Abstract
The differences in efficacy and molecular mechanisms of platinum based anti-cancer drugs cisplatin (CP) and oxaliplatin (OX) have been hypothesized to be in part due to the differential binding affinity of cellular and damage recognition proteins to CP and OX adducts formed on adjacent guanines in genomic DNA. HMGB1a in particular exhibits higher binding affinity to CP-GG adducts, and the extent of discrimination between CP- and OX-GG adducts is dependent on the bases flanking the adducts. However, the structural basis for this differential binding is not known. Here, we show that the conformational dynamics of CP- and OX-GG adducts are distinct and depend on the sequence context of the adduct. Molecular dynamics simulations of the Pt-GG adducts in the TGGA sequence context revealed that even though the major conformations of CP- and OX-GG adducts were similar, the minor conformations were distinct. Using the pattern of hydrogen bond formation between the Pt–ammines and the adjacent DNA bases, we identified the major and minor conformations sampled by Pt–DNA. We found that the minor conformations sampled exclusively by the CP-GG adduct exhibit structural properties that favor binding by HMGB1a, which may explain its higher binding affinity to CP-GG adducts, while these conformations are not sampled by OX-GG adducts because of the constraints imposed by its cyclohexane ring, which may explain the negligible binding affinity of HMGB1a for OX-GG adducts in the TGGA sequence context. Based on these results, we postulate that the constraints imposed by the cyclohexane ring of OX affect the DNA conformations explored by OX-GG adduct compared to those of CP-GG adduct, which may influence the binding affinities of HMG-domain proteins for Pt-GG adducts, and that these conformations are further influenced by the DNA sequence context of the Pt-GG adduct.
INTRODUCTION
Cisplatin (CP) and Carboplatin are platinum (Pt) based anticancer drugs used in the treatment of testicular, ovarian, head, neck and nonsmall cell lung cancer (1). Oxaliplatin (OX) is a third generation Pt drug that has been approved for treatment of colorectal cancer and cisplatin-resistant tumors (1). The main mode of action of these drugs is through formation of Pt adducts on genomic DNA, primarily on adjacent guanines. These Pt–DNA adducts have been shown to stall replication and transcription and recruit DNA damage recognition proteins leading to apoptosis (1,2). The Pt–DNA adducts are similar for CP and OX except for their carrier ligand, which is diammine for CP and diaminocyclohexane for OX (3–5).
A number of cellular proteins, especially those containing the HMG-domain have been shown to bind specifically to Pt-GG intrastrand DNA adducts (6–13). HMG-domain proteins that bind to Pt-GG adducts fall into two classes: structure-specific, abundant, chromatin architectural proteins like HMGB1 (9–11) and sequence-specific, cell-type specific transcription factors like LEF-1 (12,13) and SRY (12). The binding of structure-specific HMG-domain proteins to Pt–DNA adducts has been shown to inhibit nucleotide excision repair, which may increase the longevity of Pt–DNA adducts (14,15). Pt–DNA adducts have also been postulated to sequester some HMG-domain containing transcription factors that are essential for maintaining the uncontrolled cell growth characteristic of tumor cells (16,17). Thus, the binding of HMG-domain proteins to Pt–DNA could significantly sensitize cells to CP and OX.
Cells and tumors that are resistant to CP are generally not cross resistant to OX. Since CP and OX form the same adducts at the same sites on DNA, this lack of cross-resistance is thought to be in part due to the effects downstream of formation of CP- and OX-DNA adducts. The differential binding affinities of HMG-domain proteins to CP- and OX-DNA adducts (13,16,18) have been proposed to contribute to the distinctive downstream effects of CP- and OX-DNA adducts in the cell. Hence, understanding the basis of the differential binding of HMG-domain proteins to CP- and OX-DNA could lead to a better understanding of the differential efficacies of CP and OX.
The binding of HMGB1 to Pt–DNA adducts has been characterized in great detail. HMGB1 contains two HMG domains: domain A (HMGB1a) and domain B (HMGB1b). HMGB1a binds more strongly than HMGB1b to most Pt-GG DNA adducts (18). Full-length HMGB1 binds with lower affinity compared to HMGB1a mainly due to its highly acidic C-terminal tail, which when removed results in the tandem HMG domains having same affinity as HMGB1a (19). However, HMGB1a binds to Pt-GG adducts with the same specificity as full length HMGB1 (19). Further, footprinting studies combined with site-directed mutagenesis have shown that only domain A of full-length HMGB1 binds to DNA containing the Pt-GG DNA adduct (19,20). Thus, most of the structural and mechanistic studies have been performed with HMGB1a alone.
Like most structure-specific HMG-domain proteins HMGB1 has been shown to recognize and bind to pre-bent/distorted DNA and bend it further (12). A crystal structure of HMGB1a in complex with CP-GG DNA in the TGGA sequence context has been reported (21). That structure, coupled with site-directed mutagenesis (20), has provided considerable insight into the mechanism of HMGB1a binding to Pt–DNA adducts. There are a number of hydrophobic and hydrogen bond interactions between HMGB1a and the minor groove of DNA on the 3′ side of the CP-GG adduct. Among these interactions, the most significant appears to be the intercalation of Phe37 between the two Gs of the Pt-GG adduct, which allows a π–π stacking interaction between Phe37 and the 3′G. Mutation of Phe37 to Ala resulted in more than 667-fold decrease in binding affinity (20). The crystal structure of HMGB1a bound to Pt–DNA in the TGGA sequence context also shows a hydrogen bond between Ser41 and the N3 atom of Adenine 3′ of the Pt-GG adduct. A Ser41Ala mutation resulted only in a 3.9-fold decrease in binding affinity, a much weaker effect than the Phe37Ala mutation (20). The contributions of other interactions between HMGB1a and the minor groove of Pt–DNA adducts to the binding affinity of the protein–DNA complex have not been characterized, but are likely to be less than or equal to the Ser41-3′A-N3 interaction.
Given that HMG-domain proteins bind Pt–DNA adducts along the minor groove and do not directly contact the drug (which is covalently bound in the major groove), the carrier ligand does not have a direct role to play in the binding of HMG-domain proteins. However, if the carrier ligand were to influence the extent of distortion of Pt–DNA to a conformation that favors binding of HMG-domain proteins, the binding affinities of HMG-domain proteins to CP- and OX-DNA could be influenced in that fashion. For example, in the case of HMGB1a, conformations that favor bending of the DNA, stacking of Phe37 with the 3′G or formation of a hydrogen bond between Ser41 and the 3′A in the TGGA sequence context would be expected to enhance binding.
The differential binding affinity of HMGB1a towards CP- and OX-DNA is also dependent on the identity of bases flanking the central intrastrand GG (11,18). The binding affinity of HMGB1a for CP-DNA adducts has been shown to be ∼53 times greater than that for OX-DNA adducts in the TGGA sequence context (18), while in the AGGC (18) and the CGGA (11) sequence contexts, the binding affinity for CP-DNA is only three times greater than that of OX-DNA adducts (Supplementary Text S1). In the TGGT sequence contest, the binding affinity of HMGB1a for CP-GG and OX-GG adducts is almost the same (11). Most of the sequence-dependent variations in differential binding affinity of HMGB1a towards CP- and OX-DNA adducts is due to sequence dependent effects for binding to OX-DNA adducts since the binding affinity of HMGB1a for CP-GG adducts is roughly comparable in all three sequence contexts (11,18). This sequence dependent effect has also been shown for HMGB1b and TBP, but the trends are slightly different (18). Thus, the increased binding affinity of HMGB1a towards CP-GG adducts compared to OX-GG adducts in most sequence contexts suggests that the structures of CP- and OX-GG adducts should exhibit significant differences which would lead to differential recognition. The sequence context based effects would also suggest that the preferential binding conformations are highly dependent on the bases flanking the Pt–GG adduct.
Surprisingly, the crystal and solution structures of both CP- and OX-GG adducts in the TGGT and the AGGC sequence contexts do not reveal significant structural differences (1,22–25) that can explain the differences in binding affinities of HMG-domain proteins for CP- and OX-DNA. Hence, we have postulated that the differences in conformational dynamics of CP- and OX-DNA adducts might be responsible for the differential recognition of these adducts by HMG-domain proteins. To test this hypothesis, we performed all-atom molecular dynamics simulations on undamaged 12-mer DNA and CP- and OX-GG 12-mer DNA adducts both free in solution and in complex with HMGB1a. All of these simulations were performed in the TGGA sequence context in which the discrimination of HMGB1a between CP- and OX-GG adducts is 53-fold. These simulations were compared with previous simulations in the AGGC sequence context (26) in which the discrimination between CP- and OX-GG adducts by HMGB1a is 3-fold.
Initial studies involving all-atom, fully solvated MD simulations of CP- and OX-DNA in the AGGC sequence context had revealed the formation of hydrogen bonds between Pt-amine hydrogens and the surrounding DNA bases (26). CP- and OX-GG adducts in the AGGC sequence context differed in the pattern of hydrogen bond formation between Pt-amine hydrogens and the surrounding DNA bases and these differences in hydrogen bond patterns correlated with differences in DNA conformational dynamics that might be important for binding of HMG-domain proteins.
