Abstract
7,8-Dihydro-8-oxoguanine (8-oxoG) is an abundant and mutagenic DNA lesion. In Saccharomyces cerevisiae, the 8-oxoG DNA N-glycosylase (Ogg1) acts as the primary defense against 8-oxoG. Here, we present evidence for cooperation between Rad18–Rad6-dependent monoubiquitylation of PCNA at K164, the damage-tolerant DNA polymerase η and the mismatch repair system (MMR) to prevent 8-oxoG-induced mutagenesis. Preventing PCNA modification at lysine 164 (pol30-K164R) results in a dramatic increase in GC to TA mutations due to endogenous 8-oxoG in Ogg1-deficient cells. In contrast, deletion of RAD5 or SIZ1 has little effect implying that the modification of PCNA relevant for preventing 8-oxoG-induced mutagenesis is monoubiquitin as opposed to polyubiquitin or SUMO. We also report that the ubiquitin-binding domain (UBZ) of Pol η is essential to prevent 8-oxoG-induced mutagenesis but only in conjunction with a functional PCNA-binding domain (PIP). We propose that PCNA is ubiquitylated during the repair synthesis reaction after the MMR-dependent excision of adenine incorporated opposite to 8-oxoG. Monoubiquitylation of PCNA would favor the recruitment of Pol η thereby allowing error-free incorporation of dCMP opposite to 8-oxoG. This study suggests that Pol η and the post-replication repair (PRR) machinery can also prevent mutagenesis at DNA lesions that do not stall replication forks.
INTRODUCTION
The integrity of DNA in the cell is under constant threat from physical and chemical agents of endogenous and exogenous origin (1–4). Reactive oxygen species (ROS) that escape the cellular metabolism are an important source of DNA damage, which has been involved in pathological processes (5,6). An oxidized guanine, 7,8-dihydro-8-oxoguanine (8-oxoG), is an abundant and mutagenic lesion in DNA (4,7–11). In vitro studies show that 8-oxoG can be bypassed by eukaryotic RNA polymerases or DNA polymerases via incorporation of dCMP or dAMP opposite to 8-oxoG (9,12–18). The efficiency of the bypass reaction and the relative incorporation of dCMP vs dAMP depend upon the DNA polymerase used and the accessory proteins (PCNA and RPA) present in the reaction mixture (9,12–18). In vivo, 8-oxoG is thought to be mutagenic, yielding GC to TA transversions, which is consistent with an incorporation of adenine opposite to 8-oxoG (8,11). To counteract the deleterious effects of 8-oxoG, living organisms have evolved robust DNA repair mechanisms. In Escherichia coli, the repair of 8-oxoG mostly relies on two DNA N-glycosylases, Fpg and MutY. Fpg is an 8-oxoG DNA N-glycosylase/AP lyase that excises 8-oxoG opposite to a cytosine, whereas MutY is an adenine DNA N-glycosylase that excises adenine opposite to 8-oxoG (8,9). Inactivation of both, Fpg and MutY results in a strong GC to TA spontaneous mutator phenotype (8).
In Saccharomyces cerevisiae, the main defense against 8-oxoG is also the base excision repair (BER) pathway initiated by the Ogg1 protein (10,19,20). Yeast Ogg1 is a functional-albeit not structural-homologue of Fpg (21,22). Ogg1 is a DNA N-glycosylase/AP lyase that excises 8-oxoG and 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyG) from γ-irradiated DNA (23,24). Finally, inactivation of the Ogg1 protein results in GC to TA spontaneous mutator phenotype (25–28). In addition to BER, DNA mismatch repair (MMR) and DNA polymerase η (the RAD30 gene product) also play a critical role in the prevention of 8-oxoG-induced mutagenesis (26–28). The ogg1 msh6 and ogg1 rad30 double mutants have a synergistic increase in the rate of GC to TA mutations compared with that of the single mutants (26–28). The data suggest that MMR acts as a functional homologue of the MutY protein of E. coli. The Msh2–Msh6 complex recognizes the A.8-oxoG pair with high affinity and initiates the excision of adenine opposite to 8-oxoG (26). DNA polymerase η is thought to efficiently promote the error-free incorporation of cytosine opposite to 8-oxoG in the course of the DNA gap-filling reaction at the end of the MMR process (27,28). The RAD18 and RAD6 genes also play a prominent role in the network that prevents 8-oxoG-induced mutagenesis in yeast (28). The ogg1 rad6 and ogg1 rad18 double mutants have a synergistic increase in the rate of GC to TA mutations compared with that of the single mutants (28). The Rad18–Rad6 complex possesses an ubiquitin-conjugating activity that catalyzes the monoubiquitylation of PCNA at lysine K164, which is required for all branches of the post-replication repair (PRR) pathway (29,30).
In the present study, we explored the molecular events that link PRR and DNA Polymerase η (Rad30/Pol η) to 8-oxoG-induced mutagenesis in S. cerevisiae. We investigated the impact on spontaneous CanR mutation rate of a panel of mutants affected in the formation and the recognition of the modified forms of PCNA. The following mutants were tested: rad5Δ, siz1 Δ, pol30-K164R, pol32Δ, rad30-D570A, rad30-HH-568/572AA, rad30-FF627/628AA and rad30-(D570A, FF627/628AA). Structural studies of the ubiquitin-binding zinc finger domain (UBZ) of human Pol η suggest that mutation D570A and HH568/H572AA should primarily impair the binding of ubiquitin and the formation of the zinc-finger structure in Pol η from S. cerevisiae, respectively (31). On the other hand, F627 and F628 are essential components of the PCNA-binding domain (PIP) (30). Our results point to the monoubiquitylation of PCNA at K164 (PCNA-Ub1) as a prerequisite for Pol η's ability to prevent 8-oxoG-induced mutagenesis in vivo. They also suggest that Pol η's antimutator activity requires both the binding of PCNA-Ub1 via its PIP-domain and the binding of the ubiquitin moiety of PCNA-Ub1 via its UBZ-domain. Finally, this study proposes that Pol η and the PRR machinery can also prevent mutagenesis at DNA lesions such as 8-oxoG that do not stall replication forks.
MATERIALS AND METHODS
Media and growth conditions
Yeast strains were grown at 30°C in YPD medium (1% yeast extract, 1% bactopeptone and 2% glucose, with 2% agar for plates), YNBD medium (0.7% yeast nitrogen base without amino acids and 2% glucose, with 2% agar for plates) supplemented with appropriate amino acids and bases or YNBGal medium (YNB with 2% galactose). Supplemented YNBD medium lacking arginine but containing l-canavanine (Sigma) at 60 mg/l was used for the selective growth of canavanine-resistant (CanR) mutants on plates. Pre-sporulation and sporulation media have been described (32).
Yeast strains, plasmids and microbiological methods
Saccharomyces cerevisiae strains used in the present study are listed in Table 1. All strains are haploid and isogenic to wild type (WT) strains FF18733 (MATa, leu2-3-112, trp1-289, his7-2, ura3-52, lys1-1, CAN1) or FF18734 (MATα, leu2-3-112, trp1-289, his7-2, ura3-52, lys1-1, CAN1). Gene deletions were performed by a PCR-mediated one-step replacement technique (33,34). All disruptions were confirmed by PCR on genomic DNA. Strains were also obtained after genetic crossing and tetrad analysis. Micromanipulation and dissection of asci were performed using a Singer MSM System (35). For biochemical detection of the modified forms of PCNA, the POL30 open reading frame was replaced at its endogenous locus by a POL30-His6-tagged allele (36). For genetic analysis of mutants in the RAD30 gene, rad30 or ogg1 rad30 deletion strains were transformed with a shuttle vector based on YIplac128, which carried WT or mutated RAD30 gene under the control of the endogenous RAD30 promoter, for integration into the LEU2 locus (37). The empty vector YIplac128 served as control. Plasmids, p990 (YIplac128-Rad30), p991 (YIplac128-rad30-D570A), p1080 (YIplac128-rad30-FF627/628AA) and p1081 (YIplac128-rad30-D570A, FF627/628AA), were as described (37). Plasmid pBU001 (YIplac128-rad30-HH568/572AA) was derived from p990. Plasmids were restricted at EcoRV, transformed into rad30Δ or ogg1Δ rad30Δ cells and Leu+ transformants were selected and accurate integration was controlled by PCR analysis (Table 1). The mutY gene of E. coli was PCR-amplified from a WT strain (AB1157) and cloned into the pRS415-GAL1 (CEN ARS LEU2) plasmid. Mutations were introduced by PCR-based mutagenesis. Details of primers, strains and plasmids are available upon request.
