Abstract
Motivation: Ever increasing amounts of biological interaction data are being accumulated worldwide, but they are currently not readily accessible to the biologist at a single site. New techniques are required for retrieving, sharing and presenting data spread over the Internet.
Results: We introduce the DASMI system for the dynamic exchange, annotation and assessment of molecular interaction data. DASMI is based on the widely used Distributed Annotation System (DAS) and consists of a data exchange specification, web servers for providing the interaction data and clients for data integration and visualization. The decentralized architecture of DASMI affords the online retrieval of the most recent data from distributed sources and databases. DASMI can also be extended easily by adding new data sources and clients. We describe all DASMI components and demonstrate their use for protein and domain interactions.
Availability: The DASMI tools are available at http://www.dasmi.de/ and http://ipfam.sanger.ac.uk/graph. The DAS registry and the DAS 1.53E specification is found at http://www.dasregistry.org/.
Contact: mario.albrecht@mpi-inf.mpg.de
Supplementary information: Supplementary data and all figures in color are available at Bioinformatics online.
1 INTRODUCTION
Molecular interactions are of fundamental importance to many biological processes (BPs). In recent years, the amount of interaction data has increased substantially due to growing attention by the scientific community as well as the widespread use of high-throughput techniques that afford screening of vast numbers of molecules. Nowadays, large-scale protein interaction maps are available for model organisms like yeast, fly and worm (Goll and Uetz, 2007), and the current research focus is shifting towards interaction screens for human (Cusick et al., 2005; Stelzl and Wanker, 2006). In addition, computational methods have been developed for predicting molecular interactions, some of which reach prediction quality comparable to that of experimental high-throughput data (Ramírez et al., 2007; Shoemaker and Panchenko, 2007). However, this rapid accumulation of interaction data makes it difficult for scientists to keep track of all available information and data sources. The unification of heterogeneous and decentralized interaction data is thus a prerequisite for an effective study of interactomes (Brazma et al., 2006).
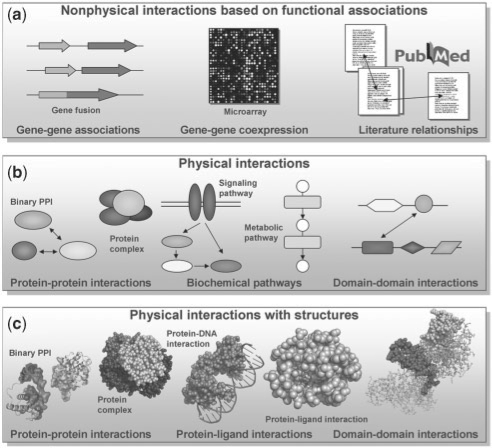
Molecular interactions can be studied at different levels of detail (Fig. 1). In general, physical and non-physical interaction types can be distinguished. While a physical interaction implies a real contact between the interacting molecules (interactors), the other interaction type denotes a purely functional association between them. For instance, such associations can be based on similar genomic contexts, coexpression analyses or literature relationships (Jensen et al., 2006). Physical interactions between proteins may involve two and more proteins, forming binary interactions and protein complexes (Frishman et al., 2009). In particular, protein–protein interactions are formed by the physical contact of binding sites, which are frequently evolutionarily conserved in domains of protein families (Finn et al., 2008). Further protein interactions exist with other ligands, for instance, nucleic acids, lipids and certain small molecules in signaling or metabolic pathways. Techniques like X-ray crystallography, NMR spectroscopy or 3D structure modeling can provide even more molecular details by identifying the interacting atoms or residues in the protein binding sites (Aloy and Russell, 2006; Finn et al., 2005).
Fig. 1.
Levels of molecular interactions. Physical and non-physical interaction types can be distinguished: (a) Non-physical interactions are based on functional associations. (b) Physical interactions imply a direct physical contact between the interacting molecules like proteins, domains or small ligands. (c) Raising the level of detail to 3D structural data, the interacting atoms or residues of binding sites can be identified.
A number of databases keep track of experimentally determined protein interactions (Bader et al., 2003; Breitkreutz et al., 2008; Chatr-Aryamontri et al., 2007; Güldener et al., 2006; Kerrien et al., 2007a; Keshava Prasad et al., 2009; Salwinski et al., 2004). Such databases are essential components of interactomics, however, each of them contains information not found in other databases (Mathivanan et al., 2006). The IMEx consortium formed by eight major interaction data providers aims at overcoming the fragmentation by sharing the curation effort and exchanging curated protein interaction records among its members (Orchard et al., 2007). However, IMEx and its member databases are restricted to experimentally determined protein interactions, which cover only a fraction of the estimated interactomes (Stumpf et al., 2008). Voluminous data on predicted protein and domain interactions that have been made publicly available by different research groups (Schelhorn et al., 2008; Schlicker et al., 2007; Shoemaker and Panchenko, 2007) are not included.
