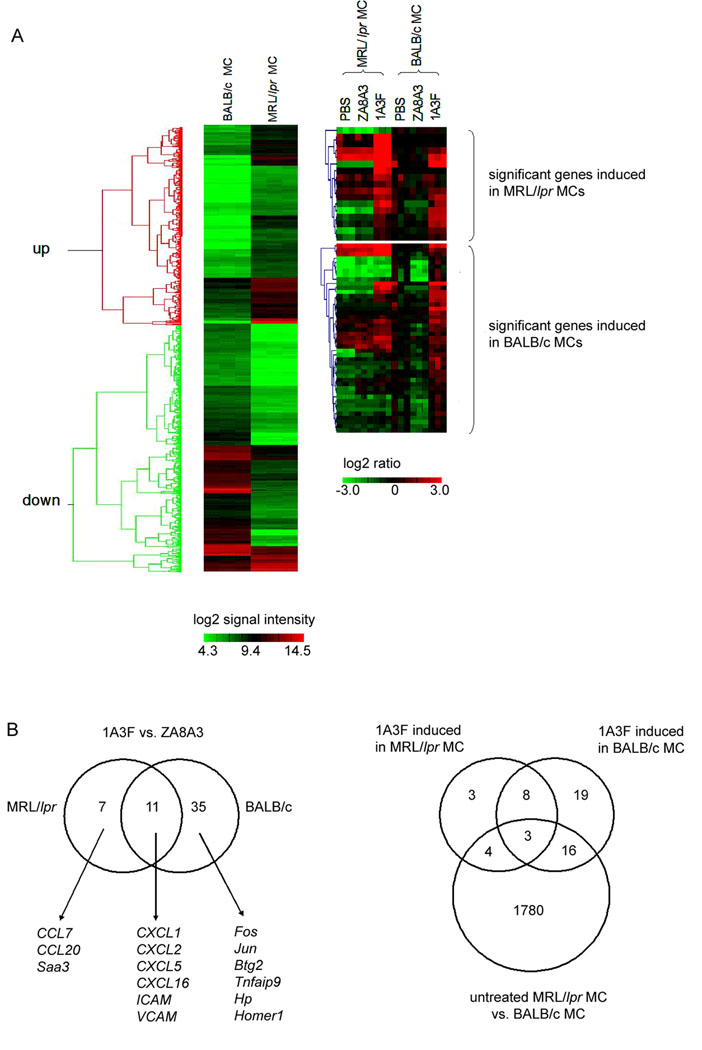
Genetic predisposition plays an essential role in the pathogenesis of SLE [46]. In order to test if the genetic background contributes to antibody induced gene modulation in MC, we examined gene expression profiles in MC from the lupus prone MRL/lpr and non-autoimmune BALB/c mice, prior to and following antibody stimulation. Initially, MCs without antibody treatment were compared between the two strains. Over 700 out of 14,000 genes were identified, with 341 upregulated and 370 downregulated in MRL/lpr MC compared to BALB/c MC (p<0.001, fold change >3), as shown by hierarchical clustering (Figure 6A, left panel). According to the Kyoto Encyclopedia of Genes and Genomes (KEGG) classification, these differentially expressed gene clusters are related to or involved in apoptosis, cell cycle, integrin-mediated cell adhesion, Wnt and G-protein signaling pathways, and matrix metalloproteinases.
Fig. 6.
MCs from lupus prone MRL/lpr and normal BALB/c mice respond differently to pathogenic antibody stimulation. (A) Hierarchical clustering analysis of differentially expressed genes identified by microarray. Left panel shows the comparison of untreated cells. Significantly differentially expressed genes were selected for hierarchical clustering analysis (p<0.001, fold change >3). Colors in the picture represent log2 transformed intensity of signals. Right panel illustrates transcripts induced by the nephritogenic antibody 1A3F in MRL/lpr MCs versus BALB/c MCs. Cells were treated with 1A3F or the control antibody ZA8A3 at 50 µg/ml or PBS, for 6 h. Differentially expressed genes were selected for hierarchical clustering analysis (p<0.05, fold change >2). Colors in the right panels represent log2 transformed ratio of signals (antibody against PBS treatment). There are three biological replicates (independent experiments) in each group. (B) Venn diagram of transcripts induced by 1A3F in MRL/lpr MC and BALB/c MC (left). In the right panel, a Venn diagram of transcripts induced by 1A3F in MRL/lpr MC and BALB/c MC compared to differentially expressed transcripts in untreated MCs is shown.
Next, genes induced by 1A3F were compared between the two backgrounds. MC derived from MRL/lpr and BALB/c mice were treated with the pathogenic antibody 1A3F, the control antibody ZA8A3, or PBS. The same numbers of cells from both strains were seeded in order to minimize the variation in cell to antibody ratios. In MRL/lpr MC, 1A3F induced 18 genes including CXCL1, CXCL2, CXCL5, CXCL16, CCL7, CCL20, Saa3, VCAM-1, and ICAM-1, many of which are inflammatory response genes such as chemokines. In contrast, more genes were identified in BALB/c MC treated with 1A3F (46 genes upregulated), including several chemokines as well (CXCL1, CXCL2, CXCL5, CCL2 and VCAM-1). Remarkably, many genes upregulated in BALB/c MC encode for transcription factors or proteins involved in signaling transduction including c-FOS, JUNB, ERG2, ATF2, BTG2 and NR4A1. In total, 11 genes were induced by 1A3F in both MRL/lpr and BALB/c MCs, as illustrated by hierarchical clustering and the Venn diagram (Figure 6, A and B). It is also interesting to note that at least in BALB/c MC there was no significant difference in the gene expression profile of MC treated with the non-pathogenic ZA8A3 antibody and PBS. Among the 18 transcripts induced by 1A3F in MRL/lpr MC, 7 genes were also differentially expressed between untreated MRL/lpr MC and BALB/c MC, while 19 out of 46 transcripts induced by 1A3F in BALB/c MC were found in the latter group. Three transcripts were found shared by all three groups (Figure 6B). Put differently, about 60% of 1A3F induced genes were originally expressed at similar levels in MC from MRL/lpr and BALB/c.
