Abstract
Mitochondrial DNA (mtDNA) is located in close proximity of the respiratory chains, which are the main cellular source of reactive oxygen species (ROS). ROS can induce oxidative base lesions in mtDNA and are believed to be an important cause of the mtDNA mutations, which accumulate with aging and in diseased states. However, recent studies indicate that cumulative levels of base substitutions in mtDNA can be very low even in old individuals. Considering the reduced complement of DNA repair pathways available in mitochondria and higher susceptibility of mtDNA to oxidative damage than nDNA, it is presently unclear how mitochondria manage to maintain the integrity of their genetic information in the face of the permanent exposure to ROS. Here we show that oxidative stress can lead to the degradation of mtDNA and that strand breaks and abasic sites prevail over mutagenic base lesions in ROS-damaged mtDNA. Furthermore, we found that inhibition of base excision repair enhanced mtDNA degradation in response to both oxidative and alkylating damage. These observations suggest a novel mechanism for the protection of mtDNA against oxidative insults whereby a higher incidence of lesions to the sugar–phosphate backbone induces degradation of damaged mtDNA and prevents the accumulation of mutagenic base lesions.
INTRODUCTION
Mutations in mitochondrial DNA (mtDNA) are an underlying factor in many mitochondrial diseases (1) and have been associated with cancer (2,3), neurodegenerative disorders (4,5), diabetes (6–8) and aging (9,10). Therefore, mechanisms of mtDNA mutagenesis are of considerable interest. Mutations in mtDNA can arise following exposure to environmental mutagens, from DNA polymerase errors during replication, from failure of the mtDNA repair machinery, and from defects in the mechanisms of degradation of damaged mtDNA. However, perhaps the most recognized source of mtDNA mutagenesis is reactive oxygen species (ROS), which are produced by the mitochondrial electron transport chain (ETC). Rates of oxygen reduction to the principal ROS, the superoxide anion (O2.−), have been reported to be as high as 1–5% of total consumed oxygen (11,12). The charged and relatively unstable O2.− is membrane-impermeable and, depending on the site of production, can be released into either the mitochondrial matrix or the intermembrane space. There, it is quickly converted to hydrogen peroxide (H2O2), the principal cellular mediator of oxidative stress. This conversion occurs either spontaneously or enzymatically, with the help of superoxide dismutases. The relative stability and membrane permeability of H2O2 allows it to freely diffuse throughout the cell, where it can generate, by means of the Fenton reaction, the extremely reactive hydroxyl radical, which can efficiently damage DNA (13,14). It has been reported that the main products of mtDNA base damage are thymine glycol among pyrymidines (15) and 7,8-dihydro-8-oxo-2′-deoxyguanosine (8-oxoG) among purines (16–18). The former has low mutagenicity, whereas the latter upon replication can cause characteristic G→T transversions (15).
Mitochondrial production of ROS and the ensuing damage inflicted by these ROS on biological macromolecules, including DNA, constitutes the basis of the free radical/mitochondrial theory of aging (19–21). According to this theory, the production of ROS by mitochondria leads to mtDNA damage and mutations which in turn lead to progressive respiratory chain dysfunction and to a further increase in ROS production as a consequence of this dysfunction. The exponential escalation of these processes is commonly referred to as a ‘vicious cycle’, and the theory predicts that the rise in mtDNA mutations and ROS eventually reach levels that are incompatible with life.
In this study, we examine the effects of oxidative stress on the integrity of mtDNA and conclude that oxidative stress can result in the loss of damaged mtDNA molecules. This phenomenon is unique to the mitochondrial compartment, and is enabled by the high redundancy of mtDNA.
MATERIALS AND METHODS
Cell growth and treatment
HCT116 cells (a kind gift of Drs Vogelstein and Kinzler) were grown in Dulbecco's modified Eagle medium (DMEM) containing 10% Fetal Bovine Serum and 50 µg/ml gentamycin in a humidified atmosphere containing 5% CO2 at 37°C. For oxidative damage and repair experiments cells were grown in 100 mm dishes until 50–80% confluence. Then cultures were washed with warm HBSS and exposed to the indicated concentrations of H2O2 in HBSS for the indicated periods of time at 37°C in a humidified atmosphere containing 5% CO2. Control cultures were mock-treated with HBSS alone. After incubation, cells were either lysed immediately or allowed to repair for the indicated periods of time in complete growth medium. Cell treatment with hypoxanthine/xantine oxidase (XO) was performed for 1h using indicated concentrations of enzyme and 0.5 mM hypoxanthine in PBS. Methyl methane sulfonate (MMS) treatments were performed in PBS for 30 min at 37°C in a humidified atmosphere containing 5% CO2. Where indicated, methoxyamine (30 mM) was included both during treatment and recovery periods.
For long-term rotenone treatment, cells were seeded in three T-75 flasks, allowed to attach for 20 h, and the medium was replaced with fresh medium containing 200 nM rotenone. From this point on, the medium was replaced every 48 h, and cells were split as necessary as they approached confluency. At the end of the experiment, cultures were allowed to undergo at least two cell divisions to allow for the fixation of mtDNA mutations prior to DNA extraction.
Mitochondrial ROS measurement
To measure mitochondrial ROS production in response to rotenone, cells were collected by trysinization, washed with PBS, and loaded with 5 μM of MitoSOX (Invitrogen, Carlsbad, CA) in the presence of indicated concentrations of rotenone for 30 min at 37°C in a humidified atmosphere containing 5% CO2 followed by flow cytometry.
Quantitative Southern blotting
Quantitative alkaline Southern blotting was performed essentially as described earlier (22). Neutral Southern blots were performed similarly, except that there was no alkaline pretreatment, and NaOH was not included in the loading dye, agarose gel or the electrophoresis buffer. When blotting BamHI-digested total DNA, after transfer the membrane was cut at the level of the 9-kb band of lambda/HindIII marker. The upper portion was then hybridized with the mtDNA probe (16.5-kb fragment), and the lower portion was hybridized with the 18S rDNA probe (5-kb fragment). After hybridization, membranes were exposed to an imaging screen to determine band intensity. The number of pixels per band was determined by encompassing bands with identical rectangular regions of interest and subtracting the background.
The break frequency was determined using the Poisson expression (s = –ln P0, where s is the number of breaks per fragment and P0 is the fraction of fragments free of breaks).
