SUMMARY
The HBO1 HAT protein is the major source of histone H4 acetylation in vivo and has been shown to play critical roles in gene regulation and DNA replication. A distinctive characteristic of HBO1 HAT complexes is the presence of three PHD finger domains in two different subunits: tumor suppressor proteins ING4/5 and JADE1/2/3. Biochemical and functional analyses indicate that these domains interact with histone H3 N-terminal tail region, but with a different specificity towards its methylation status. Their combinatorial action is essential in regulating chromatin binding and substrate specificity of HBO1 complexes, as well as cell growth. Importantly, localization analyses on the human genome indicate that HBO1 complexes are enriched throughout the coding regions of genes, supporting a role in transcription elongation. These results underline the importance and versatility of PHD finger domains in regulating chromatin association and histone modification cross-talk within a single protein complex.
INTRODUCTION
The protruding tails of nucleosomal histones are substrates for a large number of post-translational modifications such as acetylation, methylation, phosphorylation, ubiquitination and sumoylation. Different combinations of these modifications regulate each other and establish an epigenetic signature which can be read by specific protein domains present in nuclear effector proteins, such as chromodomains, bromodomains, tudor domains, MBT domains and PHD fingers (Kouzarides, 2007).
The HBO1 acetyltransferase is responsible for the bulk of histone H4 acetylation at lysines 5, 8 and 12, is implicated in gene regulation and is a key regulator of DNA replication (Avvakumov and Cote, 2007; Miotto and Struhl, 2008). Human tumor suppressor proteins ING4 and ING5 (INhibitor of Growth; (Soliman and Riabowol, 2007)) each associate with HBO1 in distinct tetrameric complexes with JADE1/2/3 paralogs and hEaf6 (Doyon et al., 2006)(Fig. 1A). Here we report the molecular dissection of functional domains within the HBO1 HAT complexes and describe how their combinatorial action allows the complex to target distinct loci within gene coding regions.
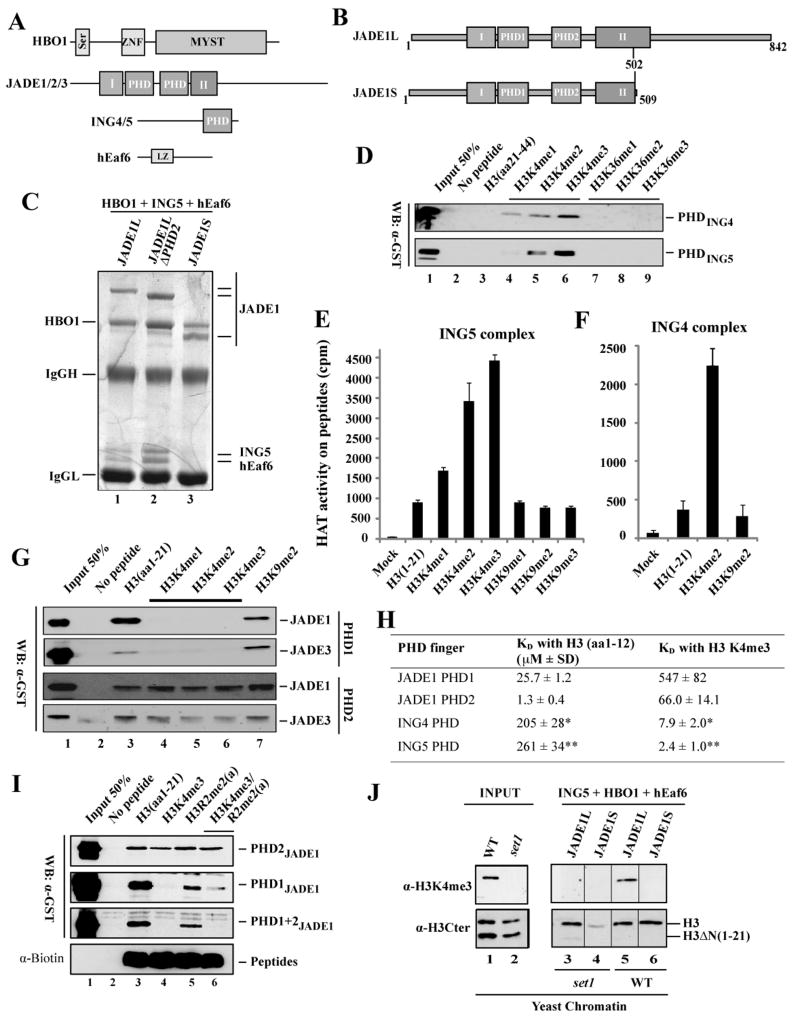
Figure 1. The interplay between the three PHD fingers within the HBO1 HAT complex.
(A) Schematic representation of functional domains found within subunits of the HBO1 complex. Domains: MYST, MYST-family HAT domain; I and II, regions conserved in all MYST-ING complexes; Ser, serine-rich; ZNF, zing-finger; LZ, leucine zipper;.
(B) Isoforms of the JADE1 protein and domains contained therein.
(C) Association of ING5 and hEaf6 with the HBO1 complex requires full-length JADE1 protein (lane 1), while the JADE1S isoform only associates with HBO1 (lane 3). The PHD finger domain of JADE1 does not affect the composition of the complex (lane 2). Immunopurified JADE1 complexes from the indicated co-transfected cells were analyzed for subunit associations by gel staining. Subunit identifications were confirmed by western blot (not shown).
(D) ING4/5 PHD fingers preferentially bind H3 peptides trimethylated on K4. Peptide pull-down assays with the indicated biotinylated peptides and recombinant PHD fingers fused to GST were analyzed by western blot with anti-GST.
(E–F) Native ING4 and ING5 complexes preferentially acetylate histone H3 tails that are methylated on K4. Purified native ING4/5 complexes were tested in HAT assays with the indicated histone H3 peptides with different levels of methylation on K4 or K9. Reactions were spotted on membranes and counted by liquid scintillation. Values are based on duplicate assays with standard deviation.
(G) JADE PHD finger domains interact with histone H3 tail peptides in vitro, but with different effect of K4 methylation. H3 peptide pull-down assays as in (D).