In the TGGA sequence context, the pattern of hydrogen bond formation between the Pt-amine hydrogens and the surrounding bases is different than that seen in the AGGC sequence context. These different patterns of hydrogen bond formation correlate with certain conformations that could favor binding to HMG-domain protein that are sampled only by CP-DNA, while the conformations favorable for HMG-domain protein binding had been sampled by both CP- and OX-DNA to different extents in the AGGC sequence context (26).
METHODS
Starting structures
We performed simulations on a 12-mer DNA sequence (shown in Scheme 1), which was either undamaged or covalently bound to CP or OX at the N7 of G6 and G7. The initial structures for the CP- and OX-TGGA simulations were obtained by modifying the NMR structure of OX-DNA in the TGGT sequence context (unpublished results). We chose this structure because, of the NMR solution structures of Pt–DNA adducts that we have obtained, it was the closest in sequence to the TGGA sequence context. Using INSIGHT II (Accelrys Inc., CA, USA), the T8-A17 base pair was replaced by the A8-T17 base pair in the TGGT NMR structure to obtain the initial structure of OX-DNA in TGGA sequence context. Further, OX was modified to CP to obtain CP-DNA structure in the TGGA sequence context. INSIGHT II was used to generate canonical B-DNA structure, which was used as the starting structure for simulations of the undamaged DNA.
Scheme 1.
Dodecamer sequences used in simulations.
For the simulations of HMGB1a-Pt–DNA complex, we used the crystal structure of rat HMGB1a bound to CP-DNA in the TGGA sequence context as the starting structure for the CP-TGGA simulations [PDB ID 1ckt (21)]. This structure was modified appropriately using Insight II to generate starting structures for OX-TGGA, CP-AGGC and OX-AGGC simulations.
Molecular dynamics simulations
We performed 5 sets of 10 ns simulations for each of CP-, OX- and undamaged DNA. The five sets had the same starting structures but the initial velocities were randomized. We employed simulation protocols identical to our published work on the AGGC sequence context (26). We performed a single MD production run of 50 ns for the HMGB1a-Pt–DNA complex in two sequence contexts and with CP and OX to yield four trajectories. The details of the force field and simulation protocols are described in Supplemental Material S2.
Analysis: hydrogen bonds
All the trajectories were analyzed for the presence of hydrogen bonds between all possible donors and acceptors based on a distance cut-off of 3.5 Å between the donor and acceptor and an angular cut-off of 135° between donor-H-acceptor. In addition to the Watson–Crick interactions, hydrogen bonds were formed for a significant amount of time between Pt-amines and the surrounding base pairs. All the frames of the trajectory were classified based on the type of hydrogen bonds formed between Pt-amines and the adjacent bases.
Centroid structures
We calculated the average structures for both the complete ensembles of CP-, OX- and undamaged DNA and also for the ensembles forming different hydrogen bonds. Since average structures are not actual structures from simulations and may feature some abnormal bond lengths/angles, we assigned the structure from each ensemble that had the lowest mass-weighted root-mean-square deviation (RMSD) to the average structure as the centroid structure of that ensemble (26).
Comparisons of centroid structures using RMSD
We compared the centroid structures of CP- and OX-DNA adducts in TGGA and AGGC sequence contexts respectively. In these comparisons, we used only the atoms from the DNA part of the molecule in calculating the mass-weighted RMSD.
Helical parameters
Helical parameters were calculated for each snapshot of the trajectory using the CURVES program, version 5.3 (27). The following CURVES parameters were extracted: global inter base-pair parameters: shift, slide, rise, tilt, roll and twist; global base–base parameters: shear, stretch, stagger, buckle, propeller and opening. Histograms were then constructed for each of these parameters as frequency of occurrence versus discrete units of each DNA helical parameter. The discrete units (bin width) were set as 0.2 Å for all the distance parameters and 2° for all the angular parameters. Histograms were plotted for parameters corresponding to the central 4 bp of the DNA oligomer (the adduct is formed on central 2 bp). These distributions were also clustered on the basis of the pattern of hydrogen bond formation between the DNA and the drug.
To calculate the P-value of the difference between helical parameters corresponding to CP-, OX-DNA and undamaged DNA and also between different hydrogen bonded species in CP- and OX-DNA, we used the Kolmogorov–Smirnov (KS) test (28). The lower cut-off threshold of significant differences was calculated by dividing each data set into five equal subsets and performing KS test comparing all possible combinations of the five subsets of divided data. The highest –log(P) among the subsets of each data set was set as the threshold for that data set. When two data sets were compared, the difference between them was significant only if –log(P) of the comparison of the data sets was higher than the threshold. The ratio of –log(P) of the comparison of the data sets to their thresholds was calculated and then this ratio for all the parameters are represented as colors in a diagram (heat map). The ratios transition from white (lowest ratios) to blue then red (highest ratios) in the heat map. The range of KS ratios and their corresponding color are shown as a scale in the bottom of the heat maps.
In order to compare the helical parameters of free Pt–DNA in solution to those of Pt–DNA bound to HMGB1a, we calculated the helical parameters by considering only the central 12 bp of the longer (16 bp) DNA present in the HMGB1a-Pt-DNA complex.
Bend angle
Overall bend angles of the snapshots from the simulations were calculated using the program MADBEND (29). MADBEND uses the local tilt and roll parameters from the CURVES (version 5.3) output to calculate the overall bend angle. The bend angle was calculated for the central 10 bp of the oligomer centered at G6-G7 where the Pt adduct is formed. The normalized histograms were plotted for bend angle versus percent occupancy with a bin width of 2°. The bend angle values were also clustered on the basis of the type of hydrogen bond formed.
RESULTS
We performed simulations of CP- and OX-GG adducts both with and without bound HMGB1a to probe for conformational differences that would explain the increased affinity of HMGB1a for CP-GG adducts compared to OX-GG adducts in general and the much greater affinity of HMGB1a for CP-GG adducts compared to OX-GG adducts in the TGGA sequence context. The simulations attained equilibrium within the first few nanoseconds, with the all-atom mass weighted RMSD of OX- and undamaged DNA remaining <3 Å and that of CP-DNA remaining <4 Å throughout the simulation (Supplementary Figure S1). The all-atom mass weighted RMSDs of protein and DNA in simulations of HMGB1a-Pt–DNA equilibrated within 5 ns and did not exceed 5 Å (S. Ramachandran, B. Temple, S. G. Chaney and N. V. Dokholyan; manuscript in preparation).
The centroid structures of CP- and OX-DNA do not reveal major differences
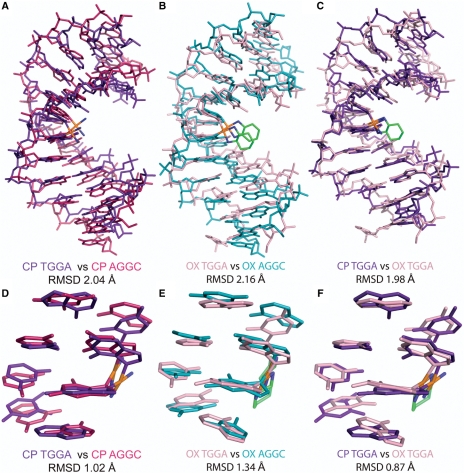
We first compared the centroid structures of CP- and OX-DNA simulations in the TGGA and the AGGC sequence contexts (Figure 1). The all-atom RMSD was ∼2 Å when we compared either CP-TGGA with CP-AGGC or OX-TGGA with OX-AGGC, indicating lack of global structural differences in the major conformations sampled by CP- and OX-DNA adducts in the two sequence contexts. Even when we compared the centroid structures of CP- and OX-DNA in the TGGA sequence context (Figure 1C, RMSD = 1.98 Å), there were no large conformational differences. The same comparisons for the central 4 bp of the centroid structures (Figure 1D–F) yielded the same conclusion: the centroid structures did not display any major structural differences that would warrant high differential binding affinities between CP- and OX-DNA adducts.
Figure 1.
Comparison of centroid structures. Structural alignment of the centroid structures using only the atoms from the DNA part of the molecule from the simulations of CP-DNA in AGGC and TGGA sequence contexts (A), OX-DNA in AGGC and TGGA sequence context (B) and CP- and OX-DNA in TGGA sequence context (C). The structural alignment of the central four base-pairs of the centroid structures of CP-DNA in AGGC and TGGA sequence contexts (D), OX-DNA in AGGC and TGGA sequence context (E) and CP- and OX-DNA in TGGA sequence context (F).