To provide broader data access to currently available interactomes, several integration frameworks have appeared recently. We will refer to the methodology underlying these frameworks as static data integration because either the user is assisted in building a local data warehouse (Aragues et al., 2006; Lee et al., 2006b; Shah et al., 2005; Shannon et al., 2006) or the software facilitates access to central data repositories via web interfaces (Birkland and Yona, 2006; Breitkreutz et al., 2008; Cerami et al., 2006; Chaurasia et al., 2009; Goll et al., 2008; Hoffmann and Valencia, 2004; Jensen et al., 2009; Pagel et al., 2008; Prieto and Rivas, 2006; Raghavachari et al., 2008; Tarcea et al., 2009) or by software plugins, for instance, as available for Cytoscape (Avila-Campillo et al., 2007; Cerami et al., 2006; Hernandez-Toro et al., 2007; Shannon et al., 2003; Tarcea et al., 2009). However, static integration has the drawback of providing only a snapshot of a fixed number of data sources at a certain point of time. Once the data have been included into the central repository, curation efforts are required to keep them up to date and in sync with the original data source. This permanent update problem can be aggravated by possible format changes of the source, which hampers further data processing. Apart from that, these integration frameworks are rather rigid because the inclusion of additional datasets like new experimental data or the results of a novel prediction method can normally be accomplished solely by the central authority.
A data fragmentation situation similar to the current diversity of interaction data arose with genomic data several years ago. One possible solution to the integration of genomic data and their annotations was the Distributed Annotation System (DAS) (Dowell et al., 2001). In general, it is anticipated that decentralization will become an important data-sharing concept in the future (Murray-Rust, 2008; Stein, 2008; Thorisson et al., 2009). DAS is based on a client–server architecture in which numerous decentralized servers offer annotations of a reference entity provided by another server. The combination and visualization of a reference entity and its annotations is performed in a DAS client.
We aim to overcome the shortcomings of the aforementioned static data integration frameworks by adopting and extending a DAS-based approach for the exchange of molecular interaction data and their annotations. Instead of unifying all available interaction data into a central database, the interaction data remain with their original providers and are retrieved and integrated online on request. This eliminates the issue of centralized data maintenance and ensures that the interaction data are always kept up to date. Our system, named DAS for Molecular Interactions (DASMI), is sufficiently generic to support all types of interaction data described above. It is not restricted to protein interactions and considers both experimentally determined and predicted interactions. This is the main distinguishing feature from interaction repositories like HPRD (Keshava Prasad et al., 2009) or IntAct (Kerrien et al., 2007a). Instead of competing with them, we want to supplement their data with additional sources and help the user to assess the available information.
In the following section, we will introduce the distributed architecture of DASMI and describe its components: the specification of a DAS extension for the exchange of interaction data and their annotations as well as the software libraries that implement the new specification in servers and clients. Finally, we will demonstrate the exemplary use of DASMI for protein and domain interactions.
2 METHODS
2.1 Distributed architecture
DAS (Dowell et al., 2001) is a data integration approach with the main goal of replacing central data repositories with distributed storage systems. DAS is built on a client–server architecture, consisting of two types of servers, namely, reference and annotation servers, and a client for visualization purposes. Reference servers provide the biological reference entity, for example, a nucleotide or peptide sequence. Annotation servers make additional information available that is related to the reference sequence, for instance, information on exons or protein domains. Coordinate systems are used to define the entities that a DAS server provides or annotates, for example, chromosomes, genes, protein sequences or protein structures (Prlić et al., 2007). A data exchange specification handles the communication between DAS clients and DAS servers by prevalent techniques, namely HTTP URL requests and XML responses.
Originally, DAS was designed for the exchange of annotations of DNA sequences. In recent years, several extensions to the protocol have widened its use to other areas (Jenkinson et al., 2008): Protein DAS affords the exchange of protein sequence annotations and alignments (Jones et al., 2005), 3D-DAS utilizes DAS for the annotation of protein structure alignments (Prlić et al., 2005) and 3D-EM DAS for electron microscopy (Macías et al., 2007). In addition to these DAS extensions, a registry has been developed that maintains a list of available DAS servers and thus allows DAS clients finding suitable servers (http://www.dasregistry.org/).
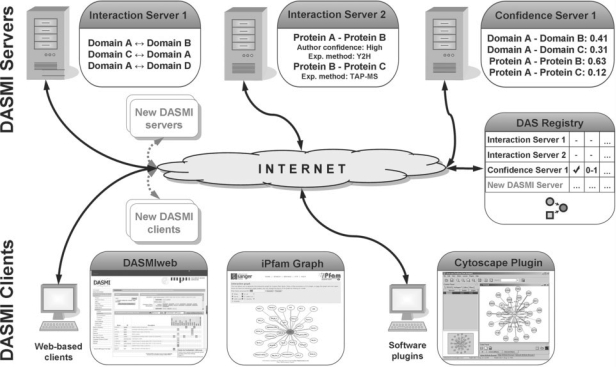
DASMI aims at resolving the problems of current integration frameworks for interaction data by transferring the idea of DAS into the field of molecular interactions. This includes the specification of a DAS extension defining the data exchange between servers and clients as well as reference implementations of servers and clients (Fig. 2).
Fig. 2.
Schematic DASMI system architecture. The DASMI architecture is similar to the original DAS architecture (Dowell et al., 2001). Interaction servers provide interactions (Interaction Server 1) and, optionally, additional information like experimental conditions (Interaction Server 2). Confidence servers provide confidence scores for interactions (Confidence Server 1). DASMI clients query interaction servers and combine their results. The DAS registry maintains a list of available DAS servers.
As in the original DAS architecture, there are different server types in the DASMI framework. The majority of servers provide interaction data and optionally additional information; these are the equivalents of DAS reference servers. Examples of additional information, which can be associated specifically with an interaction, include the known or predicted interaction regions, the strength and type of the interaction, or the conditions in which the interaction occurs. Confidence servers are comparable with DAS annotation servers and provide reliability scores for potential interactions. Notably, the HUPO-PSI community wants to utilize our distributed scoring architecture for a common confidence scoring system for protein–protein interactions (Orchard et al., 2008).