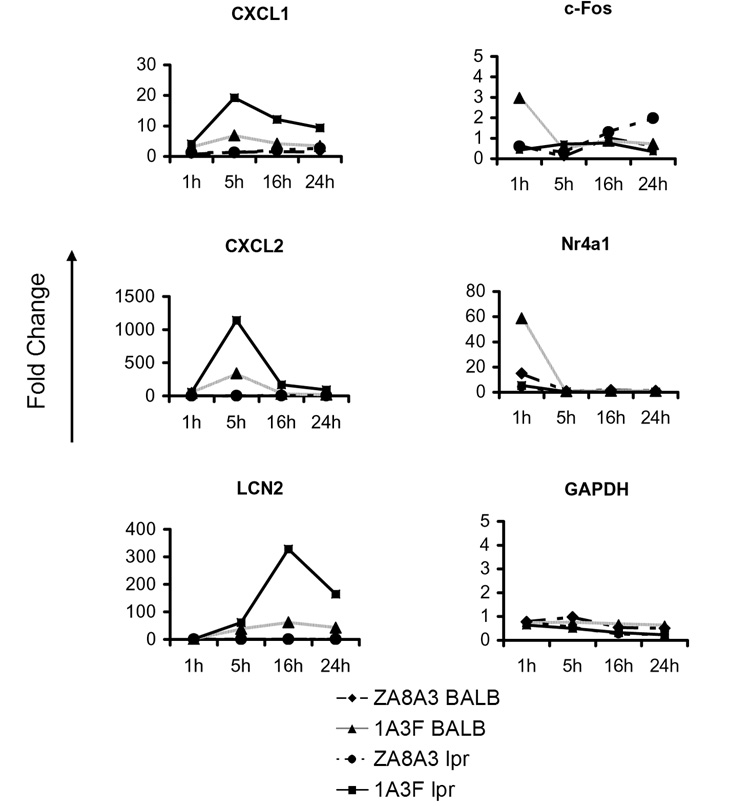
As some of the apparent differences in MC gene expression between the strains may reflect single nucleotide polymorphisms in the probes used for the array, the distinct patterns of 1A3F induced genes in MRL/lpr and BALB/c MC were further confirmed by real-time PCR. Since the microarray analysis sampled the gene expression profile only at a single time point (following 6 h of antibody treatment), a time course analysis was performed (Figure 7). Most of the chemokines studied were induced as early as 1 h upon antibody treatment, peaking at 5 h, and lasting over 24 h. Although both strains express chemokine genes following a similar kinetic pattern, it is clear that those in MRL/lpr MC reached much higher levels than in BALB/c MC at each time point (Figure 7). In contrast, several of the genes responding early (c-FOS and NR4A1) were induced at a higher level in BALB/c MC (Figure 7).
Fig. 7.
Real-time PCR confirmation of differentially expressed genes induced by pathogenic antibody in MRL/lpr MCs and BALB/c MCs. Cells were treated with PBS, ZA8A3 or 1A3F at 50 µg/ml for 1, 5, 16, and 24 h. Gene expression was analyzed by real-time PCR. GAPDH is shown as a control gene. CXCL5, CCL7, CCL20, VCAM-1, and MCP-1 also displayed up to 50 fold higher gene expression levels at the 5 or 16 hour time points in MRL/lpr as compared to BALB/c MC following stimulation by 1A3F (data not shown). Data displayed here shows one representative experiment of at least 3 independent repeats with similar results.
