Abstract
Select changes in microRNA (miRNA) expression correlate with estrogen receptor α (ERα) expression in breast tumors. miR-21 is higher in ERα positive than negative tumors, but no one has examined how estradiol (E2) regulates miR-21 in breast cancer cells. Here we report that E2 inhibits miR-21 expression in MCF-7 human breast cancer cells. The E2-induced reduction in miR-21 was inhibited by 4-hydroxytamoxifen (4-OHT), ICI 182 780 (Faslodex), and siRNA ERα indicating that the suppression is ERα-mediated. ERα and ERβ agonists PPT and DPN inhibited and 4-OHT increased miR-21 expression. E2 increased luciferase activity from reporters containing the miR-21 recognition elements from the 3′-UTRs of miR-21 target genes, corroborating that E2 represses miR-21 expression resulting in a loss of target gene suppression. The E2-mediated decrease in miR-21 correlated with increased protein expression of endogenous miR-21-targets Pdcd4, PTEN and Bcl-2. siRNA knockdown of ERα blocked the E2-induced increase in Pdcd4, PTEN and Bcl-2. Transfection of MCF-7 cells with antisense (AS) to miR-21 mimicked the E2-induced increase in Pdcd4, PTEN and Bcl-2. These results are the first to demonstrate that E2 represses the expression of an oncogenic miRNA, miR-21, by activating estrogen receptor in MCF-7 cells.
INTRODUCTION
Although the precise sequence of events leading to breast tumors are not understood, lifetime exposure to estrogens is widely accepted as a major risk factor for the development of breast cancer. Estrogens promote cell replication by binding to the estrogen receptors α and β (ERα and ERβ). Ligand-activated ER acts genomically by binding directly to estrogen response elements (EREs) or by a ‘tethering mechanism’, e.g. by interacting with AP-1 (1) or Sp1 (2). These interactions recruit coregulators to initiate chromatin remodeling resulting in increased gene transcription (3). ER can also suppress target gene transcription, although the mechanisms involved are unresolved (4). In addition to its ER-mediated, genomic activity, E2 also has ‘non-genomic’ or ‘membrane-initiated’ effects, i.e. independent of ER-mediated transcription, that occur within minutes after estradiol (E2), or other ER ligand, administration (5,6).
Inhibition of estrogen action is used as the adjuvant therapy of choice to treat both pre- and post-menopausal women with breast cancer. The anti-estrogen/Selective ER Modulator (SERM) tamoxifen (TAM) is the ‘gold standard’ of treatment of women with ER positive tumors (7). TAM is a SERM because it has mixed agonist/antagonist activity in a cell- and gene-specific manner whereas Faslodex (Fulvestrant, ICI 182 780) has pure antiestrogen activity (8). Ablation of endogenous estrogen production using aromatase inhibitors (AIs, e.g. anastrozole, letrozole and exemestane) has an efficacy greater than TAM in preventing disease recurrence in post-menopausal breast cancer patients (9). Together, these data demonstrate the importance of endogenous estrogens in promoting breast cancer recurrence.
MicroRNAs (miRNAs) are a class of naturally occurring, small, non-coding RNA molecules distinct from small interfering RNAs (siRNAs) (10–12). miRNA genes are mostly transcribed by RNA polymerase II, processed by Drosha into short hairpin RNAs that are exported from the nucleus, and processed by Dicer to form mature 21–25 nucleotide miRNAs which are transferred to Argonaute proteins in RISC. miRNAs bind to the 3′-untranslated region (3′ UTR) of target mRNAs and either block the translation of the message or target the mRNA transcript to be degraded (13). miRNAs may also increase translation of select mRNAs in a cell cycle-dependent manner (14).
The human genome contains >700 miRNAs (15) and miRNAs are expressed in a tissue-specific manner (16). Each miRNA targets ∼200 transcripts directly or indirectly (17). Aberrant patterns of miRNA expression have been reported in human breast cancer (16–40). A number of genes involved in breast cancer progression have been identified by in silico analysis to be targets of miRNAs that are deregulated in breast cancer (41) and some, e.g. AIB1 have been experimentally proven (42). We recently reported that miR-21 downregulates the translation of human PDCD4, a tumor suppressor in MCF-7 cells (43). Although miR-21 was identified as an ‘oncomiR’, was the most significantly up-regulated miRNA in breast tumor biopsies (37), and was significantly higher in ERα+ than ERα– breast tumors (40), no one has examined whether E2 or SERMs regulate miR-21 expression in human breast cancer cells.
In this study, we tested the hypothesis that miR-21, an ‘oncomiR’, is regulated by E2 in MCF-7 breast cancer cells. Although E2 increases proliferation of MCF-7 cells, we found that E2 inhibits miR-21 expression. Experiments were performed to test the effect of E2 on targets of miR-21. In silico analysis identified miR-21 seed elements in six target genes and these miRNA recognition elements (MREs) were cloned into the 3′UTR of a Renilla reporter for subsequent transcriptional evaluation and examination of the effect of antisense to miR-21 on Renilla luciferase. Antisense to miR-21 was used to confirm the importance of miR-21-MRE interaction in response to E2. Importantly, the E2-mediated decrease in miR-21 correlated with increased expression of miR-21-targets PDCD4, PTEN and Bcl-2 at the protein level. These results identify miR-21 as an E2-ER- regulated miRNA in MCF-7 cells.