PCR-cloning-sequencing
The isolation of total cellular DNA was described previously (22). The amplification of mtDNA was performed in triplicate or quadruplicate 100 µl reactions per each independent experimental condition. Each reaction contained 50 µl of 2× high fidelity PCR master mix (New England Biolabs, Ipswich, MA), 500 ng of total DNA, 1 µM of each primer (forward AATGTCTGCACAGCCACTTTCCAC and reverse AGGGTTTGCTGAAGATGGCGGTAT), and the balance of water. The PCR was performed for 25 cycles (denaturation for 10 s at 98°C, annealing for 5 s at 55°C and extension for 30 s at 55°C). The PCR product was directly purified from the reaction mixture using PCR purification kit (Qiagen, Valencia, CA) and ligated to the EcoRV− digested plasmid pBluescriptII SK+ for 4 h at 25°C. The ligated mixture was introduced into Escherichia coli cells, allowed to incubate for 1 h at 37°C and then 25% of the total mixture was diluted 1:4 and 100 µl aliquots of this dilution were plated on LB agar plates containing 200 µg/ml ampicillin, 40 µg/ml X-gal and 1 mM IPTG. Typically, no more than 16% of electroporated cells were plated. White colonies were picked, inoculated into TB medium (1.2% Tryptone, 2.4% yeast extract, 0.4% glycerol, 72 mM K2HPO4 and 17 mM KH2PO4), and grown overnight to OD600 = 5–8. At this point, bacterial cells were pelleted by centrifugation, and plasmid DNA was extracted using Qiaprep spin kit (Qiagen). 0.5–1.0 µg of this DNA was used in 10 µl cycle-sequencing reaction (BigDye v3.1., Applied Biosystems) in 96-well PCR plates using T3 and T7-sequencing primers. Sequencing products were purified using 96-well DTR plates (Edge Biosystems, Gaithersburg, MD), and submitted to high throughput capillary sequencing runs at either the DNA-sequencing facility at the Iowa State University, or Functional Biosciences (Madison, WI). The resulting chromatograms were aligned against the consensus sequence of amplified fragments.
RESULTS
Validation of PCR-cloning-sequencing assay
Several methods have been described for the mutational analysis of mtDNA. Among those are a single-molecule PCR (23), a random mutation capture (RMC) assay (24) and PCR-cloning-sequencing (25). The first of these methods is difficult to adapt for the detection of low levels of heteroplasmy, while the second samples mutation frequencies in very short palindromic sequences and may underreport mutation frequency due to the relaxed specificity of the restriction enzyme used to eliminate wild type restriction sites. Therefore, we have adopted a PCR-cloning-sequencing strategy for our studies. To experimentally determine the level of PCR-induced mutations in our system, we amplified and cloned an 1111-bp DNA fragment of the mtDNA control region in a kanamycin-resistant derivative of pBluescriptII SK+, extracted this test plasmid DNA, and subjected it to PCR-cloning-sequencing. To approximate the starting template quantity, we assumed that mtDNA constitutes 1% of the total cellular DNA [or 4000 copies per cell (26)]. In PCR-cloning-sequencing we used 500 ng of total DNA for the amplification, which corresponds to 5 ng of mtDNA (1% of total DNA). The size of the template plasmid (4.1 kb) is roughly 25% of that of the mtDNA molecule (16.5 kb). Therefore, 1.25 ng of plasmid DNA was used as the template. The template was subjected to 25 cycles of PCR with Phusion DNA polymerase, and the PCR product was ligated to ampicillin-resistant pBluescriptII SK+ (this manipulation excludes contamination with kanamycin resistant template plasmid DNA). The resulting clones were subjected to sequencing as described in the ‘Materials and methods’ section. The level of PCR-induced mutagenesis under these experimental conditions was 5.6 × 10−6 ± 5.6 × 10−6 (n = 3), or a little less than 1 mutation per 10 mitochondrial genomes (Figure 1A). This is in very good agreement with the calculated upper limit of PCR-induced mutagenesis under these conditions of 1 × 10−5 (see below).
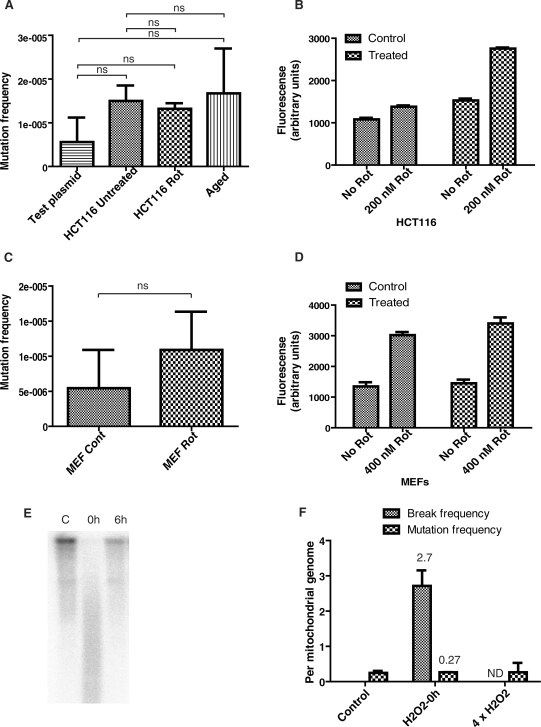
Figure 1.
Levels of mtDNA mutations in response to ROS. (A) mtDNA cloned in a plasmid vector (Test plasmid) as well mtDNA from very old individuals (98–101 years old) and mtDNA from HCT116 cells, which were either left untreated or were treated for 30 days with the ROS generator rotenone, were subjected to PCR-cloning-sequencing analysis. No significant increase was detected under any of the conditions tested (n = 3, one-way ANOVA with Tukey post-test). (B and D) Prolonged treatment of HCT116 and MEF cells with rotenone does not result in the selection of cells unable to generate ROS. Control and rotenone-treated (30 days, 200 nM, see ‘Materials and Methods’ section) HCT116 cells were loaded with 5 µM of the mitochondrial superoxide radical indicator MitoSOX, treated with 200 nM rotenone for 30 min, and subjected to FACS analysis; (C) chronic treatment of MEFs with rotenone (400 nM, 30 days) does not cause a significant increase in the mutations frequency (n = 3, two-tailed Student's t-test assuming unequal variances). (E) A representative quantitative Southern blot illustrating both mtDNA damage immediately after treatment (0 h), and repair over 6-h period (6 h). HCT116 cells were treated with 400 µM H2O2 for 60 min, and total BamHI-digested genomic DNA was subjected to both quantitative Southern blotting under the alkaline conditions and PCR-cloning-sequencing. (F) Relative frequency of combined SSBs and abasic sites versus mutagenic base lesions in mtDNA of control (untreated) cells, in mtDNA of cells from (E) (H2O2—0 h), and in mtDNA of cells treated with H2O2 four times with 24 h intervals between treatments (4 × H2O2). The frequency is expressed on a per mtDNA molecule basis (n = 3).
