Abstract
Mutational analysis of the histone H3 N-terminal region has shown it to play an important role both in chromatin function in vivo and nucleosome dynamics in vitro. Here we use a library of mutations in the H3 N-terminal region to investigate the contribution of this region to the action of the ATP-dependent remodelling enzymes Chd1, RSC and SWI/SNF. All of the enzymes were affected differently by the mutations with Chd1 being affected the least and RSC being most sensitive. In addition to affecting the rate of remodelling by RSC, some mutations prevented RSC from moving nucleosomes to locations in which DNA was unravelled. These observations illustrate that the mechanisms by which different ATP-dependent remodelling enzymes act are sensitive to different features of nucleosome structure. They also show how alterations to histones can affect the products generated as a result of ATP-dependent remodelling reactions.
INTRODUCTION
Chromatin presents an obvious impediment to nuclear processes that require access to DNA. To counter this obstruction the eukaryotic cell has evolved mechanisms that alter chromatin structure and/or dynamics. An assortment of enzymes have been found to participate in gene regulation that act to post-translationally modify histone proteins (1). Several of these modifications, including phosphorylation, acetylation and methylation have been shown at least in some cases to create epitopes that are specifically recognized by chromatin-binding proteins (2). However, it is also possible that a subset of chromatin modifications act to directly alter the biophysical properties of chromatin (3–5). A second class of enzyme acting to manipulate chromatin structure during gene regulation non-covalently reconfigures chromatin in energy-consuming reactions (6). These ATP-dependent remodelling enzymes share homology with the superfamily 2 group of helicase related proteins (7) and have been referred to as Snf2 family proteins as they share additional homology within the catalytic subunit of the SWI/SNF complex (8,9). There is likely to be interplay between these two classes of enzyme as many ATP-dependent remodelling enzymes are associated with protein motifs capable of recognizing histone modifications.
To investigate this interplay we previously investigated the effects of histone truncation on ATP-dependent remodelling (10). This pointed to an important role for the histone H3 tail in nucleosome dynamics. A peptide ligation approach was used to show that H3 K14 acetylation acted to direct recruitment of RSC complexes to chromatin (11). Consistent with this H3 K14 acetylation is important for RSC function (12). In addition to this specific effect on recruitment of RSC, point mutations in histone H3 were found to have a range of distinct effects on the dynamic properties of nuclesomes (10). In particular, it was found that mutation of the histone H3 αN helix could alter different aspects of the dynamic properties of nucleosomes such as inherent mobility, site exposure and histone dimer stability (10).
Here we take advantage of this designer library of mutant nucleosomes to investigate how the dynamic properties of nucleosomes influence the action of ATP-dependent remodelling enzymes. We find that modifications affecting different aspects of the nucleosome have specific effects on different remodelling enzymes, and that these can act to alter either the rate of remodelling or the products generated.
MATERIALS AND METHODS
Chromatin assembly
Xenopus laevis histones were bacterially expressed, purified and assembled into tetramers and octamers, as appropriate, as described (13). Site-directed mutagenesis was performed using the Stratagene QuikChange Kit. Nucleosomes were assembled by salt dialysis using an equilmolar ratio of octamer and purified PCR-amplified DNA fragments (14). The nomenclature used to define DNA fragments is aBc, with a and c are numbers that describe the upstream and downstream bp extensions, respectively. B is the nucleosome positioning sequence source, with A representing the MMTV nucleosome A (15). Fluorescently labelled oligos were from Eurogentec (Belgium) and unlabelled oligos from the Oligonucleotide Synthesis Laboratory (University of Dundee, UK). The oligo sequences to amplify the 54A18 fragment are 5′TATGTAAATGCTTATGTAAACCA and 5′TACATCTAGAAAAAGGAGC; for the 54A0 fragment 5′TATGTAAATGCTTATGTAAACCA and 5′ATCAAAACTGTGCCGCAG. The PCR was purified by ion exchange chromatography using a 1.8-ml SOURCE 15Q (GE Healthcare) column.
Purification of chromatin-remodelling enzymes
Yeast strains TAP tagged for RSC (16) and SWI/SNF (17) were purified as described previously (11).