MATERIALS AND METHODS
Cells and treatments
MCF-7 cells were purchased from ATCC and maintained as previously described (44). 17β-estradiol (E2), 4-hydroxytamoxifen (4-OHT), Actinomycin D (ActD, a transcriptional inhibitor) and cycloheximide (CHX, a protein synthesis inhibitor) were purchased from Sigma; ICI 182 780 (ICI), 4,4′,4′′-(4-propyl-[1H]-pyrazole-1,3,5-triyl)trisphenol (PPT, an ERα-selective agonist) and 2,3-bis(4-hydroxyphenyl)-propionitrile (DPN, an ERβ-selective agonist) were purchased from Tocris. Prior to ligand treatment, the medium was replaced with phenol red-free IMEM supplemented with 5% dextran charcoal-stripped FBS (DCC-FBS) for 48 h (serum-starved). Where indicated, MCF-7 cells were pre-treated with 10 μg/ml ActD or 10 μg/ml CHX, for 1 h before ligand treatment. Cells were treated with ethanol (EtOH, the vehicle control) 0.01% final volume, 10 nM E2, 100 nM 4-OHT, 10 nM PPT, or 10 nM DPN, alone or in combination with 100 nM ICI for 6 h. For the indicated experiments, cells were pretreated with 100 nM ICI for 6 h prior to EtOH or E2 treatment.
miRNA microarray
RNA was isolated from MCF-7 cells treated with EtOH or 10 nM E2 for 6 h using the mirVana miRNA Isolation Kit from Ambion (Austin, TX) and was sent to LC Sciences (Houston, TX) (http://lcsciences.com/) where the RNA samples were labeled either with Cy3 or Cy5 and were hybridized with two identical, dual-color miRNA microarray chips (MRA-1001, LC Sciences). The array contains probes to detect mature miRNA sequences as well as pre-miRNAs in the Sanger miRNA registry (http://microrna.sanger.ac.uk/sequences/). Each human miRNA on the chip contains seven redundancies for each sequence to increase sensitivity. Microarray analysis was performed by LCS including background subtraction and data normalization to the statistical median of all detectable transcripts. Two lists of differentially expressed transcripts (based on a P-value < 0.01) from two chips were merged into one list and a statistical correlation between the two sets of data was calculated.
Constructs of miRNA-recognition elements (MREs)
For MRE sequences, synthetic DNA oligonucleotides (∼35 bp) containing the MRE sequence (Supplementary Table 1) and ∼5 bp adjacent sequences from each end were annealed and ligated into the NotI/XhoI sites located in the 3′UTR region of the pRL-TK Renilla luciferase reporter from Promega. Full-length (FL) 3′-UTRs of PDCD4 and RASA1 were amplified by PCR and inserted into the phRL-TK vector, similarly. All constructs were confirmed by DNA sequencing.
Quantitative real-time PCR (Q-PCR) analysis of miRNA and mRNA expression
miRNA-enriched total RNA was extracted from MCF-7 cells using the mirVana miRNA isolation kit (Ambion). Quantification of miRNAs was performed using TaqMan MicroRNA Assays (Applied Biosystems). U6 RNA was used for normalization of miRNA expression. For analysis of PTEN, PDCD4, BCL2 and TMEM49 mRNA expression, RNA was extracted using Trizol and quantitation was performed using TaqMan primers and probes from ABI using 18S for normalization. Analysis and fold change were determined using the comparative threshold cycle (Ct) method. The change in miRNA or mRNA expression was calculated as fold-change, i.e. relative to EtOH-treated (control).
Western blot
Cells were treated as indicated in individual figure and whole cell extracts (WCE) were prepared in modified RIPA buffer as described (22). Western analysis was performed and quantitated as described (19). Membranes were probed with ERα antibodies AER320 from NeoMarkers or HC-20 from Santa Cruz Biotechnology, ERβ antibody H150 (Santa Cruz Biotechnology), polyclonal PDCD4 antibody from Genetex, monoclonal PTEN antibody from Cell Signaling, or monoclonal Bcl-2 antibody from Assay Designs. Membranes were stripped and re-probed for β-actin (Sigma).
Transient transfection
MCF-7 cells were plated in 24-well plates at a density of 1.5 × 104 cells/well in phenol red-free OPTI-MEM I reduced serum medium (GIBCO/Invitrogen) supplemented with 10% DCC-FBS. Transient transfection was performed using FuGene6 (Roche). For experiments in Figures 2 and 3A, each well received 10 ng of pGL3-pro-luciferase reporter (Promega) as a control and 10 ng of pRL-TK, Renilla luciferase reporter (Promega) containing the indicated MRE or 3′-UTR of miR-21 target genes. For some experiments, cells were also co-transfected with 2′-O-Me-anti-miR-21 [antisense (AS)-miR-21] and the control used was the negative control #1 from Ambion: a random-sequence 2′-O-Me modified RNA molecule that has been extensively tested in many human cell lines and tissues and validated to not produce any identifiable effect on known miRNA function (23). For Figure 3A, MCF-7 cells were transfected with 250 ng of pmiR-21s-luc or pmiR-21as-luc reporters described in (45) and 5 ng pRL-TK (control). Twenty-four hours after transfection, triplicate wells were treated with EtOH (vehicle control), E2, 4-OHT or ICI 182 780 as indicated in the figure legend. The cells were harvested 30 h post-treatment using Promega's Passive Lysis buffer. Luciferase and Renilla luciferase activities were determined using Promega's Dual Luciferase assay. For Figure 2, Renilla luciferase was normalized by Firefly luciferase to correct for transfection efficiency. For Figure 3A, Firefly luciferase was normalized to Renilla luciferase. Fold induction was determined by dividing the averaged normalized values from each treatment by the EtOH value for each transfection condition within that experiment. Values were averaged from multiple experiments as indicated in the figure legends.
