Abstract
DNA composition in general, and codon usage in particular, is crucial for understanding gene function and evolution. CodonExplorer, available online at http://bmf.colorado.edu/codonexplorer/, is an online tool and interactive database that contains millions of genes, allowing rapid exploration of the factors governing gene and genome compositional evolution and exploiting GC content and codon usage frequency to identify genes with composition suggesting high levels of expression or horizontal transfer.
Contact: rob@spot.colorado.edu
1 INTRODUCTION
Nucleotide, codon and amino acid preferences vary greatly among genes and organisms. Codon usage preferences can occur because there are 64 codons but only 20 amino acids (with exceptions), so some amino acids are encoded by multiple codons. The earliest studies of nucleotide frequencies (Sueoka, 1961) suggested that organisms have different biases towards certain synonyms. Even the first gene sequences confirmed that codon usage varies greatly in different organisms. These systematic biases can be exploited to perform many analyses of great theoretical and practical interest.
Many factors affect codon usage. For example, the Codon Adaptation Index (CAI) (Sharp and Li, 1987) measures similarity to codon usage of known highly expressed genes and correlates with overall expression. However, despite the effects of selection for amino acid and codon usage, strong linear trends relate the GC content of the whole genome to the GC content at each codon position across many organisms, suggesting that mutational biases also play a crucial role in shaping codon usage (Muto and Osawa, 1987). Indeed, multivariate analyses typically identify expression levels and genomic GC content as the principal factors structuring composition of individual genes (Gupta and Ghosh, 2001). Thus selection and mutation are key codon usage determinants.
Codon usage also allows insight into mutational processes operating in different genomes (e.g. Sueoka, 2002), or in different parts of the same genome where regions of compositional heterogeneity such as isochores exist (Bernardi, 1993). Codon usage can also suggest horizontal gene transfer (HGT), the movement of genes between different genomes, because different genomes have different characteristic compositions (Karlin et al., 1998).
CodonExplorer, built using PyCogent (Knight et al., 2007), provides a platform for rapid testing of hypotheses about codon usage in sequenced genes and genomes. By precomputing statistics from whole-genome databases, using thousands of CPU-hours, CodonExplorer provides graphical summaries of vast datasets consisting of millions of genes and hundreds of genomes in seconds.
CodonExplorer is especially effective for revealing patterns associated with gene expression changes, mutational biases and HGT. It allows users to conveniently retrieve genes by genome, function or orthology, and then to visualize the composition of these genes. Analyses include:
P1 and P2 versus P3 GC: the effects of selection for amino acid usage and mutational bias can be contrasted by plotting GC content at the first or second codon positions (P1 and P2) against that at the third position (P3) (Sueoka, 1995).
Codon fingerprint and PR2 bias plots: the ratio of different kinds of codons can provide insights into whether deamination or oxidation contributes more to the pattern of codon usage in a specific organism via techniques such as the fingerprint plot and the PR2 bias plot (Sueoka (2002) has examples of both: see references contained therein to prior work). Chargaff's second parity rule (PR2) states that within each DNA strand, the frequency of A ≈ T and the frequency of G ≈ C (Rudner et al., 1969). In the fingerprint plot, circles representing different amino acids are plotted such that the location of each circle on the y-axis represents the frequency of A at position 3 in codons of that amino acid relative to the frequencies of A and T at that position A3/(A3+T3), while the position on the x-axis represents G3/(G3+C3). The radius of each circle is proportional to the relative frequency of that amino acid. PR2 bias plots show the extent of bias relative to P3.
Histograms: histograms can be constructed of CAI values, GC content at the third codon position (P3), gene length or peptide hydrophobicity, and can include a customizable Monte Carlo analysis to test whether observed differences between the properties of a selected subset of genes and the rest of the genome are statistically significant.
2 EXAMPLES OF CODONEXPLORER USAGE
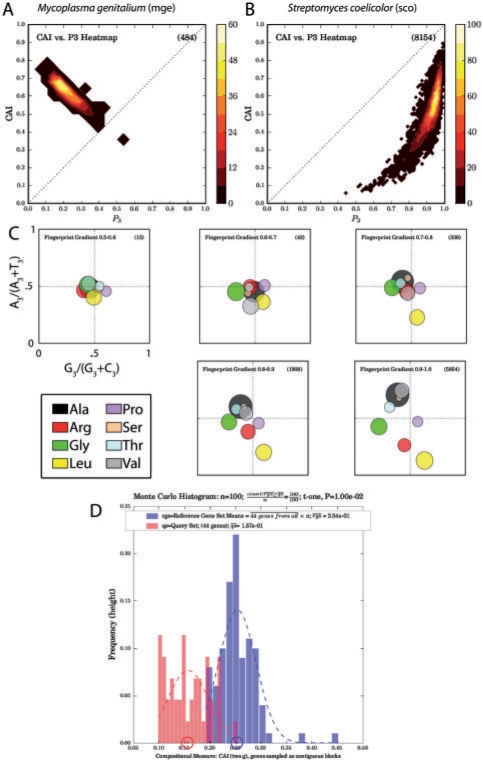
Histograms of CAI values, and plots of CAI against GC content at the third codon position (P3), can provide insight into selection for translational efficiency or mutational effects on codon usage. Figure 1a and b shows how two different genomes, Mycoplasma genitalium (low-GC) and Streptomyces coelicolor (high-GC), differ in compositional evolution: genes that fit each genome's overall GC preference are more likely to be highly expressed with high CAI values. Figure 1c shows fingerprint plots from different GC ranges of the S.coelicolor genome: at lower (non-preferred) GC, codon usage is relatively unbiased, whereas at higher GC distinct preferences for specific codons are apparent.