(H) Affinities of JADE and ING PHD finger domains for histone H3 peptides and the effect of K4 trimethylation (measured by tryptophan fluorescence). * Taken from (Hung et al., 2009); ** taken from (Champagne et al., 2008).
(I) JADE1 PHD1 is dominant over PHD2 for blocking interaction with H3K4me3 in the absence of ING PHD. H3 peptide pull-down assays as in (D).
(J) Histone H3 tail is essential for HBO1 complexes to associate with native chromatin. The indicated JADE1L/S complexes (immunopurified from co-transfected cells as in (C)) were tested for binding to native chromatin purified from wild type or set1 (no H3K4me) yeast cells. Bound histones were visualized by western blot with the indicated H3 antibodies. The presence of the N-terminal truncation of H3 is shown on the right (lines separate samples because some lanes were removed).
Interestingly, alternative splicing of the JADE1 tumor suppressor regulates ING4/5 association with HBO1, modifying its chromatin binding characteristics and HAT activity. A characteristic of ING4/5-HBO1 complexes is the presence of three distinct PHD finger domains within a single complex (Fig. 1A). We show that, while ING4/5 PHD binds H3K4me3-containing chromatin (Pena et al., 2006), the two PHD domains in JADE proteins have distinct specificities. The second PHD (PHD2) is essential for chromatin interaction irrespective of the methylation status, for chromatin acetylation by HBO1 and for tumor suppressor activity. However, the first PHD (PHD1) is blocked by H3K4me in the absence of ING4/5, allowing binding and acetylation of different chromatin regions. In agreement with this, high-resolution localization analyses of HBO1 complexes on the human genome indicate their enrichment over the entire coding regions of genes, supporting an important role during transcription elongation.
Our data unravel how different PHD finger domains within a single HAT complex cooperate to regulate binding to distinct chromatin regions and modulate histone acetylation. The combinatorial specificity of chromatin-binding PHD domains also adds a new layer in the intricate relationship between acetylation and methylation of histone tails during nuclear processes and tumor suppressor function. Our discovery that the HBO1 HAT complex localizes to coding regions of genes is of particular note. It is direct demonstration of an acetyltransferase complex functioning outside gene promoters, likely in support of transcription elongation.
RESULTS AND DISCUSSION
JADE1 protein is the platform for assembly of HBO1 complexes
JADE1 is a candidate renal tumor suppressor that has been physically linked to VHL function, transcription regulation and apoptosis (Zhou et al., 2005). While our purification of HBO1 complexes identified a full-length JADE1 protein of 842 amino acids (Doyon et al., 2006), functional data in the literature characterized a natural smaller 509 amino acid-protein produced by an abundant mRNA splice variant (Fig. 1B, JADE1L vs JADE1S). Mass spectrometry analysis of a ~63KDa protein band reproducibly obtained in our purification of native HBO1 complexes supports association of JADE1S with HBO1 in vivo (Fig. S1A and see (Doyon et al., 2006)).
To study HBO1 complex structure and function we affinity-purified JADE1 proteins after cotransfection of all subunits. As shown in figure 1C, immunopurified JADE1L associates with HBO1, ING5 and hEaf6, producing a native-like complex. JADE1 PHD finger region is not required for this assembly (lane 2). In contrast, the JADE1S isoform is unable to associate with ING5 or hEaf6 but retains its interaction with HBO1 (lane 3). Thus, the JADE1S isoform does not associate with ING proteins in vivo, suggesting a mechanism to regulate the presence of ING4/5 proteins in the HBO1 complex.
ING4 and ING5 subunits stimulate HAT activity on histone H3 tails methylated at K4
As shown in figure 1D and S1B, pull-down assays confirm that ING4 and ING5 PHD fingers show strong and specific binding to histone H3 peptides carrying di- and trimethylated forms of histone H3 K4, as shown previously (Pena et al., 2006). To determine whether this association has in impact on HAT activity of HBO1-ING complexes, purified native ING4 and ING5 complexes were tested in HAT assays with histone H3 tail peptides methylated on K4 or K9, or unmodified. The HAT activities of ING4 and ING5 complexes were greatly stimulated by methylation of K4 but not K 9 (Fig. 1E and F). The stimulatory effect of H3K4me was dependent on the level of methylation as native ING5 complexes showed the strongest stimulation with H3K4me3 (Fig. 1E). We also determined that an HBO1/JADE dimer is unaffected by K4me2, contrary to what is seen with the HBO1/JADE/ING trimer (Fig. S1C and D). These results support a role for ING subunits in stimulating HBO1 HAT activity towards H3K4me-containing chromatin, a mark highly enriched around transcription start sites of active genes.
HBO1-ING complexes contain PHD finger domains that bind histone H3 with different specificities towards K4 methylation status
We were also interested to see if the two PHD domains of JADE (Fig. 1A) play a role in binding to chromatin. Peptide pull-down assays using recombinant JADE PHD fingers also detected strong binding to histone H3 tail peptides, but in their unmodified form (Fig. 1G). While methylation of K4 or K9 had no effect on the binding of JADE’s second PHD finger (PHD2), any methylation of K4 completely disrupted binding by the first PHD finger (PHD1). Binding affinities (KD) for these different interactions measured by fluorescence spectroscopy were strong and fell within a similar range (Fig. 1H). Removal of K4 methylation decreases ING PHD affinity towards H3 by almost 100-fold, while gradual addition of methyl groups decreases JADE1 PHD1’s affinity to almost millimolar range (Fig. 1H and Table SI). Almost identical KD values were obtained for JADE3 PHDs (Table SII).
We also tested a recombinant protein corresponding to the double PHD finger region of JADE1 in peptide pull-down assays (Fig. 1I). The double PHD1+2 protein behaved similarly to PHD1 alone (Fig. 1I, lanes 1–4). Thus, in contrast to their individual affinities, JADE1 PHD1 is dominant over PHD2 in blocking interaction with histone H3 tail containing K4me3. In parallel, we also tested the impact of H3R2 methylation on the interactions detected for each PHD since this modification is mutually exclusive with H3K4me2/3 (Klose and Zhang, 2007) (Fig. 1I, lanes 5 and 6, and Fig. S2A/B). The results indicate that the presence H3R2me has no strong effect on H3 binding by JADE or ING PHDs.