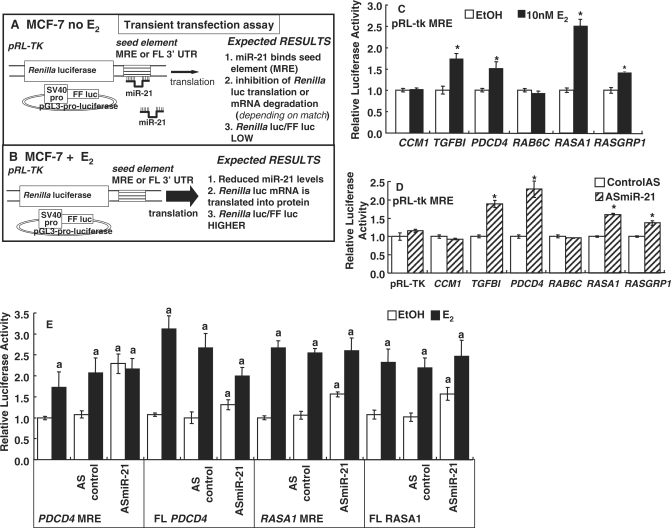
Figure 2.
Luciferase reporter assay of putative miR-21 target genes and the effect of antisense (AS) to miR-21 on reporter activity. (A) Model of transient transfection assays in MCF-7 cells. MCF-7 cells were transiently transfected with pGL3-pro-luciferase and pRL-TK parental or pRL-TK containing putative miR-21 MREs from target genes (Supplementary Table 1) cloned in the 3′UTR as described in ‘Materials and methods’ section. Expected results are indicated without E2 (A) and when cells are treated with E2 (B). (C) MCF-7 cells were transfected as indicated and treated with EtOH or 10 nM E2 for 24 h. Renilla luciferase was normalized by firefly luciferase to correct for transfection efficiency. Values are the average ± SEM of triplicate determinations. *Significantly different from EtOH control, P < 0.01. (D) MCF-7 cells were transfected with 2′-O-Me-antisense-miR-21 (ASmiR-21). Renilla luciferase reporter gene expression from the indicated gene MREs was determined and data analyzed as described in ‘Materials and Methods’ section. The control was a random-sequence 2′-O-Me modified RNA (control AS) as described in Materials and methods section. Values are the average ± SEM of triplicate determinations. *Significantly different from control AS, P < 0.05. (E) MCF-7 cells were transfected with the pRL-tk-MREs or FL 3′-UTRs as indicated. Indicated cells were co-transfected with ASmiR-21 or a control AS. Cells were treated with EtOH or 10 nM E2 as indicated for 24 h. Dual luciferase reporter assays were performed and data quantitated as described in ‘Materials and Methods’ section. Values are the average ± SEM of triplicate determinations normalized to EtOH for each construct except that cells transfected with the ASmiR-21 were normalized against the control AS-EtOH value. aSignificantly different from EtOH control, P < 0.01. bSignificantly different from control AS transfected values, P < 0.01.
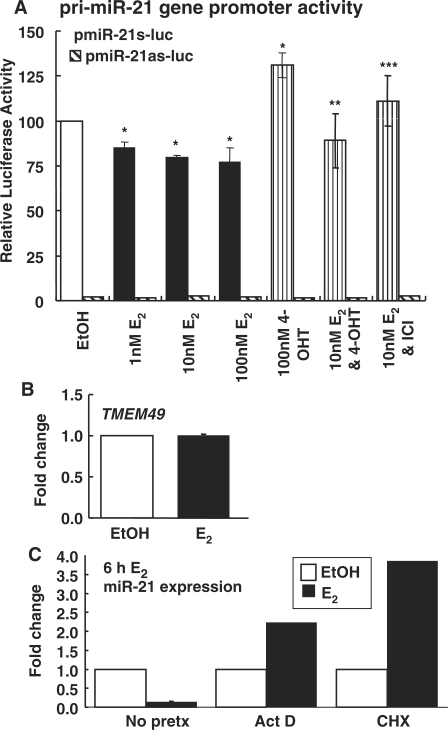
Figure 3.
Regulation of miR-21 transcription in MCF-7 cells. (A) Effects of E2, 4-OHT and ICI 182 780 (ICI) on the primary miR-21 (pri-miR-21) gene promoter in the sense (pmiR-21s-luc) or antisense (as) pmiR-21as-luc orientation. MCF-7 cells were transfected with pri-miR-21s-luc or pri-miR-21as-luc (hatched bars, values were very low) (45) and Renilla luciferase as an internal control. Cells were treated with the indicated concentrations of E2, 4-OHT, or ICI for 24 h. Dual luciferase assays were performed and luciferase values were divided by Renilla values in the same sample. Values are the average ± SEM of triplicate determinations normalized to EtOH for the pmiR-21s-luc construct. *Significantly different from EtOH control, P < 0.05. **Significantly different from 4-OHT alone, P < 0.05. ***Significantly different from 10 nM E2, P < 0.05. (B) E2 does not affect TMEM49 transcription in MCF-7 cells. miR-21 is encoded within the 10th intron of the TMEM49 gene (56). MCF-7 cells were treated with EtOH or 10 nM E2 for 6 h, total RNA was reverse transcribed and Q–PCR was performed. TMEM49 was normalized to 18S. Values are the average ± SEM of triplicate determinations normalized to EtOH. (C) The E2-induced decrease in miR-21 expression in MCF-7 cells is mediated in a primary transcriptional/genomic and secondary estrogen-target-dependent manner. MCF-7 cells were pre-treated with stripped medium or stripped medium containing 10 μg/ml ActD or CHX for 1 h before treatment with vehicle control (EtOH), or 10 nM E2 for 6 h as described in ‘Materials and Methods’ section. miR-21 expression was determined using Q-PCR as described in ‘Materials and Methods’ section. The bar graph summarizes the fold change in miR-21 expression relative to no pretreatment (No pretx)-EtOH-treated cells.