There are three methodological concerns associated with the PCR-cloning-sequencing approach:
The presence of nuclear ‘pseudogenes’ for the mitochondrial genome. The amplification and sequencing of these pseudogenes results in overreporting mutation frequencies (27). Therefore, the putative PCR product was homology-searched for its absence in nuclear DNA (nDNA).
Sequencing multiple copies of the same PCR fragment. This is only relevant for DNA ligations that produce a low yield of bacterial colonies. Indeed, during a 1-h incubation in the medium for the expression of a selective marker following electroporation, E. coli cells can divide at least once. Therefore, exhaustive sequencing of all clones would result in sequencing of two to four copies of each cloned PCR fragment. To control for this problem, no more than 16% of electroporated bacteria were spread on selective plates and then only a fraction of white colonies was sequenced.
PCR-induced mutations. DNA-polymerase-mediated errors accumulate in PCR fragments as a function of the cycle number and the fidelity of the DNA polymerase. With each PCR cycle new mutations are introduced by the DNA polymerase. Also, PCR errors introduced in earlier cycles are faithfully copied in each subsequent cycle so that their number grows exponentially. To model this process, Lin et al. (25) have derived a formula for the estimation of an average number of mutations introduced into each strand of PCR product. <k> = cfLη/(1 + η), where <k> is the average number of errors per strand, c is the number of PCR cycles, f is the fidelity of DNA polymerase (4 × 10−7 for Phusion polymerase), L is the length of the amplified fragment and η is a fraction of template strands amplified in each cycle. Making substitutions for a 1111-bp fragment amplified with Phusion polymerase, we obtain <k> = 25 × 4 × 10−7 × 1111 × η/(1 + η) = 0.01111 × η/(1 + η). Assuming that the fraction of template strands amplified in each cycle η is somewhere between 0.3 and 1, we obtain <k> values 0.0026 and 0.0056, respectively, for each strand, or for a double-stranded PCR fragment 0.005 and 0.01, respectively. This means that PCR-introduced errors will be present at the most in 1 fragment per every 100 clones sequenced, or an error rate of 10−5 per base pair.
Chronic superoxide exposure and aging do not result in significant mtDNA mutagenesis
The mitochondrial ETC complex I is an important site for ROS production. When inhibited with rotenone, this complex releases superoxide to the matrix side of the inner mitochondrial membrane, in close proximity to mtDNA (28,29). Chronic systemic rotenone infusion has been utilized to induce symptoms in one of the most faithful animal models of Parkinsonism, a disease associated with mtDNA mutations and advanced age (30). Therefore, to approximate exposure of mtDNA to ROS over a lifetime, HCT116 cells and immortalized mouse embryonic fibroblasts (MEFs) were treated with 200 nM and 400 nM rotenone, respectively, for 30 days. 200 nM and 400 nM have been determined to be the maximum concentrations tolerated by HCT116 and MEF cells, respectively, without induction of apoptosis. These treatments did not result in a statistically significant increase in the rate of mtDNA mutagenesis over untreated controls (Figure 1A and C). If, during the treatment period, a selection of cells unable to produce O2.− in response to rotenone inhibition had occurred, this artifact could have manifested itself as a lack of mtDNA mutagenesis in response to rotenone treatment. To explore this possibility, the production of O2.− in response to rotenone treatment was measured in both HCT116 and MEF cells before and after the completion of experiments. Both types of cells preserved their ability to respond to rotenone treatment with superoxide production (Figure 1B and D).
It is often postulated that a significant accumulation of ROS-induced mtDNA mutations should be observed at the end of the lifespan. Accordingly, mtDNA from the lymphocytes of two female centenarians and one male non-agenarian (98 years old), available as a panel of samples from the NIA repository at the Coriell Institute (samples NG11426, NG11467 and NG11716), was subjected to PCR-cloning-sequencing. The frequency of mtDNA mutations in these samples was not statistically different from that observed in the control experiment with amplified plasmid or in untreated HCT116 cells (Figure 1A).
H2O2 has low mutagenicity in HCT116 cells
Several factors can underlie the apparent lack of a significant accumulation of mtDNA mutations in aging cells, including the low mutagenicity of ROS in mitochondria. This possibility is especially interesting in the light of recent reports that susceptibility of mtDNA to the accumulation of oxidative base lesions may have been overestimated (31,32).
Hydrogen peroxide is a universal intracellular mediator of oxidative stress. Therefore, we studied the effect of H2O2 on mtDNA mutagenesis in HCT116 cells. HCT116 cells were treated with 400 µM of H2O2 for 1 h, after which they were either lysed or allowed to repair for 6 h, and total DNA was extracted and subjected to a quantitative alkaline Southern blotting and PCR-cloning-sequencing analysis.
H2O2 treatment induced strand breaks and abasic sites in mtDNA (Figure 1E). The prevalence of abasic sites is impossible to evaluate because they are readily converted into single-strand breaks (SSBs) by mitochondrial apurinic/apyrimidinic endonuclease (APE/Ref-1) and therefore manifest themselves as SSB in quantitative Southern blotting under denaturing conditions. Importantly, mtDNA analyzed immediately after H2O2 treatment contained ten times more combined strand breaks and abasic sites than mutagenic base lesions (Figure 1F). This ratio does not take into account the background of PCR-induced mutagenesis (0.09 mutations per mitochondrial genome), and therefore represents a conservative estimate. Indeed, subtraction of the PCR-induced mutagenesis rate from experimentally observed values would result in a ratio of 14.