The overall conformation of CP- and OX-GG adducts are similar
To further characterize the conformations sampled by CP- and OX-DNA adducts in the vicinity of platinum in the TGGA sequence context, we calculated the helical parameters of the central 4 bp of the dodecamer (T5G6G7A8) and the 3-bp steps between them. With six parameters describing each base pair and six more for each base-pair step, there were a total of 42 helical parameters corresponding to the central 4 bp. We used the KS ratio (as described in Methods section) as a test for significant difference in comparing any two distributions of a particular helical parameter. To identify the parameters that showed major differences across different comparisons, we plotted the KS ratio of all the 42 parameters as a heat map (described in Methods section) of KS ratios of different comparisons. From the heat map, we selected the parameters that exhibited significant differences. When we compared the overall distributions of the helical parameters for CP-, OX- and undamaged DNA in the TGGA sequence context, both CP- and OX-DNA had significantly different conformations compared to undamaged DNA, especially in the G6-G7 base-pair step, G6-C19 base pair and G7-C18 base pair (Supplementary Figure S2). When comparing CP- and OX-DNA, we observed differences in T5-G6 slide, A8-T17 buckle, T5-A20 opening and G6-C19 opening (Supplementary Figure S2). However, the differences between CP- and OX-DNA were minor, yielding KS ratios of ≤2. Thus, the CP- and OX-TGGA adducts appear to sample the same major DNA confirmations when examined by this criteria.
The pattern and frequency of hydrogen bond formation between the drug and DNA depend both on the sequence context and carrier ligand
An earlier study had reported the formation of hydrogen bonds between platinum amines and adjacent bases in the AGGC sequence context (26), which correlated with minor conformational differences between CP- and OX-AGGC adducts that could influence the binding of HMGB1a to these adducts. We examined the trajectories of CP- and OX-TGGA adducts for hydrogen bond formation between all possible donors and acceptors using distance and angle criteria (as described in Methods section). In addition to the Watson–Crick hydrogen bonds in the DNA, we also observed the formation of hydrogen bonds between Pt-amines and adjacent base pairs. When we compared the hydrogen bond patterns in the two sequence contexts, we observed that the bases involved and the frequency of formation of these hydrogen bonds were dictated both by the sequence context and the carrier ligand (Table 1).
Table 1.
Frequency of formation of different hydrogen bonds between Pt-amines and adjacent bases
| TGGA | |||
|---|---|---|---|
| CP |
OXa |
||
| Hydrogen bond type | Frequency (%) | Hydrogen bond type | Frequency (%) |
| G7-O6 | 32 | G7-O6 | 55 |
| A8-N7 | 13 | T17-O4 | 15 |
| T5-O3′ | 6 | ||
| None | 54 | None | 23 |
| AGGCb | |||
| CP |
OXc |
||
| Hydrogen bond type | Frequency (%) | Hydrogen bond type | Frequency (%) |
| G7-O6 | 13 | G7-O6 | 34 |
| A5-N7 | 40 | A5-N7 | 14 |
| A5-N7 + G7-O6 | 34 | A5-N7 + G7-O6 | 45 |
| None | 13 | None | 8 |
aOnly the equatorial hydrogen of the OX-amine was involved in hydrogen bonding on the 3′ side of the adduct.
bFrom ref. 26.
cBoth equatorial and axial hydrogens of the OX-amine were involved in hydrogen bonds on the 5′ side, and only equatorial hydrogen of the OX-amine was involved in hydrogen bonding on the 3′ side of the adduct.
In the AGGC sequence context, although the same hydrogen bonds were formed by both CP- and OX-Pt-amines, the frequency of hydrogen bond formation was different (26). The hydrogen bond to A5-N7 was formed more frequently for CP-DNA while the hydrogen bond to G7-O6 was formed more frequently for OX-DNA (26). In the TGGA sequence context, the G7-O6 hydrogen bond was formed by both CP- and OX-DNA and like in AGGC sequence context, the G7-O6 hydrogen bond was formed more frequently by OX-DNA (Table 1). Other hydrogen bonds seen in the TGGA sequence context were unique to either CP- or OX-DNA. OX-DNA formed hydrogen bonds on the 5′ side of the adduct, to T5-O3′ but only for a small fraction of time (6%, Table 1). On the 3′ side, CP formed hydrogen bond with A8-N7 with a frequency of 13% while OX formed hydrogen bonds with T17-O4 with a frequency of 15%. These hydrogen bond patterns in the TGGA sequence context that were unique to CP- and OX-DNA adducts (A8-N7 for CP-DNA, T17-O4 and T5-O3′ for OX) all represented relatively minor conformations (occurring ≤15% of the time) of the Pt–DNA adduct. Even though these conformations were transient, it was thought possible that the different hydrogen bonded conformations could influence protein binding.
Platinum amines in OX are more constrained compared to CP
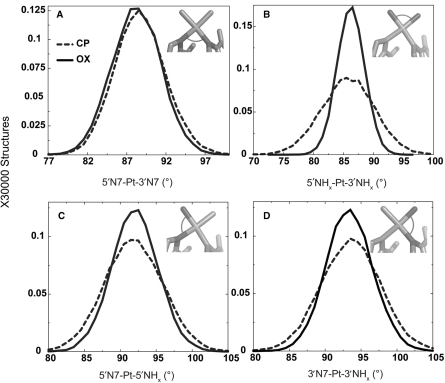
To determine the origin of differences between CP- and OX-DNA in the frequency of hydrogen bond formation with flanking bases on both sides of the Pt-GG adduct and their occupancy, we calculated the distributions of geometrical parameters of Pt from the simulations. As expected from the constraints imposed by the cyclohexane ring in OX, we observed that the 5′NHx-Pt-3′NHx bond angle had a significantly greater range for the CP-DNA adduct compared to the OX-DNA adduct (Figure 2B) and the greater range for CP-DNA was observed in both TGGA and AGGC sequence contexts (data not shown). The 5′N7-Pt-5′NHx and the 3′N7-Pt-3′NHx bond angles also showed a slightly greater range for CP-DNA compared to OX-DNA (Figure 2C–D). However, there was essentially no difference between CP- and OX-DNA in the 5′N7-Pt-3′N7 bond angle, presumably because of the constraints imposed by the DNA (Figure 2A). Thus, overall, CP-DNA was more flexible compared to OX-DNA with respect to the Pt-amines. The greater flexibility of CP compared to OX (especially with respect to the 5′NHx-Pt-3′NHx bond angle) may have resulted in the differences in local structure of the adduct, leading to differences in formation of hydrogen bonds between the drug and the adjacent bases.
Figure 2.
Geometrical parameters of platinum. The distributions of the four angles around the square-planar platinum atom are plotted from the CP- and OX-TGGA simulations: 5′N7-Pt-3′N7 (A), 5′NHx-Pt-3′NHx (B), 5′N7-Pt-5′NHx (C), 3′N7-Pt-3′NHx (D). The Pt-amines at the 5′ and 3′ side of the adduct are denoted as 5′NHx and 3′NHx, respectively. The N7s of G6 and G7 that are involved in covalent bonds with Pt are denoted as 5′N7 and 3′N7, respectively. The frequency distribution histograms were calculated from the structures obtained at every picosecond over the final 6 ns of each equilibrated MD simulation, resulting in a total of 30 000 structures each for CP-, OX- and undamaged DNA. The distribution is plotted against the number of structures in the trajectory. The frequency distributions for CP- and OX-TGGA adducts are shown as a dashed line and solid line, respectively.
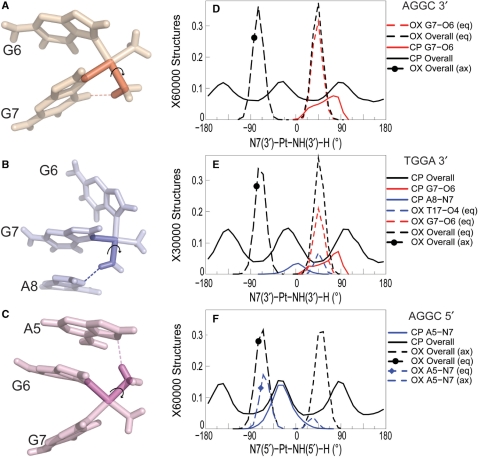
To further characterize the differences in the flexibility of CP- and OX-amines, we examined the conformational flexibility of the N7-Pt-N-H dihedral angles for CP and OX on both 5′ and 3′ side of the adduct. The N7-Pt-N-H dihedral angle (Figure 3A–C) determines the orientation of the amine hydrogens, which influences the ability of those hydrogens to form hydrogen bonds with the adjacent bases. The N7-Pt-N-H dihedral angle for the CP-GG adduct has three broad peaks on both the 3′ and 5′ side of the adduct because all the hydrogens are equivalent and sample three different orientations. In contrast, the N7-Pt-N-H dihedral angle for OX has two relatively narrow peaks in distribution, with the one between −100° and −20° representing the equatorial hydrogen and the one between 20° and 80° representing the axial hydrogen in the 5′ amine. The peaks are reversed for the 3′ amine. In both the AGGC sequence context (Figure 3D) and the TGGA sequence context (Figure 3E), the 3′N7-Pt-3′N-H dihedral angle positions the equatorial hydrogen of OX for the formation of the G7-O6 hydrogen bond, while the CP hydrogens are less frequently in the right orientation (compared to OX) for the formation of the G7-O6 hydrogen bond. Similarly in the TGGA sequence context, only the CP hydrogens spend a significant amount of time in a conformation favorable for the formation of the A8-N7 hydrogen bond and only the OX equatorial hydrogen spends a significant amount of time in a conformation favorable for the formation of the T17-O4 hydrogen bond (Figure 3E). Finally, in the AGGC sequence context, the 5′N7-Pt-N-H dihedral angle of CP allows one of the positions of the ammine hydrogens to be highly favorable for the formation of the A5-N7 hydrogen bond (Figure 3F), while the equatorial hydrogen in 5′ side of OX has an orientation that is not as favorable as CP for the formation of the A5-N7 hydrogen bond. Thus, the allowable N7-Pt-N-H dihedral angles (in part determined by the carrier ligand) provides a structural basis for the type and frequency of hydrogen bond formed in CP- and OX-DNA adducts in the TGGA and AGGC sequence contexts.