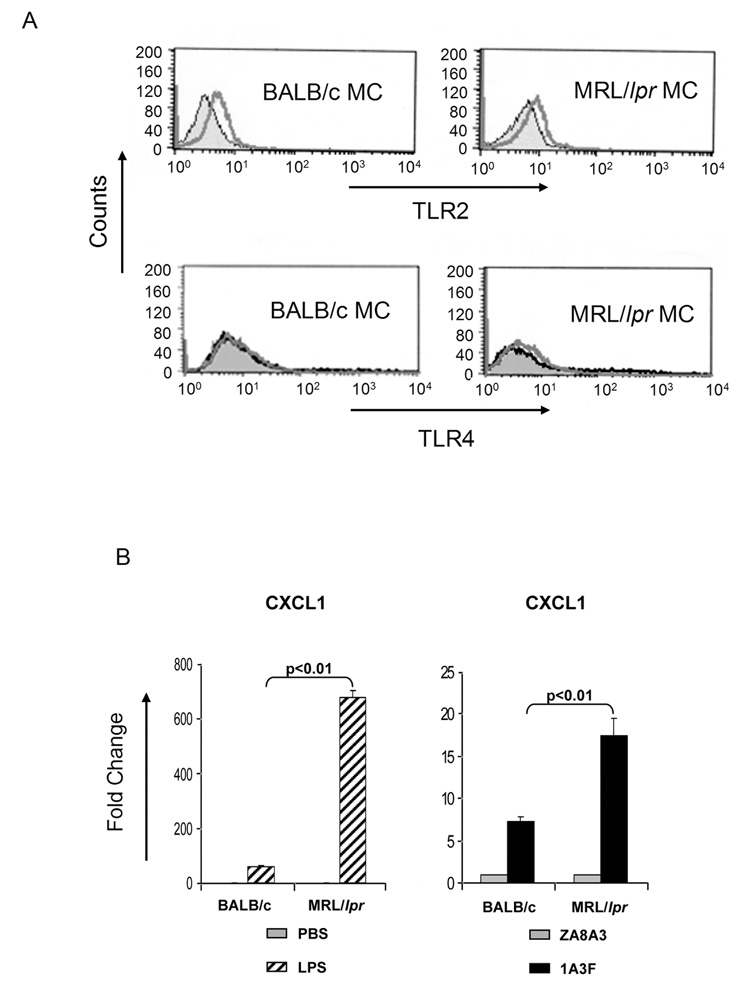
To investigate whether differential TLR2 and TLR4 expression or function can contribute to this enhanced susceptibility of MRL/lpr MC to stimulation by pathogenic antibody, we investigated the relative expression of these receptors in MC. We found that while TLR2 was expressed at a similar level on MRL/lpr and BALB/c MC, TLR4 was only detected on MRL/lpr MC, but not on BALB/c MC (Figure 8A). Nevertheless, this does not exclude the possibility that TLR4 can be induced in BALB/c MC when cells are activated. To determine if increased TLR4 expression levels on MRL/lpr MC are physiologically relevant, we stimulated MC with LPS, a TLR4 ligand. Consistent with the higher TLR4 expression levels observed in MRL/lpr MC, LPS stimulated a significantly enhanced inflammatory response in these cells (as measured by CXCL1 levels) as compared to BALB/c MC (Figure 8B, left panel). While we did not observe dramatic upregulation of TLR4 on MRL/lpr derived mesangial cells as compared to BALB/c derived cells, the hypersensitivity of MRL/lpr derived mesangial cells to LPS as compared to BALB/c derived cells (Figure 8B) is strongly supportive of a biologically significant difference in TLR4 expression levels between these strains. No significant differences between the two strains were found in the MC expression of other TLRs, including TLR 1, 3, 5, 6, 7, 9, and 11 (data not shown).
Fig. 8.
Differential expression of TLRs between MRL/lpr and BALB/c MCs. (A) Both TLR2 and TLR4 were detected in unstimulated MRL/lpr MCs, while only TLR2 was found on the cell surface of BALB/c MCs. Filled histogram, PBS; open lines, corresponding antibody. (B) MCs from the lupus prone MRL/lpr background are hyperresponsive to stimulation by LPS (left panel) and pathogenic antibody (right panel) as compared to MC from normal BALB/c mice. Cells were treated with LPS at 10 µg/ml or mAb at 50 µg/ml for 6 h. The induction of CXCL1 was analyzed by real-time PCR. Data displayed here shows one representative experiment of at least 2 independent repeats with similar results. Mean and SD are shown.
As HMGB1 is bound by 1A3F and is a ligand for TLR4, and we found that TLR4 is overexpressed by MRL/lpr MC, we predicted that MRL/lpr MC would be more sensitive than BALB/c MC to stimulation by pathogenic antibody. Indeed, stimulation of MC with 1A3F, but not the non-pathogenic mAb ZA8A3, resulted in significantly greater upregulation of CXCL1 expression in MRL/lpr than in BALB/c MC (Figure 8B, middle panel). In support of TLR4 as a mediator of antibody induced gene regulation was the very similar pattern of responses to antibodies and LPS in MC derived from the different strains (Figure 8B).
5. Discussion
Initiation of kidney damage by pathogenic anti-DNA antibodies involves glomerular deposition of anti-DNA antibodies in the kidney, leading to in situ immune complex formation, cross linking of Fc receptors and subsequent activation of complement. We recently reported that anti-DNA antibodies with mesangial cell reactivity can also modulate gene expression upon binding to MC through both Fc-dependent and Fc-independent mechanisms [33]. Among genes induced by the pathogenic antibodies are several proinflammatory chemokines and cytokines. In the present study, we further addressed the mechanisms of renal gene regulation by pathogenic anti-DNA antibodies. We found that pathogenic antibodies can mediate inflammatory injury via binding to HMGB1 and engagement of the HMGB1 receptors, in particular TLR2 and RAGE. Although we not able to definitively implicate TLR4 in inflammatory gene modulation by anti- DNA antibodies in MC, the studies comparing the differential responses of MRL/lpr and BALB/c MC are consistent with involvement of this HMGB1 receptor as well.
TLRs, and in particular TLR7 and TLR9, have been intensively studied in recent years for their involvement in the pathogenesis of lupus nephritis and other renal diseases (reviewed in [47]). In this study, we focused on the role of cell surface expressed TLRs. Several groups had previously reported the involvement of TLR2 in antibody related organ damage [5,14,21,48]. The nephrotoxic serum nephritis model requires TLR2 expression on both bone-marrow derived cells and intrinsic renal cells [14]. Deficiency in TLR2, but not TLR4, completely abrogated the inflammatory response induced by anti-phospholipid antibodies in mouse fibroblasts [48]. TLR2 recognizes Gram-positive and Gram-negative bacteria as well as mycoplasma and yeast. In a pathogen-free system, damaged cells may be a source for endogenous TLR2 ligands, including HMGB1 [19–21]. Here, we demonstrated that pathogenic anti-DNA antibodies bind to HMGB1. Interestingly, antibodies to α-actinin, a major cross-reactive specificity in the pathogenic anti-DNA response in human and murine lupus, also bind to a shared epitope on HMGB1 [41].