Table 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| FF 18733 | MATa, leu2-3-112, trp1-289, his7-2, ura3-52, lys1-1 | F. Fabre |
| FF 18734 | MATα, leu2-3-112, trp1-289, his7-2, ura3-52, lys1-1 | F. Fabre |
| CD 138 | FF 18733 with ogg1Δ::TRP1 | This study |
| BPS 1002 | FF 18733 with, rad18Δ::kanMX6 | This study |
| BPS 1119 | FF 18733 with ogg1Δ::TRP1, rad18Δ::kanMX6 | This study |
| BG 177 | FF 18734 with rad51Δ::LEU2 | This study |
| BA 009 | FF 18734 with ogg1Δ::TRP1, rad51Δ::LEU2 | This study |
| BPS 1029 | FF 18733 with pol32Δ::kanMX6 | This study |
| BPS 1056 | FF 18733 with ogg1Δ::TRP1, pol32Δ::kanMX6 | This study |
| BS 4 | FF 18733 with rad5Δ::URA3 | This study |
| BA 007 | FF 18733 with ogg1Δ::TRP1, rad5Δ::URA3 | This study |
| BA 004 | FF 18734 with siz1Δ::URA3 | This study |
| BA 005 | FF 18734 with ogg1Δ::TRP1, siz1Δ::URA3 | This study |
| FF182028 | FF 18734 with pol30-K164R | F. Fabre |
| BPS 1117 | FF 18734 with ogg1Δ::TRP1, pol30-K164R | This study |
| BPS 1059 | FF 18734 with ogg1Δ::TRP1, rad18Δ::kanMX6, pol32::kanMX6 | This study |
| BPS 1011 | FF 18733 with rad30Δ::kanMX6 | This study |
| BPS 1063 | FF 18733 with ogg1Δ::TRP1, rad30Δ::kanMX6 | This study |
| BU001a | FF 18733 with POL30-His6-tag | This study |
| BU002 | FF 18733 with ogg1Δ::TRP1 POL30-His6-tag | This study |
| BU003 | FF 18733 with msh6Δ::kanMX6 POL30-His6-tag | This study |
| BU004 | FF 18733 with ogg1Δ::TRP1 msh6Δ::kanMX6 POL30-His6-tag | This study |
| BU005 | FF 18733 with apn1Δ::LEU2 apn2Δ::kanMX6 POL30-His6-tag | This study |
| BU006 | FF 18733 with rad30Δ::kanMX6 LEU2 | This study |
| BU007 | FF 18733 with rad30Δ::kanMX6 LEU2 RAD30 | This study |
| BU008 | FF 18733 with rad30Δ::kanMX6 LEU2 rad30-D570A | This study |
| BU009 | FF 18733 with rad30Δ::kanMX6 LEU2 rad30-HH568/572AA | This study |
| BU010 | FF 18733 with rad30Δ::kanMX6 LEU2 rad30-FF627/628AA | This study |
| BU011 | FF 18733 with rad30Δ::kanMX6 LEU2 rad30-(D570A, FF627/628A) | This study |
| BU012 | FF 18733 with ogg1Δ::TRP1 rad30Δ::kanMX6 LEU2 | This study |
| BU013 | FF 18733 with ogg1Δ::TRP1 rad30Δ::kanMX6 LEU2 RAD30 | This study |
| BU014 | FF 18733 with ogg1Δ::TRP1 rad30Δ::kanMX6 LEU2 rad30-D570A | This study |
| BU015 | FF 18733 with ogg1Δ::TRP1 rad30Δ::kanMX6 LEU2 rad30-HH568/572AA | This study |
| BU016 | FF 18733 with ogg1Δ::TRP1 rad30Δ::kanMX6 LEU2 rad30-FF627/628AA | This study |
| BU017 | FF 18733 with ogg1Δ::TRP1 rad30Δ::kanMX6 LEU2 rad30-(D570A,FF627/628A) | This study |
aConstruction of strains BU001 to BU017 is detailed in the ‘Materials and Methods’ section.
Spontaneous mutation rates
For each strain, 11 independent cultures were inoculated with about 5 × 102 cells in 2 ml of YPD, YNBD or YNBGal medium and grown at 30°C for 3 days. Cell density was measured by plating dilutions on YPD or YNBD agar plates and counting the colonies after 3 days at 30°C. The quantification of canavanine-resistant mutants (CanR) was determined after plating on selective medium (YNBD agar plates containing 60 mg/l l-canavanine) (38). Colonies were counted after 4–5 days at 30°C. All experiments were repeated independently 2–5 times. Mutation rates were determined from the number of CanR colonies by the method of the median (39).
Mutation spectra
For each strain, 32 independent cultures were grown and plated onto l-canavanine-containing plates (one culture per plate). After 3 days at 30°C, a single CanR mutant colony per plate was isolated and streaked onto selective canavanine-containing plates. Genomic DNA was extracted from saturated 1.5 ml YPD cultures obtained from individual colonies isolated from the canavanine-containing plates using ZymolyaseR (ICN) and the DNeasy Tissue Kit (Qiagen). The CAN1 gene was amplified by PCR from genomic DNA with 5-CAN-40 (5′CAGACTTCTTAACTCCTG3′) and 3-CAN1880 (5′GAAATGTGATCAAAGGTA-ATAAAACG3′). Sequencing of the CAN1 gene (1773 base pairs) was performed using three primers as previously described (28,40). Sequence alignment and analysis were performed using the DNA-STRIDER program.
MutY activity assay
A 34-mer oligodeoxyribonucleotide containing a single 8-oxoG at position 16 [5′-GGCTTCATCGTTGT-8-oxoG-CAGACCTGGTGGATACCG-3′] (a kind gift of Dr J. Cadet, CEA-Grenoble, France) and its complementary sequence with an adenine (A) at position 19 were used to generate a double stranded DNA substrate with a single A.8-oxoG pair. Before annealing the A-containing complementary strand was [32P]-labeled at the 5′-end as described (41). The assay mixture (15 μl final volume) contained 25 mM Tris–HCl pH 7.6, 100 mM NaCl, 50 fmol [32P]-labeled A.8-oxoG duplex and limited amounts of cell free extract of WT cells hosting pRS415-GAL1 or pRS415-GAL1-mutY plasmid. To express the MutY protein, cells were grown at 30°C in YNBGal medium in absence of leucine until OD600 = 1.0. Cell free extracts were prepared as described (42). The reactions were performed at 37°C for 15 min. Then, 6 μl of formamide dye was added before heating at 95°C for 5 min. The product of the reaction (P) was separated from the substrate (S) by 20% PAGE containing 7 M urea and quantified using a Molecular Dynamics PhosphorImager (41).
Detection of PCNA modifications
The POL30 open reading frame was replaced at its endogenous locus by a His6-tagged allele in WT or msh6, ogg1, msh6 ogg1 and apn1 apn2 deletion mutant strains (Table 1). Exponentially growing cultures, about 109 cells, were prepared. Cells were either untreated or exposed to 0.02% methyl-methane sulfonate (MMS) for 90 min at 30°C. Cells were harvested and processed essentially as previously described (36). Briefly, cells were lysed under denaturing conditions, and HisPCNA and its modified forms were isolated by Ni-NTA affinity chromatography from the total extracts. Samples were separated on NuPAGE™ 4–12% Bis-Tris gradient gels (Invitrogen), followed by Western blotting with polyclonal antibodies against yeast PCNA and a commercial monoclonal anti-ubiquitin antibody, P4D1 (Cell Signaling Technologies) (36).