AS-control and AS-miR-21 transfection
MCF-7 cells were transfected with AS- duplexes and control-nonspecific siRNA obtained from Ambion using Lipofectamine RNAiMAX from Invitrogen according to the manufacturer's protocol. Twenty-four hours post-transfection, the medium was replaced with phenol red-free IMEM with 5% DCC for 48 h and the cells were treated with ethanol (EtOH) vehicle control, 10 nM E2, 10 nM PPT or 10 nM DPN for 24 h prior. Total RNA was isolated for Q-PCR analysis and WCEs were prepared and stored for 24 h at −80°C until western blot analysis. Each experiment was repeated for a total of three biological replicates. Western blots were quantified as above and the ratio of each protein/β-actin in the AS-control in EtOH-treated samples was set to 1 in each experiment.
ERα and ERβ knockdown by siRNA
MCF-7 cells were transfected with siRNA duplexes and control-nonspecific siRNA obtained from New England Biolabs (44). Forty-eight hours post-transfection, the cells were treated with 10 nM E2, 10 nM PPT or 10 nM DPN for 6 h for mRNA analysis, or 24 h for protein analysis. Total RNA was isolated for Q-PCR analysis and WCEs were prepared and stored for 24 h at −80°C until western blot analysis.
Statistics
Statistical analyses were performed using Student's t-test or one-way ANOVA followed by Student–Newman–Keuls or Dunnett's post-hoc tests using GraphPad Prism (San Diego, CA).
RESULTS
E2 regulates miR-21 expression in MCF-7 breast cancer cells
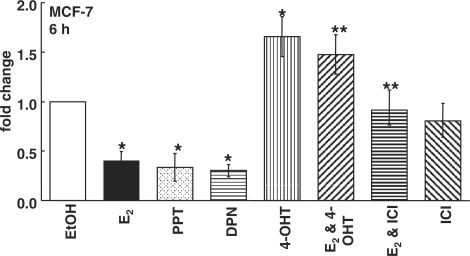
Estrogens promote breast tumor development by increasing transcription of protooncogenes and growth factors (46) and by negatively modulating the expression or functional activity of tumor suppressors (47). To determine the identity of primary E2-regulated miRNAs in estrogen-responsive human breast cancer cells, ERα-positive MCF-7 human breast cancer cells were treated with 10 nM E2 or EtOH (vehicle control) for 6 h. Among the E2-down-regulated miRNAs, we selected miR-21 for further evaluation because miR-21 is an oncomiR and its expression is higher in ERα positive versus negative tumors (40). Furthermore, no one has examined if E2 regulates miR-21 expression in breast cancer cells. Q-PCR using the TaqMan primer/probe sets from ABI indicated a ∼60% reduction in mature miR-21 by E2 (Figure 1). To determine the mechanism by which E2 reduces miR-21, MCF-7 cells were pre-incubated with 100 nM ICI 182 780 (ICI, Faslodex), a pure antagonist of ER genomic action (48,49), or 100 nM 4-OHT, the active metabolite of the antiestrogen tamoxifen, and then treated with E2. The effect of 4-OHT or ICI alone was also examined. If E2 represses miR-21 expression by binding ER, then ICI should block the decrease. Because 4-OHT has mixed ER agonist/antagonist activity in a gene- and cell-specific manner, its effect on miR-21 expression could either mimic or oppose the E2 effect, reflecting its selective ER modulator (SERM) agonist/antagonist activity. ICI reduced ERα protein by ∼30–50% in MCF-7 cells (Supplementary Figure 1), but had no effect on basal miR-21 expression (Figure 1). 4-OHT increased miR-21, indicating that 4-OHT opposes E2-induced miR-21 repression through ER binding. Since both 4-OHT and ICI relieved E2 suppression of miR-21, this reduction is ER-mediated.
Figure 1.
E2 inhibits miR-21 expression. Summary of Q-PCR data on (mature) miR-21 expression. MCF-7 cells were treated with EtOH, 10 nM E2, 10 nM PPT (ERα-selective), or 10 nM DPN (ERβ-selective) for 6 h. as indicated by the different fills. Where indicated MCF-7 cells were pretreated with 100 nM ICI 182 780 [ICI, an ER antagonist termed a ‘selective ER disrupter’ (SERD)] or 100 nM 4-OHT for 6 h and then ethanol or 10 nM E2 was added for an additional 6 h. Values are fold increase compared to EtOH for each miRNA and were calculated as described in ‘Materials and Methods’ section. Values are the average of three to eight separate experiments ± SEM. *Significantly different from the EtOH control, P < 0.05. **Significantly different from E2, P < 0.05.