Fig. 1.
Examples of CodonExplorer output. (a and b) Plots of CAI (a measure correlated with expression) against third position GC content for two genomes with extreme codon bias, M.genitalium and S.coelicolor. (c) Fingerprint plots broken down by ranges of per-gene GC content: codon usage is more biased at higher GC content. (d) Monte Carlo histogram of CAI values for the Salmonella pathogenicity island 2 (SPI-2), which differs significantly (P=0.01) in average CAI values from the genome as a whole using a t-test to compare the actual mean to a distribution of means of other genes: these should be approximately normally distributed due to the Central Limit Theorem.
CodonExplorer can also employ Monte Carlo techniques for testing the statistical significance of differences in codon usage or nucleotide sequence composition between putatively transferred sets of genes and the genome as a whole. Figure 1d shows unusually low CAI for the SPI-2 pathogenicity island in the genome of Salmonella enterica serovar Typhimurium LT 2, consistent with the hypothesis that this region underwent HGT.
By allowing users to rapidly perform a wide array of compositional analyses on customizable gene collections, CodonExplorer provides a powerful platform for investigating many phenomena.
ACKNOWLEDGEMENTS
We thank Justin Kuczynski and other members of the Knight lab for comments on drafts of the manuscript. James Michaelis and Brice Pelle contributed to interface design in the course CSCI4838. CodonExplorer is hosted on the W.M. Keck Foundation Bioinformatics Facility at CU Boulder.
Funding: NIH/CU Molecular Biophysics and Signaling & Cell Regulation Training Programs (T32GM065103 and T32GM08759, in part); NSF EAPSI fellowship (OISE0812861, in part); National Institutes of Health (P01DK078669, in part); CU URAP and HHMI/UROP scholarships (in part).
Conflict of Interest: none declared.
REFERENCES
- Bernardi G. The vertebrate genome: isochores and evolution. Mol. Biol. Evol. 1993;10:186–204. doi: 10.1093/oxfordjournals.molbev.a039994. [DOI] [PubMed] [Google Scholar]
- Gupta SK, Ghosh TC. Gene expressivity is the main factor in dictating the codon usage variation among the genes in Pseudomonas aeruginosa. Gene. 2001;273:63–70. doi: 10.1016/s0378-1119(01)00576-5. [DOI] [PubMed] [Google Scholar]
- Karlin S, et al. Codon usages in different gene classes of the Escherichia coli genome. Mol. Microbiol. 1998;29:1341–1355. doi: 10.1046/j.1365-2958.1998.01008.x. [DOI] [PubMed] [Google Scholar]
- Knight R, et al. PyCogent: a toolkit for making sense from sequence. Genome Biol. 2007;8:R171. doi: 10.1186/gb-2007-8-8-r171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobry JR, Sueoka N. Asymmetric directional mutation pressures in bacteria. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-10-research0058. RESEARCH0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A, Osawa S. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc. Natl Acad. Sci. USA. 1987;84:166–169. doi: 10.1073/pnas.84.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner R, et al. Separation of microbial deoxyribonucleic acids into complementary strands. Proc. Natl Acad. Sci. USA. 1969;63:152–159. doi: 10.1073/pnas.63.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li WH. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka N. Compositional correlation between deoxyribonucleic acid and protein. Cold Spring Harb. Symp. Quant. Biol. 1961;26:35–43. doi: 10.1101/sqb.1961.026.01.009. [DOI] [PubMed] [Google Scholar]
- Sueoka N. Intrastrand parity rules of DNA base composition and usage biases of synonymous codons. J. Mol. Evol. 1995;40:318–325. doi: 10.1007/BF00163236. [DOI] [PubMed] [Google Scholar]
- Sueoka N. Wide intra-genomic G+C heterogeneity in human and chicken is mainly due to strand-symmetric directional mutation pressures: dGTP-oxidation and symmetric cytosine-deamination hypotheses. Gene. 2002;300:141–154. doi: 10.1016/s0378-1119(02)01046-6. [DOI] [PubMed] [Google Scholar]