Oxidative stress induces in DNA premutagenic base lesions rather than mutations. A fraction of these lesions (mutagenic lesions) is fixed as mutations only upon replication by DNA polymerase. Since different DNA polymerases have different propensities for misincorporation of wrong nucleotides across from the damaged base during replication, there exists a formal possibility that low mutation rates observed in mtDNA immediately after H2O2 treatment are due to a higher fidelity of Phusion polymerase as compared to mtDNA polymerase γ (Pol γ). This possibility was explored by subjecting cells to four consecutive treatments with H2O2 with 24 h intervals between treatments. At the end of experiment, cultures were allowed to undergo at least five cell divisions prior to DNA extraction to allow for the fixation of mutations with Pol γ. This mode of treatment did not result in the increase in the frequency of mtDNA mutations over untreated control or over mtDNA isolated immediately after treatment (n = 3, Figure 1F), suggesting that differences in the propensities of Phusion and Pol γ to induce mutations at the site of oxidative DNA damage do not confound our results.
mtDNA repair and degradation pathways compete for the H2O2-damaged mtDNA
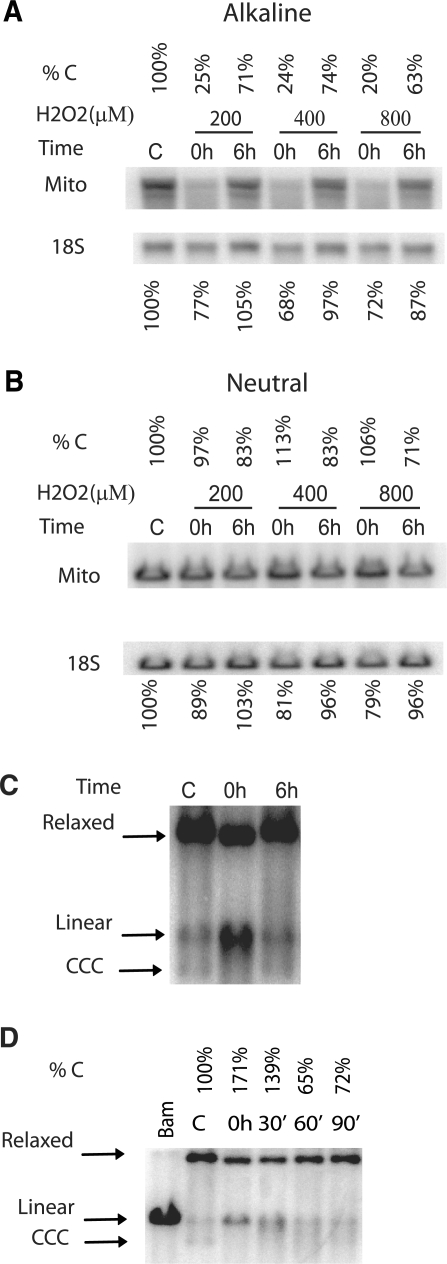
If accumulation of abasic sites and strand breaks, rather than oxidative base lesions in mtDNA is the predominant consequence of exposure to H2O2, the main cellular mediator of oxidative stress, then what is the fate of damaged mtDNA? This question was addressed by following the fate of H2O2-damaged mtDNA by Southern blotting under denaturing (alkaline) and non-denaturing (neutral) conditions. H2O2 induced lesions in both nDNA and mtDNA (Figure 2A). However, specific patterns observed in mtDNA under the denaturing and non-denaturing conditions were markedly different in that under the denaturing conditions, the amount of intact DNA was the lowest immediately after the treatment, and grew over the 6 h allowed for repair (Figure 2A). In contrast, under the non-denaturing conditions, the greatest amount of intact DNA was observed immediately after treatment, and then it decreased over the repair period (Figure 2B). This trend was preserved between different concentrations of H2O2 (Figure 2A and B), in different experiments, and in HeLa and A549 cell lines (Supplementary Figures 1 and 2). The differences observed in the same samples of mtDNA separated under the denaturing and non-denaturing conditions are attributable to the fact that sample preparation and electrophoresis under the alkaline conditions results in the separation of the two DNA strands and in the conversion of abasic sites into SSBs. Therefore, mtDNA containing oxidative damage in the form of abasic sites or strand breaks will be fragmented resulting in the decrease in intensity of the band corresponding to intact mtDNA. In contrast, neither SSBs, nor abasic sites will affect the quantity of intact mtDNA under non-denaturing conditions, as under these conditions the complementary strand will bridge the nick thus preventing fragmentation. Under the non-denaturing conditions, only double-strand breaks (DSBs) will result in mtDNA fragmentation and in decrease in the quantity of full-length mtDNA. The repair of DSBs in mtDNA has not been described, and introduction of DSBs in mtDNA of mammalian cells by restriction endonucleases results in mtDNA elimination through degradation (33,34). Therefore, trends in the quantity of the full length mtDNA observed under the denaturing and non-denaturing conditions following oxidative stress can be interpreted as follows. If a cell is unable to repair all the damage inflicted by H2O2 in the mitochondrial genome, a fraction of mtDNA molecules (presumably, those which received greater damage) receives DSBs and is degraded (hence the reduction in the amount of total mtDNA seen on a neutral gel, Figure 2B), while moderately damaged genomes are repaired, which is reflected in the increase in mtDNA quantity between 0 h and 6 h seen in alkaline gels (Figure 2A).
Figure 2.
Oxidative damage results in the degradation of mtDNA. (A and B) HCT116 cells were either left untreated (C, control) or treated with the indicated concentrations of H2O2 for 1 h, after which total genomic DNA was either extracted immediately (0 h), or allowed to repair for 6 h (6 h) prior to extraction. The DNA was digested with BamHI, which has a single recognition site in human mtDNA, quantitated and separated in 0.6% agarose gel under either alkaline (A) or neutral (B) conditions. After Southern blotting with mitochondrial (Mito) and nuclear (18S) DNA probes, the intensity of bands corresponding to intact mtDNA was expressed as a percent of control (untreated) values. (C) Oxidative DNA damage is accompanied by accumulation of linear mtDNA intermediates. HCT116 cells were treated with 200 µM H2O2, total DNA was extracted, digested with Bgl II, and subjected to Southern blotting under the neutral conditions. Bgl II has no recognition sites in mtDNA, and therefore the banding pattern is reflective of the native state of mtDNA. The positions of markers corresponding to covalently closed circular (CCC), linear and relaxed mtDNA molecules are indicated by arrows. (D) A time course of degradation of linear mtDNA intermediates. HCT116 cells were either left untreated (C) or treated with 400 μM H2O2 for 30 min, and then either lysed immediately, or allowed to recover for 30, 60, or 90 min. The percentage of linear mtDNA as compared to untreated control is indicated.