A pGEX-6p plasmid containing the Saccharomyces cerevisiae CHD1 gene made by Daniel Ryan was transformed into Rosette 2 cells (Novagen). 6 l of cells were grown in 2YT media (1.6% bactotryptone, 1.0% yeast extract, 0.5% NaCl, pH 7.5) to an A600 of ∼0.6, induced with 0.4 mM IPTG and expressed at 25°C overnight. All subsequent steps were performed at 4°C. The cells were pelleted in a Beckman JS 4.2 rotor at 4000 r.p.m. for 25 min, washed with 1 X PBS and re-suspended in 120 ml of lysis buffer [20 mM Tris pH 7.5, 350 mM NaCl, 0.1% (v/v) Tween, 0.05% (v/v) 2-mercaptoethanol, 2 µM E-64, 1 mM AEBSF, 1 µM pepstatin, 2.6 mM aprotinin], before freezing at –80°C. The thawed cells were homogenized with a 30-cm3 dounce, followed by sonication with a Branson S450 digital sonicator at 30% intensity using a 10-min cycle of 20 s ON, 30 s OFF and centrifugation in a Beckman JA-25.50 rotor at 31 000 g for 15 min. The supernatant was added to 300 μl TALON metal affinity resin (Clontech Laboratories Incorporated), followed by rotation for 2 h, before centifugation in a Heraeus Megafuge 1.0 centrifuge (Thermos Fischer Scientific) at 300 g for 1 min. The beads were re-suspended in lysis buffer and added to a 10-ml Poly-Prep column (BioRad). The column was washed with 20 ml of lysis buffer, followed by elution in 4 ml of lysis buffer with 250 mM imidazole. The eluate was added to 600 µl of glutathione Sepharose 4B beads (GE Healthcare) in a 10-ml Poly-Prep chromatography column and washed with 10 ml of glutathione elution buffer [20 mM Tris pH 7.5, 350 mM NaCl, 0.1% (v/v) Tween, 0.5 mM EDTA, 1 mM DTT]. The beads were rotated overnight with 2 ml glutathione elution buffer containing 5 μg of precision protease. The eluate collected and the column washed with a further ml of glutathione elution buffer. The eluate was concentrated with a YM-50 Microcon spin concentrator (Millipore) at 14 000 g in a Eppendorf benchtop centrifuge to a 100 µl volume. The sample was dialysed for 2 h against storage buffer [20 mM Tris pH 7.5, 350 mM NaCl, 1 mM DTT, 0.1% (v/v) Tween, 10% (v/v) glycerol] in a microdialysis apparatus (15) using 12 000–14 000-Da MWCO membrane (SpectraPor 2.1). The dialysed sample was aliquoted in 10 µl volumes and stored at –80°C.
Nucleosome repositioning assays
Nucleosomes were assembled on 54A18 DNA fragments for RSC and SWI/SNF repositioning and 54A0 DNA fragments for Chd1 repositioning. Each 10 µl reaction contained 1 pmol of wild-type and mutant nucleosomes assembled on cy3 and cy5 labelled DNA, respectively, 50 mM NaCl, 50 mM Tris pH 7.5, 3 mM MgCl2, 1 mM ATP and the quantity of remodeller specified in figures. Samples were incubated in 0.2-ml thin walled PCR tubes (ABgene, UK) in an Eppendorf mastercycle with heated lid at 30°C for the times specified in figures, before reaction termination by transferal to ice and addition of 500 ng of HindIII-digested bacteriophage lambda competitor DNA (Promega, USA) and 5% (w/v) sucrose. Samples were resolved on a native polyacrylamide gel [5% acrylamide:bis acrylamide (49:1 ratio), 0.2 × TBE buffer (0.5 mM EDTA, 22.3 mM Tris-borate, pH 8.3), 0.1% APS and 0.1% TEMED]. Gels were cast horizontally between 20 by 20-cm glass plates using 1.5-mm Teflon spacers, before mounting vertically in the gel apparatus (Thermo Fischer Scientific, USA) and pre-running at 300 V for 3 h with continuous pump recirculation of 0.2 × TBE buffer between the upper and lower compartments at 4°C. Gels were run at 300 V for 3.5 h and imaged using a Phosphoimager FLA-5100 (Fujifilm, Japan). Gel band intensities were quantitated using AIDA software (Raytest, Germany). The proportion of nucleosomes repositioned was calculated as the intensity of the sum of all detectable products over the sum of the major initial position plus products. In many assembly reactions a proportion of nucleosomes are assembled at a minor assembly location marked as the P position (e.g. *in Figure 2A) (18). Including this band in the calculations was found not to significantly affect the determination of initial rates. In some histone mutants the proportion of nucleosomes assembled at the P position was altered, and some additional less abundant sites of deposition were also observed. However, again inclusion nucleosomes assembled at these locations was not found to significantly affect the comparison of initial rates. As a result, in the analysis presented here nucleosomes deposited at locations other than the major NucA locations are not in included in quantitation. For some mutations a small proportion of nucleosomes were observed to undergo thermal repositioning in control reactions lacking remodelling enzyme. However, this was generally detected after long incubations and the proportion of nucleosomes repositioned at early time points was low enough not to affect the calculation of initial rates. The reproducibility of the assay when carried out in this manner was found to be high (Supplementry Figure 1).
Figure 2.
