Abstract
Increasing experimental evidence shows a prominent role of histone modifications in the coordinated control of gene expression in the human malaria parasite Plasmodium falciparum. The search for the histone-mark-reading machinery that translates histone modifications into biological processes, such as formation of heterochromatin and antigenic variation is of foremost importance. In this work, we identified the first member of a histone modification specific recognition protein, an orthologue of heterochromatin protein 1 (PfHP1). Analysis of the PfHP1 amino-acid sequence revealed the presence of the two characteristic HP1 domains: a chromodomain (CD) and a chromo shadow domain (CSD). Recombinant CD binds to di- and tri-methylated lysine 9 from histone H3, but not to unmodified or methylated histone H3 in lysine 4. PfHP1 is able to interact with itself to form dimers, underlying its potential role in aggregating nucleosomes to form heterochromatin. Antibodies raised against PfHP1 detect this molecule in foci at the perinuclear region. ChIP analysis using anti-PfHP1 shows that this protein is linked to heterochromatin of subtelomeric non-coding repeat regions and monoallelic expression of the major virulence var gene family. This is the first report implicating an HP1 protein in the control of antigenic variation of a protozoan parasite.
INTRODUCTION
Plasmodium falciparum, the causal protozoan agent of the most severe form of human malaria, has a complex life cycle with two different hosts, the Anopheles mosquito and humans. In order to complete its life cycle, P. falciparum invades different types of cells and self-propagates in very distinct environments in the mosquito (gut, hemolymph and salivary glands) as well as in the human host (skin, liver and erythrocytes). Each of these distinct environments exerts selective pressure related to morphological changes that force P. falciparum to exhibit differential gene expression during its life cycle (1,2). A surprising finding was the identification of a relatively low number of genes encoding transcription factors in the parasite, although the basal core transcriptional machinery and the protein-coding genes involved in nucleosome assembly and regulation of chromatin structure were found to be conserved (3–5). To date, no specific DNA-binding proteins have been identified other than PfMyb1 (6) and the ApiAp2 gene family (7). This suggested that epigenetic mechanisms play a significant role in controlling gene expression in Plasmodium (3,8–10). The importance of reversible chromatin modifications involving histone modifications and the chromatin-associated protein PfSir2 was first demonstrated for a subtelomeric gene family (var gene family) involved in the control of antigenic variation in P. falciparum (10). Knock out of PfSir2 resulted in the de-repression of a fraction of the members of the var gene family (11). A screen for histone modifications revealed that H3K9me3 plays a particular role in var gene repression (12,13) and H3K4me2/3 in transcriptional activity and epigenetic memory (12). These dynamic histone modifications occur mainly in the 5′-UTR regions of var genes. It was shown that histone modifications also play a major role in the regulation of non-var genes (12,14–16). The identification of factors able to ‘read’ specific histone marks and translate them into changes in gene activity remains elusive in Plasmodium.
In this work, we identified the first plasmodial protein that displays features of the histone-reading machinery that binds specifically to H3K9me2/3. The identified protein is an orthologue to heterochromatin protein 1 (PfHP1) and shows a specific location to perinuclear foci. Chromatin immunoprecipitation (ChIP) assays reveal that the PfHP1 protein is a component of subtelomeric chromatin. Furthermore, our data links PfHP1 to epigenetically silenced var genes, since an active var gene is devoid of PfHP1 in its 5′-UTR.
MATERIALS AND METHODS
Parasites
Plasmodium falciparum FCR3 strain was cultivated according to standard culture conditions (17). Panning assays for selection of FCR3 parasites that transcribe var genes associated with CD36 and CSA binding have been done as previously described (18).
Recombinant proteins
A 801 bp DNA fragment of the PfHP1 gene was obtained by RT–PCR. The sequences of direct and reverse oligonucleotides were: PfHP1EcoRI 5′-CGGAATTCATGACGGGTCAGATGAAGAA-3′ and PfHP1XhoI 5′-CCGCTCGAGTTAAGCTGTACGGTATCTTAG-3′, respectively. The PCR fragment was digested, retrieved and inserted into the EcoRI-XhoI-digested pGEX4T1 vector (Amersham) fused to a GST-tag. The resulting construct was sequenced and called GST-PfHP1. The primers PfHP1-CDEcoRI 5′-CGGAATTCATGACAGGGTCAGATGAAGAA-3′ and PfHP1-CDXhoI 5′-CCGCTCGAGCTCATTAGCTTTCGATAAAAAATT-3′ were used to amplify a DNA fragment of 204 bp that contains PfHP1 chromodomain (CD) (PfHP1-CD). Integrity of all recombinants clones was confirmed by sequencing with 5′ and 3′ primers to the pGEX cloning site (Pharmacia). All of them were transformed into Escherichia coli DH5α strain. Expression of GST fusion protein was induced with 0.5 mM IPTG at 30°C for 4 h. GST fusion proteins were purified with glutathione sepharose (Pharmacia) by standard methods. The PfHP1 DNA fragment of 801 bp was cloned into BamHI 5′-CGGAATTCATGACGGGTCAGATGAAGAA-3′ and NotI sites 5′-ATAAGAATGCGGCCGCTTAAGCTGTACGGTATCTTAG-3′ of the pROEX™ HTb vector (Invitrogen). 6HisPfHP1 protein was induced in BL21 bacterial cells by addition of 0.6 mM IPTG. Recombinant proteins were purified using nickel-nitriloacetic acid resin (TALON) according to manufacturer protocol. Purified recombinant proteins were verified by Western blot.
Production of PfHP1 antibodies
The GST-PfHP1 recombinant protein was purified as recommended by the manufacturers (see above). For polyclonal antibodies against PfHP1, two rabbits were inoculated i.c. with 100 μg of GST-PfH1 protein (first dose). The fusion protein was emulsified with complete adjuvant (Sigma). Following the inoculation series, animals were sacrificed and serum was collected. Immunoglobulins were purified from serum by protein A-sepharose (Pharmacia) chromatography, following standard procedures.