Each interaction or confidence server belongs to a certain coordinate or identifier system, which specifies how interactions can be requested and how they are returned. For instance, a data source with the Entrez Gene identifier system may be queried for interactions by using gene identifiers, another data source with the UniProtKB identifier system by using protein identifiers. DASMI clients thus need to transform the results of servers from different identifier systems in order to unify them.
2.2 DASMI data exchange specification
Data exchange between interaction servers and clients requires a DAS URL and XML specification. An advantage of a well-defined data exchange specification is the resulting modularity and extensibility of the system. New servers and clients can be readily incorporated if they follow the specification and thus communicate with the existing parts in a well-defined manner.
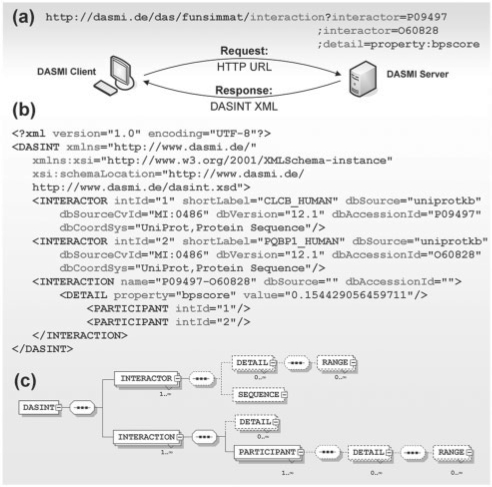
We extend the DAS specification by the new interaction command and the associated DASINT XML response format. This extension is part of the DAS 1.53E specification (Jenkinson et al., 2008). Figure 3 shows an interaction request and the associated DASINT response for an exemplary protein–protein interaction.
Fig. 3.
Exemplary interaction request and DASINT response. The communication between DASMI server and client is performed using formatted URL requests and XML responses. (a) Interactions are requested using the interaction command. In the example shown here, all protein interactions involving the proteins P09497 and O60828 and annotated with a BPscore would be retrieved. (b) The server response is a DASINT XML format. (c) Overview of the DASINT XML Schema Definition. Mandatory elements are marked using solid frames, optional elements have dashed frames.
Requests to a DASMI server are issued in the same form of a formatted URL request as those to a standard DAS server (Fig. 3a). The new command for requesting interactions is interaction and offers additional query parameters of three types: interactor, operation and detail. Please refer to Supplementary Data File 1 for the full data exchange specification.
The response of a DASMI server to an interaction request is a DASINT XML document (Fig. 3b). In contrast to the widely adopted PSI-MI XML2.5 format (Kerrien et al., 2007b), which provides an extensive specification with numerous elements and a deeply branched hierarchy, DASINT uses a concise and flexible document format. PSI-MI XML2.5 and DASINT can thus be regarded as complementary approaches: whilst PSI-MI XML2.5 has the goal of describing experimentally determined interactions in detail, naturally resulting in very complex documents, DASINT provides a lightweight intermediate exchange format, which facilitates fast communication between clients and servers. In this regard, DASINT is comparable with MITAB2.5 (Kerrien et al., 2007b), the simplified tabular version of PSI-MI XML2.5. However, DASINT is more versatile because it supports, for example, the representation of protein complexes without the need of transforming the data into a spoke or matrix model. A more detailed differentiation of DASINT from alternative data exchange formats can be found in Supplementary Data File 1. Figure 3c shows an illustration of the DASINT XML Schema Definition. The complete definition of the proposed DASINT XML format can be found in Supplementary Data File 2.
2.3 DASMI server
A DASMI server responds to an interaction request by providing interaction data in the DASINT XML format defined above. One of the objectives of DAS and thus of DASMI is to make the setup of DAS servers easy. To achieve this aim, three versatile open-source DAS server libraries are available, Dazzle (http://www.biojava.org/wiki/Dazzle) and MyDas (http://code.google.com/p/mydas/) for Java, and ProServer (Finn et al., 2007) for Perl (http://www.sanger.ac.uk/Software/analysis/proserver/). We provide DASMI reference server implementations by extending Dazzle and ProServer, while MyDAS is being extended by the DAS community.
The stand-alone servers Dazzle and ProServer both work in a modular fashion. They consist of a server core, which provides basic functionalities like handling requests and responses, and components, which manage specific DAS commands and data storage formats. Existing DataSource classes (Dazzle) and Transport modules (ProServer) act as brokers between the underlying interaction data and the modules that build the DASINT XML response. This simplifies access to a range of data storage formats, for instance, PSI-MI XML2.5 documents (Kerrien et al., 2007a) or flat files in the Simple Interaction Format (SIF) defined for Cytoscape (Cline et al., 2007; Shannon et al., 2003). In order to set up a new server, data providers need only to use an existing or implement a new module that is tailored to the specifics of their molecular interaction data.