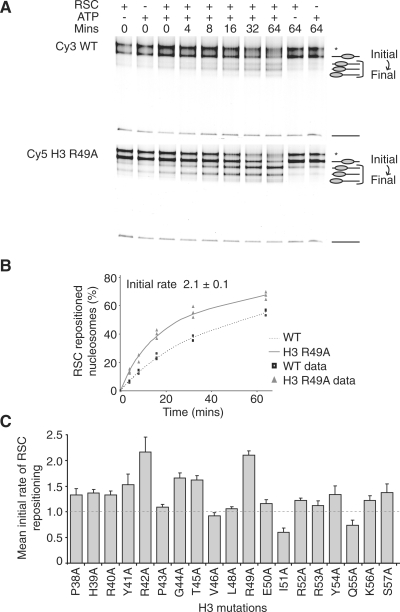
Histone H3 mutations differentially affect RSC repositioning. (A) Native polyacrylamide gel displaying repositioning of wild-type and H3 R49A nucleosomes assembled on 54A18 fragments by 0.44 fmol RSC at 30°C for the indicated times. Asterisk denotes P position. (B) RSC repositioned H3 R49A nucleosomes 2.1-fold ± 0.1 faster than wild-type nucleosomes. The raw data of the repositioning assays are shown as points, the curved line represents the average hyperbolic equation fit. (C) The initial rates of RSC repositioning for the histone H3 mutations. The dashed line indicates an initial rate of 1. Error bars represent the standard error of the mean. WT, wild-type.
In this way the proportion of nucleosomes repositioned is calculated independently of the total signal detectable in each lane. The initial rate was calculated as previously described (11). Each initial rate was repeated at least three times using chromatin prepared in separate assembly reactions.
High-resolution nucleosome mapping
Nucleosomes for high-resolution site-directed mapping (H3 C110A, H4 S47C) were prepared as previously described (18). Each 10 µl reaction contained 4 pmol mapping nucleosome, 50 mM Tris pH 7.5, 50 mM KCl, 1 mM MgCl2 and the RSC quantities specified in Figure 5. The samples were incubated at 30°C for 2 h, before termination by the addition of 500 ng of HindIII-digested bacteriophage lambda competitor DNA (Promega) and incubation on ice. One microlitre of 33 nM and 333-nM ammonium ferrous sulphate was added to the no RSC control and RSC repositioned samples, respectively and incubated on ice for 10 min. One microlitre of the reaction was removed and resolved on a 5% native polyacrylamide gel. To the remaining sample, 5 µl of 19.2-mM ascorbic acid and 0.05% (v/v) hydrogen peroxide was added to each sample, followed by incubation on ice for 90 min. The reactions were terminated by phenol:chloroform:isoamyl alcohol (25:24:1) extraction and ethanol precipitation. Each sample was re-suspended in 5 µl of mapping loading buffer (5 mM EDTA, 0.1% bromophenol blue in formamide), heated at 90°C for 3 min and loaded onto a sequencing gel [8% acrylamide:bis acrylamide (49:1 ratio), 8 M urea, 1 × TBE, 0.1% APS and 0.1% TEMED]. The gels were pre-run using 1 L of boiling 1 × TBE buffer until the temperature probe reached 50°C. The gel was loaded immediately and run at 50°C temperature, for 1.5–2 h. Fluorescent DNA markers for the 54A18 DNA fragment were created using a CycleReader Auto DNA Sequencing kit (Fermentas, Canada). The dideoxyribonucleotide sequencing products migrate 1 bp slower relative to the mapping cleavage products, as determined by examining the previously characterised +70 cleavage site for nucleosomes assembled on the 54A18 DNA fragments (19).
Figure 5.
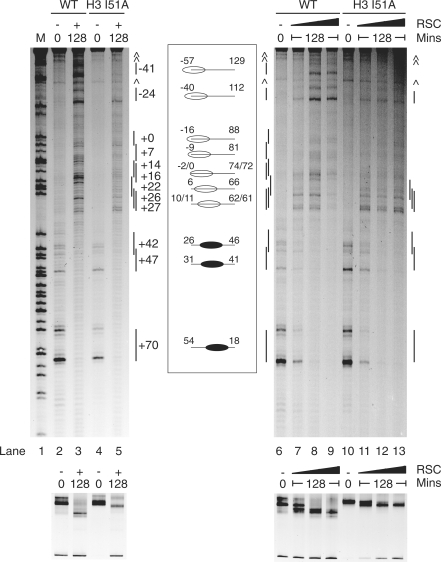
RSC redistributes H3 I51A nucleosomes to a subset of locations. The base-pair locations of RSC repositioned nucleosomes on the 54A18 fragment were determined by site-directed hydroxyl radical mapping. At top, sequencing gels. M, guanine marker of the 54A18 fragment, lane 1. No RSC controls, lanes 2, 4, 6 and 10. Repositioning of nucleosomes at 30°C for 128 min using 10 fmol RSC, lanes 7, 11; 83 fmol RSC, lanes 8, 12; and 660 fmol RSC, lanes 3, 5, 9 and 13. The cleavage products and an illustration of nucleosome locations (black ovals initial locations, clear ovals RSC redistributed locations) on the 54A18 DNA fragment are shown alongside the sequencing gels. At bottom, the corresponding native polyacrylamide gel. ⁁ (caret sign), non-specific bands. WT, wild-type mapping nucleosomes.