Nuclear and cytoplasmic extracts preparation
Nuclear and cytoplasmic extracts were prepared as previously described (10,19) with some modifications. Briefly, 5 × 109 parasites of an asynchronous culture of FCR3 P. falciparum strain were isolated from infected erythrocytes by saponin lysis, resuspended in 1 ml of lysis buffer (10 mM HEPES, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT, 0.65% NP-40) and incubated 30 min a 4°C. Then, parasites were lysed by 200 strokes on a prechilled douncer homogenizer. The nuclei were collected by centrifugation. The supernatant containing the cytoplasmic fraction was recovered and kept at −80°C. The nuclei were purified by sucrose gradient centrifugation. For this purpose, the nuclei were resuspended in 1 ml of lysis buffer and the resulting suspension was layered on 3 ml of lysis buffer containing 0.34 M sucrose. The nuclei were resuspended in 100 µl of extraction buffer (20 mM HEPES, pH 7.9, 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA and 1 mM DTT). Following 15 min of vigorous shaking at 4°C, the extract was centrifuged and the supernatant containing nuclear proteins was collected. All buffers used in this protocol contained protease inhibitors (Complete, Roche).
Immunofluorescence microscopy
Immunofluorescence assays were performed as previously described (20). The parasites were fixed in suspension with 4% paraformaldehyde solution for 15 min on ice. Next, the fixed parasites were incubated with the primary antibody for 60 min at room temperature followed by incubation for 30 min with a secondary antibody conjugated with fluorochrome and deposited on microscope slides. The final antibody dilutions were rabbit anti-PfHP1 1:500, rat anti-PfSir2 1:50, rat anti-PfNop1 1:50, Alexa Fluor 568 goat anti-rabbit highly cross-absorbed 1:500 and Fluorescein-conjugated goat anti-rat 1:500.The samples were analysed in a Nikon microscope.
ChIP and dot-blots
The ChIP assay was performed as previously described (10) with minor modifications. The parasites were treated with saponin and cross-linked in 1% formaldehyde for 10 min at 37°C. Chromatin fragments were incubated overnight at 4°C with 2.5 µl of rabbit polyclonal antibody anti-PfHP1 or preimmune serum. The immunoprecipitated DNA was radioactively labelled and hybridized with a Hybond N+ membrane dot-blotted with 35 ng of Telomeric, TARE1, TARE2, TARE2-3, TARE3, TARE6 repeats as previously published (20). The 5′-UTR region of var genes (UpsB, UpsC and UpsE) were obtained as previously described (10). For input DNA samples, an aliquot of lysate used in the immunoprecipitation was processed along with the rest of the samples. Quantification of the signal was done with the ImageQuant software (Molecular Dynamics, Sunnyvale). The amount of DNA immunoprecipitated in each ChIP corresponding to the rate of the bound and input (B/I) values and represent percentage of input.
In vitro binding assays
The His-PfHP1 protein was denatured and renatured by dialysis against 8 M urea, followed by 6 M, 4 M, 2 M, 1 M and 0.5 M urea and finally with nickel-binding buffer without urea. 40 μg of purified His-PfHP1 fusion protein was incubated with Nickel-nitriloacetic acid resin (TALON) (50 μl) according to manufacturer protocol. Once the His-PfHP1 protein was immobilized on TALON resin, it was mixed with 2 mg/ml total bacterial lysate that over-expressed GST-PfHP1 protein fusion and incubated for 2 h at 4°C with 1 ml binding buffer (20 mM Tris–HCl pH 8.0, 100 mM NaCl). Next, the beads were washed three times with 1 ml washing buffer (20 mM Tris–HCl pH 8.0, 100 mM NaCl, 15 mM Imidazol pH 8.0). Bound proteins were eluted with sample buffer and boiled before loading on a SDS–PAGE gel. After electrophoresis, proteins were transferred to Hybond ECL nitrocellulose membrane (Amersham) as previously described (21) and the blot was probed with anti-GST and Anti-6His-Tag–specific antibodies. Rabbit polyclonal antibodies against GST and polyhistidine domain 6His-probe were purchased from Santa Cruz Biotechnology, Inc.
Dot-blot overlay assays
The dot-blot assays were performed as described by Sambrook et al., 1989 (22). Two hundred nanograms of the GST-CD protein (GST-PfHP1-CD) were dotted to Hybond ECL nitrocellulose (Amersham) and blocked using 4°C fat-free dried milk in TBS-Tween (10 mM Tris–HCl, 150 mM NaCl, 1% Tween-20, pH 7.5). Membranes were incubated overnight at 4°C in BC100 buffer (25 mM HEPES, pH 7.6; 100 mM NaCl, 1 mM MgCl2, 0.5 mM EGTA, 0.1 mM EDTA, 10% glycerol, 1 mM DTT and 0.2 mM PMSF) with 2 μg or different amounts of biotynilated peptides: H3K9me3 (Upstate), H3K9me2 (Washington Biotechnology), H3K9ac (Upstate), H3K4me3 (Upstate), H3K4me2 (Washington Biotechnology) and non-modified H3 (Washington Biotechnology). The membranes were washed twice (10 min each) with BC-100 plus 0.05% NP40 and twice with BC200 (25 mM HEPES, pH 7.6; 200 mM NaCl, 1 mM MgCl2, 0.5 mM EGTA, 0.1 mM EDTA, 10% glycerol, 1 mM DTT, 0.2 mM PMSF and 0.05% NP-40). Membranes were incubated with streptavidin, coupled to HRP (1:10 000 dilutions in TBS plus 1% BSA) 30 min at 37°C. Finally the membranes were washed seven times with TBS plus 1% Tween-20 and bound peptides were detected by chemiluminescence using an ECL kit. Quantification of the signal was done with the ImageQuant software (Molecular Dynamics, Sunnyvale).