HMGB1 was initially identified as a ubiquitous non-histone DNA-binding protein. It is now well established that HMGB1, via TLR2/4 or RAGE [19–21,23,24,49–52], also possesses cytokine activity important to inflammation and immune responses [53,54]. Our results provide the first evidence to link HMGB1 with the renal effects of pathogenic antibodies in lupus nephritis. HMGB1 not only activated MC in vitro and the kidney in vivo in a similar pattern as pathogenic antibody, but also could amplify antibody effects. We should note that although the level of HMGB1 complexing spontaneously with the pathogenic anti-DNA antibody (0.3 µg/ml) was lower than the concentration needed alone to stimulate MC (>5 µg/ml; data not shown), HMGB1 concentrations in local environments in vivo, as we have found in the kidney, may be higher. Moreover, recent studies suggest that factors in serum may lead to the underestimation of HMGB1 concentrations by ELISA [55]. In any case, HMGB1 in complex with pathogenic antibody had a synergistic proinflammatory effect on MC. However, several questions remain to be clarified. First, is antibody binding mediated cooperatively between HMGB1 and FcR? We have previously shown that FcR signaling is important in gene regulation induced by 1A3F in MC [33]. TLR2/4 and RAGE are expressed on the cell surface; HMGB1 should be able to access its receptor without delivery by FcR, as opposed to nucleic acids which require FcR to reach TLR3/7/9 located in the endosomes. It is possible that anchoring of HMGB1 complexed antibody by FcR may facilitate closer contact between HMGB1 with its receptor, or that the antibody-antigen interaction may modify the active site on HMGB1. Second, what is the source of HMGB1 bound by pathogenic antibodies in vivo? Inconsistent with a previous report [55], HMGB1 was undetectable in the serum from lupus mice or SLE patients (data not shown). Nevertheless, we did find increased expression of HMGB1 in the kidneys from lupus patients with advanced renal pathology, raising the possibility that locally accumulated HMGB1 can be bound to deposited antibodies.
We clearly demonstrated that the pathogenic anti-dsDNA antibody 1A3F binds to HMGB1, which then activates a renal inflammatory response by binding to its receptors RAGE, TLR2 and TLR4. However, it is then important to reconsider whether 1A3F truly recognizes dsDNA, or rather binds to this antigen via HMGB1. Indeed, we showed that the purified antibody which stimulates kidney cells contains HMGB1 (albeit in small amounts). To address this question directly, we carried out high performance liquid chromatography to separate 1A3F complexed with HMGB1 from 1A3F alone. However, despite multiple attempts we were not successful in isolating non-complexed from complexed antibody since the size difference was not sufficiently large to precisely separate these fractions. We also considered testing whether the binding of 1A3F to dsDNA would be inhibited in the presence of a specific anti-HMGB1 Ab (not recognizing dsDNA); if inhibition is demonstrated, the conclusion would be that the binding of 1A3F to DNA is mediated by HMGB1. However, the only non-dsDNA anti-HMGB1 antibody available to us is HAP46.5, which we had shown becomes DNA binding when preincubated with HMGB1 (Figure 1C). This latter finding would suggest that there are different binding sites on HMGB1 for dsDNA and HAP46.5, precluding the use of HAP46.5 as a blocking antibody for the binding of HMGB1 to dsDNA. Nevertheless, we recently discovered that several other pathogenic, but not non-pathogenic, anti-DNA mAbs also bind to HMGB1 [41], indicating that: 1) binding to HMGB1 is not limited to 1A3F alone; and 2) that binding to DNA is not necessarily associated with HMGB1 binding. Indeed, we found that HMGB1 contains a common epitope present in other cross-reactive autoantigens bound by anti-DNA antibodies [41]. Therefore, it appears that pathogenic anti-DNA antibodies can bind HMGB1, although excluding the unlikely possibility that any DNA binding of these antibodies is mediated by HMGB1 will need to be further addressed in future studies.
Interestingly, the anti-DNA/HMGB1/RAGE dependent mechanism that we have shown here for pathogenic anti-DNA antibody stimulation of MC has some similarities to that recently described for activation of plasmacytoid dendritic cells by immune complexes. Tian et al demonstrated that HMGB1 binds to CpG oligodeoxynucleotides and enhances α-interferon secretion in dendritic cells, through a mechanism dependent on RAGE and TLR9 [56]. However, in contrast to Tian et al, in our studies we did not find evidence that MC activation is dependent on DNA contained in the antibody binding site.
As a cytokine, HMGB1 can amplify the inflammatory response [53]. However, the fact that pathogenic anti-DNA antibodies react with HMGB1 suggests that it may also be immunogenic. We also tested other reported endogenous TLR2/4 ligands, like heat shock proteins and fibrinogen [18,57,58]. Unfortunately, commercially available HSP70 and fibrinogen were both contaminated with endotoxin (data not shown). Nevertheless, our data is very consistent with a recent report by Yokota et al that a combination of HSP70 with an anti-HSP70 mAb enhanced the proinflammatory effect induced by HSP70 in monocytes, requiring both FcR and TLR4 [59].