Although ERα expression is higher than ERβ in MCF-7 cells, both ER subtypes are expressed (44). To examine the contributions of ERα and ERβ to the E2-induced reduction in miR-21, MCF-7 cells were treated with 10 nM PPT or 10 nM DPN, concentrations at which each is an ERα- or ERβ- selective agonist, respectively (50). PPT and DPN, like E2, reduced miR-21 (Figure 1). E2 did not regulate miR-21 expression in ERα+/ERβ+ T47D cells (Supplementary Figure 2), indicating cell-line-specific differences, similar to previous reports that E2 responses differ between MCF-7 and T47D cells (51–54). Together, these data indicate that both ERα and ERβ contribute to miR-21 repression by E2.
Effect of E2 on miR-21 target gene reporter activity in MCF-7 cells
The biological activity of miRNAs is primarily mediated by interaction with matching recognition sequences in the 3′ UTRs of target genes and reducing translation. A ∼33-bp region from the 3′UTR centering on the putative miR-21 miRNA regulatory element (miRNA recognition elements (MREs), also called a ‘seed element’, 5′-ATAAGCTA-3′), and minimally 4 bp flanking this sequence from the six genes listed in Supplementary Table 1 were cloned into the 3′UTR of pRL-TK Renilla reporter plasmid. The pRL-TK-MRE or pRL-TK parental plasmids were transiently transfected into MCF-7 cells with pGL3-pro-luciferase as a control and cells were treated with EtOH or E2 (Figure 2A and B). If E2 reduces miR-21, we would expect an increase in the expression of Renilla but not Firefly luciferase activity since repression would be relieved. Figure 2C shows that E2 specifically increased the expression of the Renilla luciferase protein from the pRL-TK- transforming growth factor β 1 (TGFB1), Programmed Cell Death 4 (PDCD4), RAS p21 Protein Activator 1 (RASA1) and RAS Guanyl Nucleotide-Releasing Protein 1 (RASGRP1) reporters in MCF-7 cells, data consistent with miR-21 downregulation by E2. In contrast, E2 did not alter luciferase expression from the putative miR-21 MREs in Cerebral Cavernous Malformations 1 (CCM1) or a member of the RAS oncogene family (RAB6C). Thus, the E2-mediated decrease in miR-21 expression (Figure 1) resulted in lower amounts of miR-21 available to bind the MRE sequences from the TGFB1, PDCD4, RASA1 and RASGRP1 genes, in turn reducing the targeting of these reporter transcripts for degradation/translational inhibition and thus increasing the amount of Renilla protein and luciferase activity. In contrast, the lack of change in Renilla activity from CCM1 and RAB6C indicates that the MREs in these genes do not appear to be targets of E2-induced reduction of miR-21 expression in MCF-7 cells under our assay conditions.
Effect of antisense to miR-21 target gene reporter activity in MCF-7 cells
If the E2-induced increase in Renilla luciferase from the MREs of the TGFB1, PDCD4, RASA1 and RASGRP1 genes seen in Figure 2C is due to reduced levels of endogenous miR-21, then transfection of MCF-7 cells with antisense (AS)-miR-21 should have the same effect on luciferase activity. MCF-7 cells were transiently transfected with 2′-O-Me-anti-miR-21 (AS-miR-21) (Figure 2D). A 92% knockdown of miR-21 expression was achieved (Figure 5A). AS-miR-21 resulted in a significant increase in Renilla activity from pRL-TK reporters bearing the miR-21 MREs from the TGFB1, PDCD4, RASA1 and RASGRP1 genes. In contrast, AS-miR-21 did not affect luciferase activity from the putative miR-21 MREs in CCM1 or RAB6C (Figure 2D). These data are in agreement with the E2 responses (Figure 2C), although E2 induced higher activity from the RASA1 reporter compared to the ASmiR-21. Overall, these data indicate that these MREs are bone fide targets of miR-21 regulation.
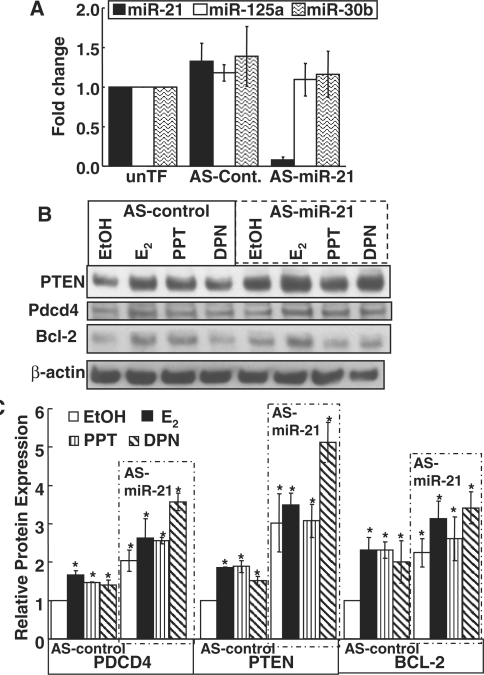
Figure 5.
AS-miR-21 increases the expression of PTEN, PDCD4 and Bcl-2. (A) The specificity of AS-miR-21 to decrease miR-21was examined by Q-PCR in parallel with miR-125a and miR-30b as negative controls. MCF-7 cells were not transfected (unTF) or were transfected with AS-control or AS-miR-21 for 48 h prior to RNA harvest. Q-PCR was performed for the indicated miRs. The values are the average of three separate experiments, each run in triplicate, ± SD. (B) MCF-7 cells were transfected with AS-control or AS-miR-21 for 24 h prior to serum deprivation for 48 h and then 24 h treatment with EtOH, 10 nM E2, PPT or DPN, as indicated. WCE were used for western blot for the indicated proteins as described in ‘Materials and methods’ section. The membrane was stripped and reprobed for β-actin for normalization as described in Materials and Methods section. The blot shown is representative of three separate biological replicates. (C) The values graphed are the mean ± SEM of the normalized western data (each protein was normalized to β-actin input and then the ratio of each protein/β-actin in the AS-control in EtOH-treated samples was set to one in each experiment) in three separate experiments. *Significantly different from the EtOH AS-control for each protein, P < 0.05.