Two-hybrid analysis
Analysis of protein–protein interactions in the two-hybrid system was performed in PJ64-4A, as described previously (37). Bait and prey plasmids carrying PCNA* (KK127/164RR) and fusions thereof to ubiquitin* (KKK29/48/63RRR) as well as WT and mutant (D570A) versions of Rad30 (Pol η), either full-length (1–632) or truncated (538–632), have been described (37). The mutant (HH568/572AA) of Rad30 was constructed analogously. Selection for the presence of the bait and prey plasmids was carried out on synthetic medium lacking tryptophan and leucine (-LW), and positive interactions were scored by growth on medium further lacking histidine (-HLW) or histidine and adenine (-AHLW).
RESULTS
Monoubiquitylation of PCNA at K164 is required to prevent high spontaneous mutation rates in Ogg1-deficient strains of S. cerevisiae
We recently reported that the RAD18 and RAD6 genes are important components of the cellular network that prevents 8-oxoG-induced mutagenesis in Ogg1-deficient strains of S. cerevisiae (28). The Rad18–Rad6 complex possesses an ubiquitin-conjugating activity that catalyzes the monoubiquitylation of PCNA at lysine K164, which is required for all branches of the post-replication repair process (29,30,43,44). Here, we investigated the impact of the K164R mutation of PCNA (POL30 gene product) that abolishes the conjugation of ubiquitin to PCNA and greatly reduces that of SUMO (29,30,43,44). The results show that the ogg1 pol30-K164R double mutant, like ogg1 rad18 and ogg1 rad30, exhibits a synergistic increase in CanR mutation rate compared to that of the single mutants (Table 2). In contrast, the ogg1 rad5 and ogg1 siz1 double mutants do not exhibit a synergistic increase in CanR mutation rates compared to that of the single mutants (Table 2). These results suggest that neither Rad5-dependent polyubiquitylation nor Siz1-dependent sumoylation of PCNA is involved in the prevention of 8-oxoG-induced mutagenesis in Ogg1-deficient cells. Table 2 also shows that homologous recombination does not play a critical role in the antimutagenic process at 8-oxoG, since the spontaneous mutation rate in the ogg1 rad51 double mutant corresponds to the sum of the rates in the two single mutants. These results are in agreement with the notion that only monoubiquitylation of PCNA (PCNA-Ub1) at K164 plays an important role in the process that prevents high spontaneous mutation rates in S. cerevisiae.
Table 2.
Spontaneous CanR mutation rate in strains that cannot undergo ubiquitin- or SUMO-conjugation of PCNA in S. cerevisiae
| Relevant genotype | CanR mutation rate × 10−7 | Ratioa |
|---|---|---|
| Wild type | 4.4 (3.8–5.0) | 1.0 |
| Δogg1 | 17.4 (13.4–24.8) | 4.0 |
| Δrad18 | 11.4 (8.2–18.6) | 2.6 |
| Δrad5 | 12.8 (10.4–16.2) | 2.9 |
| Δsiz1 | 2.4 (2.2–3.2) | 0.5 |
| Δrad51 | 51.8 (39.2–74.6) | 11.7 |
| Δrad30 | 6.0 (5.0–9.4) | 1.4 |
| pol30-K164R | 3.6 (2.8–4.8) | 0.8 |
| Δogg1Δrad18 | 70.6 (61.4–95.0) | 16.0b |
| Δogg1Δrad5 | 29.8 (24.4–40.2) | 6.8 |
| Δogg1Δsiz1 | 17.2 (13.8–20.4) | 3.9 |
| Δogg1Δrad51 | 66.6 (53.6–69.8) | 15.1 |
| Δogg1Δrad30 | 57.0 (43.2–70.6) | 13.0b |
| Δogg1 pol30-K164R | 58.0 (52.2–68.9) | 13.2b |
Rates of mutation at the CAN1 locus were determined from the number of CanR mutant colonies by the method of the median (39). The numbers in parentheses indicate the low and high values for the 95% confidence interval for each rate.
aRatio is relative to wild type.
bIndicate synergism (more than 2-fold the additivity) between ogg1 and another mutation.
Spectrum of CanR mutations in pol30-K164R and ogg1 pol30-K164R strains
If 8-oxoG is at the origin of the high spontaneous CanR mutation rate measured in the ogg1 pol30-K164R double mutant, the spectrum of mutations should be strongly biased in the favor of GC to TA (28). To test this hypothesis, sequence analysis of CanR mutations was performed in the pol30-K164R and ogg1 pol30-K164R mutant strains (Table 3). The results show that the spectrum of CanR mutations in the ogg1 pol30-K164R double mutant is nearly exclusively composed of GC to TA (96.9%), resulting in a 74-fold increase compared to the WT (Table 3). This spectrum is very similar to those obtained with the ogg1 rad18 and ogg1 rad30 double mutants (28), which points to 8-oxoG as the cause of spontaneous GC to TA in the ogg1 pol30-K164R strain. The results also show that the overall CanR mutation rate in the pol30-K164R mutant is not significantly different from that of the WT (Table 2). However, we observe a 2.2-fold increase in GC to TA in the pol30-K164R compared to the WT (Table 3). Similarly, a 1.8-fold increase in GC to TA was observed in the rad30 mutant compared to the WT (28). Although modest, these increases in GC to TA in pol30-K164R and rad30Δ suggest that a minor fraction of 8-oxoG escape the vigilance of Ogg1 and reveal the other pathways even in Ogg1-proficient cells.
Table 3.
Spectrum of CanR mutations
| Genotype | Mutation | Occurrence | Mutation rate × 10−7a |
|---|---|---|---|
| Wild type | GC to TA | 5/29 (17.2%) | 0.76 (1.0)b |
| GC to CG | 5/29 (17.2%) | 0.76 | |
| GC to AT | 8/29 (27.6%) | 1.21 | |
| AT to TA | 1/29 (3.4%) | 0.15 | |
| AT to CG | 3/29 (10.3%) | 0.45 | |
| AT to GC | 1/29 (3.4%) | 0.15 | |
| (-1) deletion | 3/29 (10.3%) | 0.45 | |
| Insertion | 1/29 (3.4%) | 0.15 | |
| Complexc | 2/29 (6.8%) | 0.30 | |
| ogg1 | GC to TA | 25/30 (83.3%) | 14.49 (19.1) |
| GC to AT | 3/30 (10.0%) | 1.74 | |
| AT to TA | 1/30 (3.3%) | 0.57 | |
| Deletion | 1/30 (3.3%) | 0.57 | |
| pol30-K164R | GC to TA | 15/32 (46.8%) | 1.68 (2.2) |
| GC to CG | 3/32 (9.4%) | 0.34 | |
| GC to AT | 9/32 (28.1%) | 1.01 | |
| AT to TA | 2/32 (6.3%) | 0.23 | |
| AT to CG | 1/32 (3.1%) | 0.11 | |
| AT to GC | 1/32 (3.1%) | 0.11 | |
| Insertion | 1/32 (3.1%) | 0.11 | |
| ogg1 pol30-K164R | GC to TA | 31/32 (96.9%) | 56.2 (73.9) |
| GC to CG | 1/32 (3.1%) | 1.80 |
aCanR mutation rates are the product of the proportion of a specific class of mutation and the total mutation rate for each strain (Table 2).
bNumber in the brackets is the fold induction of GC to TA relative to the WT value.
cComplex mutation is a mutation that is composed of more than one molecular event.