If mtDNA is indeed degraded in response to oxidative stress, then one of the initial steps in this process is likely to be the conversion of the circular mtDNA molecule into a linear intermediate. The half-life (and therefore, the steady-state level) of this intermediate is likely to be determined by a balance between formation and degradation. In the absence of knowledge of these processes, it is difficult to predict or interpret specific steady-state levels of linear mtDNA intermediates. However, the mere presence of such intermediates would support the notion of mtDNA degradation in response to oxidative stress. We therefore sought to determine whether oxidative stress is accompanied by an accumulation of linear mtDNA degradation intermediates. Indeed, H2O2 treatment resulted in an increase in the steady-state content of linear mtDNA (Figure 2C). This increase was transient, and linear intermediates disappear (degrade) within 1 h after the removal of a stressor (Figure 2D).
mtDNA degradation can be induced by enzymatically generated ROS
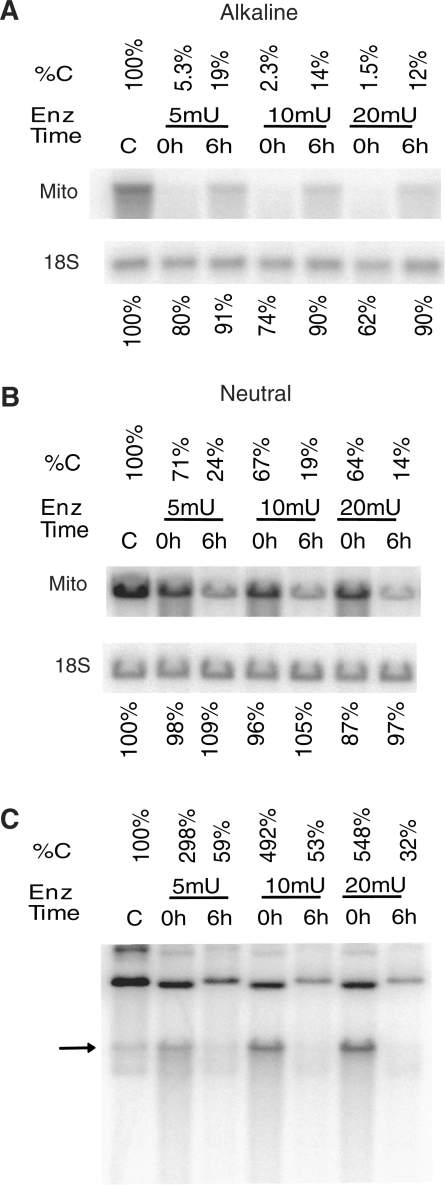
To see if degradation of oxidatively damaged mtDNA can potentially be induced by a physiologically relevant enzyme system, HCT116 cells were treated with 5, 10 or 20 mU of XO in the presence of hypoxanthine (0.5 mM). XO is released into a bloodstream upon liver damage, and XO/hypoxanthine reaction produces both H2O2 and superoxide (35). This treatment has faithfully recapitulated trends observed upon H2O2 treatment in that mtDNA repair was observed under the denaturing conditions (Figure 3A), which occurred simultaneously with mtDNA degradation observed under the non-denaturing conditions (Figure 3B), and that mtDNA degradation was accompanied by the accumulation of linear mtDNA intermediates (Figure 3C). Importantly, all three phenomena appear to be dose-dependent suggesting their specificity (Figure 3).
Figure 3.
The effect of enzymatically induced oxidative stress on mtDNA. HCT116 cells were either left untreated (C), or treated with 5, 10, or 20 mU of xanthine oxidase (XO) in the presence of hypoxanthine (0.5 mM). Cells were then either lysed immediately after treatment (0 h) or allowed for repair (6 h). Total DNA was extracted and digested with BamHI, and (A), separated under denaturing (alkaline) conditions, and subjected to Southern blotting with nuclear (18S) and mtDNA probe. %C indicates percent intact mtDNA as compared to untreated control. (B) The same samples as in A were run under non-denaturing (TBE buffer) conditions. Annotation is the same as in (A). This blot shows intact and SSBs containing mtDNA and nDNA. (C) Total DNA samples from the same experiment as in (A) and (B) were digested with BglII and run under the non-denaturing conditions to detect linear mtDNA intermediates. Annotation is the same as in (A) and (B). The arrow indicates the position of the linear mtDNA species.
Inhibition of base excision repair (BER) of oxidative DNA damage enhances mtDNA degradation
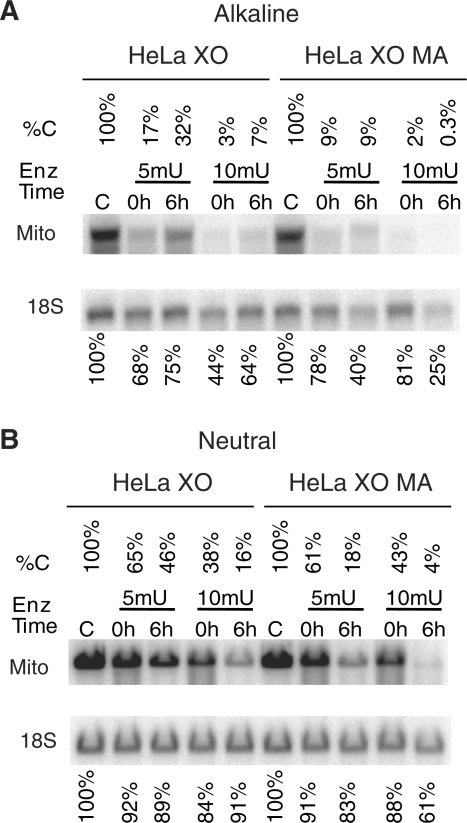
To study the effect of BER inhibition on mtDNA degradation, HeLa cells were subjected to oxidative stress in the presence or absence of the BER inhibitor methoxyamine (MA). This compound covalently modifies abasic sites generated by lesion-specific DNA glycosylases and prevents their incision by APE/Ref-1 (36). Since APE/Ref-1 is a bi-functional enzyme (AP endonuclease and a redox factor, which maintains the activity of numerous transcription factors), methoxyamine treatment is preferred over knockdown because it does not affect the redox function of APE/Ref-1. Methoxyamine treatment abrogated BER of oxidative damage in both mtDNA and nDNA (Figure 4A) and enhanced mtDNA degradation (Figure 4B).
Figure 4.
Effect of APE1 inhibition on mtDNA degradation in response to oxidative stress. HeLa cells were treated with XO in the presence of hypoxanthine and in the presence or absence of methoxyamine (MA). Cells were then either lysed immediately after treatment (0 h) or allowed for repair (6 h) in the presence or absence of MA. Total DNA was extracted and digested with BamHI, and (A), separated under denaturing (alkaline) conditions, and subjected to Southern blotting with nuclear (18S) and mtDNA (Mito) probes. %C indicates percentage intact mtDNA as compared to untreated control. (B) The same samples as in (A) were run under non-denaturing (TBE buffer) conditions. Annotation is the same as in (A). This blot shows both intact and SSB-containing nDNA and mtDNA.