The histone H3 tail is essential for HBO1 complexes to bind chromatin
In order to extend these observations to a more physiological context, we tested the interaction of immunopurified JADE1L and JADE1S complexes (Fig. 1C) with purified native chromatin. Native chromatin from yeast cells is a powerful tool due to the high level of H3K4 methylation present in this organism. Furthermore, histones or chromatin purified from yeast often contain a significant proportion of truncated histone H3 that has lost its first 21 amino acids (Lacoste et al., 2002). Histones or chromatin completely lacking H3K4 methylation can also be obtained from set1 mutant cells (Kouzarides, 2007). As shown in figure 1J, each complex bound a significant amount of chromatin, but only the population containing full-length histone H3, indicating that the first 20 amino acids of histone H3 are strictly required for binding to occur. This tail effect is specific for binding to chromatin since it is not seen with free histones (Fig. S2C). In addition, while H3K4me3 is not required for binding by both complexes (Fig. 1J, lanes 3, 4), only JADE1L complex associates with chromatin containing this mark (lane 5). These results demonstrate that binding to chromatin is efficient in the absence of ING subunit or H3K4 methylation, but that ING PHD is required to bind H3K4me-containing chromatin. Altogether, these data show that HBO1 complexes interact with chromatin through the histone H3 N-terminal domain, presumably via binding by JADE PHD fingers, and that the presence of ING can orient this binding towards H3K4me-containing chromatin regions.
The PHD finger domains of JADE1 are essential for HBO1 complexes to bind chromatin
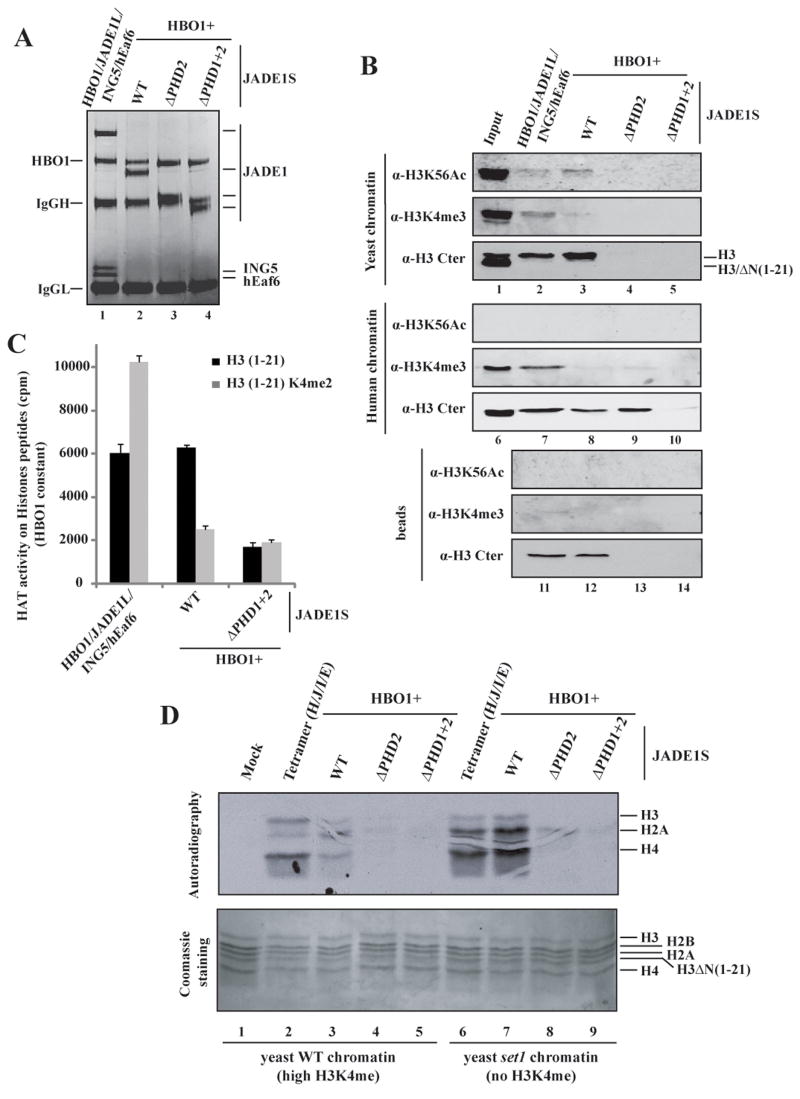
Using similar amounts of immunopurified wild type and ΔPHD JADE1S complexes along with the full tetrameric JADE1L complex (Fig. 2A), we performed chromatin pull down assays using both yeast and human native chromatin. As above, wild type tetrameric (JADE1L) and dimeric (JADE1S) complexes efficiently pulled down yeast chromatin, in an H3-tail dependent manner (Fig. 2B, lanes 1–3). A yeast-specific histone modification, H3K56ac, was also analyzed since a significant amount of endogenous human histone H3 was found to copurify with these complexes (Fig. 2B, lanes 11 and 12). Pull down assays with human chromatin and wild type complexes gave similar results (lanes 6–8). Deletion of both PHDs of JADE1 completely eliminated the interaction with both chromatin samples, demonstrating a key role of these PHD finger domains in the association of HBO1 complexes with chromatin (lanes 5 and 10). Importantly, this double deletion also eliminated the co-purification of endogenous histone H3 with the complex, indicating that this function is physiological (lane 14).
Figure 2. Histone H3 tail-dependent binding to chromatin and HBO1 nucleosomal HAT activity depend on JADE1 PHD finger domains, while ING5 is required to target H3K4 methylated regions and modulates HAT specificity.
(A) Immunopurified complexes from cells co-transfected with the indicated constructs were analyzed on gel to normalize relative amounts.
(B) Binding assays with purified native chromatin from yeast and human cells. Chromatin binding with wild type and mutant JADE1 complexes immunopurified in (A) was analyzed as in Fig. 1J.
(C) Efficient acetylation of H3 peptides by HBO1 requires JADE1 PHD fingers. HAT assays on H3 peptides as in Fig. 1E were performed with wild type and mutant JADE1 complexes immunopurified in (A).