Molecular modelling of PfHP1 protein
Homology modeling of the PfCD and PfCSD complexes was performed with MOE package (Molecular Operating Environment, http://www.chemcomp.com) using the crystallographic structure of CD from mouse HP1β (19–73) and CSD from S. pombe SpSwi6 (261–320) as template (PDB ID 1APO and 1E0B, respectively). Search for templates and sequence alignments were done with BLAST, with minor adjustments in the alignment based on template structure inspection to facilitate indel modelling. For the search of the conformation of replaced side chains we used a library of rotamers included in MOE; for indel modelling, a loop library of this package was used. By random combination of these libraries we constructed 1000 alternative models and minimized them to reach a gradient of 0.1 kcal/(mol A°). The model with the best packing was subjected to more extensive minimization, until an RMS gradient lower than 0.05 kcal/(mol A°) was obtained, and used thereafter for the structural analysis. Minimizations used the CHARMM22 force field with its partial-charge assignments for protein atoms. The stereochemical quality of the model was verified with MOE and with the Mol-Probity server (http://kinemage.biochem.duke.edu/molprobity).
RESULTS
Identification and modelling of the HP1 orthologue in P. falciparum
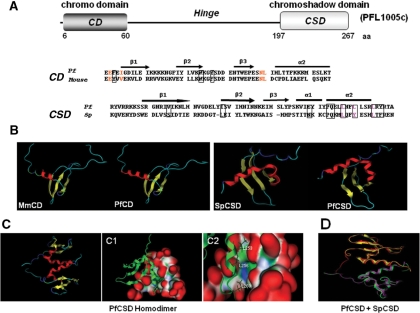
The HP1 protein is highly preserved through evolution from Schizosaccharomyces pombe to human, and participates in the formation of highly condensed chromatin by recognizing the di- or tri-methylated histone H3 lysine 9 (H3K9me2 or H3K9me3) (23). In order to determine the existence of a protein similar to HP1 and its possible participation in heterochromatin formation in P. falciparum, we performed a search in the P. falciparum databank (www.plasmodb.org/plasmo). This analysis allowed us to identify the PFL1005c gene, which is predicted to encode a 30-kDa protein. A detailed analysis of its amino-acid sequence with the SMART program revealed the presence of the two characteristic HP1 domains defined to date: a CD located between amino acids 6 and 60, and a low-homology chromo shadow domain (CSD) that spans from amino acids 197–267. Hereafter, this protein encoded by PFL1005c gene is called PfHP1.
The P. falciparum CD's highest identity (66%) is with the mouse CD, while the CSD's highest identity is with S. pombe CSD (37%) (Figure 1A). Similar to what has been reported in other eukaryotes, the PfHP1 CD has three of the most conserved aromatic cage residues (F9, W30 and Y33); in mice, these residues recognize the H3K9me3, and abrogate silencing when they are mutated (24).
Figure 1.
Structural modelling of the chromo domain (CD) and chromoshadow domain (CSD) of PfHP1. (A) Schematic diagram showing the complete ORF and location of CD and CSD of PfHP1 separated by the hinge. Numbers correspond to residue positions for each domain. The primary sequence alignment of CD from mouse HP1ß (19–73 aa) and PfHP1 (6–60 aa) was used for the modelling of the CD domain. The boxes indicate the aromatic cage residues potentially involved in the recognition of the H3K9me peptide. Conserved residues that form a complementary surface responsible for H3 peptide recognition in the CD from Drosophila are indicated in red (24). The primary sequence alignment of CSD from S. pombe (261–320 aa) and from PfHP1 (197–267 aa) was used to perform the model of the CSD domains. Residues implicated in the formation of the dimer interface are enclosed in boxes. Residues that have shown to be important for dimer formation in S. pombe (25) are indicated with pink letters. Both alignments were performed with ClustalW and used for modelling with the program MOE (www.chemcomp.com). The secondary structure elements are shown above the alignment: bars represent α-helices (α-1 and α-2) and arrows represent β-strands (β1–β3). (B) Tertiary structure of CD and CSD domains of PfHP1. The structure is conserved between mouse and P. falciparum CD domains, and between S. pombe and P. falciparum CSD domains. (C) Cartoon representation of the CSD dimer. The two monomers are indicated in red. The molecular surface of the putative PfHP1-CSD dimer is shown. (C1) A putative monomer is shown by cartoon and the other one by molecular surface. Surface convex zones are shown in red (protuberances); concave zones that correspond to binding or recognition sites are shown in green (hydrophobic regions) and the polar surface is denoted in blue. (C2) The hydrophobic region and the crucial L residues that contact the other subunit in the formation of the dimer are indicated. (D) Superposition of the 3D models of the CSD domains homodimers from Swi6 (magenta–yellow) and PfHP1 homodimers (green–red). The CSD architecture is strongly conserved between S. pombe and P. falciparum.
Secondary structure analysis of the PfHP1 CD showed high similarity to that described in mouse, since it presents the three ß chains, as well as the α2 helix structures (Figure 1A). This suggests that PfHP1 protein might be able to recognize the H3K9me2 and/or H3K9me3, similarly to the HP1 protein in higher eukaryotes. With respect to the CSD, PfHP1 consists of three beta chains and two α helixes (α1 and α2), as described in S. pombe and other eukaryotes. It also presents some of the residues reported to be important for dimerization, which in turn are necessary for heterochromatin formation (Figure 1A).
In order to determine whether the CD and CSD domains of the PfHP1 protein have a tertiary structure similar to that described in S. pombe and other eukaryotes, we made homology models, taking as a reference the mouse HP1 CD domain and Swi6 protein CSD of S. pombe. The tertiary structure obtained showed that PfHP1 CD and CSD domains have a folding similar to that of the mouse CD and S. pombe CSD (Figure 1B).
Dimer formation in S. pombe is centred on helix 2, which interacts symmetrically with the helix 2 of the other monomer, and the primary contacts involve amino acids L161, Y164 and L168. In PfHP1, L161 and L168 are present, but Y164 has been replaced by a L. Nevertheless, in mouse Y164 has also been changed and that does not affect dimer formation (25). Therefore, amino acids L161, L164 and L168 could play a role in PfHP1 dimer formation (Figures 1C–C2). In the case of S. pombe, it has been found that the bonding of the two monomers through the alpha 2 helix generates a hydrophobic groove that favours protein–protein interactions (25). As seen in Figure 1C, the PfHP1 CSD folding generates a hydrophobic groove through which the PfHP1 monomers can interact with themselves. Finally, the overlap of S. pombe CSD homodimers (magenta–yellow) and CSD of PfHP1 (red–green) showed similar structural organization (Figure 1D). In conclusion, the predicted tertiary structure suggests that PfHP1 might interact with histone H3 lysine 9 methylated, as well as participate in the formation of homodimers.