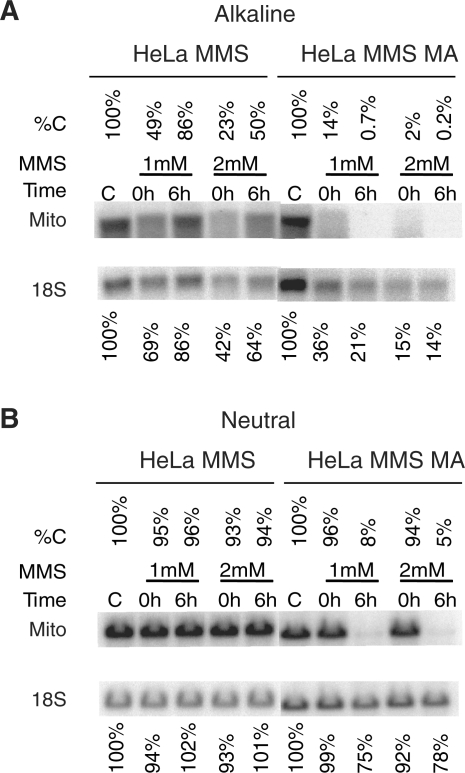
Inhibition of BER of alkylating DNA damage induces mtDNA degradation
To test whether mtDNA degradation can only be induced by oxidative DNA damage, or this is a more general phenomenon, we studied the repair of alkylation mtDNA lesions in the presence or absence of MA. At low MMS doses, no mtDNA degradation was observed in the absence of MA. However, inhibition of BER with MA both impaired the repair (Figure 5A) and dramatically induced mtDNA degradation (Figure 5B). In the presence of MA, nDNA repair was also impaired (Figure 5A). However, induction of DSBs in nDNA was much less pronounced compared to mtDNA under these conditions (Figure 5B).
Figure 5.
Effect of APE1 inhibition on mtDNA degradation in response to alkylating damage. HeLa cells were treated with indicated concentrations of MMS in the presence or absence of methoxyamine (MA). Cells were then either lysed immediately after treatment (0 h) or allowed for repair (6 h) in the presence or absence of MA. Total DNA was extracted and digested with BamHI, and (A), separated under denaturing (alkaline) conditions, and subjected to Southern blotting with nuclear (18S) and mtDNA (Mito) probes. %C indicates percentage intact mtDNA as compared to untreated control. (B) The same samples as in A were run under non-denaturing (TBE buffer) conditions. Annotation is the same as in (A). This blot shows both intact and SSB-containing nDNA and mtDNA.
DISCUSSION
While considerable progress has been achieved towards unraveling the process of nDNA repair, our current understanding of mtDNA repair and mutagenesis is much less detailed.
Here, we present evidence that the frequency of oxidative stress-induced base substitutions in mtDNA can be very low. This holds true for both human adenocarcinoma and mouse embryonic fibroblast cell lines. This is also true for oxidative stress induced by superoxide production from mitochondrial complex I and by H2O2 treatment.
The finding that the combined frequency of H2O2-induced strand breaks and abasic sites exceeds that of base substitutions by at least a factor of 10 is rather unexpected and indicates that handling of these types of mtDNA damage constitute an important aspect of the maintenance of overall mtDNA integrity. There are two reasons to believe that this observation is not likely to be confounded by an inability to amplify mtDNA fragments containing SSBs and abasic sites by PCR. First, in our experiments H2O2 induced an average of 2.71 ± 0.44 strand breaks/abasic sites per mtDNA molecule as measured immediately after the treatment (Figure 1E and F). This means an average of one strand break/abasic site per 6135 bp. Therefore, one can not expect a skewed amplification to substantially affect quantitation of mutagenic base lesions as there is only 16% probability that the amplified 1000-bp fragment would contain a strand break or abasic site. Second, if ROS-induced strand breaks were to interfere with detection of point mutations by PCR using DNA isolated immediately after treatment with ROS, then one would expect an increase in the frequency of base substitutions in samples which were allowed for repair of strand breaks and abasic sites (thus eliminating the amplification bias) as in the experiment where HCT116 cells were treated repeatedly with H2O2 (Figure 1F). However, this increase was not observed (Figure 1F). Importantly, in cells treated repeatedly with H2O2 the initial very substantial oxidative stress may be expected to be further amplified through a ‘vicious cycle’ thus enhancing mtDNA mutagenesis.
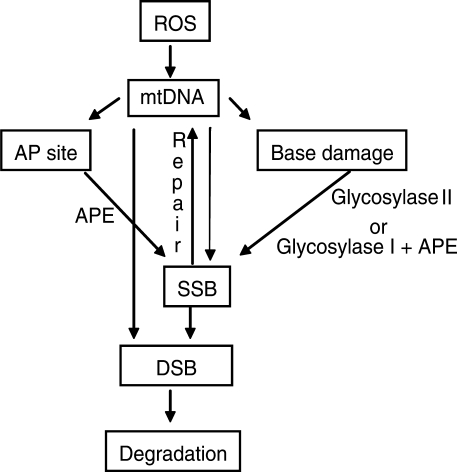
Our data provide evidence that both DNA repair and degradation processes operate on oxidatively damaged mtDNA (Figure 2A and B). The elimination of damaged mtDNA is preceded by the accumulation of linear mtDNA molecules, which may represent degradation intermediates (Figure 2C). These intermediates, unlike undamaged circular mtDNA molecules, are susceptible to exonucleolitic degradation thus ensuring the specificity of the process. mtDNA degradation can be induced by a physiologically relevant enzyme system, and in this case the amount of linear intermediates is proportional to enzyme dose (Figure 3C). Inhibition of abasic site processing by APE/Ref-1 stimulated mtDNA degradation following oxidative stress (Figure 4) suggesting that inability to repair mtDNA damage may be the signal for its degradation. This notion was further supported by induction of mtDNA degradation by inhibition of BER at levels of alkylation damage, which do not normally induce degradation (Figure 5). Importantly, inhibition of BER enhanced degradation of mtDNA containing both oxidative and alkylation lesions suggesting that degradation of mtDNA molecules, which failed to repair is a general mechanism. The higher propensity of ROS to induce mtDNA strand breaks and abasic sites, as compared to mutagenic base lesions, provides a new mechanism whereby mitochondria maintain the integrity of their genetic information (Figure 6). According to this mechanism, under conditions of oxidative stress, the generation of a single steady-state premutagenic lesion in mtDNA is accompanied by generation of as many as 10 strand breaks and abasic sites, which lead to elimination of mtDNA molecules containing such premutagenic lesions. Mitochondrial BER pathway components such as lesion-specific DNA glycosylases may aid in generation of abasic sites, which if left unrepaired, stimulate mtDNA degradation (Figures 4 and 5). The formation of DSBs is a commitment step, which may be mediated by stalled DNA or RNA polymerase, or by another