Figure 3.
Dihedral angle involved in hydrogen bond formation. The dihedral angle describing the Pt-amine hydrogen orientation while forming hydrogen bond with adjacent DNA bases is shown for the G7-O6 hydrogen bond in CP-DNA in the AGGC sequence context (A), the A8-N7 hydrogen bond in CP-DNA in the TGGA sequence context (B) and the A5-N7 hydrogen bond in CP-DNA in the AGGC sequence context (C), respectively. OX has two hydrogens for each Pt-amine—equatorial (eq) and axial (ax). The distribution of the 3′N7-Pt-NHx-H dihedral angle is shown for the AGGC sequence context (D) and the TGGA sequence context (E). The distribution of the 5′N7-Pt-NHx-H dihedral angle is shown for the AGGC sequence context (F). The distribution of dihedral angles involving all the structures of an adduct are plotted in black, distribution of dihedral angles for structures with the G7-O6 hydrogen bond are plotted in red and that for structures with the hydrogen bond to adjacent base are plotted in blue. Similarly, distributions for OX-DNA are plotted as a dashed line while those for CP-DNA are plotted as a solid line. The frequency distribution histograms for the overall distributions were calculated from the final 30 000 structures (taken every picosecond) for the TGGA and 60 000 structures for the AGGC sequence contexts, for the CP-, OX- and undamaged DNA simulations as described in Figure 2. [Simulations in the AGGC sequence context were performed from two starting structures each as described in Sharma et al. (26).] The frequency distribution for a particular hydrogen bonded species was obtained from structures that formed that particular hydrogen bond. However, the normalization was performed over the full 30 000 structures to show the relative abundance of different hydrogen bonded species.
Hydrogen bond formation is associated with distinctive distortions near the base to which the hydrogen bond is formed
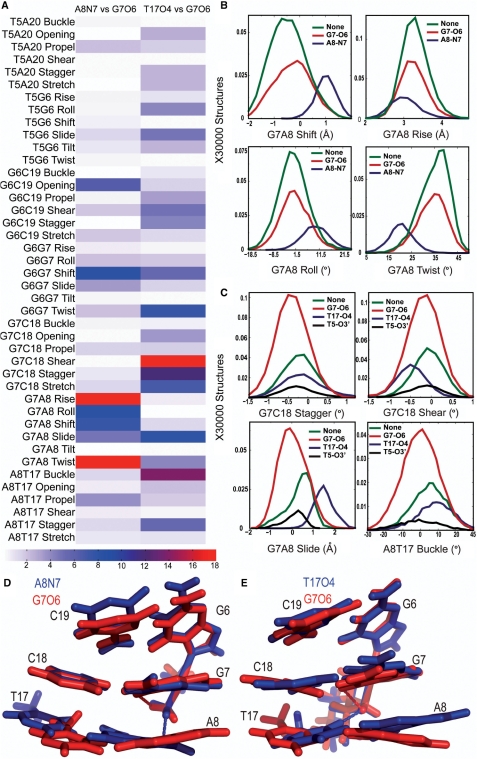
To examine if hydrogen bond formation correlated with unique conformations in the central 4 bp in the TGGA sequence context, we calculated the KS ratios for the helical parameters of the central 4 bp associated with the different hydrogen bonded species. For both CP- and OX-DNA, structures forming G7-O6 hydrogen bond did not have major differences when compared to structures forming no hydrogen bonds to the Pt-amines (Supplementary Figure S3). However, we did observe significant differences when comparing the helical parameters of structures with CP-A8-N7 hydrogen bond to structures with the CP-G7-O6 hydrogen bond and in comparing structures with OX-T17-O4 hydrogen bond to structures with OX-G7-O6 hydrogen bond (Figure 4). From the KS ratio map of comparison of structures with the CP-A8-N7 hydrogen bonds to the structures with the CP-G7-O6 hydrogen bond (Figure 4A), we observed the greatest differences in the G7-A8 base pair step, especially in rise, roll, shift, twist and slide parameters. Significant differences were also observed for G6-G7 shift and G6-C19 opening. The parameters with major differences between structures with CP-A8-N7 hydrogen bond and all other species in CP-DNA yielded KS ratios of ∼8–15 indicating that the CP-A8-N7 hydrogen bonded species had a unique conformation compared to all other species in CP-DNA. Similarly, we observed from the KS ratio map of comparison of structures with OX-T17-O4 hydrogen bond to structures with OX-G7-O6 hydrogen bond (Figure 4A) that the formation of OX-T17-O4 hydrogen bond resulted in unique conformations at G7-C18 base pair, G7-A8 base-pair step and A8-T17 base pair, as seen in G7-C18 shear, stagger and stretch; G7-A8 slide and A8-T17 buckle parameters. A significant difference was also observed for the G6-G7 twist. The same differences were also seen when the helical parameters were compared for structures with the OX-T17-O4 hydrogen bond to structures with no hydrogen bonds (data not shown).
Figure 4.
Helical parameters in the TGGA sequence context. The conformational differences in the central 4 bp for different hydrogen bonded species in the TGGA sequence context are represented at three different levels. The differences between two hydrogen bonded species in all the 42 parameters are shown in a heat map (A). For CP-DNA, we compare structures with G7-O6 hydrogen bond to structures with the A8-N7 hydrogen bond and for OX-DNA we compare structures with G7-O6 hydrogen bond to structures with the T17-O4 hydrogen bond. We represent the differences as the KS ratio, which is color-coded (The KS ratio decreases in the order of Red > Blue > White). Plots of histograms of the four helical parameters showing significant differences for different hydrogen bonded species of CP-DNA adduct (B) and OX-DNA adduct (C), are shown with the frequency distribution calculated as described before. The distribution for structures with no hydrogen bond is plotted in green, for structures with G7-O6 hydrogen bond in plotted in red and for structures with hydrogen bond to the adjacent base is plotted in blue. Alignment of 5′G6G7A8 3′ base pairs of the centroid structures forming the CP-G7-O6 hydrogen bond (red) and the CP-A8-N7 hydrogen bond (blue) (D) are shown. Alignment of 5′G6G7A8 3′ base pairs of the centroid structures forming the OX-G7-O6 hydrogen bond (red) and the OX-T17-O4 hydrogen bond (blue) (E) are also shown. The structures are aligned based on Pt-G6G7.
The distributions of helical parameters of the hydrogenbonded species showing major differences in CP-DNA and OX-DNA are plotted in Figure 4B and C, respectively. The G6G7A8 portion of the centroid structures forming the CP-A8-N7/OX-T17-O4 hydrogen bond is compared with CP-G7-O6/OX-G7-O6 hydrogen bond in Figure 4D and E, respectively. In summary, we found that the formation of the minor hydrogen bonds on the 3′ flanking base pair in CP- and OX-DNA was associated with unique conformations on the 3′ side of the adduct. We do not describe here the helical parameters of structures with T5-O3′ hydrogen bond in OX-DNA since this hydrogen bond was formed only in 6% of the structures and because binding of HMGB1a to Pt–DNA is primarily through interactions on the 3′ side of the adduct.