RESULTS
Histone H3 mutations have specific effects on nucleosome repositioning by Chd1 and RSC
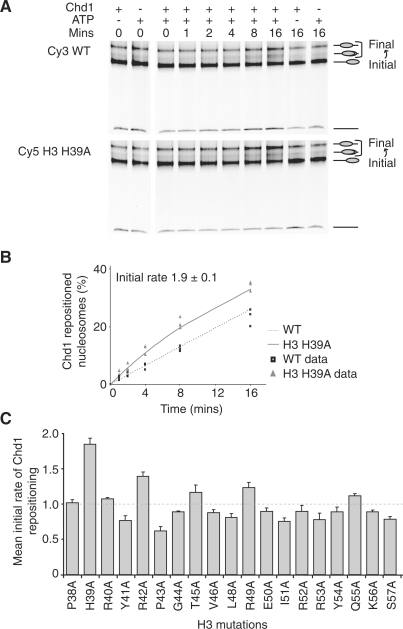
A competitive repositioning assay was used to compare the initial rates at which mutated and wild-type nucleosomes are moved by RSC and Chd1 (10,11). This involves mixing equimolar amounts of wild-type and mutated nucleosomes assembled on cy3 and cy5 labelled DNA fragments, ATP and the appropriate amount of remodelling enzyme at 30°C, before reactions were stopped by the addition of excess competitor DNA. The reaction products were analysed on native polyacrylamide gels and the extent to which the wild-type and mutant nucleosomes are repositioned established through the use of selective filters. As Chd1 repositions mononucleosomes most efficiently from positions close to DNA ends to more centrally located positions (18), we used a DNA fragment that positions nucleosomes such that 54 bp is located on one side and none on the other (54A0). Native gel electrophoresis can be used to track the loss of nucleosomes form their initial location and accumulation at destinations (Figure 1A). Quantitation enables the proportion of nucleosomes moved to be tracked over a time course (Figure 1B). Approximately three-quarters of the histone mutations tested caused relatively minor changes of 25% or less to the initial rate (Figure 1C). The H3 H39A mutation caused the greatest effect on Chd1 repositioning with a 1.9-fold ± 0.1 (mean ± standard error of the mean) increase to the initial rate relative to wild-type controls (Figure 1A and B). These results suggest that Chd1 is not greatly affected by mutations in this region of H3.
Figure 1.
Competitive repositioning of histone H3 mutations by Chd1. (A) A native polyacrylamide gel for a repositioning assay using wild-type and H3 H39A nucleosomes assembled on 54A0 fragments, 6.9 fmol Chd1 and incubation at 30°C for the indicated times. (B) Chd1 repositioned H3 H39A nucleosomes 1.9-fold ± 0.1 (mean ± standard error of the mean) faster than wild-type nucleosomes. The raw data from the repositioning assays are plotted as points, the average hyperbolic fit of the data is shown as a line. (C) The initial rate of Chd1 repositioning is plotted for the histone H3 mutations. The dashed line marks an initial rate of 1. The error bars represent the standard error of the mean. WT, wild-type.
To determine whether the above effects were a property shared among all chromatin-remodelling enzymes we tested the ability of the RSC complex to reposition the H3 mutated nucleosomes. RSC and related SWI/SNF complexes reposition nucleosomes from central locations towards and up to 50 bp beyond the end of an MMTV nucA DNA fragment (19) and similar behaviour has been observed on other DNA fragments (20,21). For this reason, RSC repositioning was performed using nucleosomes assembled on 54A18 DNA fragments. The H3 R42A and R49A mutations were observed to have the greatest effects with ∼2-fold increases in the initial rate of RSC repositioning relative to wild-type nucleosomes (Figure 2A and B). Comparison of the effects of mutations on the initial rates of RSC and Chd1 repositioning revealed many differences (Table 1). For example, the H3 P43A mutation caused a 1.6-fold reduction to the initial rate of Chd1 repositioning, whereas RSC repositioned this same nucleosome equivalently to wild-type controls. In contrast, the H3 I51A and Q55A mutations caused 1.7- and 1.4-fold decreases to the initial rate of RSC repositioning, but had far smaller effects on Chd1 action. Collectively, these results show that single alanine mutations within the histone H3 N-terminal region cause different effects on the repositioning capabilities of the Chd1- and RSC-remodelling enzymes.
Table 1.
Effects of histone mutations on repositioning by RSC and Chd1
| Mutation | Initial rate of Rsc respositioninga | Initial rate of Chd1 respositioninga |
|---|---|---|
| H3 P38A | 1.3 ± 0.1 | 1.0 ± 0.05 |
| H3 H39A | 1.4 ± 0.05 | 1.9 ± 0.1 |
| H3 R40A | 1.3 ± 0.1 | 1.1 ± 0.02 |
| H3 Y41A | 1.5 ± 0.2 | 0.8 ± 0.05 |
| H3 R42A | 2.2 ± 0.3 | 1.4 ± 0.1 |
| H3 P43A | 1.1 ± 0.05 | 0.6 ± 0.05 |
| H3 G44A | 1.7 ± 0.1 | 0.9 ± 0.01 |
| H3 T45A | 1.6 ± 0.1 | 1.2 ± 0.1 |
| H3 V46A | 0.9 ± 0.05 | 0.9 ± 0.05 |
| H3 L48A | 1.1 ± 0.05 | 0.8 ± 0.05 |
| H3 R49A | 2.1 ± 0.1 | 1.2 ± 0.05 |
| H3 E50A | 1.2 ± 0.1 | 0.9 ± 0.05 |
| H3 I51A | 0.6 ± 0.1 | 0.8 ± 0.05 |
| H3 R52A | 1.2 ± 0.1 | 0.9 ± 0.1 |
| H3 R53A | 1.1 ± 0.1 | 0.8 ± 0.1 |
| H3 Y54A | 1.3 ± 0.2 | 0.9 ± 0.05 |
| H3 Q55A | 0.7 ± 0.1 | 1.1 ± 0.05 |
| H3 K56A | 1.2 ± 0.1 | 0.9 ± 0.05 |
| H3 S57A | 1.4 ± 0.2 | 0.8 ± 0.05 |
| H3 K56Q | 1.3 ± 0.1 | ND |
aEach value represents the mean ± SEM. ND, not determined.