In vitro binding of the PfHP1 CD to H3K9me2 and H3K9me3 peptides
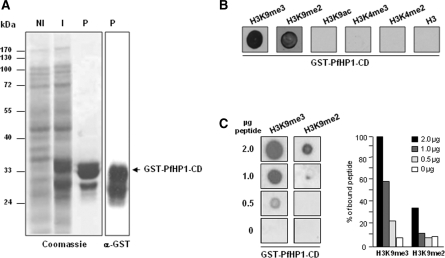
The CD in eukaryotes is responsible for recognizing H3K9me2 or H3K9me3 (26,27). To determine whether the PfHP1 protein is able to perform the same function in malaria parasites, a soluble GST fusion protein corresponding to the CD was produced. This protein was purified and immunoblotted with anti-GST antibodies (Figure 2A). A band of the predicted size (33 kDa) was observed together with minor bands, which are probably degradation products often seen for GST fusion proteins. The GST-PfHP1-CD protein was then used to investigate its binding specificity to different histone modifications. The GST-PfHP1-CD protein was dotted on nylon membranes and incubated with the following biotinylated histone H3 peptides: H3K9me3, H3K9me2, H3K9ac, H3K4me3, H3K4me2 and unmodified histone H3. Binding of the peptides to the CD of PfHP1 was detected using streptavidin conjugated to HRP. Signal was observed only with H3K9me2 and H3K9me3 (Figure 2B). To show that the modified histone H3 peptides did not bind the recombinant GST-CD protein through GST, the GST protein was used as a negative control (data not shown). These results demonstrate that the CD from PfHP1 protein is able to interact to H3K9me2 and H3K9me3, but not to H3K9ac, H3K4me2, H3K4me3 or to unmodified histone H3.
Figure 2.
PfHP1 binds to histone H3 peptide methylated at lysine 9. (A) Total proteins from DH5α strain expressing the CD of PfHP1 before (NI) and after IPTG induction (I), as well as the purified GST-PfHP1-CD fusion protein (P), were separated by SDS–PAGE and stained with Coomassie blue (left). An anti-GST antibody was used to detect the presence of the fusion protein (right). (B) Purified protein GST-PfHP1-CD was assayed for binding to histone H3 peptides. 200 ng of GST-CD protein (GST-PfHP1-CD) were dotted to a nitrocellulose membrane and incubated with 2 µg of the following biotinylated peptides: H3K9me3, H3K9me2, H3K9ac, H3K4me3, H3K4me2 and unmodified H3. (C) Two hundred nanograms of GST-PfHP1-CD fusion protein were dotted to nitrocellulose and incubated with decreasing amounts of H3K9me3 and H3K9me2 biotinylated peptides (left panels). A representative result of two independent experiments is shown. The right panel shows a quantitative presentation of the GST-PfHP1-CD affinity to the peptides.
To determine the affinity of the GST-PfHP1-CD protein for both H3K9me3 and H3K9me2 peptides, dot blot binding-assays were performed using a fixed amount of GST-PfHP1-CD fusion protein with different concentrations of peptides. Densitometry analysis showed that in vitro GST-PfHP1-CD binds three times more efficiently to H3K9me3 than to H3K9me2 (Figure 2C).
Recombinant PfHP1 forms homodimers
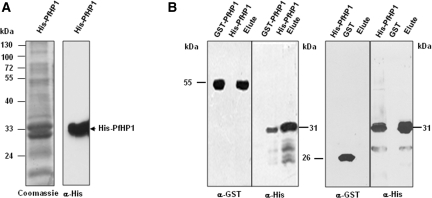
Cowieson et al. demonstrated that recombinant CSD in its unmodified form is able to dimerize in solution through the alpha helixes, forming a non-polar cavity where proteins that have the P×V×L pentapeptide can bind (28). To demonstrate that PfHP1 is able to form dimers as suggested by the tertiary structure, we generated recombinant PfHP1 proteins fused to GST (GST-PfHP1) and to six histidines (His-PfHP1). Inclusion bodies containing His-PfHP1 were incubated in the presence of urea, renatured and incubated with Nickel-beads. These beads were washed and the bound protein was eluted, analysed by SDS–PAGE, blotted and tested with the anti-histidine antibodies. These antibodies recognized a 33 kDa protein, which corresponds to the monomeric form of PfHP1 (Figure 3A).
Figure 3.
In vitro dimerization of the PfHP1 protein. (A) His-PfHP1 protein purified using nickel-nitriloacetic acid resin (left). Western blot analysis with anti-histidine antibodies identified the monomer of PfHP1 (right). (B) Interaction between His-PfHP1 (∼30 kDa) and GST-PfHP1 fusion proteins (∼55 kDa). Purified His-PfHP1 was mixed with total bacterial lysate overexpressing GST-PfHP1 fusion protein (left panels) or only GST (right panels). Protein complexes were recovered, separated by SDS–PAGE and detected by Western blot with anti-histidine and anti-GST antibodies.
We next incubated lysates from bacterial cells over-expressing the GST-PfHP1 fusion protein (55 kDa) with the renatured His-PfHP1 fusion protein bound to the nickel beads. Bound proteins were eluted, separated by SDS–PAGE and immunoblotted with anti-histidine and anti-GST antibodies (Figure 3B). The Western blot analysis confirmed that His-PfHP1 interacts specifically with GST-PfHP1. GST alone (26 kDa) did not bind to His-PfHP1 under the same conditions (Figure 3B). Similar results were obtained when we used purified GST-PfHP1 and His-PfHP1 fusion proteins in the pull-down assays (data not shown). In summary, our in vitro experiments suggest the ability of PfHP1 to form dimers, which appears to be important for HP1-mediated heterochromatin formation (see model in Figure 6A).