Suppression of 8-oxoG-induced mutagenesis by the MutY protein of E. coli
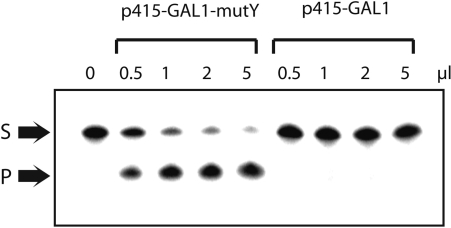
The synergistic increase in spontaneous GC to TA observed in ogg1 rad18 and ogg1 pol30-K164R is most probably due to 8-oxoG. However the role of another DNA lesion with the same coding properties cannot be completely ruled out. To further confirm 8-oxoG as the cause of CanR mutations in various yeast mutants, we explored the capacity of the MutY protein of E. coli to suppress spontaneous mutagenesis in S. cerevisiae. The MutY protein of E. coli is an adenine DNA N-glycosylase that specifically recognizes A.8-oxoG pairs in DNA (8). Amongst eukaryotes, most organisms including man possess a MutY-homolog with the considerable exception of S. cerevisiae (11). Genetic and biochemical data point to MMR as the functional homolog of MutY in S. cerevisiae (26). Here, we cloned the mutY gene of E. coli in the pRS415-GAL1 plasmid yielding pRS415-GAL1-mutY, which was used to transform the WT strain of S. cerevisiae. Figure 1 shows that cell free extract of WT/pRS415-GAL1 does not exhibit detectable MutY activity. This result is in agreement with the notion that S. cerevisiae does not possess a known or unknown DNA N-glycosylase able to excise adenine opposite to 8-oxoG. In contrast, cell free extract of WT/pRS415-GAL1-mutY possesses a robust MutY activity (Figure 1). Control experiments show that the MutY activity in extracts is undetectable when WT/pRS415-GAL1-mutY cells are grown in the presence of glucose instead of galactose (data not shown).
Figure 1.
Expression of the MuY protein of E. coli in S. cerevisiae. WT strain of S. cerevisiae harboring p415-GAL1 or p415-GAL1-mutY was grown at 30°C in YNBGal supplemented medium until OD600 = 1.0. Cell free extracts were prepared and assayed for MutY activity using a 34-mer DNA duplex that contained a single A*.8-oxoG pair. Total protein concentration in extracts was as follows: WT/p415-GAL1 (4.2 mg/ml) and WT/p415-GAL1-mutY (3.8 mg/ml). S: 34-mer Substrate, P: 19-mer Product. A*: the A-containing strand was [32P]-labeled at the 5′-end.
Table 4 shows that MutY has no significant effect on the overall CanR mutation rate in WT, rad18 or pol30-K164R strains. Although biased in favor of GC to TA, the spontaneous mutator phenotype of rad18 is most likely not due to 8-oxoG but to other endogenous DNA damage(s) or structure(s) (28,45). In contrast, the MutY protein greatly reduces spontaneous CanR mutation rate in the ogg1 single mutant, which is expected if CanR mutations are due to 8-oxoG (Table 4). Table 4 also shows that the expression of MutY only partially suppresses spontaneous mutagenesis (to the rad18 level) in the ogg1 rad18 double mutant, which confirms the impact of another class of damage at the origin of mutations in Rad18-deficient cells. Finally, the expression of MutY completely suppresses (to the WT level) spontaneous mutagenesis in the ogg1 pol30-K164R double mutant (Table 4). Therefore, both the mutation spectrum and the suppression of the mutator phenotype by MutY point to 8-oxoG as the major cause of mutations in the ogg1 pol30-K164R double mutant.
Table 4.
The MutY protein of E. coli suppresses 8-oxoG-induced mutagenesis in S. cerevisiae
| Relevant genotype | CanR mutation rate × 10−7 | Ratioa |
|---|---|---|
| Wild type/p415-GAL1 | 3.6 (3.2–7.5) | 1.0 |
| Wild type/p415-GAL1-mutY | 4.1 (2.4–6.2) | 1.1 |
| Δogg1/p415-GAL1 | 19.5 (15.2–22.6) | 5.4 |
| Δogg1/p415-GAL1-mutY | 2.9 (2.3–3.5) | 0.8 |
| Δrad18/p415-GAL1 | 10.1 (6.8–11.9) | 2.8 |
| Δrad18/p415-GAL1-mutY | 9.4 (7.7–11.2) | 2.6 |
| pol30-K164R/p415-GAL1 | 5.1 (4.0–6.7) | 1.4 |
| pol30-K164R/p415-GAL1-mutY | 3.9 (3.4–4.6) | 1.1 |
| Δogg1Δrad18/p415-GAL1 | 60.6 (49.9–72.6) | 16.8 |
| Δogg1Δrad18/p415-GAL1-mutY | 15.5 (12.4–22.3) | 4.3 |
| Δogg1 pol30-K164R/p415-GAL1 | 68.1 (51.7–84.5) | 21.7 |
| Δogg1 pol30-K164R/p415-GAL1-mutY | 4.9 (4.2–5.9) | 1.4 |
Rates of forward mutation at the CAN1 locus were determined from the number of CanR mutant colonies by the method of the median (39). The numbers in parentheses indicate the low and high values for the 95% confidence interval for each rate.
aRatio is relative to wild type.
Ubiquitylation of PCNA in strains unable to repair 8-oxoG or AP sites
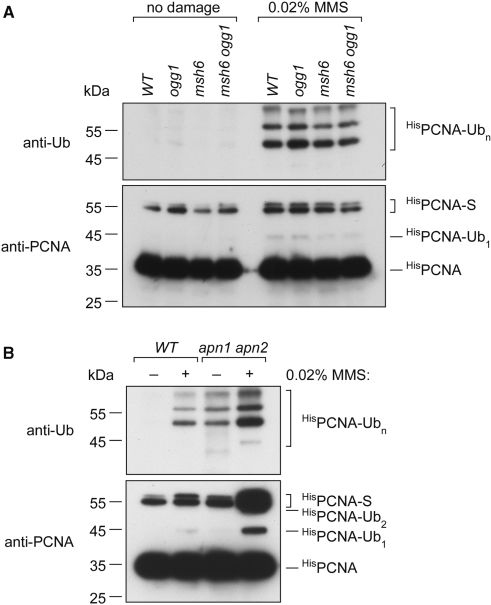
Our genetic analysis shows that monoubiquitylated PCNA (PCNA-Ub1) is an important player in the error-free processing (TLS) of 8-oxoG in S. cerevisiae. These results point to the formation of PCNA-Ub1 in untreated Ogg1-deficient cells at presumably low but potentially detectable levels. Since 8-oxoG is not thought to be a strong block to DNA replication, the formation of PCNA-Ub1 could occur in the course of the gap-filling synthesis after MMR-dependent excision of adenine paired with 8-oxoG (26–28). Therefore, if detectable under normal growth condition, the level of PCNA-Ub1 should be higher in an ogg1 single mutant than in an ogg1 msh6 double mutant. To identify the ubiquitylated forms of PCNA (PCNA-Ub) in yeast cells, the POL30 (PCNA) open reading frame was replaced at its endogenous locus by a His6-tagged allele. The presence of PCNA-Ub was monitored using affinity chromatography-purified PCNA and Western blotting with polyclonal antibodies against yeast PCNA and monoclonal anti-ubiquitin antibody (36). Figure 2A shows that PCNA-Ub cannot be detected in untreated WT, ogg1, msh6 or ogg1 msh6 cells. This is probably due to the fact that the signal is below the limit of detection of the western blotting assay. Control experiments show the formation of mono- and polyubiquitylated PCNA (PCNA-Ubn) in all four strains after exposure to 0.02% MMS (Figure 2A). SUMO-PCNA is also observed in all strain tested either untreated or exposed to 0.02% MMS (Figure 2). Therefore, the absence of detectable PCNA-Ub in Ogg1-deficient cells does not allow conclusions about the role of MMR in the formation of PCNA-Ub during the processing of 8-oxoG.
Figure 2.