Histone mutations alter the products generated following remodelling with RSC
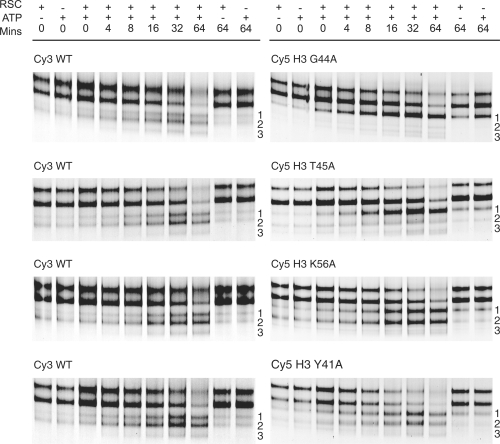
Typically, RSC repositions nucleosomes assembled on 54A18 DNA fragments to three faster migrating bands on native PAGE gels (11), denoted as 1, 2 and 3 in order of increasing mobility (Figure 3). For wild-type nucleosomes the majority of nucleosomes accumulate with the intensity in band 2 > band 1 > band 3 (Figure 3). Marked differences to this banding pattern were observed following repositioning of H3 mutated nucelosomes. For example, H3 G44A or T45A mutated nucleosomes are largely repositioned to band 1, with minimal nucleosomes present in either band 2 or band 3, whereas other H3 mutations such as H3 Y41A or K56A showed a pattern more similar to wild-type controls (Figure 3).
Figure 3.
Histone H3 mutations alter banding patterns following remodelling with RSC. When nucleosomes that have been remodelled by RSC (0.44 fmol) are separated on a native polyacrylamide gel three faster migrating species can be detected. These are labelled 1, 2 and 3. Nucleosomes H3 G44A and T45A were predominantly relocated to band 1 after a 64-min incubation at 30°C, in contrast wild-type nucleosomes were relocated equally to bands 1 and 2. Repositioning of mutations H3 K56A and Y41A produced a similar banding pattern relative to wild-type. Note band 3 represents a doublet in some gels. WT, wild-type.
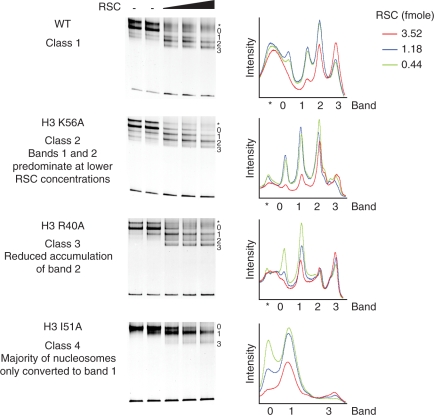
In principle, these differences to the products observed could reflect either a slower rate of remodelling and the accumulation of intermediates or a genuine difference in the products generated. To investigate this further, reactions were performed with higher concentrations of RSC over extended time courses. We observed that wild-type nucleosomes were initially converted to band 2, but at completion equivalent amounts of band 2 and band 3 accumulated (Figure 4). Importantly, histone mutants showed distinct effects following prolonged remodelling. These effects were assigned into four main categories. Class 1 mutations, generated a similar distribution to wild-type. Class 2 mutations, such as H3 K56A, displayed increased accumulation of band 1. Class 3 mutations, such as H3 R40A, were under-represented for band 2 and class 4 mutations, such as H3 I51A, showed the most marked differences to wild-type controls with the majority of nucleosomes only converted to band 1 (Figure 4).
Figure 4.
The effect of H3 mutations on the end products of RSC directed remodelling. To drive remodelling reactions towards completion, reactions were performed for 2 h at 30°C with increasing concentrations of RSC as indicated. The products were separated into one of four classes dependent on the distribution pattern of the repositioned products (bands 1, 2 and 3). Wild-type nucleosomes represent class 1, H3 K56A nucleosomes class 2, H3 R40A nucleosomes class 3 and H3 I51A nucleosomes class 4. The band intensities of the RSC remodelled nucleosomes are plotted in the graph to the right of each respective native polyacrylamide gel. Band 0, main initial nucleosome position. Asterisk denotes P position.