2.4 Interaction datasets
DASMI has been developed to support molecular interactions at different levels (Fig. 1). To demonstrate its use, we set up DASMI servers for a collection of protein–protein interaction datasets: two large-scale experimental datasets [CCSB-HI1 (Rual et al., 2005) and MDC (Stelzl et al., 2005)], four curated experimental datasets [DIP (Salwinski et al., 2004), HPRD (Keshava Prasad et al., 2009), IntAct (Kerrien et al., 2007a) and MINT (Chatr-Aryamontri et al., 2007)] and six predicted datasets [Bioverse (McDermott et al., 2005), HiMAP (Rhodes et al., 2005), HomoMINT (Persico et al., 2005) OPHID (Brown and Jurisica, 2005), POINT (Huang et al., 2004) and Sanger (Lehner and Fraser, 2004)]. More information on these datasets is found in Ramírez et al. (2007). In addition, several domain–domain interaction datasets are offered by DASMI servers: three experimental datasets derived from 3D structures obtained by X-ray crystallography or NMR spectroscopy [3did (Stein et al., 2009), iPfam (Finn et al., 2005) and PiNS (Bordner and Gorin, 2008)] and 11 predicted datasets (Chen and Liu, 2005; Guimarães et al., 2006; Jothi et al., 2006; Lee et al., 2006a; Liu et al., 2005; Ng et al., 2003; Pagel et al., 2008; Riley et al., 2005; Schelhorn et al., 2008; Wang et al., 2007; Wuchty, 2006), see Supplementary Data File 3. Moreover, we set up two confidence servers, FunSimMat (Schlicker and Albrecht, 2008; Schlicker et al., 2006) and Domain support (Finn et al., 2005; Ramírez et al., 2007), which can be used to assess the reliability of protein interactions.
Of course, our current selection of data sources, with the majority of them temporarily maintained at our institute, serves only as a prototype for the capabilities of our system because it necessitates the replication of some interaction datasets, resulting in the same update problem the aforementioned central repositories are facing. However, for the near future, we already know from other scientists that new sources for interactions and confidence measures will be made available at other institutions.
2.5 DASMI client
A DASMI client offers the user an easy way of communicating with various DASMI servers without having to know any data exchange specification details. Subsequent to a user request, a DASMI client will contact all DASMI servers, retrieve and unify the interaction data and present the results to the user. A list of all publicly available DASMI servers is provided by the DAS registry (Prlić et al., 2007).
To facilitate the development of new DASMI clients, the two existing open-source DAS client libraries, Dasobert (http://www.spice-3d.org/dasobert/) in Java and Bio-Das-Lite (http://search.cpan.org/dist/Bio-Das-Lite/) in Perl, have been upgraded to support our interaction extension.
2.5.1 Identifier mapping
Proteomics affords a substantial diversity of object identifiers, ranging from RefSeq and Entrez Gene identifiers to UniProtKB accession numbers. Accordingly, protein interaction datasets use different identifier systems to describe their interactions. In order to unify them, a DASMI client has to convert between various systems to incorporate servers that have different identifier systems. This mapping procedure can produce considerable computational overhead. For instance, while there is usually a one-to-one mapping from UniProtKB to Entrez Gene identifiers, mapping in the opposite direction may produce multiple results as one gene can be responsible for several protein variants or fragments. Therefore, a DASMI client might need to issue multiple queries to retrieve all protein–protein interactions for one gene. Furthermore, the mapping procedure implemented by a DASMI client determines if it is able to distinguish splice variants. In contrast, the identifier diversity for domain interaction datasets is less problematic as stable Pfam identifiers (Finn et al., 2008) are predominantly used.
3 RESULTS
On the basis of the client libraries Dasobert and Bio-Das-Lite, two DASMI clients have been developed to illustrate the potential of our new system: the DASMIweb client as an entry gate to various protein–protein and domain–domain interaction datasets and the iPfam graphical domain interaction browser that uses DASMI to incorporate predicted domain–domain interactions into its results.
3.1 DASMIweb for protein and domain interactions
The DASMI client DASMIweb is publicly accessible at http://www.dasmi.de/web/. The aim of DASMIweb is to establish a starting point for interactome studies by consolidating protein and domain interaction data from various sources.
3.1.1 User interface
The DASMIweb user interface is designed to be clear and intuitive (Fig. 4a). The screen window is divided into several panels; permanent panels are the Query Panel in the top left corner of the window and the Information Panel in the top right corner. Interactions are presented within the Interaction Panel, located in the central part of the window. The configuration of DASMIweb can be managed in the optional Source Configuration Panel. The DASMIweb user interface relies heavily on the use of Asynchronous JavaScript and XML (AJAX) (Jimenez et al., 2008; Sagotsky et al., 2008). This technique is required to present interactions to the user as soon as they are received from a DASMI server. The asynchronous communication is provided by the Java framework Direct Web Remoting (DWR, http://getahead.org/dwr/).
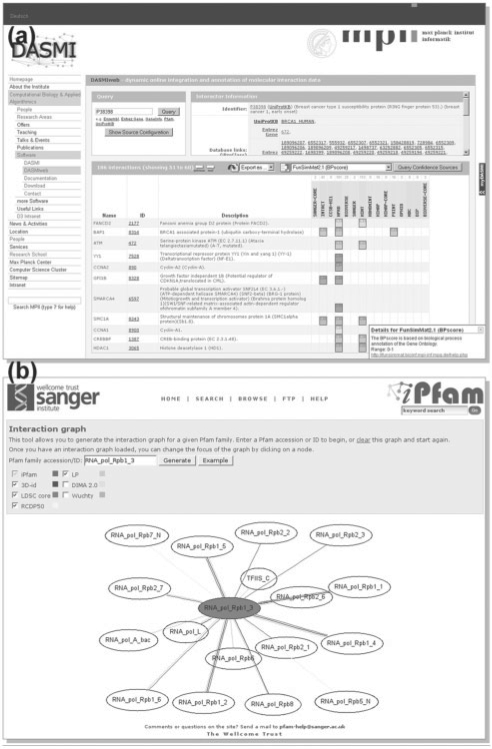
Fig. 4.