(D) HBO1 requires JADE1 PHDs to acetylate chromatin, while ING5 modulates histone tail specificity in the presence of H3K4me. HAT assays with the same complexes on yeast native chromatin with (WT) or without (set1) H3K4me mark. The reactions were loaded and acetylation revealed by fluorography. Coomassie staining of the gel is shown to ensure equivalent loading.
Essential and regulatory roles of different PHD fingers in chromatin acetylation by HBO1 complexes
Using equivalent amounts of wild type and mutant HBO1 complexes, we performed HAT assays using peptides, core histones and native chromatin. In contrast to our earlier observations (Fig. 1E), K4me2 peptides inhibited HBO1 activity in the absence of ING5 (JADE1S in Fig. 2C), confirming our previous result that JADE1 PHD1 blocks interaction with H3 tails containing K4me (Fig. 1G–I). Deletion of JADE1S PHDs greatly diminished HBO1 HAT activity on both peptides, irrespective of methylation status. When using native wild type chromatin in HAT assays, the JADE1S complex is much less active than the JADE1L complex (Fig. 2D, lanes 2 vs 3). This weak acetylation is clearly due to the high level of H3K4me since the JADE1S complex is highly active on chromatin lacking this mark (set1, compare lanes 3 to 7). In this case, JADE1L and JADE1S complexes show equivalent acetyltransferase activity (compare lanes 6 and 7). These results strongly support our binding data, with ING PHD allowing acetylation of H3K4me-containing chromatin regions and JADE PHD1 blocking it. Since JADE PHD1 is always present in HBO1 complexes, this implies that the association of ING PHD masks its function, modifying the way HBO1 interacts with H3K4me-containing chromatin. This is supported by a slight change in acetylation specificity where histone H3 tail is now more efficiently acetylated (compare lane 2 to lanes 3, 6 and 7).
In addition, HAT assays shown in figure 2D clearly indicate that the PHD2 domain of JADE1S is essential for HBO1 to acetylate chromatin substrates (lanes 4 and 8). Similar results were obtained with human native chromatin but not with free core histones (Fig. S2D/E), indicating that the essential role of PHD2 is specific to chromatin substrates. These results indicate that three different PHD domains within HBO1 complexes play distinct roles in chromatin acetylation, from being essential (PHD2) to reading the substrate methylation status (ING PHD and PHD1).
JADE1 PHD2 is essential for binding to chromatin in vivo and for JADE1L tumor suppressor activity
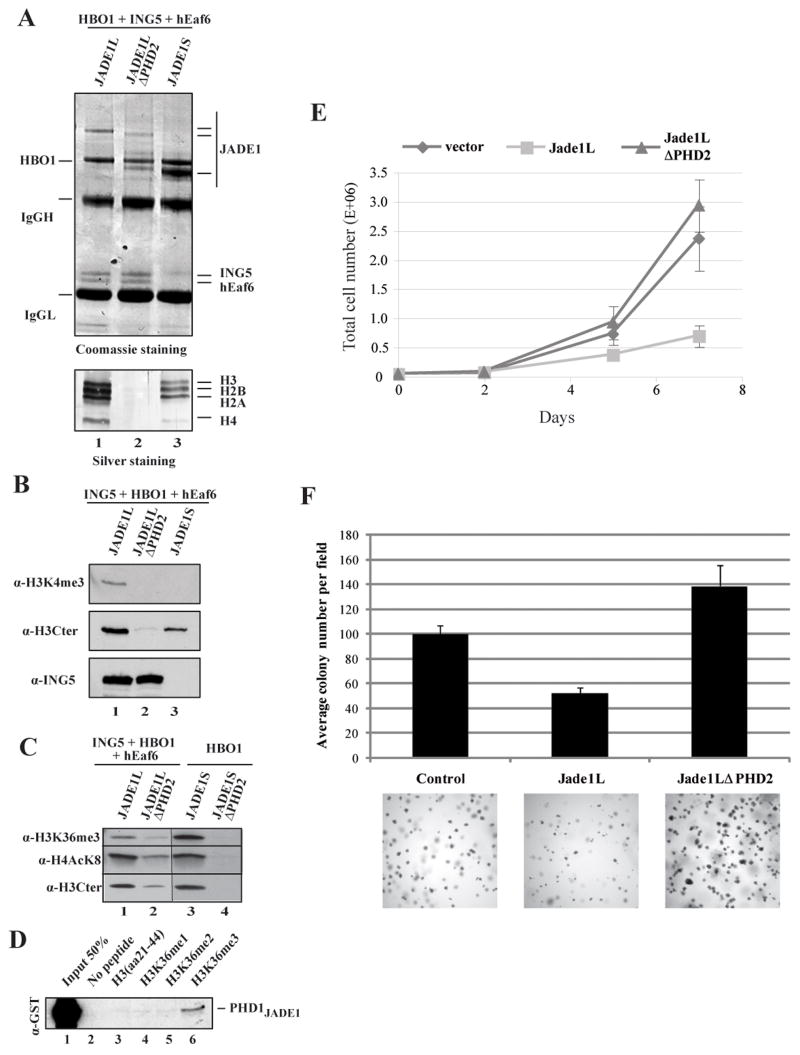
To determine the functional requirement of PHD2 in an HBO1 complex containing ING4/5, we immunopurified wild type and ΔPHD2 JADE1 complexes. Gel staining of the samples demonstrated that a full set of core histones co-purified with wild type JADE1L and JADE1S complexes, indicating native association with endogenous chromatin (Fig. 3A, lanes 1 and 3). Strikingly, no histones could be detected in the ΔPHD2 complex (lanes 1 vs 2). Western analysis of the bound chromatin confirmed that PHD2 is required for all HBO1 complexes (JADE1L and JADE1S) to associate with chromatin in vivo (Fig. 3B and C). It also showed the ING5-dependent presence of the H3K4me3 mark, suggesting cooperation with PHD2 (Fig. 3B). In agreement with the observation that overexpressed JADE1/HBO1 complexes make bona fide interactions with chromatin in vivo, we can detect an increase in the bulk levels of histone H4 acetylation on K8 (Fig. S3A), a stimulation fully dependent on JADE1 PHD2 (Fig. S3B).
Figure 3. JADE1 PHD2 is essential for HBO1 binding to chromatin in vivo and for tumor suppressor activity.