MRE and FL 3′-UTRs activities of PDCD4 and RASA1 in reporter assays in MCF-7 cells
Since sequences flanking the MRE affect miRNA binding and activity (55), it is important to compare the effect of E2 and AS-miR-21 in reporters bearing the MRE versus the FL 3′UTR of PDCD4 and RASA1 genes (Figure 2E). E2 induced greater luciferase activity from the FL than the PDCD4 MRE. AS-miR-21 increased reporter activity more from the MRE than the FL PDCD4. The AS-miR-21-induced increase in basal luciferase activity was comparable for the MRE and FL RASA1 reporters. AS-mR-21 transfection reduced the fold E2-induction for the MRE and FL PDCD4 and RASA1 reporters. The miR-21 knockdown data are consistent with E2-ER downregulation of miR-21 increasing reporter activity.
Regulation of primary (pri)-miR-21 promoter activity by E2, 4-OHT and ICI 182,780 in MCF-7 cells
miR-21 is located in the 10th intron of the TMEM49 gene (56). To test whether E2 regulates miR-21 gene expression through the ∼−1 kb 5′flanking region previously reported to function as a promoter for miR-21 (45), transient transfection assays were performed using two constructs: pmiR-21s-luc and pmiR-21as-luc, corresponding to the sense (s) and antisense (as) orientations of this ∼1 kb region cloned in front of the Firefly luciferase gene (45) (Figure 3A). The activity from the pmiR-21as-luc reporter was ∼2% of that of the pmiR-21s-luc construct, indicating orientation-dependent promoter activity. If E2 represses miR-21 expression by an interaction of ER with the 5′ promoter, we should detect a decrease in luciferase reporter activity. E2 reduced luciferase activity ∼25% whereas 4-OHT increased pmiR-21 activity by ∼25% (Figure 3A). ICI abrogated the inhibition by E2, indicating that ER is responsible for reduction in reporter activity. E2 did not alter TMEM49 transcription (Figure 3B). To our knowledge, this is the first examination of the effect of E2 on TMEM49 transcription. These data are consistent with the independent regulation of TMEM49 and miR-21 in HL-60 cells (56). Overall, these data agree with the direction, although not magnitude, of changes in endogenous miR-21 expression in response to E2, 4-OHT and ICI in MCF-7 cells (Figure 1) and indicate that the −1 kb promoter of miR-21 mediates in part, the observed reduction in miR-21 expression by E2.
Actinomycin D (ActD) and cycloheximide (CHX) block E2-mediated miR-21 expression
To determine whether the E2-mediated reduction in miR-21 expression is a direct effect of ER at the genomic level or requires synthesis of a secondary estrogen-responsive protein, MCF-7 cells were pretreated with the transcriptional inhibitor ActD or the protein synthesis inhibitor CHX prior to EtOH or E2 treatment (Figure 3C). Pretreatment with ActD and CHX blocked E2-mediated miR-21 repression, indicating that E2-repression is mediated by both transcriptional (primary genomic) and secondary mechanisms.
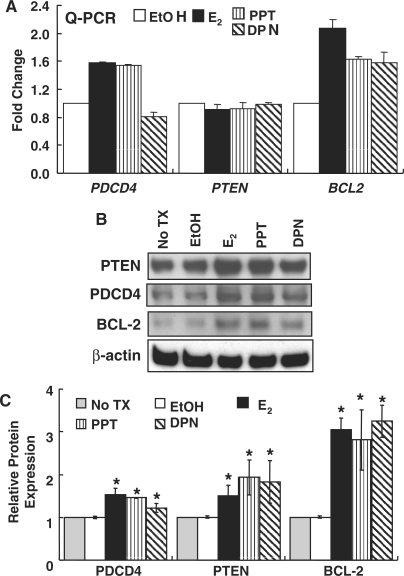
Effect of E2, PPT and DPN on endogenous miR-21 target genes in MCF-7 cells
Since E2 reduced miR-21 expression in MCF-7 cells and increased the expression of miR-21 target reporter gene activity, the effect of E2 on the mRNA and protein levels of endogenous miR-21-target genes PDCD4, PTEN and BCL2 was examined by Q-PCR (Figure 4A) and western blot (Figure 4B and C). To determine the relative contribution of the two ER subtypes to these effects, MCF-7 cells were treated with 10 nM PPT or 10 nM DPN, concentrations at which each is an ERα- or ERβ-selective agonist, respectively (50). As expected based on the reporter assay data for PDCD4 in Figure 2, E2 increased mRNA (Figure 4A) and protein (Figure 4B and C) levels of PDCD4, results reflecting reduced miR-21 levels (Figure 1), thus increased transcript stability. Similar results were observed for BCL2. PPT also increased PDCD4 and BCL2 mRNA and protein levels, whereas DPN reduced PDCD4 and increased BCL2 mRNA levels (Figure 4A) while increasing protein amounts (Figure 4B). E2, PPT and DPN increased PTEN protein but not RNA levels (Figure 4A and C), suggesting translational inhibition. Overall, these data indicate roles for both ERα and ERβ in mediating the effects of E2 on miR-21 target gene expression, consistent with results shown in Figure 1.