Another important question addressed in this work is the contribution of genetic background to the susceptibility of MC to antibody stimulation. Studies in various mouse models of SLE have repeatedly showed that the genetic background significantly affects the onset or outcome of the disease. Liu et al recently performed gene profiling in MRL/lpr, MRL/+ and C57BL/6 mice. A few interferon-regulated and cell surface proteins were upregulated in kidneys from 6 wk old MRL/lpr mice compared to MRL/+ mice. The majority of genes encoding interferon-responsive proteins, chemokines and cytokines were elevated starting at week 12 in MRL/lpr kidneys, in parallel with increasing autoantibody titers and inflammatory infiltrates [60]. Our experiments used MC derived from very young mice (3–4 wk), therefore minimizing the likelihood that the cells have been ‘primed’ in an inflammatory milieu. In our study, hyperresponsiveness to pathogenic antibody as well as LPS was found in MRL/lpr MC, as compared to MC derived from non-autoimmune BALB/c (current paper) and C57Bl/6 mice (data not shown). The increased production of proinflammatory mediators by lupus derived MC that we demonstrate here is consistent with the recent observations of Ka et al, who reported that mesangial cells of lupus prone NZB × NZW F1 mice produce higher chemokine levels in cell culture than MC derived from DBA × NZW F1 mice, an MHC class II matched non-autoimmune strain [61]. Furthermore, following LPS stimulation, NZB × NZW F1 MC had significantly increased TLR4 and MyD88 mRNA and augmented MCP-1 and osteopontin production, when compared to control DBA × NZW F1 MC.
Our results reported above lend further support to the conclusion that genetic susceptibility in the target organ (kidney) contributes to the pathogenesis of lupus nephritis. However, the exact mechanisms of enhanced MRL/lpr MC responsiveness still need to be further clarified. We demonstrated at least 2 possible elements. Although TLR2 seems to be essential in activation of MRL/lpr MC by the pathogenic antibody, no significant difference in TLR2 expression was found between MRL/lpr and BALB/c MC. As for TLR4, its upregulation on MRL/lpr MC nicely explains not only hyperresponsiveness to 1A3F, but also to LPS. Interestingly, upregulation of TLR4 has recently been shown to be sufficient to induce lupus like disease in a transgenic mouse model [62]. A possible contribution of RAGE as well to differential sensitivity to antibody stimulation will need to be explored in future studies. Finally, a large number of genes were identified that are differentially expressed in MC between the two strains. Variability in as yet uncharacterized downstream pathways may also contribute to the hyperresponsiveness of MC from lupus prone mice.
In conclusion, our findings reveal a novel mechanism by which pathogenic anti-DNA antibodies may contribute to renal damage, implicating the endogenous TLR2/4 and RAGE ligand HMGB1. Furthermore, these studies provide an additional rationale for targeting TLRs as a potential novel approach for the development of therapeutics for lupus. Finally, the enhanced sensitivity of lupus MC to inflammatory stimuli such as anti-DNA antibodies or LPS lends further support to recent studies from our laboratory showing the importance of the genetic background of the kidney target organ in the development of antibody induced nephritis [40]. Further investigation of this unique mechanism for renal injury may yield promising new therapeutic targets for lupus, which remains a clinically challenging disease.
Acknowledgement
This work was supported by NIH grants RO1 AR48692 and PO1 AI51392 (to C.P.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Foster MH, Cizman B, Madaio MP. Nephritogenic autoantibodies in systemic lupus erythematosus: immunochemical properties, mechanisms of immune deposition, and genetic origins. Lab Invest. 1993;69:494–507. [PubMed] [Google Scholar]
- 2.Hahn BH. Antibodies to DNA. N Engl J Med. 1998;338:1359–1368. doi: 10.1056/NEJM199805073381906. [DOI] [PubMed] [Google Scholar]
- 3.Liang Z, Xie C, Chen C, Kreska D, Hsu K, Li L, et al. Pathogenic profiles and molecular signatures of antinuclear autoantibodies rescued from NZM2410 lupus mice. J Exp Med. 2004;199:381–398. doi: 10.1084/jem.20030132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clynes R, Dumitru C, Ravetch JV. Uncoupling of immune complex formation and kidney damage in autoimmune glomerulonephritis. Science. 1998;279:1052–1054. doi: 10.1126/science.279.5353.1052. [DOI] [PubMed] [Google Scholar]
- 5.Fu Y, Xie C, Chen J, Zhu J, Zhou H, Thomas J, et al. Innate stimuli accentuate end-organ damage by nephrotoxic antibodies via Fc receptor and TLR stimulation and IL-1/TNF-alpha production. J Immunol. 2006;176:632–639. doi: 10.4049/jimmunol.176.1.632. [DOI] [PubMed] [Google Scholar]