Ubiquitylation of PCNA in mutator strains of S. cerevisiae. Asynchronous exponentially growing untreated (no damage) cells were allowed to grow at 30°C until OD600 = 1.0. MMS-treated cells were exposed to 0.02% MMS for 90 min at 30°C. Crude extracts from untreated and MMS-treated cells were prepared under denaturing conditions. HisPCNA was isolated and its modifications were detected by Western blotting with anti-PCNA and anti-ubiquitin antibodies. Migration of the ubiquitylated forms of PCNA (PCNA-Ub1, PCNA-Ub2 and PCNA-Ubn) and sumoylated forms of PCNA (PCNA-S) are indicated (Figure 2, right). Migration of molecular weight markers (kDa) is indicated (Figure 2, left). (A) WT, ogg1, msh6, msh6 ogg1 His-PCNA strains untreated and MMS (0.02%) treated. (B) WT and apn1 apn2 His-PCNA strains untreated and MMS (0.02%) treated.
For comparison, we also His-tagged PCNA in the apn1 apn2 double mutant, another spontaneous mutator strain in S. cerevisiae (3,46). Figure 2B shows the presence of PCNA-Ubn in untreated apn1 apn2 double mutant, which is deficient in the repair of abasic (AP) sites (3). Interestingly, the CanR mutation rate in apn1 apn2 [9.4 x 10−7 (6.6–15.2)] is comparable to that observed in ogg1 (Table 2). Therefore, the comparison of ogg1 and apn1 apn2 reveals that spontaneous mutation rates and levels of PCNA-Ub in untreated cells are not correlated. This result probably reflects the properties of the DNA lesions, 8-oxoG versus AP site, at the origin of the mutational events. These data support the notion that 8-oxoG does not act as a block to replication forks in vivo whereas AP site does.
UBZ and PIP domains of DNA polymerase η are required to prevent 8-oxoG-induced mutagenesis
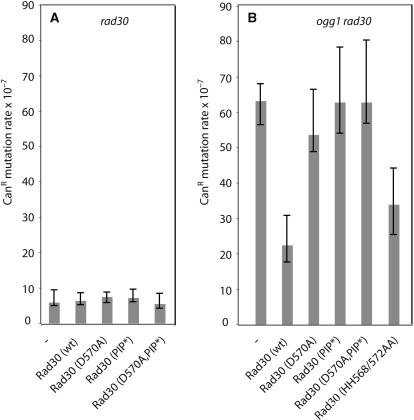
Our genetic data point to a model that involves Rad18–Rad6 and PCNA-Ub1 in a cellular process that prevents 8-oxoG-induced mutagenesis. PCNA-Ub1 could be used to specifically recruit Pol η to perform the translesion synthesis (TLS) with error-free incorporation of dCMP opposite to 8-oxoG (27,28). Indeed, the recruitment of specialized DNA polymerases by PCNA-Ub1 is a widely accepted mechanism to explain DNA synthesis across DNA lesions that stall replication forks such as pyrimidine dimer (29,30). In order to investigate the impact of the interactions between Pol η and PCNA on 8-oxoG-induced mutagenesis, we used mutant versions of Pol η that are affected in the PCNA-interacting domain (PIP) or in the ubiquitin-binding zinc finger domain (UBZ) (37,47). The PIP-domain mutant used here (FF627/628AA) has already been demonstrated to be deficient in its TLS function after UVC-irradiation in vivo (48). Based on structural information, we used two types of mutant versions of Pol η affected in the UBZ-domain (31). According to this study, mutation D570A should primarily affect the interaction between Pol η and ubiquitin, whereas HH568/572AA should abrogate the formation of the putative zinc-finger structure, respectively (31). Whereas the D570A mutation had previously been demonstrated to be deficient in TLS and ubiquitin binding (37,47), no measurable UVC sensitivity was found for HH568/572AA indicating functional TLS (47). Yeast strains that express WT and mutant versions of Pol η were constructed and spontaneous CanR mutation rates were determined. Figure 3A shows that the expression of Pol η WT and mutants in rad30 cells does not significantly impact on CanR mutation rates. Figure 3B shows that the expression of Pol η-WT and Pol η-HH568/572AA in ogg1 rad30 cells greatly reduces, to the level of an ogg1 single mutant, the spontaneous CanR mutation rates. In contrast, the expression of Pol η-D570A, Pol η-FF627/628AA and Pol η-(D570A, FF627/628AA) does not result in a significant reduction of CanR mutation rates in ogg1 rad30 cells (Figure 3B). These results strongly suggest a critical role of both the UBZ and the PIP domain of Pol η to prevent 8-oxoG-induced mutagenesis in S. cerevisiae. They also correlate with the UVC sensitivities of the respective mutants reported previously (37,47).
Figure 3.
The UBZ- and PIP-domains of Pol η are required to prevent 8-oxoG-induced mutagenesis. Strains bearing a rad30 deletion (A) or ogg1 rad30 deletions (B) were complemented by expressing a chromosomal version of Rad30-WT, Rad30-D570A, Rad30-FF627/628AA (PIP*), Rad30-(D570A, PIP*) and Rad30-HH568/572AA placed under the control of its natural promoter. Strains are described in ‘Materials and methods’ section and Table 1. CanR mutation rates were determined as described (Table 2). Δ ogg1 CanR mutation rate value: 17.4 × 10−7 (13.4–24.8).
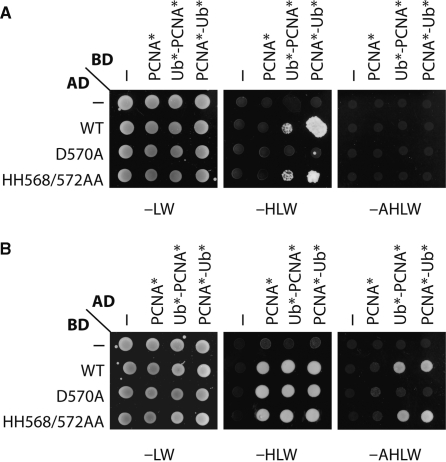
Although essential, the precise role of the UBZ domain of Pol η in the TLS process remains unclear. Indeed, the mutational inactivation of the zinc binding ability of Pol η-HH568/572AA was expected to abolish its ubiquitin-binding ability raising the question of whether ubiquitin binding was in fact required for TLS (Figure 3 and ref. 47). Here, we compared the capacity of Pol η-WT, Pol η-D570A and Pol η-HH568/572AA to interact with PCNA and PCNA-Ub in the two-hybrid system. Figure 4 shows the interactions between the different versions of Pol η, PCNA* and linear fusions of ubiquitin to PCNA* (Ub*-PCNA* and PCNA*-Ub*). As previously reported (37), fusion of ubiquitin to PCNA significantly enhances interaction with Pol η in an UBZ-dependent manner. Hence, Pol η-WT fused to GAL4-AD in combination with Ub*-PCNA* or PCNA*-Ub* fused to GAL4-BD activates the HIS3 reporter gene, whereas interactions with PCNA* alone or those involving the Pol η-D570A mutation are too weak to give a signal (Figure 4A). Reversal of the orientation in the two-hybrid system results in coherent but overall stronger signals, where Pol η fused to GAL4-BD in combination with PCNA* alone fused to GAL4-AD activates the HIS3 reporter (Figure 4B). Preferential binding of Pol η-WT to the ubiquitin fusions is now observed on plates selecting for activation of the HIS3 and ADE2 reporters. Under this assay condition (-AHLW), Pol η-D570A does not generate a detectable signal with Ub*-PCNA* or PCNA*-Ub*, whereas Pol η-HH568/572AA and Pol η-WT efficiently bind Ub*-PCNA* or PCNA*-Ub* (Figure 4B). We also performed these two-hybrid assays with the full-length proteins, Rad30(1–632), with identical results (ref. 37 and data not shown). These results led us to conclude that mutation D570A in the UBZ domain of Pol η adversely affects its capacity to bind PCNA-Ub, whereas mutation HH568/572AA does not. Taken together, our data suggest that Pol η's capacity to suppress 8-oxoG-induced mutagenesis correlates with its capacity to directly bind the ubiquitin moiety on linear Ub-PCNA fusions. They also suggest that the putative Zn-finger structure in the UBZ domain of Pol η is not required for ubiquitin binding and for TLS of 8-oxoG in S. cerevisiae.
Figure 4.