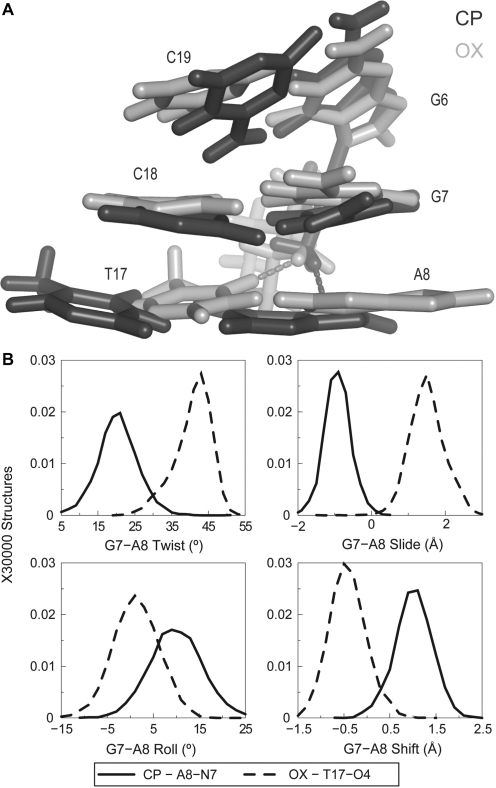
OX T17-O4 and CP A8-N7 are conformationally distinct with respect to the minor groove
Formation of unique hydrogen bonds by CP- and OX-DNA to the drug suggested that these hydrogen-bonded species could lead to structural differences between CP- and OX-DNA. When we compared structures forming the CP-A8-N7 hydrogen bond to structures forming the OX-T17-O4 hydrogen bond, we did observe significant differences. When seen facing the minor groove, the formation of CP-A8-N7 hydrogen bond shifted the A8-T17 base pair towards the left, while the formation of OX-T17-O4 hydrogen bond shifted the A8-T17 base pair to the right (Figure 5A). Thus, the structural distortions associated with the formation of these hydrogen bonds are in opposite directions with reference to the minor groove. Analyzing these differences quantitatively using helical parameters, we observed that the mean value of G7-A8 slide was −1 Å for the CP-A8-N7 hydrogen bond, while it was +1.8 Å for the OX-T17-O4 hydrogen bond (Figure 5B). Similarly, the shift, twist and roll of the G7-A8 base-pair step were clearly different for structures with CP-A8-N7 hydrogen bond compared to those with the OX-T17-O4 hydrogen bond (Figure 5B). As might be expected, these conformational differences correlated with the T17-O4 atom being closer to the Pt-amine hydrogen of OX and the A8-N7 atom being closer to the Pt-ammine hydrogen of CP. In summary, when comparing CP-DNA to OX-DNA, the minor conformations had distinct distributions of helical parameters in the vicinity of hydrogen bond formation that were unique to either CP- or OX-DNA.
Figure 5.
Differences in DNA conformation and helical parameters of A8-N7 and T17-O4 hydrogen bonded species in CP- and OX-DNA, respectively. (A) Alignment of the 5′G6G7A8 3′ base pairs of the centroid structures forming A8-N7 hydrogen bond in CP-DNA adduct and the T17-O4 hydrogen bond in the OX-DNA adduct is shown. The structures are aligned based on Pt-G6G7. (B) Plots of histograms of the four helical parameters showing greatest differences between the A8-N7 hydrogen bonded species in CP-DNA and T17-O4 hydrogen bonded species in OX-DNA are shown. The distributions of twist, slide, roll and shift parameters of the G7-A8 base-pair step are plotted because the greatest differences are in this base-pair step (data not shown). The solid line represents distribution of structures with CP-A8-N7 hydrogen bond while the dashed line represents distribution of structures with OX-T17-O4 hydrogen bond. The frequency distributions are calculated and normalized as described earlier.
In the crystal structure of the HMGB1a-CP-DNA complex in the TGGA sequence context, Ser41 of HMGB1a forms a hydrogen bond with N3 atom of the adenine 3′ to Pt-GG adduct (21) (which would correspond to A8 in our simulations of free Pt–DNA). Mutagenesis experiments suggest that this hydrogen bond has a minor (∼4-fold) effect on the affinity of HMGB1a for CP-DNA (20). The formation of the Ser41-A8N3 hydrogen bond is observed in our simulations of the HMGB1a complexes with both CP-DNA and OX-DNA (data not shown). In the HMGB1a-Pt–DNA complexes, structures containing the Ser41-A8 hydrogen bond have a G7-A8 slide and twist that is intermediate between that observed in free CP- and OX-DNA simulations. However, the HMGB1a-Pt–DNA conformers containing the Ser41-A8 hydrogen bond have a G7-A8 roll of 0–7.5° and a G7-A8 shift of −1-0 Å, which is very close to the conformation of the G7-A8 base-pair step seen in structures with T17-O4 hydrogen bond in free Pt–DNA simulations (Supplementary Figure S4). Thus, structures with the OX-T17-O4 hydrogen bond in free Pt–DNA would aid the formation of Ser41 hydrogen bond during complex formation, while structures with CP-A8-N7 hydrogen bonds have the A8 in a position unfavorable for hydrogen bonding with Ser41 during binding to HMGB1a.
Hydrogen bond formation to N7 of an adjacent adenine is associated with higher bend angles
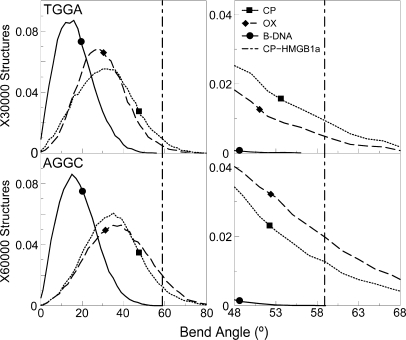
HMGB1a, whose differential binding affinity to CP- and OX-DNA has been experimentally characterized (11,30), binds specifically to bent or distorted DNA and bends it further (31,32). The formation of the Pt-GG adduct induces a sharp bend in the DNA towards the major groove and results in the formation of a wide and shallow minor groove which provides multiple sites for interaction with HMGB1a (21). The higher bend angle of the Pt–DNA adduct has been postulated to make the binding of HMGB1a to Pt–DNA energetically more favorable compared to undamaged DNA (31), and hence the distribution of the bend angles could influence the preferential binding of HMG-domain proteins to Pt–DNA. We calculated the overall bend angle of the central 10 bp of the different hydrogen-bonded conformations using the program MADBEND (29).
The overall distribution of bend angles indicated that the platinated DNA is more bent than undamaged DNA as expected (Figure 6). When we compared CP- and OX-GG adducts in the TGGA sequence context, we observed a higher population of structures in CP-GG adduct compared to OX-GG adduct having bend angles close to 58.9°, which is the bend angle of CP-DNA bound to HMGB1a found in its crystal structure (21) (represented as a dashed vertical line in Figure 6). When we computed the percentage of species having bend angles ±10° of the bend angle of CP-DNA bound to HMGB1a, we found that CP-GG had 10.25% of the population in this region, while OX-GG adduct had only 5.86% of its population in this region. The situation was reversed in the AGGC sequence context, where OX had 22.15% of the population within ±10° of the bend angle of CP-DNA bound to HMGB1a [in its crystal structure (21)], while CP-GG adduct had only 15.24% of the population in the higher bend angle region of the distribution.
Figure 6.
Bend angle distributions of hydrogen-bonded species. The bend angle distributions of CP-, OX-GG adducts and undamaged DNA in the TGGA and the AGGC sequence context are plotted. The distribution for undamaged DNA is plotted as a solid line, the distribution for CP-DNA is plotted as a dotted line and that for OX-DNA is plotted as a dashed line. The bend angle of DNA in the crystal structure of CP-DNA bound to HMGB1a (21) is plotted as a vertical dashed line. The region of bend angle ±10° of the bend angle in the crystal structure of CP-DNA bound to HMGB1a is plotted alongside to highlight differences between CP- and OX-GG adducts.
Upon clustering the distribution of bend angles based on hydrogen bond formation, we observed significant correlation between hydrogen bond pattern and bend angle (Supplementary Figure S5, Table 2). In the TGGA sequence context, the A8-N7 hydrogen bond formed by the CP-GG adduct was associated with higher bend angles (mean bend angle of 44.3°) compared to species with no hydrogen bonds (29.8°) and with the G7-O6 hydrogen bond (29.9°). For the OX-GG adduct, no single species stood out with higher mean bend angle. In the AGGC sequence context, the formation of A5-N7 hydrogen bond was associated with a higher bend angle compared to the structures with the G7-O6 hydrogen bond or no hydrogen bonds with the drug for both CP- and OX-GG adducts. In summary, higher bend angles were associated with hydrogen bond formation to adjacent adenines in both TGGA and AGGC sequence contexts, which gave a significant advantage to the CP-GG adduct in the TGGA sequence context and a slight advantage to the OX-GG adduct in the AGGC sequence context in terms of interaction with HMGB1a.
Table 2.