- 6.Kaneko Y, Nimmerjahn F, Madaio MP, Ravetch JV. Pathology and protection in nephrotoxic nephritis is determined by selective engagement of specific Fc receptors. J Exp Med. 2006;203:789–797. doi: 10.1084/jem.20051900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsumoto K, Watanabe N, Akikusa B, Kurasawa K, Matsumura R, Saito Y, et al. Fc receptor-independent development of autoimmune glomerulonephritis in lupus-prone MRL/lpr mice. Arthritis Rheum. 2003;48:486–494. doi: 10.1002/art.10813. [DOI] [PubMed] [Google Scholar]
- 8.Tarzi RM, Davies KA, Robson MG, Fossati-Jimack L, Saito T, Walport MJ, et al. Nephrotoxic nephritis is mediated by Fcgamma receptors on circulating leukocytes and not intrinsic renal cells. Kidney Int. 2002;62:2087–2096. doi: 10.1046/j.1523-1755.2002.00687.x. [DOI] [PubMed] [Google Scholar]
- 9.Barrat FJ, Meeker T, Gregorio J, Chan JH, Uematsu S, Akira S, et al. Nucleic acids of mammalian origin can act as endogenous ligands for Toll-like receptors and may promote systemic lupus erythematosus. J Exp Med. 2005;202:1131–1139. doi: 10.1084/jem.20050914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau CM, Broughton C, Tabor AS, Akira S, Flavell RA, Mamula MJ, et al. RNA-associated autoantigens activate B cells by combined B cell antigen receptor/Toll-like receptor 7 engagement. J Exp Med. 2005;202:1171–1177. doi: 10.1084/jem.20050630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leadbetter EA, Rifkin IR, Hohlbaum AM, Beaudette BC, Shlomchik MJ, Marshak-Rothstein A. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416:603–607. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- 12.Pisitkun P, Deane JA, Difilippantonio MJ, Tarasenko T, Satterthwaite AB, Bolland S. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science. 2006;312:1669–1672. doi: 10.1126/science.1124978. [DOI] [PubMed] [Google Scholar]
- 13.Subramanian S, Tus K, Li QZ, Wang A, Tian XH, Zhou J, et al. A Tlr7 translocation accelerates systemic autoimmunity in murine lupus. Proc Natl Acad Sci U.S.A. 2006;103:9970–9975. doi: 10.1073/pnas.0603912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown HJ, Lock HR, Sacks SH, Robson MG. TLR2 stimulation of intrinsic renal cells in the induction of immune-mediated glomerulonephritis. J Immunol. 2006;177:1925–1931. doi: 10.4049/jimmunol.177.3.1925. [DOI] [PubMed] [Google Scholar]
- 15.Rifkin IR, Leadbetter EA, Busconi L, Viglianti G, Marshak-Rothstein A. Toll-like receptors, endogenous ligands, and systemic autoimmune disease. Immunol Rev. 2005;204:27–42. doi: 10.1111/j.0105-2896.2005.00239.x. [DOI] [PubMed] [Google Scholar]
- 16.Biragyn A, Ruffini PA, Leifer CA, Klyushnenkova E, Shakhov A, Chertov O, et al. Toll-like receptor 4-dependent activation of dendritic cells by beta-defensin 2. Science. 2002;298:1025–1029. doi: 10.1126/science.1075565. [DOI] [PubMed] [Google Scholar]
- 17.Kariko K, Ni H, Capodici J, Lamphier M, Weissman D. mRNA is an endogenous ligand for Toll-like receptor 3. J Biol Chem. 2004;279:12542–12550. doi: 10.1074/jbc.M310175200. [DOI] [PubMed] [Google Scholar]
- 18.Smiley ST, King JA, Hancock WW. Fibrinogen stimulates macrophage chemokine secretion through toll-like receptor 4. J Immunol. 2001;167:2887–2894. doi: 10.4049/jimmunol.167.5.2887. [DOI] [PubMed] [Google Scholar]
- 19.Park JS, Svetkauskaite D, He Q, Kim JY, Strassheim D, Ishizaka A, et al. Involvement of toll-like receptors 2 and 4 in cellular activation by high mobility group box 1 protein. J Biol Chem. 2004;279:7370–7377. doi: 10.1074/jbc.M306793200. [DOI] [PubMed] [Google Scholar]
- 20.Park JS, Gamboni-Robertson F, He Q, Svetkauskaite D, Kim JY, Strassheim D, et al. High mobility group box 1 protein interacts with multiple Toll-like receptors. Am J Physiol Cell Physiol. 2006;290:C917–C924. doi: 10.1152/ajpcell.00401.2005. [DOI] [PubMed] [Google Scholar]
- 21.Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–195. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- 22.Wang H, Vishnubhakat JM, Bloom O, Zhang M, Ombrellino M, Sama A, et al. Proinflammatory cytokines (tumor necrosis factor and interleukin 1) stimulate release of high mobility group protein-1 by pituicytes. Surgery. 1999;126:389–392. [PubMed] [Google Scholar]
- 23.Hori O, Brett J, Slattery T, Cao R, Zhang J, Chen JX, et al. The receptor for advanced glycation end products (RAGE) is a cellular binding site for amphoterin. Mediation of neurite outgrowth and co-expression of rage and amphoterin in the developing nervous system. J Biol Chem. 1995;270:25752–25761. doi: 10.1074/jbc.270.43.25752. [DOI] [PubMed] [Google Scholar]
- 24.Kokkola R, Andersson A, Mullins G, Ostberg T, Treutiger CJ, Arnold B, et al. RAGE is the major receptor for the proinflammatory activity of HMGB1 in rodent macrophages. Scand J Immunol. 2005;61:1–9. doi: 10.1111/j.0300-9475.2005.01534.x. [DOI] [PubMed] [Google Scholar]