Physical interactions between Rad30-WT, Rad30-D570A, Rad30-HH568/572AA and PCNA*, Ub*-PCNA* or PCNA*-Ub* fusions. Interactions were monitored in the two-hybrid system, based on fusions to the GAL4 activation (AD) and GAL4 DNA-binding (BD) domains. Truncated RAD30, comprising amino acids 538–632, either WT or mutant (D570A and HH568/572AA) were used. Mutated PCNA (PCNA*) and linear fusions of mutated ubiquitin and PCNA at the N-terminus (Ub*-PCNA*) or the C-terminus (PCNA*-Ub*) were fused to GAL4-AD and GAL4-BD [‘Materials and Methods’ section (37)]. Interactions were scored by growth on plates lacking histidine (-HLW) and plates lacking histidine and adenine (-AHLW). Plates were scored after 3 days at 30°C. (A) Rad30 (538–632) fused to GAL4-AD. (B) Rad30 (538–632) fused to GAL4-BD.
Impact of DNA Polymerase δ subunit (Pol32) on 8-oxoG-induced mutagenesis
Our model suggests a competition between DNA polymerases δ (Pol δ) and η to bind PCNA or PCNA-Ub1 during the gap-filling reaction after the MMR-dependent excision of A.8-oxoG mismatches (28). The role of Pol δ cannot be directly addressed because of its essential function in S. cerevisiae. However, one of the three subunits of Pol δ, Pol32, is non-essential and can be deleted (49,50). We therefore investigated the impact of the Pol32 protein on spontaneous CanR mutation rate in ogg1 and ogg1 rad18 backgrounds. Table 5 shows that the ogg1 pol32 double mutant does not exhibit an enhanced CanR spontaneous mutation rate, compared to an ogg1 single mutant. Interestingly, the CanR mutation rate is greatly reduced in the ogg1 rad18 pol32 triple mutant compared to the ogg1 rad18 double mutant (Table 5). We propose that in the absence of PCNA-Ub1 in the ogg1 rad18 strain, both Pol δ and Pol η are recruited at unmodified PCNA allowing significant dAMP incorporation opposite to 8-oxoG, which explains the enhanced mutagenesis in the ogg1 rad18 strain. In the ogg1 rad18 pol32 triple mutant, the recruitment of Pol η at unmodified PCNA could be favored because of the loss of affinity of Pol δ for PCNA in absence of its Pol32 subunit, which also possesses a PIP domain (50). Consequently, the shift in favor of Pol η would promote dCMP incorporation and explain the reduced CanR mutation rate in the ogg1 rad18 pol32 triple mutant.
Table 5.
Impact of Pol32 on CanR mutation rates in S. cerevisiae
| Relevant genotype | CanR mutation rate × 10−7 | Ratioa |
|---|---|---|
| Wild type | 4.4 (3.8–5.0) | 1.0 |
| Δogg1 | 17.4 (13.4–24.8) | 4.0 |
| Δrad18 | 11.4 (8.2–18.6) | 2.6 |
| Δpol32 | 4.6 (4.2–5.6) | 1.0 |
| Δogg1Δrad18 | 70.6 (61.4–95.0) | 16.0 |
| Δogg1Δpol32 | 17.8 (14.4–23.8) | 4.0 |
| Δogg1Δrad18Δpol32 | 26.6 (23.4–36.4) | 6.0 |
Rates of forward mutation at the CAN1 locus were determined from the number of CanR mutant colonies by the method of the median (39). The numbers in parentheses indicate the low and high values for the 95% confidence interval for each rate.
aRatio is relative to wild type.
DISCUSSION
The aim of our studies is to decipher the biological network that protects the genome from the deleterious action of 8-oxoG in eukaryotic cells. In S. cerevisiae, this network involves at least four components: the 8-oxoG DNA N-glycosylase (Ogg1), the Msh2–Msh6-dependent MMR, the Rad18–Rad6 complex and the DNA polymerase η (26–28). Ogg1 is the major player and the only one able to remove 8-oxoG from DNA, which may explain why the contribution of other partners is unambiguously assessed only in Ogg1-deficient cells. Recently, we proposed a model (28): (i) due to endogenous oxidative stress, 8-oxoG forms in genomic DNA, (ii) DNA polymerase δ in the presence of auxiliary proteins present at the replication fork efficiently incorporates adenine opposite to 8-oxoG, (iii) Msh2–Msh6 complex recognizes the A.8-oxoG pairs and initiates the MMR-dependent excision of the adenine paired with 8-oxoG, which results in the formation of a gapped-structure with 8-oxoG in a single-stranded DNA (ssDNA) region and (iv) During repair synthesis, the Rad18–Rad6 complex promotes the recruitment of Pol η, which incorporates a cytosine across from the lesion, preventing mutation fixation and regenerating the 8-oxoG.C pair substrate of Ogg1. Although attractive, this model presents a major caveat since it does not provide information about the molecular mechanisms that allow the specific recruitment of Pol η at the site of the lesion.
In the present study, we show that the ogg1 pol30-K164R double mutant exhibits a synergistic increase in spontaneous CanR mutation rate, compared to the single mutants. Our results unambiguously point to 8-oxoG as the primary cause for the high CanR mutation rate in the ogg1 pol30-K164R strain: (i) the spectrum of CanR mutations in ogg1 pol30-K164R is nearly exclusively composed of GC to TA and (ii) the high CanR mutation rate in ogg1 pol30-K164R is reduced to the WT level upon expression of the bacterial MutY protein. These data led us to conclude that modification of PCNA at lysine K164 is a critical component of the cellular network that prevents 8-oxoG-induced mutagenesis. In fact, the K164R mutation of PCNA abolishes the formation of all kinds of PCNA-Ub and greatly reduces that of PCNA-SUMO (29,30). Here, we show that neither rad5 nor siz1 synergizes with ogg1, leading us to conclude that neither polyubiquitylated PCNA nor SUMO-PCNA is critical to prevent mutations at 8-oxoG. On the other hand, they strongly suggest that monoubiquitylation of PCNA (PCNA-Ub1) is essential.
Our genetic analysis points to the formation of PCNA-Ub1 in the course of the error free processing (TLS) of 8-oxoG. To investigate this issue at the biochemical level, we measured by western blotting the formation of PCNA-Ub in an ogg1 deletion mutant. For comparison, we also tested an apn1 apn2 mutant, unable to repair AP sites. Unfortunately, untreated ogg1 cells do not exhibit detectable amounts of PCNA-Ub, like WT cells, which do not allow us to provide further information about the mechanisms of formation of PCNA-Ub1 during the processing of 8-oxoG. In contrast, untreated apn1 apn2 cells present detectable level of PCNA-Ubn. These results may suggest that 8-oxoG does not efficiently stall replication forks, whereas the AP site does (3). This is in favor of a model where PCNA-Ub1 forms during the gap-filling reaction after the MMR-dependent excision of adenine opposite to 8-oxoG. Indeed, this model relies on consistent but circumstantial evidence (26–28) and its remains possible that the role of PCNA-Ub1 and Pol η also occurs at the stage of replication. The impact of 8-oxoG on DNA replication is also probably modulated by the sequence context around the lesion.
However, most in vitro and in vivo studies support the notion that the majority of the error-free TLS events at 8-oxoG occur in the course of a gap-filling reaction after MMR (12–18,26–28). MMR-dependent excision of adenine opposite to 8-oxoG results in the formation of stretches of ssDNA coated with RPA that might be used to activate Rad18–Rad6 and cause the formation of PCNA-Ub1 (51). However, it is potentially deleterious (mutagenic) to trigger the formation of PCNA-Ub1 for the recruitment of Pol η during the repair synthesis after the processing of any mismatch subject to MMR (52). To reconcile these two notions, one should conclude that 8-oxoG in ssDNA specifically initiates the molecular cascade that results in the formation of PCNA-Ub1. Clearly, we suggest that Pol δ efficiently bypasses 8-oxoG in ‘replication mode’, but may stall or pause at the same lesion in ‘gap-filling-repair mode’. It should be noted that purified Pol δ poorly replicates through 8-oxoG in vitro showing two strong stall sites, one right before the lesion and the other opposite to the lesion (27). In this model, Pol δ would initiate the gap-filling reaction and stall (pause) at 8-oxoG, thus allowing the recruitment of Rad18–Rad6 and the formation PCNA-Ub1.