Bend angle distributions
| CP |
OX |
||||
|---|---|---|---|---|---|
| Hydrogen bond type | Frequency (%) | Mean bend angle (°) | Hydrogen bond type | Frequency (%) | Mean bend angle (°) |
| TGGA | |||||
| A8-N7 | 13 | 44.3 | T17-O4 | 15 | 35.3 |
| G7-O6 | 32 | 29.9 | G7-O6 | 55 | 28.5 |
| None | 54 | 29.9 | None | 23 | 29.9 |
| AGGC | |||||
| A5-N7 | 40 | 36.3 | A5-N7 | 14 | 40.2 |
| A5-N7 + G7 − O6 | 34 | 36.0 | A5-N7 + G7 − O6 | 45 | 39.7 |
| G7-O6 | 13 | 28.3 | G7-O6 | 34 | 31.2 |
| None | 13 | 28.8 | None | 8 | 33.0 |
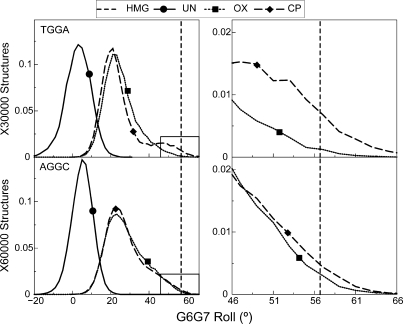
Hydrogen bond formation to N7 of an adjacent adenine is associated with higher roll in the platinated base-pair step
Binding of HMB1a to Pt–DNA involves the intercalation of Phe37 in the platinated base-pair step, resulting in the roll of this base-pair step being 56.7° (21). Thus, ability to sample higher roll could result in better binding affinity for Pt–DNA. The G6-G7 base-pair step has a mean roll of 3.3° and 5.3° in undamaged DNA in the TGGA and AGGC sequence contexts, respectively. The formation of either CP- or OX-G6G7 adducts shifted the G6-G7 roll to significantly higher values compared to undamaged DNA (Figure 7). There was also a pronounced shoulder in the distributions of G6G7 roll parameters in Pt–DNA adducts in both TGGA and AGGC sequence contexts (Figure 7). This shoulder lies in the region that is favorable for binding of HMGB1a, and differs between CP- and OX-GG adducts to different extents in different sequence contexts. In the TGGA sequence context, the CP-GG adduct explored conformations with G6G7 roll ±10° of the roll of CP-DNA bound to HMGB1a 3.2 times more frequently than OX-GG adducts, while in the AGGC sequence context, CP- and OX-GG adducts explored rolls within ±10° of the roll of CP-DNA bound to HMGB1a to about the same extent.
Figure 7.
Distribution of the G6-G7 roll. The distribution of G6-G7 roll, which is important for intercalation of Phe37 of HMGB1a between the G6-G7 base-pair step is plotted for the AGGC and the TGGA sequence contexts. The distributions of CP-, OX- and undamaged DNA are plotted in each sequence context. The distribution for undamaged DNA is plotted as a solid line, the distribution for CP-DNA is plotted as a dashed line and that for OX-DNA is plotted as a dotted line. The bend angle of DNA in the crystal structure of CP-DNA bound to HMGB1a (21) is plotted as a vertical dashed line. The region of roll ±10° of the roll in the crystal structure of CP-DNA bound to HMGB1a is plotted alongside to highlight differences between CP- and OX-DNA in the TGGA sequence context.
The distributions with higher roll were mainly comprised of structures forming the CP-A8-N7 hydrogen bond in the TGGA sequence context (Supplementary Figure S6), while in the AGGC sequence context, the regions of high roll were associated mainly with the formation of both CP- and OX-A5-N7 hydrogen bond (Supplementary Figure S6). Thus, hydrogen bonding to adjacent adenine in both sequence contexts was associated with higher roll in the G6G7 base-pair step.
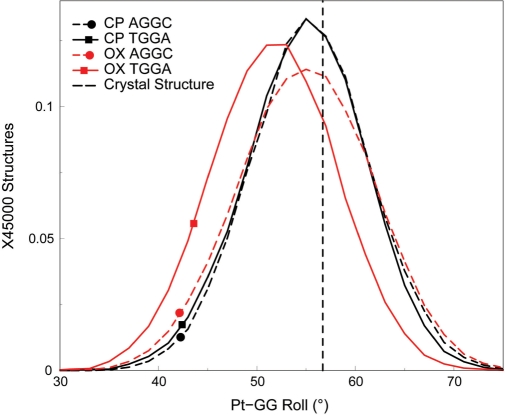
The difference between CP- and OX-DNA in the roll of the G6-G7 base-pair step may be significant for HMGB1a binding to Pt–DNA adducts. Ohndorf et al. (21) have shown that the crystal structure of the HMGB1a-CP-DNA complex has significantly greater roll (56.7°) at the platinated GG base-pair step than free CP-DNA and have postulated that the greater roll enables ideal geometry for π–π stacking of Phe37 of HMGB1a with the 3′ guanine of the platinated base-pair step. This interaction appears to be particularly important for the stability of the HMGB1a-Pt–DNA complex, since the Phe37Ala mutation reduces binding affinity >667-fold. The ability of free CP-DNA in the TGGA sequence context and both free CP- and OX-DNA in the AGGC sequence context (but not free OX-DNA in the TGGA sequence context) to sample roll at values seen in the crystal structure of the HMGB1a-CP-DNA complex would suggest that these Pt–DNA adducts have the conformational flexibility to readily form the HMGB1a-Pt–DNA complex. In contrast, OX-TGGA alone samples lower roll angles at the platinated base-pair step, suggesting that it would have more difficulty forming a complex that has optimal stacking of Phe37 to the 3′G. As might be expected, our simulations of the HMGB1a complexes with Pt–DNA show that indeed the mean platinated base-pair roll of OX-TGGA is downshifted by ∼3.2° compared to the roll seen in CP- and OX-AGGC and CP-TGGA and in the crystal structure of the HMGB1a-CP-DNA complex (Figure 8).
Figure 8.
Distributions of the Pt-GG roll in the HMGB1a-Pt–DNA simulations. The distribution of the roll of the Pt-GG base-pair step, which is important for stacking of Phe37 of HMGB1a to the 3′G is plotted for the HMGB1a-Pt–DNA complexes in the AGGC and the TGGA sequence contexts. The distributions of the roll of CP- and OX-DNA are plotted for each sequence context. Distributions of adducts in the TGGA sequence context are plotted as solid lines, while the distributions of adducts in the AGGC sequence context are plotted as dashed lines. CP-DNA is plotted in black, while OX-DNA is plotted in red. The dashed vertical line represents the roll of the G6-G7 base-pair step from the HMGB1a-CP-DNA crystal structure. The frequency distribution histograms were calculated from the structures obtained at every picosecond over the final 45 ns of each equilibrated MD simulation, resulting in a total of 45 000 structures each for CP- and OX-DNA in the TGGA and AGGC sequence contexts. The distribution is plotted against the number of structures in the trajectory.
In summary, CP-DNA in the TGGA sequence context was able to sample conformations that resulted in a high roll in the G6G7 base-pair step, which has been suggested to favor binding to HMGB1a (21), while OX-DNA in the same sequence context was unable to sample these conformations. Contrastingly, both CP- and OX-DNA sampled conformations with a high roll in the G6G7 base-pair step to the same extent in the AGGC sequence context. Furthermore, these differences in roll between CP- and OX-DNA adducts in the TGGA sequence context appear to be preserved in the complexes with HMGB1a.
DISCUSSION
The goal of this study was to uncover the conformational differences between CP- and OX-GG adducts which could explain the differential binding affinities of cellular proteins to these adducts. The molecular basis of these differential binding affinities is not clear and understanding the underlying mechanism may throw light on the contrasting downstream effects of CP- and OX-GG adducts at the cellular level. It has been hypothesized that the different downstream effects of CP- and OX-GG adducts are caused in part by the differential binding affinities of cellular damage-recognition proteins to CP- and OX-GG adducts (8,16,18,33–35). Further, it has been shown that HMGB1a binds to CP-GG adducts with approximately the same affinity in both the TGGA and AGGC sequence contexts, while it binds to OX-GG adducts with ∼3-fold lower affinity in the AGGC sequence context and 53-fold lower affinity in the TGGA sequence context (18). Thus, the binding affinity of HMGB1a to Pt-GG adducts is dependent both on the carrier ligand and the sequence context. We have postulated that the differences in binding affinity to CP- and OX-DNA by Pt–DNA binding proteins is due to the differences in their conformational dynamics, i.e. the time spent by CP- and OX-DNA adducts in various conformations favorable for binding (26).
From our simulations of free Pt–DNA in the TGGA sequence context, we were able to identify various conformations on the basis of the pattern of hydrogen bonds between Pt-amines and the DNA. The major conformations, which are characterized by either no hydrogen bonds being formed or hydrogen bond formed with G7-O6, have similar structural parameters, which explains the lack of differences in the overall centroid structures in these simulations and the lack of major conformational differences between CP- and OX-GG adducts in previous NMR and crystal structures. However, the minor conformations, which are accompanied by hydrogen bonds to bases adjacent to the Pt-GG, feature significantly different structural parameters that are unique to both CP- and OX-GG adducts. The differences in hydrogen bond patterns between CP- and OX-GG adducts appear to be due to constraints imposed by the cyclohexane ring on the amine-Pt-amine bond angle and the N7-Pt-N-H dihedral angle. If conformational dynamics influence the differences in the binding affinity of HMGB1a to CP- and OX-DNA, then we would expect these minor conformations to have distinct aspects of structural parameters that favor binding by HMGB1a.
The crystal structure of HMGB1a-Pt–DNA complex reveals a number of hydrophobic contacts and hydrogen bonds between HMGB1a and the DNA minor groove. Given that HMGB1a has been shown not to make any sequence-specific contacts with its target DNA (21), the flexibility and bending ability of free Pt–DNA could have a major influence on the binding affinity. The affinity of HMGB1a for Pt–DNA adducts appears to be strongly influenced by three conformational features of DNA containing Pt-GG adducts: overall bend angle, Phe37 stacking with the 3′G of the Pt-GG adduct and, in the TGGA sequence context, hydrogen bond formation between Ser41 and the 3′A. The overall bend angle is important because bending of the DNA towards the major groove favors formation of a widened minor groove opposite the Pt-GG adduct which serves as the binding site for HMGB1a (31). However, based on both the crystal structure and mutagenesis studies, the π–π stacking of Phe37 with the 3′G appears to be of primary importance, with the Ser41 hydrogen bond with the 3′AN3 in the TGGA sequence context being of secondary importance (20,21). The effects of the other interactions of HMGB1a with the widened minor groove Pt–DNA adducts have not been characterized, but are likely to be of less or equal significance than the Ser41 hydrogen bond.