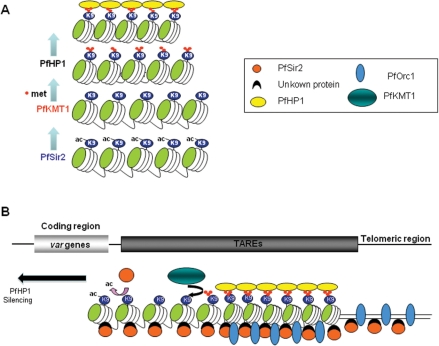
Figure 6.
Hypothetical model for heterochromatin formation at P. falciparum chromosome ends. (A) PfHP1-mediated heterochromatin model depends on the action of the histone deacetylase PfSir2 and histone methyltransferase PfKMT1 (10,15,37). (B) General view of the known chromatin components at P. falciparum subtelomeres. Spreading of heterochromatin along the different TAREs into adjacent coding regions probably involves the cooperation of PfHP1, PfSir2 and PfKMT1. The role of PfOrc1 in this process remains unknown.
PfHP1 localizes at the nuclear periphery
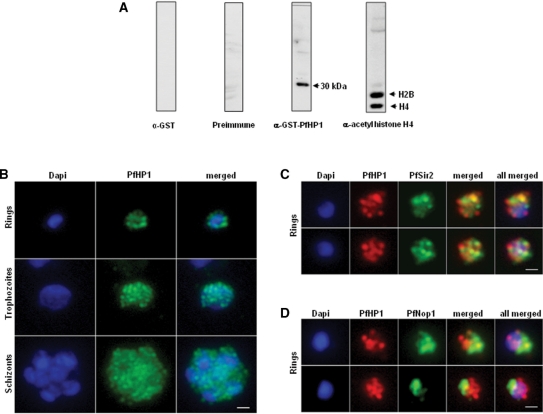
To further investigate the function of PfHP1, we raised polyclonal antibodies against the recombinant GST-PfHP1. Immunoblot analysis of parasite nuclear extracts with this anti-GST-PfHP1 antibody detected a 30 kDa protein, which has the predicted size of PfHP1. The specificity of the nuclear extracts was verified using an anti-acetyl histone H4 antibody, which recognizes both histones H2B and H4 (Figure 4A). No signal was detected in the cytoplasmic fraction (data not shown). Preimmune serum and anti-GST antibody gave no signal with the parasite's nuclear extracts (Figure 4A).
Figure 4.
Nuclear expression and cellular localization of PfHP1. (A) Western blot analysis of nuclear extracts from P. falciparum using the following antibodies: anti-GST antibody (left), preimmune serum, anti-GST-PfHP1 antibody (middle) and anti-acetyl histone 4 antibodies (right). (B) IFA analysis of PfHP1 (green) throughout the parasite asexual blood stage cycle (ring, trophozoite and schizont stages). (C) In ring stage, partial co-localization between PfHP1 (red) and PfSir2 (green) antibodies (yellow spots) is observed. (D) Co-localization with a nucleolar marker PfNop1 shows minimal overlap of PfHP1 with this compartment. Scale bars represent 1 µm.
To study the subnuclear location of PfHP1, we performed immunofluorescence assays (IFA) in different stages of the 48-h asexual cycle. PfHP1 appears to form nuclear foci during the entire cycle (Figure 4B). In ring stages (∼16 h), PfHP1 locates predominantly to perinuclear space, whereas in trophozoite stages (∼30 h) and schizonts (∼40 h) the number of foci increases and seems to redistribute in the nucleoplasm.
To test whether PfHP1 localizes to telomeric perinuclear clusters we performed dual IFA using anti-PfHP1 and anti-Sir2 antibodies. We observed that PfHP1 localizes with some PfSir2 fluorescent signals (Figure 4C). Because PfSir2 is not an exclusive marker for telomeric clusters and anti-PfSir2 also stains the nucleolus (10), we carried out a dual IFA using an antibody against a nucleolar protein (anti-PfNop1). We observed that PfHP1 did not (or only marginally) co-localize with the nucleolar marker (Figure 4D), indicating that the PfHP1 foci co-localizing with PfSir2 correspond indeed to telomeric foci.
PfHP1 binds to subtelomeric TAREs and is implicated in the control of var genes expression
The IFA suggested that the PfHP1 protein is located on the telomeric and/or subtelomeric regions of P. falciparum. To determine the in vivo distribution of PfHP1 over the chromosome ends of the parasite, we performed ChIP assays. DNA pulled down with anti-GST-PfHP1 was hybridized to different repetitive TARE elements and telomere probes in a dot blot assay. We observed that PfHP1 is enriched in the subtelomeric region and almost absent in the telomeric repeats (Figure 5B). A single copy blood-stage expressed gene (HRP1) did not immunoprecipitate with the anti-GST-PfHP1 antibody (Figure 5B). These results show that PfHP1 is associated to chromatin across subtelomeric repeat elements, most likely mediating chromatin compaction.
Figure 5.
PfHP1 is associated to constitutive and facultative heterochromatin. (A) Schematic representation of telomeric and subtelomeric regions of P. falciparum. In this parasite, chromosome ends are composed of telomeres and telomere associated repetitive elements (TAREs 1–6). The subtelomeric coding region contains members of several multigene families important for virulence, such as the var genes. The var gene repertoire is classified in different types according to the conserved upstream sequences. Ups A, B and E types are located on subtelomeric regions, whereas UpsC types are found in central chromosomal position. The positions of the probes used in ChIP assays are shown. (B) Dot blot based ChIP analysis of PfHP1 abundance at telomeric and non-coding subtelomeric regions. DNA probes corresponding to TARE 1, 2, 3, 6 and a sequence between TARE 2–3 were fixed to nylon membranes and hybridized to DNA immunoprecipitated with anti-PfHP1 antibodies and preimmune serum (PI) (left panels). The 5′-UTR region of the histidine rich protein 1 (HRP1) gene, expressed during blood stage, was used as control (left panels). (C) ChIP analysis of FCR3-CSA parasites, in which the var2CSA gene is active (UpsE ON). (D) ChIP analysis of FCR3-CD36 parasites, in which the var2CSA gene is repressed (UpsE OFF). In (C) and (D) DNA corresponding to different types of var 5′-UTR regions (UpsE, UpsC and UpsB), 5′-UTR region of HRP1 and TARE6 were fixed on nylon membranes and hybridized to DNA immunoprecipitated with anti-PfHP1 antibodies and PI serum (left panels). The right panels show a quantitative representation of PfHP1 levels at the different DNA regions. In all cases, ChIP analysis was performed in ring stage parasites and a representative result is shown. Error bars represent the standard deviation from three independent experiments each performed in duplicate. Input corresponds to DNA prepared from fragmented chromatin prior to immunoprecipitation.