mechanism. This model provides a mechanistic explanation for the observations made by Suter and Richter (37) who have found that 8-oxoG content of circular mtDNA is low and does not increase in response to oxidative insult. However, fragmented mtDNA had very high 8-oxoG content, which further increased after oxidative stress. This model is consistent with observations of Yakes and van Houten (38) who found higher incidence of polymerase-blocking lesions (i.e. SSBs and abasic sites) in mtDNA as compared to nDNA. Observations of Ikeda and Ozaki (39) who found that mitochondrial endonuclease G is more active in vitro on oxidatively modified DNA compared to undamaged DNA provide a candidate, which along with other yet unknown enzymes may be involved in the degradation of oxidatively damaged mtDNA. Finally, the notion of mtDNA's higher susceptibility to strand breaks in response to oxidative stress as a mechanism for the protection of the integrity of encoded genetic information concurs with evolutionary theory. It suggests that (in combination with the high redundancy of mtDNA) this unique mechanism may have evolved in response to mtDNA's close proximity to the main cellular source of ROS. Recent studies indicate that excessive accumulation of mtDNA mutations in genetically altered mice results in premature aging (40,41). However, levels of mutations which accumulate in mtDNA naturally do not appear to limit mammalian lifespan (42). These observations, together with mtDNA's proximity to a major site for the generation of potentially mutagenic ROS, suggest the existence of very efficient mechanisms which prevent excessive accumulation of ROS-induced mtDNA mutations. The degradation of oxidatively damaged mtDNA described in this study may represent a component of this machinery.
Figure 6.
Proposed interaction between mtDNA repair and degradation pathways. Glycosylase I and Glycosylase II, mono- and bi-functional DNA glycosylases. A bi-functional DNA glycosylase also posseses an AP-lyase activity (makes an incision at an abasic site). APE, apurinic/apyrimidinic endonuclease APE/Ref1. See ‘Discussion’ section for details.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (grant numbers PO1 HL06629907, R21RR02396101 to M.A. and RO1 ES03456 to G.L.W.). Funding for open access charge: R21RR02396101.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are thankful to Bert Vogelstein and Kenneth Kinzler for HCT116 cell line.
REFERENCES
- 1.Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, Lezza AM, Elsas LJ, 2nd., Nikoskelainen EK. Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy. Science. 1988;242:1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- 2.Chatterjee A, Mambo E, Sidransky D. Mitochondrial DNA mutations in human cancer. Oncogene. 2006;25:4663–4674. doi: 10.1038/sj.onc.1209604. [DOI] [PubMed] [Google Scholar]
- 3.Ohta S. Contribution of somatic mutations in the mitochondrial genome to the development of cancer and tolerance against anticancer drugs. Oncogene. 2006;25:4768–4776. doi: 10.1038/sj.onc.1209602. [DOI] [PubMed] [Google Scholar]
- 4.Wallace DC, Lott MT, Shoffner JM, Ballinger S. Mitochondrial DNA mutations in epilepsy and neurological disease. Epilepsia. 1994;35(Suppl. 1):S43–S50. doi: 10.1111/j.1528-1157.1994.tb05928.x. [DOI] [PubMed] [Google Scholar]
- 5.Krishnan KJ, Reeve AK, Turnbull DM. Do mitochondrial DNA mutations have a role in neurodegenerative disease? Biochem. Soc. Trans. 2007;35:1232–1235. doi: 10.1042/BST0351232. [DOI] [PubMed] [Google Scholar]
- 6.Newsholme P, Haber EP, Hirabara SM, Rebelato EL, Procopio J, Morgan D, Oliveira-Emilio HC, Carpinelli AR, Curi R. Diabetes associated cell stress and dysfunction: role of mitochondrial and non-mitochondrial ROS production and activity. J. Physiol. 2007;583:9–24. doi: 10.1113/jphysiol.2007.135871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rotig A, Bonnefont JP, Munnich A. Mitochondrial diabetes mellitus. Diabetes Metab. 1996;22:291–298. [PubMed] [Google Scholar]
- 8.Ballinger SW, Shoffner JM, Hedaya EV, Trounce I, Polak MA, Koontz DA, Wallace DC. Maternally transmitted diabetes and deafness associated with a 10.4 kb mitochondrial DNA deletion. Nat. Genet. 1992;1:11–15. doi: 10.1038/ng0492-11. [DOI] [PubMed] [Google Scholar]
- 9.Alexeyev MF, Ledoux SP, Wilson GL. Mitochondrial DNA and aging. Clin. Sci. 2004;107:355–364. doi: 10.1042/CS20040148. [DOI] [PubMed] [Google Scholar]
- 10.Kujoth GC, Bradshaw PC, Haroon S, Prolla TA. The role of mitochondrial DNA mutations in mammalian aging. PLoS Genet. 2007;3:e24. doi: 10.1371/journal.pgen.0030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loschen G, Azzi A. On the formation of hydrogen peroxide and oxygen radicals in heart mitochondria. Recent Adv. Stud. Cardiac Struct. Metab. 1975;7:3–12. [PubMed] [Google Scholar]
- 12.Boveris A, Oshino N, Chance B. The cellular production of hydrogen peroxide. Biochem. J. 1972;128:617–630. doi: 10.1042/bj1280617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henle ES, Luo Y, Gassmann W, Linn S. Oxidative damage to DNA constituents by iron-mediated fenton reactions. The deoxyguanosine family. J. Biol. Chem. 1996;271:21177–21186. [PubMed] [Google Scholar]
- 14.Henle ES, Luo Y, Linn S. Fe2+, Fe3+, and oxygen react with DNA-derived radicals formed during iron-mediated Fenton reactions. Biochemistry. 1996;35:12212–12219. doi: 10.1021/bi961235j. [DOI] [PubMed] [Google Scholar]
- 15.Wang D, Kreutzer DA, Essigmann JM. Mutagenicity and repair of oxidative DNA damage: insights from studies using defined lesions. Mutat. Res. 1998;400:99–115. doi: 10.1016/s0027-5107(98)00066-9. [DOI] [PubMed] [Google Scholar]