Finally, Pol η binds to PCNA-Ub1 and catalyzes the incorporation of dCMP opposite to 8-oxoG. Here, we show that the UBZ of Pol η is essential to prevent 8-oxoG-induced mutagenesis, but only in conjunction with a functional PIP motif. This notion is based on the analysis of the properties of the D570A mutation in the UBZ domain of Pol η (Figures 3 and 4) (37). However, this notion appeared to be at variance with the conclusions of another study reporting that mutations in the zinc-binding motif of the UBZ domain of Pol η such as CC552/553AA or HH568/572AA do not impair its TLS function in vivo (47), leading to the concept that the direct binding of the ubiquitin moiety on PCNA via the UBZ was not required for TLS by Pol η in S. cerevisiae (47). According to this study, the invalidation of the zinc-finger structure in the UBZ domain of Pol η should necessarily result in the loss of its ubiquitin-binding ability (47). Here we show that Pol η -HH568/572AA not only exhibits functional TLS of 8-oxoG, but is also capable of productively interacting with the ubiquitin moiety of PCNA-Ub1, like the WT (Figures 3 and 4). Even thought ubiquitin is not attached to its natural position (K164) on PCNA, our data strongly suggest that the zinc-finger structure of the UBZ domain of Pol η is not required for binding to the ubiquitin moiety of PCNA and hence for TLS in S. cerevisiae. It should be noted that the presence of a Zn-finger structure in the UBZ domain of Pol η from S. cerevisiae is not firmly demonstrated: (i) to the best of our knowledge, there is no structure of the UBZ domain of yeast Pol η (there is in fact one of the human UBZ domain, which does bind Zn) (31) and (ii) the sequence is poorly conserved, since the two cysteines are contiguous in S. cerevisiae (CCKY) versus (CEKC) in the human protein. Indeed, we cannot exclude the possibility that Pol η's UBZ domain in S. cerevisiae does not harbor any zinc at all, in analogy to the RING-like, but zinc-less U-box domain (53). Taken together, our results are compatible with the hypothesis that points to an essential role of PCNA-Ub1 and its recognition by the UBZ and PIP domains of Pol η to prevent UVC- and 8-oxoG-induced mutagenesis in S. cerevisiae. These conclusions may or may not be applicable to mammalian system where mutation in the Zn-finger of the UBZ domain of the human Pol η results in different phenotypes, since the H654A is TLS-deficient, whereas H650A is TLS-proficient (54).
To summarize, our current model for the late steps of the error-free bypass of 8-oxoG is as follows: (i) Pol δ in the ‘gap-filling-repair’ mode transiently stalls at 8-oxoG, (ii) Rad18–Rad6 is recruited at RPA-coated ssDNA allowing the formation of PCNA-Ub1, (iii) Pol η through its UBZ and PIP domains binds to PCNA-Ub1, (iv) Pol η preferentially incorporates dCMP opposite to 8-oxoG, (v) PCNA is deubiquitylated allowing the release of Pol η. When one of the components of the fidelity system that favors the recruitment of Pol η is missing the equilibrium is shifted in favor of Pol δ, which will finally lead to the mutagenic incorporation of dAMP opposite to the lesion. In S. cerevisiae, the complexity of the network orchestrated by Ogg1 points to 8-oxoG as a major cellular threat. In mammals, the inactivation of two major components of the network, that prevents mutation at 8-oxoG, namely Ogg1 and Myh1, results in a high incidence of cancer (55).
FUNDING
CAPES (Post-doctoral fellowship # 0438-01-4) and CEA (to M.P.); Association pour la Recherche sur le Cancer (ARC) (to G.B.-S.); Cancer Research UK (to H.D.U.). Funding for open access charge: iRCM at Commissariat á l'E;nergie Atomique (CEA).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank the Centre National de la Recherche Scientifique (CNRS), the Commissariat à l’Energie Atomique (CEA) and Cancer Research UK for their support. We thank Drs Francis Fabre and Xavier Veaute for yeast strains and helpful discussions.
REFERENCES
- 1.Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 2.Hoeijmakers JHJ. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 3.Boiteux S, Guillet M. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair. 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 5.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 6.Hussain SP, Hofseth LJ, Harris CC. Radical causes of cancer. Nat. Rev. 2003;3:276–285. doi: 10.1038/nrc1046. [DOI] [PubMed] [Google Scholar]
- 7.Cadet J, Berger M, Douki T, Ravanat JL. Oxidative damage to DNA: formation, measurement, and biological significance. Rev. Physiol. Biochem. Pharmacol. 1997;131:1–87. doi: 10.1007/3-540-61992-5_5. [DOI] [PubMed] [Google Scholar]
- 8.Michaels ML, Miller JH. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) J. Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grollman AP, Moriya M. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 1993;9:246–249. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- 10.Boiteux S, Radicella JP. The human OGG1 gene: structure, functions, and its implication in the process of carcinogenesis. Arch. Biochem. Biophys. 2000;377:1–8. doi: 10.1006/abbi.2000.1773. [DOI] [PubMed] [Google Scholar]
- 11.Boiteux S, Gellon L, Guibourt N. Repair of 8-oxoguanine in Saccharomyces cerevisiae: interplay of DNA repair and replication mechanisms. Free Rad. Biol. Med. 2002;32:1244–1253. doi: 10.1016/s0891-5849(02)00822-5. [DOI] [PubMed] [Google Scholar]
- 12.Einolf HJ, Guengerich FP. Fidelity of nucleotide insertion at 8-oxo,7,8-dihydroguanine by mammalian DNA polymerase η. J. Biol. Chem. 2001;276:3764–3771. doi: 10.1074/jbc.M006696200. [DOI] [PubMed] [Google Scholar]
- 13.Brégeon D, Doddridge ZA, You HJ, Weiss B, Doetsch PW. Transcriptional mutagenesis induced by uracil and 8-oxoguanine in. Escherichia coli. Mol. Cell. 2003;12:959–970. doi: 10.1016/s1097-2765(03)00360-5. [DOI] [PubMed] [Google Scholar]
- 14.Kathe SD, Guang-Ping S, Wallace SS. Single-stranded breaks in DNA but not oxidative DNA base damage block transcriptional elongation by RNA polymerase II in HeLa cell nuclear extract. J. Biol. Chem. 2004;279:18511–18520. doi: 10.1074/jbc.M313598200. [DOI] [PubMed] [Google Scholar]
- 15.Brieba LG, Eichman BF, Kokoska RJ, Doublié S, Kunkel T, Ellenberger T. Structural basis for dual coding potential of 8-oxoguanine by a high fidelity DNA polymerase. EMBO J. 2004;23:3452–3461. doi: 10.1038/sj.emboj.7600354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 17.Charlet-Berguerand N, Feuerhahn S, Kong SE, Ziserman H, Conaway JW, Conaway R, Egly JM. RNA polymerase II bypass of oxidative DNA damage is regulated by transcription elongation factors. EMBO J. 2006;25:5481–5491. doi: 10.1038/sj.emboj.7601403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maga G, Villani G, Crespan E, Wimmer U, Ferrari E, Bertocci B, Hubscher U. 8-oxoguanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature. 2007;447:606–608. doi: 10.1038/nature05843. [DOI] [PubMed] [Google Scholar]
- 19.Auffret van der Kemp P, Thomas D, Barbey R, de Oliveira R, Boiteux S. Cloning and expression in Escherichia coli of the OGG1 gene of Saccharomyces cerevisiae, which codes for a DNA glycosylase that excises 7,8-dihydro-8-oxoguanine and 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine. Proc. Natl Acad. Sci. USA. 1996;93:5197–5202. doi: 10.1073/pnas.93.11.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nash HM, Bruner SD, Scharer OD, Kawate T, Addona TA, Spooner E, Lane WS, Verdine GL. Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr. Biol. 1996;6:968–980. doi: 10.1016/s0960-9822(02)00641-3. [DOI] [PubMed] [Google Scholar]