DASMI clients DASMIweb and iPfam. (a) DASMIweb is an online tool for dynamically unifying protein and domain interaction data and additional annotations. The results are presented in tabular form: each column represents a DASMI server, each row contains an interaction partner, and squares at the intersections of rows and columns indicate interactions. In this figure, the interaction squares are colored according to the functional similarity between the interacting proteins, from dark blue for high to white for low similarity. (b) The iPfam client tool combines domain–domain interactions from several sources with interactions reported in the iPfam database. The results are presented in graphical form; protein domains are depicted as ovals and interactions as edges that connect ovals. Different edge colors distinguish individual data sources.
3.1.2 Querying
To make querying DASMIweb as intuitive as possible, the Query Panel contains only a single search field. There is no need for the user to specify the type of the query. The system will use internal identifier mapping tables derived from iProClass (Huang et al., 2003) and Pfam (Finn et al., 2008) to automatically determine whether the input is a gene, protein or domain identifier. If the identifier cannot be mapped unambiguously, the user is asked to refine the query. Furthermore, DASMIweb will map the query to all compatible identifier systems in order to maximize the number of data sources that can be used to answer the user query. For instance, if the user searches by an Entrez Gene identifier, DASMIweb will not only query all data sources in the Entrez Gene identifier system, but will also try to convert the identifier to UniProtKB, GeneInfo, RefSeq and Ensembl identifiers to cover all data sources in the respective identifier systems. If a mapping results in multiple identifiers, for instance, when mapping from gene to protein identifiers, all combinations of identifiers are used. A more detailed description of the mapping procedure and exemplary mappings can be found in the online documentation at http://www.dasmi.de/.
3.1.3 Presentation of results
Interactions are presented to the user in tabular form within the Interaction Panel. In the central table, columns represent data sources that have been contacted for the user query, rows correspond to different interactions, and squares in the intersections of rows and columns indicate particular interactions reported by a specific source (Fig. 4a). If an interaction is binary, the row contains the interaction partner of the query interactor, but if the interaction is complex, all interaction partners are presented in a single row that is highlighted. This tabular representation affords an intuitive, visual judgment of the results since an interaction reported by multiple sources, as shown by several squares in the same row, may be more likely to be accurate. A more detailed assessment of the interactions can be performed by applying confidence measures as described below.
The user can also request further information about an interaction, such as experimental details and confidence scores, by clicking on the associated ‘interaction square’. These annotation details are an optional feature provided by the individual data sources. Therefore, they might not be available for all interactions.
3.1.4 Source configuration and data export
In its initial configuration, DASMIweb incorporates all available data sources that are compatible with a user query. The user can change this selection in the Source Configuration Panel. This panel lists all known data sources that are registered in the DAS registry with their identifier systems. The user can integrate additional data sources that are not contained in the DAS registry by using the ‘Add new source’ tab of the Source Configuration. After providing all required information like the name, URL and identifier system of the new source, it will be included in all future queries. Another way of adding new interaction data is by creating a DASMI server from an existing PSI-MI XML2.5 file. After uploading the file into DASMIweb, the interactions will be made temporarily available as a new DASMI server. This procedure enables users to compare their own interactions with existing datasets or to assess them by confidence servers.
In order to analyze the results with external applications such as the network visualization software Cytoscape (Shannon et al., 2003), the user can export the results from the web client into file formats like the PSI-MI tabular format MITAB2.5 (Kerrien et al., 2007b) or the SIF (Cline et al., 2007).
3.1.5 Quality assessment
Current protein interaction networks are still incomplete to a large extent and are prone to bias and errors (Ramírez et al., 2007). To address this problem, DASMIweb offers useful options to assess the reliability of individual interactions. The following datasets of confidence scores can be requested and selected in the header of the Interaction Panel and are applied to color the interaction squares:
FunSimMat: Interaction partners frequently share similar functions. FunSimMat provides scores that measure the functional similarity of both partners (Schlicker and Albrecht, 2008; Schlicker et al., 2006). The BPscore is based on the BP annotation in the Gene Ontology, the CCscore on the cellular component (CC) annotation and the MFscore on the molecular function (MF) annotation.
Domain support: Some protein–protein interactions may be traced to the underlying domain–domain interactions. Domain support offers two subsets: domain interactions that have been derived from crystal structure analyses and domain interactions that have been computationally predicted by different methods (see Supplementary Data File 3).
In addition, the user can display the original confidence scores that are contained in the source datasets.
3.2 iPfam graphical domain interaction browser
iPfam (Finn et al., 2005) is a database of Pfam domain interactions derived from proteins with an experimentally determined 3D structure. Integrating information about domain interactions from various sources enables one to address several questions. For instance, datasets generated using different methods can be compared and structurally known domain interactions provided by iPfam or 3did (Stein et al., 2009) can be used to verify predicted domain interactions. To this end, the iPfam database has developed a client tool that graphically integrates domain interaction information from one or more DASMI servers (http://ipfam.sanger.ac.uk/graph) including an own server for structural interactions. For a user-selected domain, the iPfam tool retrieves data about interacting domains and represents them as a graph. Each domain is depicted as an oval node within the graph, and interactions are represented by graph edges. Different colors are used to distinguish interactions from individual data sources. Clicking on a domain will center the graph on the interactions for that domain, which supports the visual exploration of the domain interaction network.