(A–B) HBO1 complexes require JADE1 PHD2 to associate with chromatin in vivo, even in the presence of ING5. Additionally, the HBO1-JADE1S complex does not associate with chromatin containing H3K4me in vivo (B). Immunopurified JADE1 complexes from the indicated co-transfected cells were analyzed for subunit associations and endogenous histone binding by gel staining (A). The full set of stoichiometric core histones is labeled on the right. Samples were also analyzed by western blot (B).
(C) JADE1 complexes associate with K36me3-containing chromatin regions in vivo. JADE1L and JADE1S immunopurified complexes from the indicated co-transfections were analyzed by western with the indicated antibodies. The amount of co-purified chromatin is similar between JADE1L and JADE1S but there is significantly more H3K36me3 signal in JADE1S.
(D) JADE1 PHD1 shows some preferential binding to H3K36me3 peptides. Peptide pull-down assays as in Fig. 1D.
(E) Expression of JADE1L strongly inhibits HeLa cell growth while a protein lacking PHD2 does not. Indicated transduced cell lines were seeded at the same density and cells were counted over a period of 7 days. Values are based on duplicate counts of two independent cultures with standard deviation.
(F) JADE1L inhibits anchorage independent cell growth while deletion of PHD2 stimulates it. HeLa cell clones expressing the different constructs were incubated in soft agar for three weeks and colony formation was measured. Values are based on quadruplicate counts of two independent cultures with standard deviation. Examples of image fields are shown at the bottom.
To detect a direct impact of JADE1 PHD2 on cellular processes, we transduced HeLa cells with retroviruses expressing WT and the ΔPHD2 mutants of JADE1L and JADE1S. While we obtained a few clones expressing low levels of JADE1L, we were unsuccessful in obtaining clones for JADE1S (see similar problems reported in (Zhou et al., 2005)). Clones expressing similar levels of JADE1L and its PHD2 mutant (see Fig. S3C) were then analyzed for cell growth. As seen in figure 3E, JADE1L-expressing cells show much slower growth rate than the clone obtained with the empty vector. All the isolated JADE1L clones behaved similarly (data not shown). Strikingly, clones expressing the PHD2 mutant show normal growth, indicating that this chromatin-binding domain is essential for growth suppression by JADE1L. We then measured anchorage-independent cell growth of the transduced clones. As seen in figure 3F, JADE1L expressing cells form significantly fewer colonies in soft agar compared to control cells. It is also clear that the colonies are smaller. In contrast, cells expressing the PHD2 mutant can efficiently form larger colonies than control cells. Thus, recognition of the histone H3 tail by the PHD2 domain of JADE1L on chromatin in vivo is critical for its anti-tumor activity, in part via prevention of anchorage-independent growth.
HBO1 HAT complexes preferentially bind H3K36me3-containing chromatin in the absence of ING subunits
Both JADE1L tetramers and JADE1S dimers copurified with chromatin containing significant levels of H4 acetylation at K8, a direct target of HBO1, and this in vivo binding required JADE1 PHD2 domain (Fig. 3C). Western analyses did not detect co-purification of other methyl histone marks including H3K9me2/3, H3K27me3 and H4K20me2 with HBO1 complexes (data not shown). However, signals for H3K36me3 were observed (Fig. 3C) which encouraged us to directly test this interaction. Native human chromatin pull-downs were performed as in figure 2B and clearly showed that both JADE1L and JADE1S HBO1 complexes can bind H3K36me3-containing chromatin in vitro (Fig. S3D). Interestingly, the level of H3K36me3-containing chromatin bound in vivo was much higher for the JADE1S-HBO1 complex (Fig. 3C, higher signal compared to JADE1L while H4AcK8 and total H3 are equivalent, lanes 1 vs 3). Since H3K36 methylation is found on coding regions of transcribed genes, just downstream of H3K4 methylation, this suggests that HBO1-JADE1S HAT complexes are preferentially located on the coding regions of genes. In fact, peptide pull-down assays reproducibly detected low but specific binding of JADE1 PHD1 to the trimethylated form of H3K36 (Fig. 3D). This was confirmed by measuring binding affinities with shorter peptides by fluorescence spectroscopy (Table SI). On the other hand, loss of H3K36me marks from chromatin had limited impact on the HAT activity of JADE1L and JADE1S complexes in vitro (Fig. S3E). Altogether, these results strongly suggest that ING proteins allow HBO1 complexes to bind and acetylate chromatin near transcription start sites of genes, while JADE proteins do so further downstream in the coding region, likely playing a role during transcription elongation.
JADE1-HBO1 HAT complexes are located on the coding regions of a large number of genes in human cells
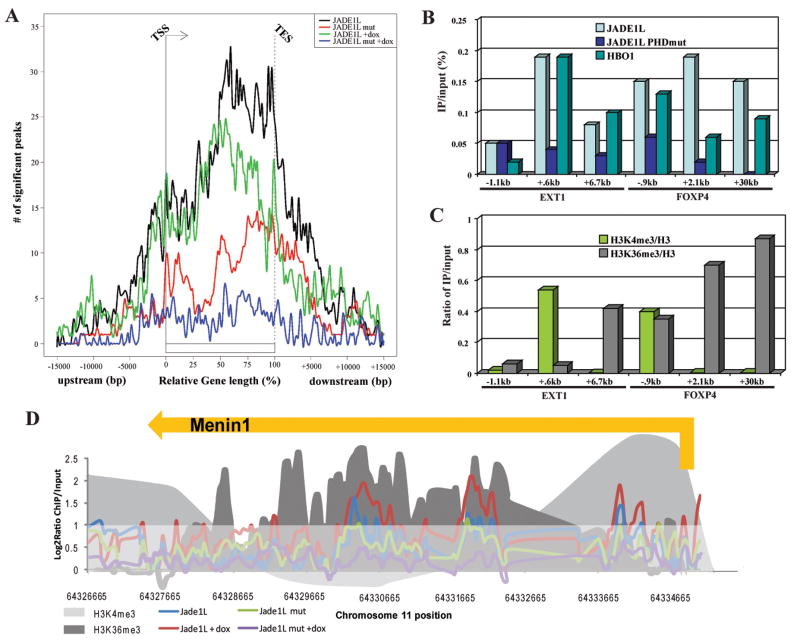
To directly analyze which genes and gene regions are bound by JADE1L complexes in near physiological conditions, we performed chromatin immunoprecipitations (ChIP) with cells expressing low levels of tagged wild-type and ΔPHD2 JADE1L. Since bona fide direct targets for JADE1 were unknown, we labeled input and immunoprecipitated DNA and hybridized to high-resolution tiled arrays covering 1% of the human genome (Consortium, 2004). Since JADE1 and INGs have been implicated in apoptosis and response to stress (Avvakumov and Cote, 2007; Zhou et al., 2005), we also prepared samples from cells treated with the genotoxic drug doxorubicin. A large number of sites with significant binding were detected in wild-type cells in the absence or presence of doxorubicin (823 and 905, respectively; Table SIII). Nearly 50% of these sites are identical between the two samples (data not shown). Importantly, the two samples from the PHD2 mutant show a large reduction of significant binding sites (125 and 298, Table SIII). Using as reference points the mapped transcription start and end sites (TSS and TES) of the known genes on the ENCODE array, compiled data indicate enrichment of binding at TSS and downstream, as well as at TES and upstream, thus placing JADE1L within the body of genes (Fig. S4).