Figure 4.
Effect of ER ligands on endogenous miR21 target gene mRNA and protein expression in MCF-7 cells. MCF-7 cells were serum-starved for 48 h and then treated with EtOH, 10 nM E2, 10 nM PPT (ERα selective), or 10 nM DPN (ERβ selective) for 6 h prior to RNA isolation (A) or 24 h prior to WCE preparation (B) as described in ‘Materials and methods’ section. (A) Q-PCR was performed for the indicated genes and fold-expression determined compared to EtOH as described in ‘Materials and Methods’ section. Values are the average of four separate determinations ± SEM. (B) Western blot for the indicated proteins. The membrane was stripped and reprobed for β-actin for normalization as described in ‘Materials and Methods’ section. The blot shown is representative of three separate biological replicates. (C) Western data are presented as relative to non-treated (No TX) MCF-7 cells. The values in C are the mean ± SEM of three separate experiments. *Significantly different from the EtOH value for each protein, P < 0.01.
AS-miR-21 inhibits endogenous miR-21 target gene protein expression in MCF-7 cells
To confirm the role of downregulation of miR-21 in the increase in protein expression of Pdcd4, PTEN and Bcl-2, MCF-7 cells were transfected with AS-control and AS-miR-21 plasmids followed by treatment with EtOH, E2, PPT and DPN for 24 h. If the ER-ligand-induced reduction in miR-21 causes an increase in target protein expression, then the AS-miR-21 should have the same effect. AS-miR-21 reduced miR-21 by 92% (Figure 5A). Specific knockdown of miR-21, and not miR-125a or miR-30b, was confirmed by Q–PCR (Figure 5A). AS-miR-21 significantly increased the basal Pdcd4, PTEN and Bcl-2 protein expression (Figure 5B and C). AS-control did not affect the observed increase in each protein in response to E2, PPT and DPN (compare Figures 4B, C and 5B, C). These data indicate that these genes are targets of repression by miR-21. No further increase in protein expression was detected with E2 or PPT treatment, but DPN significantly increased Pdcd4 and PTEN proteins (Figure 5C).
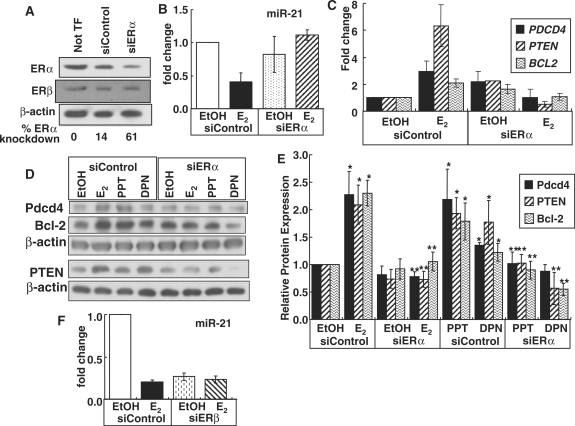
Effect of ERα knock-down on E2-induced endogenous miR-21 target gene expression in MCF-7 cells
To confirm the role of ERα in the observed decrease in miR-21 and increase in miR-21-target gene expression in response to E2 and PPT, MCF-7 cells were transfected with siRNA targeting ERα or control siRNA for 48 h and then treated with EtOH, 10 nM E2, PPT, or DPN for 6 h. Transfection of MCF-7 cells with siRNA for ERα reduced ERα mRNA expression by ∼62% (Supplementary Figure 3) and ERα protein by 61%. In contrast, ERβ protein levels were unaffected (Figure 6A, see also Supplementary Figure 4). siERα blocked the E2-induced repression of miR-21 (Figure 6B). Concordantly, knockdown of ERα reduced the E2-stimulated expression of miR-21 target genes PDCD4, PTEN and BCL2 (Figure 6C). To confirm these findings at the protein level, western blots were performed using antibodies commercially available for Pdcd4, PTEN and Bcl-2 (Figure 6D). Results confirm that ERα knockdown reduced the E2- and PPT-induced protein expression of the miR-21 target genes PDCD4, PTEN and BCL2 to basal levels (Figure 6E). siERα also reduced DPN-stimulated expression of Pdcd4, PTEN and Bcl-2 proteins suggesting that at least part of the DPN response may be ERα-mediated.
Figure 6.
ERα, but not ERβ, knockdown inhibits the E2-mediated decrease in miR-21 and thus reverses miR-21 target gene expression. (A) MCF-7 cells were not transfected (Not TF) or transfected with siControl RNA or siERα as described in ‘Materials and Methods’ section for 48 h and WCE were analyzed for ERα and ERβ by western blot as described in ‘Materials and Methods’ section. The same membrane was stripped and reprobed for β-actin for normalization. The% ERα knockdown was calculated relative to the Not TF control. (B) MCF-7 cells were transfected with siControl RNA or siERα for 48 h prior to treatment with EtOH or 10 nM E2, PPT, or DPN for 6 h. RNA and protein were extracted and Q-PCR (B and C) or western blots (D and E) were performed for the indicated miR-21 targets as described in ‘Materials and Methods’ section. The blots shown are representative of three separate biological replicates. The values in (E) are the mean ± SEM of three to four separate experiments. (F) MCF-7 cells were transfected with siControl RNA or siERβ for 48 h prior to treatment with EtOH or 10 nM E2 for 6 h. MiR-21 expression is the mean fold change ± SEM of four samples. Values are mean ± SEM. *Significantly different from the EtOH siControl for each protein, P < 0.05. **Significantly different from E2, PPT or DPN siControl value for that protein, P < 0.05.