H3 I51A prevents RSC from unravelling nucleosomes
In order to characterize the products generated following remodelling of the different histone mutations, we employed site-directed hydroxyl radical mapping (22). Briefly, this involves the incorporation of cysteaminyl EDTA at H4 S47C, a residue that is close to the DNA at the nucleosome dyad axis. Ferrous ions, ascorbic acid and hydrogen peroxide are added to produce a localized Fenton reaction at the EDTA derivative. The resulting hydroxyl radicals cleave strongly at a single proximal DNA site and more weakly at 7 and 8 bp away on each DNA strand. This characteristic cleavage pattern can be used to determine the position of the nucleosomal dyad axis at base-pair resolution (22).
Site-directed mapping showed increasing amounts of RSC progressively remove control nucleosomes (H4 S47C) away from the +70 position. Nucleosomes appear at new locations up to 111 bp along the longer arm (Figure 5, lanes 3, 7–9). This involves moving the nucleosome to locations in which DNA is removed from one side of the histone octamer as has been described previously (19–21). These results suggest that for wild-type nucleosomes the faster migrating products on native PAGE gels correspond to nucleosomes being repositioned by RSC to a series of locations on what was originally the longer linker DNA arm (Figure 5).
In contrast, repositioning of H3 I51A nucleosomes (a Class 4 mutation), resulted in accumulation at only a subset of the locations observed for wild-type nucleosomes. The major positions observed are at +27, +26 and +22, which correspond to the edge of the nucleosome being positioned at 11, 10 and 6 bp from the end of the DNA fragment (Figure 5 lanes 5, 11–13). Although wild-type nucleosomes were moved to a series of positions beyond this in which from 2 to 57 bp of DNA are removed from the edge of the nucleosome, these locations are all under-represented for I51A nucleosomes. The single predominant band on native PAGE gels observed following remodelling of H3 I51A nucleosomes corresponds to the co-migration of nucleosomes at a subset of locations in which DNA remains fully wrapped.
Comparison of remodelling by RSC and SWI/SNF
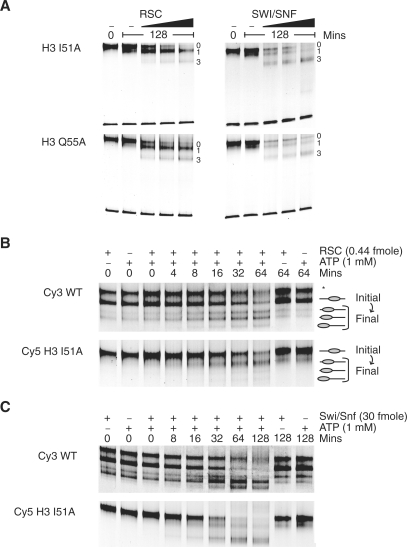
The SWI/SNF complex has a similar subunit composition to RSC. The catalytic subunits Snf2 and Sth1 share several sequence motifs as do other members of the complexes (23). Biochemical characterization of the activities of these complexes has also suggested they function similarly (19,24,25). In order to investigate whether interactions with histone H3 affect the action of these enzymes similarly, the products of remodelling reactions driven by SWI/SNF were resolved by native PAGE and assigned to one of four classes as described for RSC. Interestingly, the extent to which each of the mutations affects the two remodelling complexes differs. For instance, a marked difference is observed between the products generated following remodelling of H3 I51A nucleosomes by RSC and SWI/SNF. While RSC generates predominantly band 1 (Figure 6A), SWI/SNF remodelling results in more of band 3. A difference between RSC and SWI/SNF repositioning is also observed for the H3 Q55A mutant nucleosomes (Figure 6A). Despite the qualitative difference in the products generated by these enzymes, the rates at which SWI/SNF and RSC remodel I51A nucleosomes are comparable (Figure 6B and C) with the initial rates of remodelling being reduced by 0.6-fold in comparison to wild-type nucleosomes in both cases (data not shown).
Figure 6.
Differences observed between RSC and SWI/SNF end products. (A) Repositioning of 1 pmol of H3 I51A and Q55A mutant nucleosomes by 0.44, 1.18 or 3.52 fmol RSC showed a preferential accumulation in band 1 on native polyacrylamide gels. In contrast, repositioning of 0.5 pmol of mutant nucleosomes with 10, 20 or 60 fmol of SWI/SNF showed repositioning to band 3. Band 0, main initial position. (B) RSC and (C) SWI/SNF repositioning of H3 I51A nucleosomes. Asterisk denotes P position, not present in either mutation. WT, wild-type.
DISCUSSION
The ability to manipulate the coding sequence of histone genes together with the expression and assembly of these proteins in vitro provides an opportunity to monitor the direct effects of these mutations on chromatin dynamics. We have identified differences in the sensitivities of chromatin-remodelling enzymes to mutations affecting different aspects of nucleosome structure. This indicates there are differences in the underlying mechanisms by which even closely related enzymes act.