- 21.Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 22.Serre L, Pereira de Jesus K, Boiteux S, Zelwer C, Castaing B. Crystal structure of the Lactococcus lactis formamidopyrimidine-DNA glycosylase bound to an abasic site analogue-containing DNA. EMBO J. 2002;21:2854–2865. doi: 10.1093/emboj/cdf304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Girard PM, Guibourt N, Boiteux S. The Ogg1 protein of Saccharomyces cerevisiae: a 7,8-dihydro-8-oxoguanine DNA glycosylase/AP lyase whose lysine 241 is a critical residue for catalytic activity. Nucleic Acids Res. 1997;25:3204–3211. doi: 10.1093/nar/25.16.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karahalil B, Girard PM, Boiteux S, Dizdaroglu M. Substrate specificity of the Ogg1 protein of Saccharomyces cerevisiae: excision of guanine lesions produced in DNA by ionizing radiation- or hydrogen peroxide/metal ion-generated free radicals. Nucleic Acids Res. 1998;26:1228–1233. doi: 10.1093/nar/26.5.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas D, Scott AD, Barbey R, Padula M, Boiteux S. Inactivation of OGG1 increases the incidence of G.C→T.A transversions in Saccharomyces cerevisiae: evidence for endogenous oxidative damage to DNA in eukaryotic cells. Mol. Gen. Genet. 1997;254:171–178. doi: 10.1007/s004380050405. [DOI] [PubMed] [Google Scholar]
- 26.Ni TT, Marsischky GT, Kolodner RD. MSH2 and MSH6 are required for removal of adenine misincorporated opposite 8-oxo-guanine in S. cerevisiae. Mol. Cell. 1999;4:439–444. doi: 10.1016/s1097-2765(00)80346-9. [DOI] [PubMed] [Google Scholar]
- 27.Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase η. Nat. Genet. 2000;25:458–61. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 28.Padula M, Slezak G, Auffret van der Kemp P, Boiteux S. The post-replication repair RAD18 and RAD6 genes are involved in the prevention of spontaneous mutations caused by 7,8-dihydro-8-oxoguanine in Saccharomyces cerevisiae. Nucleic Acids Res. 2004;32:5003–5010. doi: 10.1093/nar/gkh831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ulrich HD. The Rad6 pathway: Control of DNA damage bypass and mutagenesis by ubiquitin and SUMO. Chem. Biol. Chem. 2005;6:1735–1743. doi: 10.1002/cbic.200500139. [DOI] [PubMed] [Google Scholar]
- 30.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–675. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 31.Bomar MG, Pai M-T, Tzeng S-R, Li SS-C, Zhou P. Structure of the ubiquitin-binding zinc finger domain of human DNA Y-polymerase h. EMBO Rep. 2007;8:247–251. doi: 10.1038/sj.embor.7400901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Resnick MA, Game JC, Stasiewicz S. Genetic effects of UV irradiation on excision-proficient and -deficient yeast during meiosis. Genetics. 1983;104:603–618. doi: 10.1093/genetics/104.4.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 35.Sherman F, Hicks J. Micromanipulation and dissection of asci. Methods Enzymol. 1991;194:21–37. doi: 10.1016/0076-6879(91)94005-w. [DOI] [PubMed] [Google Scholar]
- 36.Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 37.Parker JL, Bielen AB, Dikic I, Ulrich HD. Contributions of ubiquitin- and PCNA-binding domains to the activity of polymerase η in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:881–889. doi: 10.1093/nar/gkl1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whelan WL, Gocke E, Manney TR. The CAN1 locus of Saccharomyces cerevisiae: fine-structure analysis and forward mutation rates. Genetics. 1979;91:35–51. doi: 10.1093/genetics/91.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lea DE, Coulson DA. The distribution of the numbers of mutants in bacterial populations. J. Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 40.Kozmin S, Slezak G, Reynaud-Angelin A, Elie C, de Rycke Y, Boiteux S, Sage E. UVA radiation is highly mutagenic in cells that are unable to repair 7,8-dihydro-8-oxoguanine in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2005;102:13538–13543. doi: 10.1073/pnas.0504497102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Auffret van der Kemp P, Charbonnier JB, Audebert M, Boiteux S. Catalytic and DNA-binding properties of the human Ogg1 DNA N-glycosylase/AP lyase: biochemical exploration of H270, Q315 and F319, three amino acids of the 8-oxoguanine-binding pocket. Nucleic Acids Res. 2004;32:570–578. doi: 10.1093/nar/gkh224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lebedeva N, Auffret Vander Kemp P, Bjornsti MA, Lavrik O, Boiteux S. Trapping of DNA topoisomerase I on nick-containing DNA in cell free extracts of Saccharomyces cerevisiae. DNA Repair. 2006;5:799–809. doi: 10.1016/j.dnarep.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 43.Hoege C, Pfander B, Moldovan GL, Pyrovolakis G, Jentsch S. Rad6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 44.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 45.Kunz BA, Kang XL, Kohalmi L. The yeast rad18 mutator specifically increases G.C to T.A transversions without reducing correction of G-A or C-T mismatches to G.C pairs. Mol. Cell. Biol. 1991;11:218–225. doi: 10.1128/mcb.11.1.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guillet M, Boiteux S. Endogenous DNA abasic sites cause cell death in the absence of Apn1, Apn2 and Rad1/Rad10 in Saccharomyces cerevisiae. EMBO J. 2002;21:2833–2841. doi: 10.1093/emboj/21.11.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Acharya N, Brahma A, Haracska L, Prakash L, Prakash S. Mutation in the ubiquitin binding UBZ motif of DNA polymerase η do not impair its functions in translesion synthesis during replication. Mol. Cell. Biol. 2007;27:7266–7272. doi: 10.1128/MCB.01196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haracska L, Kondratick C, Unk I, Prakash S, Prakash L. Interaction with PCNA is essential for yeast DNA polymerase η function. Mol. Cell. 2001;8:407–415. doi: 10.1016/s1097-2765(01)00319-7. [DOI] [PubMed] [Google Scholar]
- 49.Huang M-E, Rio AG, Galibert D, Galibert F. Pol32, a subunit of Saccharomyces cerevisiae DNA polymerase η, suppresses genomic deletions and is involved in the mutagenic bypass pathway. Genetics. 2002;160:1409–1422. doi: 10.1093/genetics/160.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johansson E, Garg P, Burgers PMJ. The Pol32 subunit of DNA polymerase η contains separable domains for processive replication and proliferating cell nuclear antigen (PCNA) binding. J. Biol. Chem. 2004;279:1907–1915. doi: 10.1074/jbc.M310362200. [DOI] [PubMed] [Google Scholar]
- 51.Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein A. Mol. Cell. 2007;29:625–636. doi: 10.1016/j.molcel.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiricny J. The multifaceted mismatch-repair system. Nat. Rev. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 53.Ohi MD, Van der Kooi CW, Rosenberg JA, Chazin WJ, Gould KL. Structural insight into the U-box, a domain associated with multi-ubiquitination. Nat. Struct. Biol. 2003;10:250–255. doi: 10.1038/nsb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Acharya N, Yoon JH, Gali H, Unk I, Haracska L, Johnson R, Hurwitz J, Prakash L, Prakash S. Roles of PCNA-binding and ubiquitin-binding domains in human DNA polymerase h in translesion synthesis. Proc. Natl Acad. Sci. USA. 2008;105:17724–17729. doi: 10.1073/pnas.0809844105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie Y, Cunanan C, Okamoto K, Shibata D, Pan J, Barnes DE, Lindahl T, Mc Ilhatton M, Fishel R, Miller JH. Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-ras oncogene in lung tumors. Cancer Res. 2004;64:3096–3102. doi: 10.1158/0008-5472.can-03-3834. [DOI] [PubMed] [Google Scholar]