Pt–DNA adducts that can sample higher bend angles would be expected to be bound by HMGB1a with greater affinity (36). We observed that higher bend angle was associated with hydrogen bonding to adjacent adenine for both CP- and OX-GG adducts. In the TGGA sequence context, CP-GG adduct can form A8-N7 hydrogen bond and can also sample higher bend angles more frequently, while OX-GG adduct does not form the A8-N7 hydrogen bond and samples higher bend angles less frequently compared to CP-GG adducts. In the AGGC sequence context, both CP- and OX-GG adducts form the A5-N7 hydrogen bond and sample high bend angles, with OX-DNA adducts sampling at high bend angles at a slightly higher frequency than CP-DNA adducts. Thus, CP- and OX-DNA in AGGC sequence context and CP-DNA in TGGA sequence context but not OX-DNA in TGGA sequence context sample bend angles that would favor binding to HMGB1a.
In addition, the binding of HMGB1a to Pt–DNA adducts involves intercalation of Phe37 into the Pt-GG base-pair step, which is associated with an unusually large roll and dihedral angle at that base-pair step. Thus, HMGB1a would likely have a higher affinity for DNA structures that favor high roll in the G6G7 base-pair step. In our simulations, we observed that CP-DNA is able to sample structures with roll ±10° of the roll of CP-DNA bound to HMGB1a significantly more than OX-DNA in the TGGA sequence context, while in the AGGC sequence context, the populations of CP- and OX-DNA in this region were essentially the same. The differences seen in free Pt–DNA simulations are reflected in the simulations of HMGB1a-Pt–DNA simulations, where OX-DNA in the TGGA sequence context has a mean roll that is less than that of CP-DNA. The geometry of the Pt-GG base-pair step in the HMGB1a-Pt–DNA crystal structure is ideal for π–π stacking of Phe37 of HMGB1a with the 3′G, and deviations from this geometry are expected to decrease binding affinity. Decrease in the roll of Pt-GG implies decreased surface area of 3′G that is exposed for the stacking interaction. This agrees well with gel shift data, which shows appreciable binding of HMGB1a to CP- and OX-DNA in the AGGC sequence context, and to CP-DNA in the TGGA sequence context, but not to OX-DNA in the TGGA sequence context (18).
Finally, DNA helical parameters at the G7N8 base-pair step also appear to influence the affinity of HMGB1a for Pt–DNA adducts. In the TGGA sequence context, the formation of a hydrogen bond between Ser41 and the 3′A contributes to the stability of the HMGB1a-Pt–DNA complex (20), and the roll and shift of G7-A8 base-pair step appear to favor formation of this hydrogen bond between HMGB1a and OX-DNA during complex formation. In a previous comparison of the conformational dynamics of CP- and OX-GG DNA adducts in the AGGC sequence context (26), we found that the shift and slide of the G7-C8 base-pair step of CP-GG adduct favored binding of HMGB1a to the CP-GG adducts.
In summary, the specificity of binding of HMGB1a to Pt–DNA adducts is influenced by a number of complex structural features. In the case of CP- and OX-GG DNA adducts in the TGGA sequence context, the ability of CP-GG adducts to explore greater bend angles and greater roll at the G6G7 base-pair step favors Phe37 π–π stacking with G7. Both of these conformational features favor binding of HMGB1a to CP-GG adducts. In contrast, the conformations explored by the DNA at the G7A8 base-pair step favor formation of the Ser41-A8 hydrogen bond, which in turn favors binding of HMGB1a to the OX-GG adduct. The quantitative effect of DNA bending on the affinity of HMGB1a for Pt–DNA adducts has not been established, but the effect of Phe37 stacking with the 3′G is much greater than formation of the Ser41-A8N3 hydrogen bond (20), which might explain the observed 53 times higher binding affinity of HMGB1a towards CP-GG adducts compared to OX-GG adducts in the TGGA sequence context (18). For the AGGC sequence context, differences in several helical parameters at the G7A8 base-pair step favor binding of CP-GG adduct to HMGB1a, while roll at the G6G7 base-pair step does not appear to offer a significant advantage to either CP- or OX-GG adducts and the overall bend angle favors OX-GG adduct slightly, perhaps explaining the just three times higher affinity of HMGB1a towards CP-GG adduct compared to OX-GG adduct in that sequence context (11,18).
We have demonstrated here that the sequence dependent differences in conformational dynamics of CP- and OX-DNA correlate with hydrogen bond formation between the Pt-amines and adjacent bases. However, the formation of hydrogen bonds poses an interesting question in terms of cause-effect: is it the hydrogen bonds that drive these unique conformations, or is it the sequence dependent conformations that allow the formation of hydrogen bonds? Since previous binding studies with DNA which had 7-deazapurines flanking Pt-GG revealed no changes in binding affinities of HMGB1a toward CP-DNA (37) and the roll at the G6G7 base-pair step is more constrained for OX-DNA in the HMGB1a-OX-DNA complex even though hydrogen bond formation between Pt-amines and adjacent bases is minimal, we consider it likely that the hydrogen bonds do not drive these conformations. However, the association of hydrogen bond formation with conformations that are unique to CP- and OX-DNA have provided us an avenue to characterize the conformational dynamics of Pt-GG adducts.
Apart from HMG-domain proteins, other cellular proteins have also been observed to show differential binding affinity to CP- and OX-DNA adducts, which may have possible functional significance. The mismatch repair proteins, MutS and hMSH2 have been observed to bind with higher affinity to CP-DNA compared to OX-DNA and it has been shown that mismatch repair deficient cells were resistant to CP- but not to OX. Thus, mismatch repair proteins have an important role in CP toxicity but not OX toxicity. The differences in conformational dynamics could certainly contribute to the differential affinities in mismatch repair proteins, but structural information and mutational studies on hMSH2 and MutS are limited and out of the scope of this article. Similarly, error-prone polymerases are another group of proteins that have been shown to functionally discriminate CP- and OX-DNA adducts, and the conformational differences observed here might have an important role in the interactions of Pt–DNA with the error-prone polymerases as well.
To summarize, we showed that the conformational dynamics of CP- and OX-DNA are different. The differences in conformational dynamics between CP- and OX-DNA are likely determined by the constraints on the NHx-Pt-NHx bond angles and the N7-Pt-N-H dihedral angles, which in turn depended on both the sequence context and carrier ligand of the Pt-GG adduct. These differences may explain the dependence of sequence context on the extent of differential binding of HMGB1a to CP- and OX-DNA adducts. These differences in conformational dynamics may also contribute to the differential binding of mismatch repair proteins to CP- and OX-DNA adducts and also the differential translesion replication of these adducts by error-prone polymerases. Thus, the differential conformational dynamics of CP- and OX-DNA could contribute to the differences in efficacy and toxicity of these drugs.
SUPPLEMENTARY DATA
Supplementary data is available at NAR Online.
FUNDING
National Institutes of Health (CA84480); National Institute of Environmental Health Sciences (P30ES10126).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Shantanu Sharma and Dr Debadeep Bhattacharya for insightful discussions.