To determine whether PfHP1 has a role in the silencing of subtelomeric var genes, we performed ChIP assays using two isogenic parasite populations: FCR3-CSA that expresses a specific subtelomeric var gene (var2CSA), and FCR3-CD36 in which the same gene is repressed. We found that the var2CSA upstream region (UpsE) is immunoprecipitated by anti-GST-PfHP1 antibodies when this gene is inactive (FCR3-CD36 population, Figure 5D) but not when it is active (FCR3-CSA population, Figure 5C). Other types of var gene upstream regions representing subtelomeric and chromosome central var members (UpsB and UpsC, respectively), which are in a repressed state in these parasite populations, were immunoprecipitated by anti-PfHP1 antibodies. Thus, our data show that PfHP1 is linked to repression of subtelomeric but also to central var genes.
DISCUSSION
In this work, we identified and characterized PfHP1, the first protein from P. falciparum that binds specifically to a histone modification, thus demonstrating the existence of histone-mark-reading machinery in this pathogen. Our in vitro results demonstrated that PfHP1 specifically recognizes H3K9me2 and H3K9me3 peptides by its CD. The affinity of this protein seems to be greater for tri-methylated than for di-methylated peptide (Figure 2C), suggesting that the degree of methylation at histone H3K9 may lead to the formation of more or less tight chromatin. As predicted by its tertiary structure (Figure 1), PfHP1 is able to interact with itself (Figure 3B), which suggests a role in the formation and maintenance of heterochromatin, similarly to its orthologues in S. pombe, Drosophila and human (29). Furthermore, our data reinforce the idea that the machinery involved in heterochromatin formation in Plasmodium is more similar to metazoa than to S. cerevisiae. It also shows that the HP1 protein is not exclusive of metazoan, as it has been proposed (30).
Nuclear staining with anti-PfHP1 suggested that this protein is located at the perinuclear region visible as foci similar to PfSir2 and PfOrc1, two proteins known to be components of subtelomeric heterochromatin in P. falciparum (10,20). In contrast to PfSir2 and PfOrc1, our co-localization experiments demonstrated that PfHP1 does not stain the parasite nucleolus and may therefore not participate in the silencing of rDNA genes in blood stage parasites.
The notion that PfHP1 is a component of telomeric heterochromatin was further demonstrated by showing the binding of PfHP1 to subtelomeric chromatin (Figure 5). ChIP studies clearly showed that the anti-PfHP1 antibody immunoprecipitated subtelomeric regions as well as members of the var gene family, and react poorly with telomeric repeats. Like in yeast, the telomeres of P. falciparum are mostly not packed in nucleosomes whereas the subtelomeric chromatin contains nucleosomes (31). The association of PfHP1 with the subtelomeric chromatin correlates well with the nucleosomal nature of HP1 heterochromatinization function.
Importantly, our data show that PfHP1 protein is linked to monoallelic expression of the major virulence gene family in this human pathogen. The data strongly suggest that PfHP1 must be removed from the var upstream regions to allow gene expression. Our ChIP assays demonstrated that PfHP1 is able to bind not only to subtelomeric but also to central var genes (Figure 5). Consistent with these results, P. falciparum genome-wide high-resolution ChIP analysis of histone mark H3K9me3 shows a restricted localization to telomeres, subtelomeres and internal chromosomal regions that contain virulence gene families (Lopez-Rubio, JJ and Scherf A. personal communication). It is very likely that PfHP1 displays a genome-wide distribution similar to H3K9me3. The presence of PfHP1 in internal chromosomal domains may explain the partial co-localization with telomeric PfSir2 foci in the IFA analysis.
In mammals, the HP1 family is composed of three variants (alpha, beta and gamma), which are coded by distinct genes (32). These proteins are involved in different events: HP1 alpha and beta in heterochromatin formation, and HP1 gamma in euchromatin formation (29). We have found only one hp1 gene in P. falciparum genome and this one was confirmed by Southern blot analysis (data not shown). This raises the question of whether a single PfHP1 protein performs different functions. The hinge region of HP1 protein is highly amenable to post-translational modifications especially phosphorylation (29). In addition, mutations in this region have been shown to affect the location, interactions and functions of HP1 (33). For example, Ser83 phosphorylation of HP1 gamma is important for its localization only in euchromatic regions (34). In silico analysis of PfHP1 protein indicates that a number of post-translational changes, such as: sumoylation, ubiquitination and phosphorylation, may occur mainly in the hinge region (Perez-Toledo K and Hernandez-Rivas R unpublished data). It is therefore possible that different post-translational modifications may define distinct functions of PfHP1. Another component of the subtelomeric chromatin, PfSir2, has recently been shown to be sumoylated (35). It will be interesting to investigate whether mutation in the PfHP1 hinge region will affect function and nuclear location.
PfHP1 knock-out parasites would be a valuable tool to further investigate the role in gene silencing in P. falciparum. So far, all our attempts to disrupt the hp1 gene had failed. This is possibly due to the fact that only one copy exists in the genome. It may also be due to additional essential roles of PfHP1 possibly related with chromosome integrity and/or nuclear organization in this parasite. Inducible loss-of-function systems appear now to be available for P. falciparum that may allow PfHP1 phenotype analysis (36).