When the vast diversity of gene lengths is normalized to percentage of relative gene length, it becomes even clearer that JADE1L HAT complexes are highly enriched throughout the coding regions of a large number of human genes (Fig. 4A). It is also clear that the PHD2 domain is critical for this binding (Fig. 4A and S4). The shapes of the curves presented in figure 4A also suggest two distinct types of binding on genes: a large dominant one covering most of the coding region from the middle of the genes to the TES; and a smaller region of binding centered around the TSS. Overall, this high-resolution localization analysis of JADE1L on the human genome supports our hypothesis drawn from the biochemical and overexpression experiments, i.e. that JADE1-HBO1 complexes are located on the body of transcribed genes, where chromatin harbors H3K4me3 and H3K36me3 marks. These two H3 methylation marks have also been analyzed recently in the same human cell line by ChIP-chip on high resolution ENCODE arrays (Koch et al., 2007; Lian et al., 2008). We used these published datasets to compare JADE1L binding with regions containing H3K4me3 and H3K36me3. A cluster of 4 genes is shown as an example in figure S5A. A frequent overlap of JADE1L and H3K36me3 peaks is evident. Statistical analysis confirms that JADE1L and H3K36me3 colocalize significantly more frequently than would be predicted by random binding (Table SIV). There is also a notable overlap of JADE1L with H3K4me3 but it is less pronounced. Interestingly, the colocalization of K4me3 and JADE1L is more apparent when the PHD2 domain is deleted, suggesting that the remaining peaks reflect chromatin binding through the ING PHD domain present in the complex (Table SIV).
Figure 4. JADE1L HAT complexes are associated with the coding regions of genes in human cells.
(A) Graphical summary of the data obtained by ChIP-chip assay with JADE1L wild-type and ΔPHD2 proteins in the presence or absence of doxorubicin. Numbers of significant detected peaks of binding are presented in relation to relative position on the body of genes, or distance from transcription start/end sites (TSS/TES) when located outside coding regions.
(B–C) HBO1 colocalizes with JADE1L and histone H3 methylation marks in vivo. ChIP analysis of genes found to be bound by JADE1L in (A) using WT and mutant cells and also cells expressing tagged HBO1. IP/input signals obtained on the indicated loci with empty vector-transduced cells were subtracted from the signal obtained with the tagged proteins (B). H3K4me3 and H3K36me3 signal corrected for nucleosome occupancy (total H3) were also measured on the same loci (C).
(D) ChIP-chip data as in (A) obtained on the Menin1 tumor suppressor gene. H3K4me3 and H3K36me3 signals obtained by ChIP-chip on the same ENCODE sequences in HeLa cells are also depicted (Koch et al., 2007; Lian et al., 2008). Values are presented as log2 ratio of IP/input. The part of the graph below a log2 ratio of 1 is shaded to highlight regions of significant binding.
To independently confirm these results we performed classical ChIP assays with the same cell lines and with another transduced line expressing low levels of FLAG-tagged HBO1 (Doyon et al., 2006). From the genome-wide analysis we selected three distinct regions of two genes that showed significant peaks (see Fig. S5B and C) and analyzed them by realtime PCR. Again, the binding of JADE1L was detected on the TSS and coding regions of EXT1 and FOXP4 genes (Fig. 4B). This binding was similarly dependent on the PHD2 domain. These signals represent actual JADE1L HAT complexes as specific binding of HBO1 is detected at the same locations. Similar analysis of histone H3 methylation status confirmed colocalization of K4me3 and K36me3 signals with JADE1L/HBO1 complexes (Fig. 4C). Other interesting loci bound by the JADE1L complexes include every gene of the HoxA cluster (Fig. S5D) and the menin tumor suppressor gene (Fig. 4D). The latter gene is of particular interest since JADE1L binding seems to be induced by the doxorubicin treatment. Menin has been implicated in apoptosis linked to DNA damage and is an important co-factor of MLL proteins, the major H3K4 methyltranferases frequently implicated in leukemia (Yokoyama et al., 2005).