3.3 Comparison with existing interaction repositories
DASMI clients may be compared with interaction databases like HPRD (Keshava Prasad et al., 2009) or IntAct (Kerrien et al., 2007a). However, DASMI does not want to compete with such databases, but intends to complement their results with interaction data from other datasets. For example, the results of computational predictions as available in Bioverse (McDermott et al., 2005) are not included into databases of experimental protein interactions, though they might give scientists new insights into the function of proteins (Sharan et al., 2007).
Providing more interaction datasets is not only a goal of DASMI, but also the motivation for composite databases like MiMI (Tarcea et al., 2009) for protein–protein interaction or DOMINE for domain–domain interactions (Raghavachari et al., 2008). In contrast to DASMI, these composite databases combine several datasets into a central repository, which renders it difficult to ensure that the interaction data they provide is kept in sync with the original sources. IntAct, for instance, has a daily release cycle, implying daily update processes of the composite databases. DASMI avoids this problem by leaving the interaction data with its original providers.
In addition, DASMI fosters the inclusion of novel interaction data and interaction confidence scoring methods. There is no central authority that decides which data resources are included and which are not. By setting up a new DASMI server and registering it at the DAS registry, the interaction data or confidence scoring routine will automatically be available to all users (Fig. 2). Moreover, the setup of an own DASMI server without publishing the server address allows for integrating confidential data into other DASMI clients.
4 DISCUSSION AND CONCLUSION
We have introduced DASMI, a new framework for the dynamic exchange and integration of different types of interaction data. The DAS protocol extension DASMI is based on a client–server architecture and comprises three main components: data exchange specification, interaction servers and integration clients. Open source server and client libraries are available for the programming languages Java and Perl. Due to its distributed architecture, DASMI is easily extensible, for instance, by including new servers or developing additional clients. By avoiding a central interaction data repository, DASMI bypasses the problem of update cycles that static integration frameworks face.
As a prototypic application, we set up several DASMI servers and developed web clients for the exchange of protein and domain interactions. The client DASMIweb dynamically gathers interactions from various servers and integrates their results into a unified view. In addition, the reliability of interactions can be assessed by confidence measures. Furthermore, DASMI is used by an iPfam client to integrate predicted domain–domain interactions into iPfam results.
The development of DASMI will be continued and further extensions will be included. Future plans include more DASMI sources for interaction datasets and confidence measures by external providers. Additional proxies will allow incorporating servers into the DASMI system that do not use DASINT but PSI-MI XML2.5 or other XML formats. The DASMI clients DASMIweb and iPfam will be equipped with new features like a graphical network representation for DASMIweb. Additionally, new DASMI clients like a Cytoscape plugin are under development.
Supplementary Material
ACKNOWLEDGEMENTS
The work was conducted in the context of the DFG-funded Cluster of Excellence for Multimodal Computing and Interaction and the EC-funded BioSapiens Network of Excellence.
Funding: German National Genome Research Network (NGFN); German Research Foundation (DFG contract number KFO 129/1-2); European Commission (LSHG-CT-2003-503265).
Conflict of Interest: none declared.
REFERENCES
- Aloy P, Russell RB. Structural systems biology: modelling protein interactions. Nat. Rev. Mol. Cell Biol. 2006;7:188–197. doi: 10.1038/nrm1859. [DOI] [PubMed] [Google Scholar]
- Aragues R, et al. PIANA: protein interactions and network analysis. Bioinformatics. 2006;22:1015–1017. doi: 10.1093/bioinformatics/btl072. [DOI] [PubMed] [Google Scholar]
- Avila-Campillo I, et al. BioNetBuilder: automatic integration of biological networks. Bioinformatics. 2007;23:392–393. doi: 10.1093/bioinformatics/btl604. [DOI] [PubMed] [Google Scholar]
- Bader GD, et al. BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkland A, Yona G. BIOZON: a system for unification, management and analysis of heterogeneous biological data. BMC Bioinformatics. 2006;7:70. doi: 10.1186/1471-2105-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordner AJ, Gorin AA. Comprehensive inventory of protein complexes in the Protein Data Bank from consistent classification of interfaces. BMC Bioinformatics. 2008;9:234. doi: 10.1186/1471-2105-9-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazma A, et al. Standards for systems biology. Nat. Rev. Genet. 2006;7:593–605. doi: 10.1038/nrg1922. [DOI] [PubMed] [Google Scholar]
- Breitkreutz B-J, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KR, Jurisica I. Online predicted human interaction database. Bioinformatics. 2005;21:2076–2082. doi: 10.1093/bioinformatics/bti273. [DOI] [PubMed] [Google Scholar]
- Cerami EG, et al. cPath: open source software for collecting, storing, and querying biological pathways. BMC Bioinformatics. 2006;7:497. doi: 10.1186/1471-2105-7-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-Aryamontri A, et al. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaurasia G, et al. UniHI 4: new tools for query, analysis and visualization of the human protein–protein interactome. Nucleic Acids Res. 2009;37:D657–D660. doi: 10.1093/nar/gkn841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X-W, Liu M. Prediction of protein-protein interactions using random decision forest framework. Bioinformatics. 2005;21:4394–4400. doi: 10.1093/bioinformatics/bti721. [DOI] [PubMed] [Google Scholar]