In line with our data, we propose a model in which PfHP1 works in concert with a histone deacetylase and a histone methyltransferase in heterochromatin formation (Figure 6A). In our model, a histone deacetylase PfSir2 (10,37), is first recruited to chromatin and deacetylates H3K9 residues. In a subsequent step, we hypothesize that the H3K9 methyltransferase orthologue PfKMT1 (15) methylates the H3K9 residues. As a consequence, PfHP1 binds to H3K9me3, dimerizes and promotes chromatin condensation. This leads to PfHP1 spreading into the coding regions, thus contributing to the reversible silencing of virulence factor gene families immediately adjacent of TAREs (Figure 6B). What recruits PfHP1 to central var genes remains guesswork. In mammals, targeting of HP1 to chromatin requires not only K9 methylation but also a direct protein–protein interaction with SUV39 (an orthologue of PfKMT1) (38). HP1 can also interact with Orc1, as it has been demonstrated in Drosophila and Xenopus (39,40). The spreading of PfHP1 along the different TAREs is most likely controlled by complex machinery including known components such as PfSir2, PfOrc1 and PfKMT1. Further studies are necessary to determine potential protein–protein interaction between PfHP1 and other chromatin components by TAP-tag or pull down assays.
In conclusion, this work identifies a key component of the epigenetic machinery involved in heterochromatin formation. We demonstrated that PfHP1 is closely related to its counterpart in higher eukaryotes, since it is able to form homodimers and binds methylated histone H3K9. PfHP1 is the first plasmodial protein identified that recognizes a specific histone modification, which is linked gene silencing of virulence factor gene families located adjacent to TAREs and some chromosome internal regions. Upon activation of a single gene member in a mutually exclusive fashion, PfHP1 is absent of the promoter region of the activated gene. This is a major conceptual advance to our knowledge and may have a big impact on the allelic exclusion field in other pathogens and higher eukaryotes. Together with the previously identified PfSir2, PfHP1 might form the core of the subtelomeric heterochromatin machinery in P. falciparum. Our anti-PfHP1 antibodies are a new tool to investigate facultative and constitutive heterochromatin formation in malaria parasites. Telomere-linked control of phenotypic variation is an important issue in malaria pathogenesis and a better knowledge of the epigenetic factors controlling those is vital for understanding the disease and to develop new anti-parasite control strategies.
FUNDING
Consejo Nacional de Ciencia y Tecnológia [45687/A-1 to R.H.R.]; Institut Pasteur (EGIDE) and CONACYT [No 172799 to A.P.R.M.]; European Commission (BioMalPar) [contract No: LSPH-CT-2004-503578]; the ‘Fonds dédié: Combattre les Maladies Parasitaires’ Sanofi Aventis – Ministère de la Recherche; ANR [ANR-06-MIME-026-01 to A.S.] and Fundação para a Ciência e Tecnologia, Portugal [SFRH/BD/11756/2003 to L.M.S.]. Funding for open access charge: Consejo Nacional de Ciencia y Tecnología [56836-Q to LSA].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank J.J. Lopez-Rubio, L. Riviere and F. Iracheta Tovar for critical comments.
REFERENCES
- 1.Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, Haynes JD, De la Vega P, Holder AA, Batalov S, Carucci DJ, et al. Discovery of gene function by expression profiling of the malaria parasite life sycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- 3.Aravind L, Iyer LM, Wellems TE, Miller LH. Plasmodium biology: genomic gleanings. Cell. 2003;115:771–785. doi: 10.1016/s0092-8674(03)01023-7. [DOI] [PubMed] [Google Scholar]
- 4.Templeton TJ, Iyer LM, Anantharaman V, Enomoto S, Abrahante JE, Subramanian GM, Hoffman SL, Abrahamsen MS, Aravind L. Comparative analysis of apicomplexa and genomic diversity in eukaryotes. Genome Res. 2004;14:1686–1695. doi: 10.1101/gr.2615304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callebaut I, Prat K, Meurice E, Mornon JP, Tomavo S. Prediction of the general transcription factors associated with RNA polymerase II in Plasmodium falciparum: conserved features and differences relative to other eukaryotes. BMC Genomics. 2005;6:100. doi: 10.1186/1471-2164-6-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gissot M, Briquet S, Refour P, Boschet C, Vaquero C. PfMyb1, a Plasmodium falciparum transcription factor, is required for intra-erythrocytic growth and controls key genes for cell cycle regulation. J. Mol. Biol. 2005;346:29–42. doi: 10.1016/j.jmb.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 7.De Silva EK, Gehrke AR, Olszewski K, Leon I, Chahal JS, Bulyk ML, Llinas M. Specific DNA-binding by apicomplexan AP2 transcription factors. Proc. Natl Acad. Sci. USA. 2008;105:8393–8398. doi: 10.1073/pnas.0801993105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hakimi M-A, Deitsch KW. Epigenetics in apicomplexa: control of gene expression during cell cycle progression, differentiation and antigenic variation. Curr. Opin. Microbiol. 2007;10:357–362. doi: 10.1016/j.mib.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 9.Ralph SA, Scherf A. The epigenetic control of antigenic variation in Plasmodium falciparum. Curr. Opin. Microbiol. 2005;8:434–440. doi: 10.1016/j.mib.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Freitas-Junior LH, Hernandez-Rivas R, Ralph SA, Montiel-Condado D, Ruvalcaba-Salazar OK, Rojas-Meza AP, Mâncio-Silva L, Leal-Silvestre RJ, Gontijo AM, Shorte S, et al. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria Parasites. Cell. 2005;121:25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 11.Duraisingh MT, Voss TS, Marty AJ, Duffy MF, Good RT, Thompson JK, Freitas-Junior LH, Scherf A, Crabb BS, Cowman AF. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell. 2005;121:13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- 12.Lopez-Rubio JJ, Gontijo AM, Nunes MC, Issar N, Hernandez Rivas R, Scherf A. 5′ flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol. Microbiol. 2007;66:1296–1305. doi: 10.1111/j.1365-2958.2007.06009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chookajorn T, Dzikowski R, Frank M, Li F, Jiwani AZ, Hartl DL, Deitsch KW. Epigenetic memory at malaria virulence genes. Proc. Natl Acad. Sci. USA. 2007;104:899–902. doi: 10.1073/pnas.0609084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui L, Miao J, Furuya T, Li X, Su XZ. PfGCN5-mediated histone H3 acetylation plays a key role in gene expression in Plasmodium falciparum. Eukaryot. Cell. 2007;6:1219–1227. doi: 10.1128/EC.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui L, Fan Q, Miao J. Histone lysine methyltransferases and demethylases in Plasmodium falciparum. Int. J. Parasitol. 2008;38:1083–1097. doi: 10.1016/j.ijpara.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miao J, Fan Q, Cui L, Li J. The malaria parasite Plasmodium falciparum histones: organization, expression, and acetylation. Gene. 2006;369:53–65. doi: 10.1016/j.gene.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 17.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 18.Scherf A, Hernandez-Rivas R, Buffet P, Bottius E, Benatar C, Pouvelle B, Gysin J, Lanzer M. Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. EMBO J. 1998;17:5418–5426. doi: 10.1093/emboj/17.18.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruvalcaba-Salazar OK, del Carmen Ramirez-Estudillo M, Montiel-Condado D, Recillas-Targa F, Vargas M, Hernandez-Rivas R. Recombinant and native Plasmodium falciparum TATA-binding-protein binds to a specific TATA box element in promoter regions. Mol. Biochem. Parasitol. 2005;140:183–196. doi: 10.1016/j.molbiopara.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Mancio-Silva L, Rojas-Meza AP, Vargas M, Scherf A, Hernandez-Rivas R. Differential association of Orc1 and Sir2 proteins to telomeric domains in Plasmodium falciparum. J. Cell Sci. 2008;121:2046–2053. doi: 10.1242/jcs.026427. [DOI] [PubMed] [Google Scholar]
- 21.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl Acad. Sci. USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Rusell DW. Molecular Cloning a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 23.Hiragami K, Festenstein R. Heterochromatin protein 1: a pervasive controlling influence. Cell Mol. Life Sci. 2005;62:2711–2726. doi: 10.1007/s00018-005-5287-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 25.Brasher SV, Smith BO, Fogh RH, Nietlispach D, Thiru A, Nielsen PR, Broadhurst RW, Ball LJ, Murzina NV, Laue ED. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 2000;19:1587–1597. doi: 10.1093/emboj/19.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 27.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 28.Cowieson NP, Partridge JF, Allshire RC, McLaughlin PJ. Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Curr. Biol. 2000;10:517–525. doi: 10.1016/s0960-9822(00)00467-x. [DOI] [PubMed] [Google Scholar]
- 29.Lomberk G, Wallrath L, Urrutia R. The Heterochromatin Protein 1 family. Genome Biol. 2006;7:228. doi: 10.1186/gb-2006-7-7-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sautel CF, Cannella D, Bastien O, Kieffer S, Delphine A, Garin J, Belrhali Tardieux, I H, Hakimi M-A. SET8-mediated methylations of histone H4 lysine 20 mark silent heterochromatic domains in apicomplexan genomes. Mol. Cell Biol. 2007;27:5711–5724. doi: 10.1128/MCB.00482-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Figueiredo LM, Pirritt LA, Scherf A. Genomic organisation and chromatin structure of Plasmodium falciparum chromosome ends. Mol. Biochem. Parasitol. 2000;106:169–174. doi: 10.1016/s0166-6851(99)00199-1. [DOI] [PubMed] [Google Scholar]
- 32.Jones DO, Mattei MG, Horsley D, Cowell IG, Singh PB. The gene and pseudogenes of Cbx3/mHP1 gamma. DNA Seq. 2001;12:147–160. doi: 10.3109/10425170109080769. [DOI] [PubMed] [Google Scholar]
- 33.Zhao T, Heyduk T, Eissenberg JC. Phosphorylation site mutations in heterochromatin protein 1 (HP1) reduce or eliminate silencing activity. J. Biol. Chem. 2001;276:9512–9518. doi: 10.1074/jbc.M010098200. [DOI] [PubMed] [Google Scholar]
- 34.Lomberk G, Bensi D, Fernandez-Zapico ME, Urrutia R. Evidence for the existence of an HP1-mediated subcode within the histone code. Nat. Cell Biol. 2006;8:407–415. doi: 10.1038/ncb1383. [DOI] [PubMed] [Google Scholar]
- 35.Issar N, Roux E, Mattei D, Scherf A. Identification of a novel post-translational modification in Plasmodium falciparum: Protein SUMOylation in different cellular compartments. Cell Microbiol. 2008;10:1999–2011. doi: 10.1111/j.1462-5822.2008.01183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Armstrong CM, Goldberg DE. An FKBP destabilization domain modulates protein levels in Plasmodium falciparum. Nat. Methods. 2007;4:1007–1009. doi: 10.1038/nmeth1132. [DOI] [PubMed] [Google Scholar]
- 37.Merrick CJ, Duraisingh MT. Plasmodium falciparum Sir2: an unusual sirtuin with dual histone deacetylase and ADP-ribosyltransferase activity. Eukaryot. Cell. 2007;6:2081–2091. doi: 10.1128/EC.00114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blasco MA. The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 2007;8:299–309. doi: 10.1038/nrg2047. [DOI] [PubMed] [Google Scholar]
- 39.Pak DT, Pflumm M, Chesnokov I, Huang DW, Kellum R, Marr J, Romanowski P, Botchan MR. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell. 1997;91:311–323. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- 40.Huang DW, Fanti L, Pak DT, Botchan MR, Pimpinelli S, Kellum R. Distinct cytoplasmic and nuclear fractions of Drosophila heterochromatin protein 1: their phosphorylation levels and associations with origin recognition complex proteins. J. Cell Biol. 1998;142:307–318. doi: 10.1083/jcb.142.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]