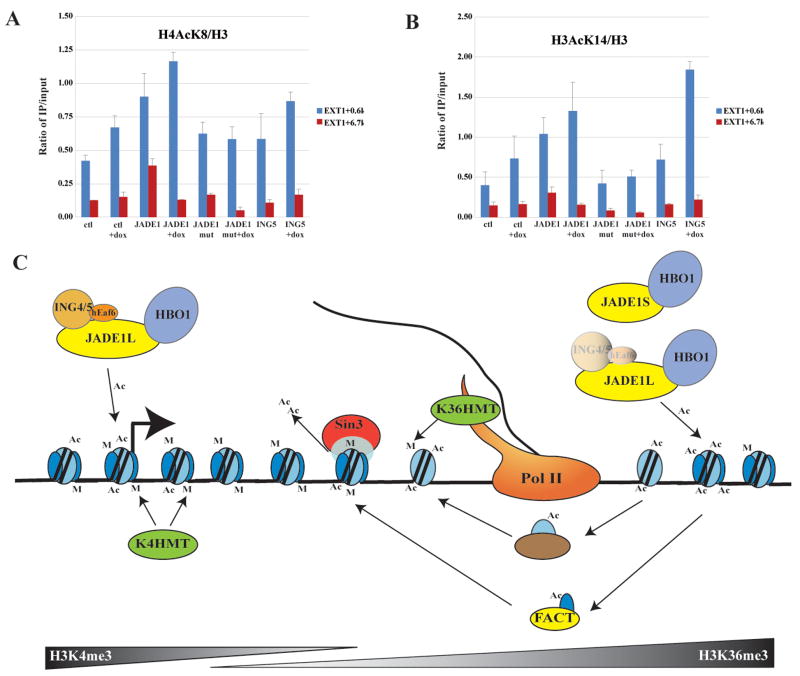
Finally, to support our model of two different mechanisms of HBO1 binding and function on transcription units, we analyzed the status of histone H3 and H4 acetylation on the EXT1 TSS region versus downstream coding region in the transduced cell lines expressing slightly higher levels of total JADE1L proteins. As shown in figure 5A, expression of JADE1L leads to a significant increase of H4K8 acetylation at both promoter and coding regions, while the PHD2 mutant has a limited effect. A transduced ING5-expressing cell line also demonstrates a limited effect but tends to show a slight increase of H4AcK8 at the promoter. The results are significantly different when H3K14 acetylation is measured (Fig. 5B). In this situation, JADE1 expression shows a similar increase of acetylation at the promoter but is much less significant in the coding region. While the PHD2 mutant has no effect, ING5 expression creates the largest increase of H3K14 acetylation in the presence of doxorubicin. This is specific to the promoter region since no increase is detected downstream. Interestingly, even JADE1L expression does not lead to an increase of H3 and H4 acetylation at the coding region in the presence of the drug (Fig. 5A & B). These results support our model proposed from the biochemical analysis, arguing that JADE1-HBO1 complexes largely target H4 in the coding regions, while association of the ING subunit boosts HBO1 action near transcription start sites, favoring H3 and H4 acetylation.
Figure 5. Differential regulation of histone acetylation by JADE1-HBO1 complexes from transcription initiation to elongation.
(A–B) ChIP analysis of histone H3 and H4 acetylation in cells stably expressing JADE1L wild type and ΔPHD2 or ING5, in the presence or absence of doxorubicin. Acetylation levels corrected for nucleosome occupancy were measured near EXT transcription start site and further downstream in the coding region. JADE1L affects acetylation at both loci while ING5 only affects the TSS region and favors H3 acetylation. Values are based on two independent experiments with standard error.
(C) Model for HBO1-dependent acetylation, depicting how different HBO1-JADE HAT complexes can target initiation versus elongation steps of transcription (see text).
A hallmark of ING-HBO1 complexes is the presence of three PHD finger domains in a single complex. We analyzed the functions of the additional PHD modules in the JADE1 protein and found that, as ING PHD, they also bind histone H3 tails with high affinity, but with distinct specificity towards varied K4 methylation status. In agreement with such multiplicity of histone H3 tail binding motifs in a single protein complex, we found that the histone H3 N-terminal region is essential for HBO1 complexes to associate with chromatin. Furthermore, our results indicate that the second PHD in JADE1 is the primary interface between HBO1 complexes and chromatin. A parallel study also confirmed the importance of JADE1 PHD fingers in allowing HBO1 to acetylate chromatin (Foy et al., 2008). Accordingly, the PHD2 domain of JADE1L is essential for its function as growth regulator and putative tumor suppressor.
Our analysis of the naturally occurring isoforms of JADE1 demonstrated that the smaller JADE1S protein forms a dimeric complex with HBO1, which only binds and acetylates chromatin that lacks H3K4me. We also observed that PHD1 of JADE1 has an intrinsic affinity for another histone mark, H3K36me3. These results uncover an intricate and essential coordination of functionally distinct PHD finger domains in a single protein complex to subtly regulate binding to different chromatin regions and the local acetylation specificity on these regions (see Fig. S6).
Our high-resolution analysis of JADE1L binding on the human genome demonstrated that JADE1-HBO1 complexes are indeed present throughout the coding regions of a large number of genes. Thus, HBO1 complexes are very likely involved in the transcription elongation process. Accordingly, we reproducibly detected co-purification of the FACT complex, an important transcription elongation factor, during our affinity purifications of native ING4/5 and HBO1 (not shown; Fig. 5C). In addition, a parallel study has mapped several ING4 target genes in vivo and found an H3K4me3-dependent interaction to be essential for transcription, stress response and apoptosis (Hung et al., 2008). Importantly, the study described a specific role of ING4 PHD in H3 acetylation by HBO1 near transcription start sites, concurring with our data.
Unfortunately, we were unable to differentially map the location of JADE1S versus JADE1L. However, our data favor a model in which two populations of JADE1L complexes interact with transcription units (Fig. 5C). We propose that a population associates with TSS regions through an interaction between the ING PHD and H3K4me3, leading to local H3 and H4 acetylation. Downstream on the bodies of the genes, JADE1L interaction with chromatin is independent of ING subunits, driven mostly by its two PHD domains interacting with the H3 tail and leading to local H4 acetylation. It is interesting to note that a physical disconnection of HBO1 and ING has been reported during initiation of replication, supporting a model of regulated association through JADE proteins (Miotto and Struhl, 2008). ING4 is also known to be targeted by the proteasome and acetylated, a process highly reminiscent of the one regulating yeast ING protein association with the NuA4 HAT complex (Lin et al., 2008; Tsai et al., 2008). Alternatively, ING4 isoforms produced by splice variants and specifically lacking their PHD finger have been described in vivo (Raho et al., 2007). It will be very interesting to decipher the exact roles of JADE and ING PHDs in HBO1 functions in transcription regulation, stress-response, apoptosis and DNA replication. Our data suggest that HBO1 complexes function ahead of the polymerase to stimulate transcription elongation by acetylating nucleosomes and allowing their disassembly (Fig. 5C). This is a highly dynamic process since nucleosomes are known to be rapidly reassembled and deacetylated behind the polymerase (Kouzarides, 2007).
EXPERIMENTAL PROCEDURES
Cell culture, plasmids and recombinant proteins
Details on the plasmid constructions, establishment of cell lines and recombinant proteins are described in the supplemental material.