- 25.Abraham E, Arcaroli J, Carmody A, Wang H, Tracey KJ. HMG-1 as a mediator of acute lung inflammation. J Immunol. 2000;165:2950–2954. doi: 10.4049/jimmunol.165.6.2950. [DOI] [PubMed] [Google Scholar]
- 26.Ek M, Popovic K, Harris HE, Naucler CS, Wahren-Herlenius M. Increased extracellular levels of the novel proinflammatory cytokine high mobility group box chromosomal protein 1 in minor salivary glands of patients with Sjogren's syndrome. Arthritis Rheum. 2006;54:2289–2294. doi: 10.1002/art.21969. [DOI] [PubMed] [Google Scholar]
- 27.Popovic K, Ek M, Espinosa A, Padyukov L, Harris HE, Wahren-Herlenius M, et al. Increased expression of the novel proinflammatory cytokine high mobility group box chromosomal protein 1 in skin lesions of patients with lupus erythematosus. Arthritis Rheum. 2005;52:3639–3645. doi: 10.1002/art.21398. [DOI] [PubMed] [Google Scholar]
- 28.Barkauskaite V, Ek M, Popovic K, Harris HE, Wahren-Herlenius M, Nyberg F. Translocation of the novel cytokine HMGB1 to the cytoplasm and extracellular space coincides with the peak of clinical activity in experimentally UV-induced lesions of cutaneous lupus erythematosus. Lupus. 2007;16:794–802. doi: 10.1177/0961203307081895. [DOI] [PubMed] [Google Scholar]
- 29.Ayer LM, Senecal JL, Martin L, Dixon GH, Fritzler MJ. Antibodies to high mobility group proteins in systemic sclerosis. J Rheumatol. 1994;21:2071–2075. [PubMed] [Google Scholar]
- 30.Bustin M, Reisch J, Einck L, Klippel JH. Autoantibodies to nucleosomal proteins: antibodies to HMG-17 in autoimmune diseases. Science. 1982;215:1245–1247. doi: 10.1126/science.6460317. [DOI] [PubMed] [Google Scholar]
- 31.Santoro P, De Andrea M, Migliaretti G, Trapani C, Landolfo S, Gariglio M. High prevalence of autoantibodies against the nuclear high mobility group (HMG) protein SSRP1 in sera from patients with systemic lupus erythematosus, but not other rheumatic diseases. J Rheumatol. 2002;29:90–93. [PubMed] [Google Scholar]
- 32.Wittemann B, Neuer G, Michels H, Truckenbrodt H, Bautz FA. Autoantibodies to nonhistone chromosomal proteins HMG-1 and HMG-2 in sera of patients with juvenile rheumatoid arthritis. Arthritis Rheum. 1990;33:1378–1383. doi: 10.1002/art.1780330910. [DOI] [PubMed] [Google Scholar]
- 33.Qing X, Zavadil J, Crosby MB, Hogarth MP, Hahn BH, Mohan C, et al. Nephritogenic anti-DNA antibodies regulate gene expression in MRL/lpr mouse glomerular mesangial cells. Arthritis Rheum. 2006;54:2198–2210. doi: 10.1002/art.21934. [DOI] [PubMed] [Google Scholar]
- 34.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 35.Deocharan B, Qing X, Lichauco J, Putterman C. Alpha-actinin is a cross-reactive renal target for pathogenic anti-DNA antibodies. J Immunol. 2002;168:3072–3078. doi: 10.4049/jimmunol.168.6.3072. [DOI] [PubMed] [Google Scholar]
- 36.Mason LJ, Ravirajan CT, Rahman A, Putterman C, Isenberg DA. Is alpha-actinin a target for pathogenic anti-DNA antibodies in lupus nephritis? Arthritis Rheum. 2004;50:866–870. doi: 10.1002/art.20103. [DOI] [PubMed] [Google Scholar]
- 37.Mostoslavsky G, Fischel R, Yachimovich N, Yarkoni Y, Rosenmann E, Monestier M, et al. Lupus anti-DNA autoantibodies cross-react with a glomerular structural protein: a case for tissue injury by molecular mimicry. Eur J Immunol. 2001;31:1221–1227. doi: 10.1002/1521-4141(200104)31:4<1221::aid-immu1221>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 38.Renaudineau Y, Croquefer S, Jousse S, Renaudineau E, Devauchelle V, Gueguen P, et al. Association of alpha-actinin-binding anti-double-stranded DNA antibodies with lupus nephritis. Arthritis Rheum. 2006;54:2523–2532. doi: 10.1002/art.22015. [DOI] [PubMed] [Google Scholar]
- 39.Zhao Z, Weinstein E, Tuzova M, Davidson A, Mundel P, Marambio P, et al. Cross-reactivity of human lupus anti-DNA antibodies with alpha-actinin and nephritogenic potential. Arthritis Rheum. 2005;52:522–530. doi: 10.1002/art.20862. [DOI] [PubMed] [Google Scholar]
- 40.Zhao Z, Deocharan B, Scherer PE, Ozelius LJ, Putterman C. Differential binding of cross-reactive anti-DNA antibodies to mesangial cells: the role of alpha-actinin. J Immunol. 2006;176:7704–7714. doi: 10.4049/jimmunol.176.12.7704. [DOI] [PubMed] [Google Scholar]
- 41.Deocharan B, Zhou Z, Antar K, Siconolfi-Baez L, Angeletti RH, Hardin J, et al. Alpha-actinin immunization elicits anti-chromatin autoimmunity in nonautoimmune mice. J Immunol. 2007;179:1313–1321. doi: 10.4049/jimmunol.179.2.1313. [DOI] [PubMed] [Google Scholar]
- 42.Emlen W, Ansari R, Burdick G. DNA-anti-DNA immune complexes. Antibody protection of a discrete DNA fragment from DNase digestion in vitro. J Clin Invest. 1984;74:185–190. doi: 10.1172/JCI111400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shuster AM, Gololobov GV, Kvashuk OA, Bogomolova AE, Smirnov IV, Gabibov AG. DNA hydrolyzing autoantibodies. Science. 1992;256:665–667. doi: 10.1126/science.1585181. [DOI] [PubMed] [Google Scholar]