REFERENCES
- 1.Chaney SG, Campbell SL, Bassett E, Wu Y. Recognition and processing of cisplatin- and oxaliplatin-DNA adducts. Crit. Rev. Oncol. Hematol. 2005;53:L3–L11. doi: 10.1016/j.critrevonc.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 2.Jamieson ER, Lippard SJ. Structure, recognition, and processing of cisplatin-DNA adducts. Chem. Rev. 1999;99:2467–2498. doi: 10.1021/cr980421n. [DOI] [PubMed] [Google Scholar]
- 3.Page JD, Husain I, Sancar A, Chaney SG. Effect of the diaminocyclohexane carrier ligand on platinum adduct formation, repair, and lethality. Biochemistry. 1990;29:1016–1024. doi: 10.1021/bi00456a026. [DOI] [PubMed] [Google Scholar]
- 4.Jennerwein MM, Eastman A, Khokhar AR. Characterization of adducts produced in DNA by isomeric 1,2-diaminocyclohexaneplatinum(II) complexes. Chem. Biol. Interact. 1989;70:39–49. doi: 10.1016/0009-2797(89)90061-6. [DOI] [PubMed] [Google Scholar]
- 5.Woynarowski JM, Chapman WG, Napier C, Herzig MCS, Juniewicz P. Sequence- and region-specificity of oxaliplatin adducts in naked and cellular DNA. Mol. Pharmacol. 1998;54:770–777. doi: 10.1124/mol.54.5.770. [DOI] [PubMed] [Google Scholar]
- 6.Pil PM, Lippard SJ. Specific binding of chromosomal protein HMG1 to DNA damaged by the anticancer drug cisplatin. Science. 1992;256:234–237. doi: 10.1126/science.1566071. [DOI] [PubMed] [Google Scholar]
- 7.Ohndorf UM, Whitehead JP, Raju NL, Lippard SJ. Binding of tsHMG, a mouse testis-specific HMG-domain protein, to cisplatin-DNA adducts. Biochemistry. 1997;36:14807–14815. doi: 10.1021/bi9717643. [DOI] [PubMed] [Google Scholar]
- 8.Trimmer EE, Zamble DB, Lippard SJ, Essigmann JM. Human testis-determining factor SRY binds to the major DNA adduct of cisplatin and a putative target sequence with comparable affinities. Biochemistry. 1998;37:352–362. doi: 10.1021/bi971675q. [DOI] [PubMed] [Google Scholar]
- 9.Dunham SU, Lippard SJ. DNA sequence context and protein composition modulate HMG- domain protein recognition of ciplatin-modified DNA. Biochemistry. 1997;36:11428–11436. doi: 10.1021/bi9709452. [DOI] [PubMed] [Google Scholar]
- 10.Farid RS, Bianchi ME, Falciola L, Engelsberg BN, Billings PC. Differential binding of HMG1, HMG2, and a single HMG box to cisplatin-damaged DNA. Toxicol. Appl. Pharmacol. 1996;141:532–539. doi: 10.1006/taap.1996.0319. [DOI] [PubMed] [Google Scholar]
- 11.Malina J, Novakova O, Vojtiskova M, Natile G, Brabec V. Conformation of DNA GG intrastrand cross-link of antitumor oxaliplatin and its enantiomeric analog. Biophys. J. 2007;93:3950–3962. doi: 10.1529/biophysj.107.116996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chow CS, Whitehead JP, Lippard SJ. HMG domain proteins induce sharp bends in cisplatin-modified DNA. Biochemistry. 1994;33:15124–15130. doi: 10.1021/bi00254a023. [DOI] [PubMed] [Google Scholar]
- 13.Chvalova K, Sari MA, Bombard S, Kozelka J. LEF-1 recognition of platinated GG sequences within double-stranded DNA. Influence of flanking bases. J. Inorg. Biochem. 2008;102:242–250. doi: 10.1016/j.jinorgbio.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Huang JC, Zamble DB, Reardon JT, Lippard SJ, Sancar A. HMG-domain proteins specifically inhibit the repair of the major DNA adduct of the anticancer drug cisplatin by human excision nuclease. Proc. Natl Acad. Sci. USA. 1994;91:10394–10398. doi: 10.1073/pnas.91.22.10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McA’Nulty MM, Lippard SJ. The HMG-domain protein Ixr1 blocks excision repair of cisplatin-DNA adducts in yeast. Mutat. Res./DNA Repair. 1996;362:75–86. doi: 10.1016/0921-8777(95)00037-2. [DOI] [PubMed] [Google Scholar]
- 16.Zhai X, Beckmann H, Jantzen HM, Essigmann JM. Cisplatin-DNA adducts inhibit ribosomal RNA synthesis by hijacking the transcription factor human upstream binding factor. Biochemistry. 1998;37:16307–16315. doi: 10.1021/bi981708h. [DOI] [PubMed] [Google Scholar]
- 17.Treiber DK, Zhai X, Jantzen HM, Essigmann JM. Cisplatin-DNA adducts are molecular decoys for the ribosomal RNA transcription factor hUBF (human upstream binding factor) Proc. Natl Acad. Sci. USA. 1994;91:5672–5676. doi: 10.1073/pnas.91.12.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei M, Cohen SM, Silverman AP, Lippard SJ. Effects of spectator ligands on the specific recognition of intrastrand platinum-DNA cross-links by high mobility group box and TATA-binding proteins. J. Biol. Chem. 2001;276:38774–38780. doi: 10.1074/jbc.M106374200. [DOI] [PubMed] [Google Scholar]
- 19.Jung Y, Lippard SJ. Nature of full-length HMGB1 binding to cisplatin-modified DNA. Biochemistry. 2003;42:2664–2671. doi: 10.1021/bi026972w. [DOI] [PubMed] [Google Scholar]
- 20.He Q, Ohndorf UM, Lippard SJ. Intercalating residues determine the mode of HMG1 domains a and B binding to cisplatin-modified DNA. Biochemistry. 2000;39:14426–14435. doi: 10.1021/bi001700j. [DOI] [PubMed] [Google Scholar]
- 21.Ohndorf UM, Rould MA, He Q, Pabo CO, Lippard SJ. Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins. Nature. 1999;399:708–712. doi: 10.1038/21460. [DOI] [PubMed] [Google Scholar]
- 22.Takahara PM, Rosenzweig A, Frederick C, Lippard S. Crystal structure of double-stranded DNA containing the major adduct of the anticancer drug cisplatin. Nature. 1995;377:649–652. doi: 10.1038/377649a0. [DOI] [PubMed] [Google Scholar]
- 23.Spingler B, Whittington DA, Lippard SJ. 2.4 Å crystal structure of an oxaliplatin 1,2-d(GpG) intrastrand cross-link in a DNA dodecamer duplex. Inorg. Chem. 2001;40:5596–5602. doi: 10.1021/ic010790t. [DOI] [PubMed] [Google Scholar]
- 24.Wu Y, Pradhan P, Havener J, Boysen G, Swenberg JA, Campbell SL, Chaney SG. NMR solution structure of an oxaliplatin 1,2-d(GG) intrastrand cross-link in a DNA dodecamer duplex. J. Mol. Biol. 2004;341:1251–1269. doi: 10.1016/j.jmb.2004.06.066. [DOI] [PubMed] [Google Scholar]
- 25.Wu Y, Bhattacharyya D, King CL, Baskerville-Abraham I, Huh SH, Boysen G, Swenberg JA, Temple B, Campbell SL, Chaney SG. Solution structures of a DNA dodecamer duplex with and without a cisplatin 1,2-d(GG) intrastrand cross-link: comparison with the same DNA duplex containing an oxaliplatin 1,2-d(GG) intrastrand cross-link. Biochemistry. 2007;46:6477–6487. doi: 10.1021/bi062291f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma S, Gong P, Temple B, Bhattacharyya D, Dokholyan NV, Chaney SG. Molecular dynamic simulations of cisplatin- and oxaliplatin-d(GG) intrastand cross-links reveal differences in their conformational dynamics. J. Mol. Biol. 2007;373:1123–1140. doi: 10.1016/j.jmb.2007.07.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavery R, Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 28.Press WH, Flannery BP, Teukolsky SA, Vetterling WT. Numerical Recipes in C: The Art of Scientific Computing. 2nd. Cambridge: Cambridge University Press; 1992. [Google Scholar]
- 29.Strahs D, Sclick T. A-tract bending: insights into experimental structures by computational models. J. Mol. Biol. 2000;301:643–663. doi: 10.1006/jmbi.2000.3863. [DOI] [PubMed] [Google Scholar]
- 30.Pil PM, Lippard SJ. Specific binding of chromosomal protein HMG1 to DNA damaged by the anticancer drug cisplatin. Science. 1992;256:234–237. doi: 10.1126/science.1566071. [DOI] [PubMed] [Google Scholar]
- 31.Pil PM, Chow CS, Lippard SJ. High-mobility-group-1 protein mediates DNA bending as determined by ring closures. Proc. Natl Acad. Sci. USA. 1993;90:9465–9469. doi: 10.1073/pnas.90.20.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bianchi ME, Beltrame M, Paonessa G. Specific recognition of cruciform DNA by nuclear protein HMG1. Science. 1989;243:1056–1059. doi: 10.1126/science.2922595. [DOI] [PubMed] [Google Scholar]
- 33.Coin F, Frit P, Viollet B, Salles B, Egly JM. TATA binding protein discriminates between different lesions on DNA, resulting in a transcription decrease. Mol. Cell Biol. 1998;18:3907–3914. doi: 10.1128/mcb.18.7.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zdraveski ZZ, Mello JA, Farinelli CK, Essigmann JM, Marinus MG. MutS preferentially recognizes cisplatin- over oxaliplatin-modified DNA. J. Biol. Chem. 2002;277:1255–1260. doi: 10.1074/jbc.M105382200. [DOI] [PubMed] [Google Scholar]
- 35.Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nat. Rev. Drug Discov. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 36.Webb M, Payet D, Lee KB, Travers AA, Thomas JO. Structural requirements for cooperative binding of HMG1 to DNA minicircles. J. Mol. Biol. 2001;309:79–88. doi: 10.1006/jmbi.2001.4667. [DOI] [PubMed] [Google Scholar]
- 37.Cohen SM, Mikata Y, He Q, Lippard SJ. HMG-domain protein recognition of cisplatin 1,2-intrastrand d(GpG) cross-links in purine-rich sequence contexts. Biochemistry. 2000;39:11771–11776. doi: 10.1021/bi001352l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.