- Cline MS, et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusick ME, et al. Interactome: gateway into systems biology. Hum. Mol. Genet. 2005;14(Spec No. 2):R171–R181. doi: 10.1093/hmg/ddi335. [DOI] [PubMed] [Google Scholar]
- Dowell RD, et al. The distributed annotation system. BMC Bioinformatics. 2001;2:7. doi: 10.1186/1471-2105-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, et al. iPfam: visualization of protein-protein interactions in PDB at domain and amino acid resolutions. Bioinformatics. 2005;21:410–412. doi: 10.1093/bioinformatics/bti011. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. ProServer: a simple, extensible Perl DAS server. Bioinformatics. 2007;23:1568–1570. doi: 10.1093/bioinformatics/btl650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frishman D, et al. Protein-protein interactions: analysis and prediction. In: Frishman D, Valencia A, editors. Modern Genome Annotation: The Biosapiens Network. Springer, Wien, Austria: 2009. pp. 353–410. [Google Scholar]
- Goll J, Uetz P. Analyzing protein interaction networks. In: Lengauer T, editor. Bioinformatics – From Genomes to Therapies. Vol. 1. Wiley-VCH, Weinheim; 2007. pp. 1121–1179. [Google Scholar]
- Goll J, et al. MPIDB: the microbial protein interaction database. Bioinformatics. 2008;24:1743–1744. doi: 10.1093/bioinformatics/btn285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarães KS, et al. Predicting domain-domain interactions using a parsimony approach. Genome Biol. 2006;7:R104. doi: 10.1186/gb-2006-7-11-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güldener U, et al. MPact: the MIPS protein interaction resource on yeast. Nucleic Acids Res. 2006;34:D436–D441. doi: 10.1093/nar/gkj003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Toro J, et al. APID2NET: unified interactome graphic analyzer. Bioinformatics. 2007;23:2495–2497. doi: 10.1093/bioinformatics/btm373. [DOI] [PubMed] [Google Scholar]
- Hoffmann R, Valencia A. A gene network for navigating the literature. Nat. Genet. 2004;36:664. doi: 10.1038/ng0704-664. [DOI] [PubMed] [Google Scholar]
- Huang H, et al. iProClass: an integrated database of protein family, function and structure information. Nucleic Acids Res. 2003;31:390–392. doi: 10.1093/nar/gkg044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T-W, et al. POINT: a database for the prediction of protein–protein interactions based on the orthologous interactome. Bioinformatics. 2004;20:3273–3276. doi: 10.1093/bioinformatics/bth366. [DOI] [PubMed] [Google Scholar]
- Jenkinson AM, et al. Integrating biological data – the Distributed Annotation System. BMC Bioinformatics. 2008;9(Suppl. 8):S3. doi: 10.1186/1471-2105-9-S8-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LJ, et al. Literature mining for the biologist: from information retrieval to biological discovery. Nat. Rev. Genet. 2006;7:119–129. doi: 10.1038/nrg1768. [DOI] [PubMed] [Google Scholar]
- Jensen LJ, et al. STRING 8 – a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez RC, et al. Dasty2, an Ajax protein DAS client. Bioinformatics. 2008;24:2119–2121. doi: 10.1093/bioinformatics/btn387. [DOI] [PubMed] [Google Scholar]
- Jones P, et al. Dasty and UniProt DAS: a perfect pair for protein feature visualization. Bioinformatics. 2005;21:3198–3199. doi: 10.1093/bioinformatics/bti506. [DOI] [PubMed] [Google Scholar]
- Jothi R, et al. Co-evolutionary analysis of domains in interacting proteins reveals insights into domain-domain interactions mediating protein–protein interactions. J. Mol. Biol. 2006;362:861–875. doi: 10.1016/j.jmb.2006.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrien S, et al. IntAct - open source resource for molecular interaction data. Nucleic Acids Res. 2007a;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrien S, et al. Broadening the horizon – level 2.5 of the HUPO-PSI format for molecular interactions. BMC Biol. 2007b;5:44. doi: 10.1186/1741-7007-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshava Prasad TS, et al. Human Protein Reference Database-2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, et al. An integrated approach to the prediction of domain-domain interactions. BMC Bioinformatics. 2006a;7:269. doi: 10.1186/1471-2105-7-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TJ, et al. BioWarehouse: a bioinformatics database warehouse toolkit. BMC Bioinformatics. 2006b;7:170. doi: 10.1186/1471-2105-7-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B, Fraser AG. A first-draft human protein-interaction map. Genome Biol. 2004;5:R63. doi: 10.1186/gb-2004-5-9-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, et al. Inferring protein-protein interactions through high-throughput interaction data from diverse organisms. Bioinformatics. 2005;21:3279–3285. doi: 10.1093/bioinformatics/bti492. [DOI] [PubMed] [Google Scholar]
- Macías JR, et al. Integrating electron microscopy information into existing distributed annotation systems. J. Struct. Biol. 2007;158:205–213. doi: 10.1016/j.jsb.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Mathivanan S, et al. An evaluation of human protein-protein interaction data in the public domain. BMC Bioinformatics. 2006;7(Suppl. 5):S19. doi: 10.1186/1471-2105-7-S5-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott J, et al. Functional annotation from predicted protein interaction networks. Bioinformatics. 2005;21:3217–3226. doi: 10.1093/bioinformatics/bti514. [DOI] [PubMed] [Google Scholar]
- Murray-Rust P. Chemistry for everyone. Nature. 2008;451:648–651. doi: 10.1038/451648a. [DOI] [PubMed] [Google Scholar]