Antibodies, peptides and native chromatin
Purification of native chromatin from yeast and human cells was described previously (Lacoste et al., 2002). A detailed list of antibodies and peptides used is in the supplemental material.
Cell growth and HAT assays, Fluorescence spectroscopy
Details on the cell growth assays are included in the supplemental material. HAT assays with histone peptides (300 ng), core histones or chromatin (500 ng) prepared from HeLa or yeast cells were performed essentially as described (Lacoste et al., 2002). For peptide binding affinity measurement, tryptophan fluorescence spectra were recorded and analyzed as described before (Champagne et al., 2008) and in the supplemental material.
Affinity purification of protein complexes
Tandem-affinity purifications of native ING complexes from mammalian cells were done as previously described (Doyon et al., 2006). Details about the protocol for affinity purification from cotransfected cells are included in supplemental material.
Peptide, chromatin and GST pull-down assays
Peptide pull-down, GST pull-down and chromatin pull-down assays with affinity-purified complexes were also performed essentially as described (Shi et al., 2006). Additional information is in the supplemental material
ChIP assays and ChIP-chip analysis
Chromatin immunoprecipitations were performed similarly to what has been described (Shi et al., 2006). ChIP-chip assays were performed on 384k human ENCODE arrays (Nimblegen) after amplification and labeling of the ChIP DNA. Hybridization and data analysis were done according to standard Nimblegen protocol. All ChIP-chip data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE14309. More details are in the supplemental material.
Supplementary Material
Acknowledgments
We apologize for omitted references due to space constrains. We thank Patricia Savard and Josée Lavoie for technical help and Nicolas Lacoste, Song Tan, Bruno Amati and Maria Panchenko for reagents. We also thank Maria Panchenko for sharing unpublished data, Rhea Utley for text editing and Dorine Rossetto for art work. This work was supported by grants from the Canadian Institutes of Health Research (CIHR) to J. C., the National Cancer Institute of Canada (NCIC) to X.-J. Y. and the National Institutes of Health (NIH) to O. G. and T. G. K.. K. S. C. holds an American Cancer Society Fellowship and N. A. a CIHR/Institute of Aging fellowship. O. G. holds Searle Scholar and Burroughs Wellcome Career Awards and J. C. is a Canada Research Chair, Tier I.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Avvakumov N, Cote J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene. 2007;26:5395–5407. doi: 10.1038/sj.onc.1210608. [DOI] [PubMed] [Google Scholar]
- Champagne KS, Saksouk N, Pena PV, Yang XJ, Johnson K, Cote J, Kutateladze TG. The crystal structure of the ING5 PHD finger in complex with an H3K4me3 histone peptide. Proteins. 2008;72:1371–1376. doi: 10.1002/prot.22140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium TEP. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- Doyon Y, Cayrou C, Ullah M, Landry AJ, Cote V, Selleck W, Lane WS, Tan S, Yang XJ, Cote J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Foy RL, Song IY, Chitalia VC, Cohen HT, Saksouk N, Cayrou C, Vaziri C, Cote J, Panchenko MV. Role of Jade-1 in the histone acetyltransferase (HAT) HBO1 complex. J Biol Chem. 2008;283:28817–28826. doi: 10.1074/jbc.M801407200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Binda O, Champagne KS, Kuo AJ, Johnson K, Chang HY, Simon MD, Kutateladze TG, Gozani O. ING4 mediates crosstalk between histone H3 K4 trimethylation and H3 acetylation to attenuate cellular transformation. Mol Cell. 2008 doi: 10.1016/j.molcel.2008.12.016. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Binda O, Champagne KS, Kuo AJ, Johnson K, Chang HY, Simon MD, Kutateladze TG, Gozani O. ING4 mediates crosstalk between histone H3 K4 trimethylation and H3 acetylation to attenuate cellular transformation. Mol Cell. 2009 doi: 10.1016/j.molcel.2008.12.016. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y. Histone H3 Arg2 methylation provides alternative directions for COMPASS. Nature structural & molecular biology. 2007;14:1058–1060. doi: 10.1038/nsmb1107-1058. [DOI] [PubMed] [Google Scholar]
- Koch CM, Andrews RM, Flicek P, Dillon SC, Karaoz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, et al. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome research. 2007;17:691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lacoste N, Utley RT, Hunter JM, Poirier GG, Cote J. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J Biol Chem. 2002;277:30421–30424. doi: 10.1074/jbc.C200366200. [DOI] [PubMed] [Google Scholar]
- Lian Z, Karpikov A, Lian J, Mahajan MC, Hartman S, Gerstein M, Snyder M, Weissman SM. A genomic analysis of RNA polymerase II modification and chromatin architecture related to 3′ end RNA polyadenylation. Genome research. 2008;18:1224–1237. doi: 10.1101/gr.075804.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YY, Qi Y, Lu JY, Pan X, Yuan DS, Zhao Y, Bader JS, Boeke JD. A comprehensive synthetic genetic interaction network governing yeast histone acetylation and deacetylation. Genes Dev. 2008;22:2062–2074. doi: 10.1101/gad.1679508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto B, Struhl K. HBO1 histone acetylase is a coactivator of the replication licensing factor Cdt1. Genes Dev. 2008;22:2633–2638. doi: 10.1101/gad.1674108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raho G, Miranda C, Tamborini E, Pierotti MA, Greco A. Detection of novel mRNA splice variants of human ING4 tumor suppressor gene. Oncogene. 2007;26:5247–5257. doi: 10.1038/sj.onc.1210335. [DOI] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman MA, Riabowol K. After a decade of study-ING, a PHD for a versatile family of proteins. Trends Biochem Sci. 2007;32:509–519. doi: 10.1016/j.tibs.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Tsai KW, Tseng HC, Lin WC. Two wobble-splicing events affect ING4 protein subnuclear localization and degradation. Exp Cell Res. 2008;314:3130–3141. doi: 10.1016/j.yexcr.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123:207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- Zhou MI, Foy RL, Chitalia VC, Zhao J, Panchenko MV, Wang H, Cohen HT. Jade-1, a candidate renal tumor suppressor that promotes apoptosis. Proc Natl Acad Sci U S A. 2005;102:11035–11040. doi: 10.1073/pnas.0500757102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.