- 44.Takeuchi O, Kaufmann A, Grote K, Kawai T, Hoshino K, Morr M, et al. Cutting edge: preferentially the R-stereoisomer of the mycoplasmal lipopeptide macrophage-activating lipopeptide-2 activates immune cells through a toll-like receptor 2- and MyD88-dependent signaling pathway. J Immunol. 2000;164:554–557. doi: 10.4049/jimmunol.164.2.554. [DOI] [PubMed] [Google Scholar]
- 45.Li Q, Xie C, Mackay M, Aranow C, Putterman C, Mohan C. Glomerular proteome assays: A novel approach to study glomerular-reactive autoantibodies in lupus. J Clin Invest. 2005;115:3428–3439. doi: 10.1172/JCI23587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harley JB, Kelly JA, Kaufman KM. Unraveling the genetics of systemic lupus erythematosus. Springer Semin Immunopathol. 2006;28:119–130. doi: 10.1007/s00281-006-0040-5. [DOI] [PubMed] [Google Scholar]
- 47.Marshak-Rothstein A. Toll-like receptors in systemic autoimmune disease. Nat Rev Immunol. 2006;6:823–835. doi: 10.1038/nri1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Satta N, Dunoyer-Geindre S, Reber G, Fish RJ, Boehlen F, Kruithof EK, et al. The role of TLR2 in the inflammatory activation of mouse fibroblasts by human antiphospholipid antibodies. Blood. 2006;109:1507–1514. doi: 10.1182/blood-2005-03-024463. [DOI] [PubMed] [Google Scholar]
- 49.Andersson U, Wang H, Palmblad K, Aveberger AC, Bloom O, Erlandsson-Harris H, Janson A, et al. High mobility group 1 protein (HMG-1) stimulates proinflammatory cytokine synthesis in human monocytes. J Exp Med. 2000;192:565–570. doi: 10.1084/jem.192.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DeMarco RA, Fink MP, Lotze MT. Monocytes promote natural killer cell interferon gamma production in response to the endogenous danger signal HMGB1. Mol Immunol. 2005;42:433–444. doi: 10.1016/j.molimm.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 51.Jaulmes A, Thierry S, Janvier B, Raymondjean M, Marechal V. Activation of sPLA2-IIA and PGE2 production by high mobility group protein B1 in vascular smooth muscle cells sensitized by IL-1beta. FASEB J. 2006;20:1727–1729. doi: 10.1096/fj.05-5514fje. [DOI] [PubMed] [Google Scholar]
- 52.Yu M, Wang H, Ding A, Golenbock DT, Latz E, Czura CJ, et al. HMGB1 signals through toll-like receptor (TLR) 4 and TLR2. Shock. 2006;26:174–179. doi: 10.1097/01.shk.0000225404.51320.82. [DOI] [PubMed] [Google Scholar]
- 53.Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat Rev Immunol. 2005;5:331–342. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- 54.Ulloa L, Messmer D. High-mobility group box 1 (HMGB1) protein: friend and foe. Cytokine Growth Factor Rev. 2006;17:189–201. doi: 10.1016/j.cytogfr.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 55.Urbonaviciute V, Furnrohr BG, Weber C, Haslbeck M, Wilhelm S, Herrmann M, et al. Factors masking HMGB1 in human serum and plasma. J.Leukoc.Biol. 2007;81:67–74. doi: 10.1189/jlb.0306196. [DOI] [PubMed] [Google Scholar]
- 56.Tian J, Avalos AM, Mao SY, Chen B, Senthil K, Wu H, et al. Toll-like receptor 9- dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat Immunol. 2007;8:487–496. doi: 10.1038/ni1457. [DOI] [PubMed] [Google Scholar]
- 57.Ohashi K, Burkart V, Flohe S, Kolb H. Cutting edge: heat shock protein 60 is a putative endogenous ligand of the toll-like receptor-4 complex. J Immunol. 2000;164:558–561. doi: 10.4049/jimmunol.164.2.558. [DOI] [PubMed] [Google Scholar]
- 58.Vabulas RM, Ahmad-Nejad P, Costa C, Miethke T, Kirschning CJ, Hacker H, et al. Endocytosed HSP60s use toll-like receptor 2 (TLR2) and TLR4 to activate the toll/interleukin-1 receptor signaling pathway in innate immune cells. J Biol Chem. 2001;276:31332–31339. doi: 10.1074/jbc.M103217200. [DOI] [PubMed] [Google Scholar]
- 59.Yokota S, Minota S, Fujii N. Anti-HSP auto-antibodies enhance HSP-induced proinflammatory cytokine production in human monocytic cells via Toll-like receptors. Int Immunol. 2006;18:573–580. doi: 10.1093/intimm/dxh399. [DOI] [PubMed] [Google Scholar]
- 60.Liu J, Karypis G, Hippen KL, Vegoe AL, Ruiz P, Gilkeson GS, et al. Genomic view of systemic autoimmunity in MRL/lpr mice. Genes Immun. 2006;7:156–168. doi: 10.1038/sj.gene.6364286. [DOI] [PubMed] [Google Scholar]
- 61.Ka SM, Cheng CW, Shui HA, Wu WM, Chang DM, Lin YC, et al. Mesangial cells of lupus-prone mice are sensitive to chemokine production. Arthritis Res Ther. 2007;9:R67. doi: 10.1186/ar2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu B, Yang Y, Dai J, Medzhitov R, Freudenberg MA, Zhang PL, et al. TLR4 up-regulation at protein or gene level is pathogenic for lupus-like autoimmune disease. J Immunol. 2006;177:6880–6888. doi: 10.4049/jimmunol.177.10.6880. [DOI] [PubMed] [Google Scholar]