- 16.De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–185. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- 17.Bohr VA. Repair of oxidative DNA damage in nuclear and mitochondrial DNA, and some changes with aging in mammalian cells. Free Radic. Biol. Med. 2002;32:804–812. doi: 10.1016/s0891-5849(02)00787-6. [DOI] [PubMed] [Google Scholar]
- 18.Christmann M, Tomicic MT, Roos WP, Kaina B. Mechanisms of human DNA repair: an update. Toxicology. 2003;193:3–34. doi: 10.1016/s0300-483x(03)00287-7. [DOI] [PubMed] [Google Scholar]
- 19.Harman D. Aging: a theory based on free radical and radiation chemistry. J. Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 20.Harman D. The biologic clock: the mitochondria? J. Am. Geriatr. Soc. 1972;20:145–147. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- 21.Miquel J. An update on the mitochondrial-DNA mutation hypothesis of cell aging. Mutat. Res. 1992;275:209–216. doi: 10.1016/0921-8734(92)90024-j. [DOI] [PubMed] [Google Scholar]
- 22.Driggers WJ, Holmquist GP, LeDoux SP, Wilson GL. Mapping frequencies of endogenous oxidative damage and the kinetic response to oxidative stress in a region of rat mtDNA. Nucleic Acids Res. 1997;25:4362–4369. doi: 10.1093/nar/25.21.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kraytsberg Y, Schwartz M, Brown TA, Ebralidse K, Kunz WS, Clayton DA, Vissing J, Khrapko K. Recombination of human mitochondrial DNA. Science. 2004;304:981. doi: 10.1126/science.1096342. [DOI] [PubMed] [Google Scholar]
- 24.Bielas JH, Loeb LA. Quantification of random genomic mutations. Nat. Methods. 2005;2:285–290. doi: 10.1038/nmeth751. [DOI] [PubMed] [Google Scholar]
- 25.Lin MT, Simon DK, Ahn CH, Kim LM, Beal MF. High aggregate burden of somatic mtDNA point mutations in aging and Alzheimer's disease brain. Hum. Mol. Genet. 2002;11:133–145. doi: 10.1093/hmg/11.2.133. [DOI] [PubMed] [Google Scholar]
- 26.Miller FJ, Rosenfeldt FL, Zhang C, Linnane AW, Nagley P. Precise determination of mitochondrial DNA copy number in human skeletal and cardiac muscle by a PCR-based assay: lack of change of copy number with age. Nucleic Acids Res. 2003;31:e61. doi: 10.1093/nar/gng060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirano M, Shtilbans A, Mayeux R, Davidson MM, DiMauro S, Knowles JA, Schon EA. Apparent mtDNA heteroplasmy in Alzheimer's disease patients and in normals due to PCR amplification of nucleus-embedded mtDNA pseudogenes. Proc. Natl Acad. Sci. USA. 1997;94:14894–14899. doi: 10.1073/pnas.94.26.14894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.St-Pierre J, Buckingham JA, Roebuck SJ, Brand MD. Topology of superoxide production from different sites in the mitochondrial electron transport chain. J. Biol. Chem. 2002;277:44784–44790. doi: 10.1074/jbc.M207217200. [DOI] [PubMed] [Google Scholar]
- 29.Muller FL, Liu Y, Van Remmen H. Complex III releases superoxide to both sides of the inner mitochondrial membrane. J. Biol. Chem. 2004;279:49064–49073. doi: 10.1074/jbc.M407715200. [DOI] [PubMed] [Google Scholar]
- 30.Betarbet R, Sherer TB, MacKenzie G, Garcia-Osuna M, Panov AV, Greenamyre JT. Chronic systemic pesticide exposure reproduces features of Parkinson's disease. Nat. Neurosci. 2000;3:1301–1306. doi: 10.1038/81834. [DOI] [PubMed] [Google Scholar]
- 31.Anson RM, Hudson E, Bohr VA. Mitochondrial endogenous oxidative damage has been overestimated. FASEB J. 2000;14:355–360. doi: 10.1096/fasebj.14.2.355. [DOI] [PubMed] [Google Scholar]
- 32.Lim KS, Jeyaseelan K, Whiteman M, Jenner A, Halliwell B. Oxidative damage in mitochondrial DNA is not extensive. Ann. NY Acad. Sci. 2005;1042:210–220. doi: 10.1196/annals.1338.023. [DOI] [PubMed] [Google Scholar]
- 33.Kukat A, Kukat C, Brocher J, Schafer I, Krohne G, Trounce IA, Villani G, Seibel P. Generation of rho0 cells utilizing a mitochondrially targeted restriction endonuclease and comparative analyses. Nucleic Acids Res. 2008;36:e44. doi: 10.1093/nar/gkn124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexeyev MF, Venediktova N, Pastukh V, Shokolenko I, Bonilla G, Wilson GL. Selective elimination of mutant mitochondrial genomes as therapeutic strategy for the treatment of NARP and MILS syndromes. Gene Ther. 2008;15:516–523. doi: 10.1038/gt.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Porras AG, Olson JS, Palmer G. The reaction of reduced xanthine oxidase with oxygen. Kinetics of peroxide and superoxide formation. J. Biol. Chem. 1981;256:9006–9103. [PubMed] [Google Scholar]
- 36.Fishel ML, Kelley MR. The DNA base excision repair protein Ape1/Ref-1 as a therapeutic and chemopreventive target. Mol. Aspects Med. 2007;28:375–395. doi: 10.1016/j.mam.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 37.Suter M, Richter C. Fragmented mitochondrial DNA is the predominant carrier of oxidized DNA bases. Biochemistry. 1999;38:459–464. doi: 10.1021/bi9811922. [DOI] [PubMed] [Google Scholar]
- 38.Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc. Natl Acad. Sci. USA. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ikeda S, Ozaki K. Action of mitochondrial endonuclease G on DNA damaged by L-ascorbic acid, peplomycin, and cis-diamminedichloroplatinum (II) Biochem. Biophys. Res. Commun. 1997;235:291–294. doi: 10.1006/bbrc.1997.6786. [DOI] [PubMed] [Google Scholar]
- 40.Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 41.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 42.Vermulst M, Bielas JH, Kujoth GC, Ladiges WC, Rabinovitch PS, Prolla TA, Loeb LA. Mitochondrial point mutations do not limit the natural lifespan of mice. Nat. Genet. 2007;39:540–543. doi: 10.1038/ng1988. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.