- Ng S-K, et al. Integrative approach for computationally inferring protein domain interactions. Bioinformatics. 2003;19:923–929. doi: 10.1093/bioinformatics/btg118. [DOI] [PubMed] [Google Scholar]
- Orchard S, et al. Submit your interaction data the IMEx way. Proteomics. 2007;7:28–34. doi: 10.1002/pmic.200700286. [DOI] [PubMed] [Google Scholar]
- Orchard S, et al. Annual spring meeting of the Proteomics Standards Initiative 23–25 April 2008, Toledo, Spain. Proteomics. 2008;8:4168–4172. doi: 10.1002/pmic.200800555. [DOI] [PubMed] [Google Scholar]
- Pagel P, et al. DIMA 2.0 - predicted and known domain interactions. Nucleic Acids Res. 2008;36:D651–D655. doi: 10.1093/nar/gkm996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persico M, et al. HomoMINT: an inferred human network based on orthology mapping of protein interactions discovered in model organisms. BMC Bioinformatics. 2005;6(Suppl. 4):S21. doi: 10.1186/1471-2105-6-S4-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto C, Rivas JDL. APID: Agile Protein Interaction DataAnalyzer. Nucleic Acids Res. 2006;34:W298–W302. doi: 10.1093/nar/gkl128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prlić A, et al. Adding Some SPICE to DAS. Bioinformatics. 2005;21(Suppl. 2):ii40–ii41. doi: 10.1093/bioinformatics/bti1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prlić A, et al. Integrating sequence and structural biology with DAS. BMC Bioinformatics. 2007;8:333. doi: 10.1186/1471-2105-8-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavachari B, et al. DOMINE: a database of protein domain interactions. Nucleic Acids Res. 2008;36:D656–D661. doi: 10.1093/nar/gkm761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, et al. Computational analysis of human protein interaction networks. Proteomics. 2007;7:2541–2552. doi: 10.1002/pmic.200600924. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, et al. Probabilistic model of the human protein-protein interaction network. Nat. Biotechnol. 2005;23:951–959. doi: 10.1038/nbt1103. [DOI] [PubMed] [Google Scholar]
- Riley R, et al. Inferring protein domain interactions from databases of interacting proteins. Genome Biol. 2005;6:R89. doi: 10.1186/gb-2005-6-10-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual J-Fo, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Sagotsky JA, et al. Life sciences and the web: a new era for collaboration. Mol. Syst. Biol. 2008;4:201. doi: 10.1038/msb.2008.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salwinski L, et al. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelhorn SE, et al. An integrative approach for predicting interactions of protein regions. Bioinformatics. 2008;24:i35–i41. doi: 10.1093/bioinformatics/btn290. [DOI] [PubMed] [Google Scholar]
- Schlicker A, Albrecht M. FunSimMat: a comprehensive functional similarity database. Nucleic Acids Res. 2008;36:D434–D439. doi: 10.1093/nar/gkm806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicker A, et al. A new measure for functional similarity of gene products based on Gene Ontology. BMC Bioinformatics. 2006;7:302. doi: 10.1186/1471-2105-7-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicker A, et al. Functional evaluation of domain-domain interactions and human protein interaction networks. Bioinformatics. 2007;23:859–865. doi: 10.1093/bioinformatics/btm012. [DOI] [PubMed] [Google Scholar]
- Shah SP, et al. Atlas – a data warehouse for integrative bioinformatics. BMC Bioinformatics. 2005;6:34. doi: 10.1186/1471-2105-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, et al. Gaggle: an open-source software system for integrating bioinformatics software and data sources. BMC Bioinformatics. 2006;7:176. doi: 10.1186/1471-2105-7-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan R, et al. Network-based prediction of protein function. Mol. Syst. Biol. 2007;3:88. doi: 10.1038/msb4100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker BA, Panchenko AR. Deciphering protein-protein interactions. Part II. Computational methods to predict protein and domain interaction partners. PLoS Comput. Biol. 2007;3:e43. doi: 10.1371/journal.pcbi.0030043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A, et al. 3did Update: domain-domain and peptide-mediated interactions of known 3D structure. Nucleic Acids Res. 2009;37:D300–D304. doi: 10.1093/nar/gkn690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD. Towards a cyberinfrastructure for the biological sciences: progress, visions and challenges. Nat. Rev. Genet. 2008;9:678–688. doi: 10.1038/nrg2414. [DOI] [PubMed] [Google Scholar]
- Stelzl U, Wanker EE. The value of high quality protein-protein interaction networks for systems biology. Curr. Opin. Chem. Biol. 2006;10:551–558. doi: 10.1016/j.cbpa.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Stelzl U, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Stumpf MP, et al. Estimating the size of the human interactome. Proc. Natl Acad. Sci. USA. 2008;105:6959–6964. doi: 10.1073/pnas.0708078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarcea VG, et al. Michigan molecular interactions r2: from interacting proteins to pathways. Nucleic Acids Res. 2009;37:D642–D646. doi: 10.1093/nar/gkn722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorisson GA, et al. Genotype-phenotype databases: challenges and solutions for the post-genomic era. Nat. Rev. Genet. 2009;10:9–18. doi: 10.1038/nrg2483. [DOI] [PubMed] [Google Scholar]
- Wang R-S, et al. Analysis on multi-domain cooperation for predicting protein-protein interactions. BMC Bioinformatics. 2007;8:391. doi: 10.1186/1471-2105-8-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuchty S. Topology and weights in a protein domain interaction network – a novel way to predict protein interactions. BMC Genomics. 2006;7:122. doi: 10.1186/1471-2164-7-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.