We observed that remodelling by the enzyme Chd1, was affected very little in terms of either the products generated or the rates at which nucleosomes were repositioned (Figure 7B; Table 1). Although the initial rate of remodelling by RSC was affected to a greater extent than Chd1 the magnitude of the effects remained low, with the maximum effect being 2.2-fold (Figure 7E). This indicates that both enzymes were able to engage and reposition nucleosomes efficiently. Consistent with this the binding of RSC to nucleosomes bearing the H3 E50A, I51A and Q55A mutations was equivalent to wild-type nucleosomes (Supplementary Figure 2). The limited effects of these mutations on the rate of nucleosome sliding by RSC and Chd1 contrasts with the more substantial effects these mutations were observed to have on spontaneous thermal nucleosome repositioning (compare Figure 7B, E and F). This lack of correlation is likely to reflect significant differences in the mechanisms for thermal and ATP-dependent nucleosome repositioning.
Figure 7.
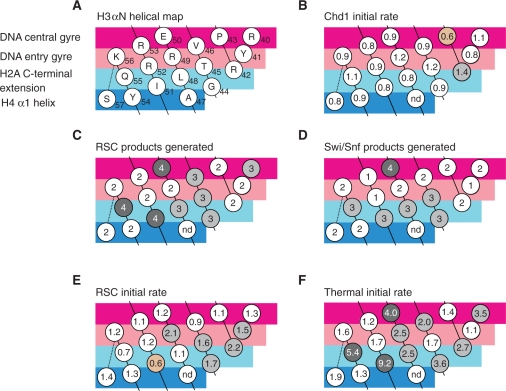
Remodelling enzymes are distinctly affected by histone H3 mutations. (A) The histone H3 αN helical map, introduced in ref. 10. This map allows the impact of each interaction surface to be compared among the different remodellers. (B) The initial rate of Chd1 repositioning. (C) The distribution of the end products after RSC repositioning, class 1–4 (see Figure 4 for description of classes). (D) The distribution of end products after SWI/SNF, classes 1–4. (E) The initial rate of RSC repositioning. (F) The initial rate of thermal repositioning, values from ref. 10, except for H3 E50A where remeasurement detected a difference from the previously published value.
In contrast to the limited effects on the initial rates of remodelling, the outcome of RSC remodelling reactions was affected dramatically with many mutations altering the spectrum of products generated (Figure 7C). A related but less dramatic effect was observed with the SWI/SNF complex (Figure 7D). While RSC repositions wild-type nucleosomes to a series of locations in which the nucleosome dyad is so close to the edge of the DNA fragment that up to 57 bp of DNA are removed from one edge of the nucleosome, these locations were under-represented when H3 I51A mutant nucleosomes were used as a substrate (Figure 5). This means that the mechanics of moving nucleosomes to locations in which DNA is unravelled differ from those involved in repositioning within the confines of a DNA fragment.
It is notable that there is a good correlation between residues that have strong effects on thermally driven nucleosome positioning and the inability of RSC to move nucleosomes into positions where DNA is unravelled (compare Figure 7C and F). At first this appears to be counterintuitive as nucleosomes that are inherently more mobile cannot be repositioned so effectively by RSC. However, it is also true that the H3 E50A, I51A and Q55A mutations reduce the stability of histone octamers (10; data not shown). One model for RSC action involves the RSC complex drawing DNA over the surface of nucleosomes while remaining bound to the histone octamer in a fixed orientation (26). The reduction to octamer stability resulting from the H3 E50A, I51A and Q55A mutations might result in the octamer rather than the DNA being distorted as a result of ATP hydrolysis. Furthermore, as RSC starts to move nucleosomes to locations in which DNA is unravelled from their surface, the overall loss of histone contacts will require the ATPase motor within the RSC complex to exert greater forces. Indeed, it has been estimated that a force of 3 pN will be required to dissociate each helical contact (27). The increased forces that are applied during DNA unravelling might drive a reconfiguration of the histone octamer that prevents any further repositioning. This could explain why RSC is able to move nucleosomes to positions where histone DNA contacts are retained, but not to locations where DNA is unravelled. In this case the reason why Chd1 is not affected in the same way could be simply that the repositioning of nucleosomes to more central locations does not involve unravelling DNA to the same extent.
Having observed that mutations to the H3 αN region of the nucleosome can affect the outcome of remodelling reactions, it will be of interest to investigate whether histone modifications exert similar effects. Our own preliminary observations made using mutations designed to mimic a selection of modifications known to occur in this region of the nucleosome revealed only subtle effects on the action of ATP-dependent remodelling enzymes (data not shown). It is possible that the use of amnio acid substitutions is not a sufficiently accurate mimic for post-translational modifications and hopefully the development of new technologies will allow the effect of histone modifications within the globular core of nucleosomes to be tested directly (28). Nonetheless, our observations illustrate a principle by which alterations to the structure of nucleosomes can selectively affect the outcome of remodelling reactions. It is of special note that the unravelling of DNA is affected as this might be anticipated to affect the ability of enzymes to catalyse octamer removal by collision (26). As a result these observations expand the repertoire of different means by which post-translational modification of histones can potentially exert effects on chromatin dynamics (3).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Todd Foundation Awards for Excellence, William Georgetti Scholarship, ORSAS, Fellow of New Zealand federation of graduate women (to J.S.); and a Wellcome Trust senior research fellowship (064414 to T.O.H.). Funding for open access charge: Wellcome Trust award to the University of Dundee.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Helder Ferreira, Andrew Flaus, Daniel Ryan and other members of the TOH lab for useful discussion.
REFERENCES
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cosgrove MS, Boeke JD, Wolberger C. Regulated nucleosome mobility and the histone code. Nat. Struct. Mol. Biol. 2004;11:1037–1043. doi: 10.1038/nsmb851. [DOI] [PubMed] [Google Scholar]
- 4.Wang XD, Hayes JJ. Acetylation mimics within individual core histone tail domains indicate distinct roles in regulating the stability of higher-order chromatin structure. Mol. Cell Biol. 2008;28:227–236. doi: 10.1128/MCB.01245-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tse C, Sera T, Wolffe AP, Hansen JC. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 7.Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 1993;3:419–429. [Google Scholar]
- 8.Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferreira H, Somers J, Webster R, Flaus A, Owen-Hughes T. Histone tails and the H3 alphaN helix regulate nucleosome mobility and stability. Mol. Cell Biol. 2007;27:4037–4048. doi: 10.1128/MCB.02229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferreira H, Flaus A, Owen-Hughes T. Histone modifications influence the action of Snf2 family remodelling enzymes by different mechanisms. J. Mol. Biol. 2007;374:563–579. doi: 10.1016/j.jmb.2007.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kasten M, Szerlong H, Erdjument-Bromage H, Tempst P, Werner M, Cairns BR. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luger K, Rechsteiner TJ, Richmond TJ. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 1999;119:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- 14.Flaus A, Richmond TJ. Base-pair resolution mapping of nucleosome positions using site-directed hydroxy radicals. Methods Enzymol. 1999;304:251–263. doi: 10.1016/s0076-6879(99)04015-x. [DOI] [PubMed] [Google Scholar]
- 15.Flaus A, Richmond TJ. Positioning and stability of nucleosomes on MMTV 3′ LTR sequences. J. Mol. Biol. 1998;275:427–441. doi: 10.1006/jmbi.1997.1464. [DOI] [PubMed] [Google Scholar]
- 16.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 2002;16:2120–2134. doi: 10.1101/gad.995002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chandy M, Gutierrez JL, Prochasson P, Workman JL. SWI/SNF displaces SAGA-acetylated nucleosomes. Eukaryot. Cell. 2006;5:1738–1747. doi: 10.1128/EC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stockdale C, Flaus A, Ferreira H, Owen-Hughes T. Analysis of nucleosome repositioning by yeast ISWI and Chd1 chromatin remodeling complexes. J. Biol. Chem. 2006;281:16279–16288. doi: 10.1074/jbc.M600682200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flaus A, Owen-Hughes T. Dynamic properties of nucleosomes during thermal and ATP-driven mobilization. Mol. Cell Biol. 2003;23:7767–7779. doi: 10.1128/MCB.23.21.7767-7779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kassabov SR, Zhang B, Persinger J, Bartholomew B. SWI/SNF unwraps, slides and rewraps the nucleosome. Mol. Cell. 2003;11:391–403. doi: 10.1016/s1097-2765(03)00039-x. [DOI] [PubMed] [Google Scholar]
- 21.Fan HY, He X, Kingston RE, Narlikar GJ. Distinct strategies to make nucleosomal DNA accessible. Mol. Cell. 2003;11:1311–1322. doi: 10.1016/s1097-2765(03)00192-8. [DOI] [PubMed] [Google Scholar]
- 22.Flaus A, Luger K, Tan S, Richmond TJ. Mapping nucleosome position at single base-pair resolution by using site-directed hydroxyl radicals. Proc. Natl Acad. Sci. USA. 1996;93:1370–1375. doi: 10.1073/pnas.93.4.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cairns BR, Kim YJ, Sayre MH, Laurent BC, Kornberg RD. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc. Natl Acad. Sci. USA. 1994;91:1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyer LA, Logie C, Bonte E, Becker PB, Wade PA, Wolffe AP, Wu C, Imbalzano AN, Peterson CL. Functional delineation of three groups of the ATP-dependent family of chromatin remodeling enzymes. J. Biol. Chem. 2000;275:18864–18870. doi: 10.1074/jbc.M002810200. [DOI] [PubMed] [Google Scholar]
- 25.Zhang YL, Smith CL, Saha A, Grill SW, Mihardja S, Smith SB, Cairns BR, Peterson CL, Bustamantel C. DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Mol. Cell. 2006;24:559–568. doi: 10.1016/j.molcel.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cairns BR. Chromatin remodeling: insights and intrigue from single-molecule studies. Nat. Struct. Mol. Biol. 2007;14:989–996. doi: 10.1038/nsmb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mihardja S, Spakowitz AJ, Zhang Y, Bustamante C. Effect of force on mononucleosomal dynamics. Proc. Natl Acad. Sci. USA. 2006;103:15871–15876. doi: 10.1073/pnas.0607526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neumann H, Peak-Chew SY, Chin JW. Genetically encoding N-epsilon-acetyllysine in recombinant proteins. Nat. Chem. Biol. 2008;4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.