Effect of ERβ knock-down on miR-21 expression in MCF-7 cells
To examine ERβ's role in mediating E2-suppression of miR-21 transcription, MCF-7 cells were transfected with siRNA targeting ERβ or control siRNA for 48 h and then treated with EtOH or 10 nM E2 for 6 h. siERβ reduced ERβ mRNA expression by ∼70% and protein by 64% (Supplementary Figure 5A and B). Knockdown of ERβ reduced basal miR-21 by 73% and E2 treatment had no further effect (Figure 6F). siERβ resulted in a commensurate increase in basal PDCD4, PTEN and BCL2 mRNA and a loss of E2, DPN and PPT-stimulated PDCD4 and BCL2 transcription (Supplementary Figure 5C and Figure 4). With ERβ knockdown, PPT and DPN increased PTEN mRNA (Supplementary Figure 5C).
DISCUSSION
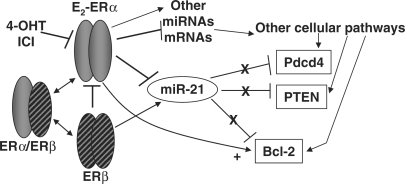
Since the oncomiR miR-21 was the most significantly up-regulated miRNA in breast tumor biopsies compared to normal breast tissue (37) and because estrogen stimulates breast tumorigenesis, the goal of this study was to determine if E2 regulates the expression of miR-21 in MCF-7 cells as an established human breast cancer model of estrogen responses. To our knowledge, this is the first report that E2 downregulates miR-21 and thus upregulates the protein expression of miR-21 target genes PDCD4, PTEN and BCL2 in MCF-7 human breast cancer cells. Furthermore, the ability of 4-OHT, ICI and siERα to block the E2 repression of miR-21 and the subsequent increase in Pdcd4, Pten and Bcl-2 proteins provide a mechanism for the E2 effect, i.e. through ERα activation. ERβ appears to regulate basal miR-21 expression in MCF-7 cells since knockdown of ERβ reduced miR-21 expression. ERβ represses/opposes ERα transcriptional activity and E2-induced cell proliferation (57–61). Stable transfection of MCF-7 cells with ERβ inhibited xenograft tumor growth, indicating that ERβ is a tumor suppressor (62). We observed that ERβ knock down reduced basal miR-21 and there was no further reduction in miR-21 expression with E2 treatment. These data appear to indicate a relief of repression of ERα's inhibition of miR-21 transcription. Figure 7 shows a schematic model illustrating ER regulation of miR-21 and miR-21 regulation of its targets. Our results showing that E2 reduces miR-21 expression in MCF-7 are in agreement with recent reports that E2 down-regulated miR-21 in endometrial stromal cells (63) and in the uterus of ovariectomized mice (64).
Figure 7.
ER regulates miR-21 expression and its downstream targets in a ligand-dependent manner. E2-ER (ERα and/or ERβ) inhibits miR-21 expression resulting in a loss of repression (indicated by the Xs) of Pdcd4, PTEN and Bcl-2 protein expression. E2-ERα directly increases BCL2 transcription (arrow, +). 4-OHT and ICI block ER-induced inhibition of miR-21 expression. E2-ER also regulates the expression of other miRNAs and mRNAs that, in turn, regulate other cellular pathways which impact the expression of PDCD4, PTEN and BCL2.
At the same time, given the established link between estrogen and breast carcinogenesis (65,66), one might expect E2 to upregulate miR-21 rather than inhibit miR-21 as shown here. Likewise, the increase in miR-21 expression by 4-OHT appears to contradict its anticipated anti-tumor role, but is consistent with 4-OHT's gene-specific SERM activity as indicated by its activity opposing E2's inhibition of miR-21 expression. For complex phenotypes including cell proliferation, genes and proteins are up- and down- regulated by a variety of interacting mechanisms that we are only beginning to understand and integrate. Our data are supported by a recent report showing that miR-21 expression was reduced in TAM-resistant MCF-7 cells (67), a finding likely reflecting the loss of ER-regulated responses in TAM-resistant cells. It is well-established that E2 and 4-OHT regulate transcription in a gene- and cell-specific manner (68–72) and the findings reported here add miR-21 to the list of ER-regulated genes. We conclude that our apparent ‘contradictory data’ of E2 down-regulating and 4-OHT increasing miR-21 expression add unexpected complexity to understanding of E2 action in breast tumorigenesis.
The reduction of miR-21 expression in response to E2 appears to be mediated, in part, by the −1kb promoter. However, because the reduction in transcription was only ∼25% in the reporter assay compared to a ∼80% reduction by Q-PCR analysis of miR-21 expression, it is possible that additional regions are also important in regulating miR-21 expression in response to E2. It has been established that E2 increases ERα binding to chromosome regions outside gene promoters (73,74). Analysis of the miR-21 promoter using TRANSFAC (http://www.gene-regulation.com/) identified a non-consensus ERE with a 2-bp spacer: 5′-AGCTGAgcTGACC-3′ located 883-bp upstream of the TATA-binding site. Previous studies showed no binding of ERα to an ERE with a 2-bp spacer in vitro (75). However, in addition to direct ERE binding, ERα regulates gene transcription by tethering to other transcription factors. Genes repressed by E2-ERα in MCF-7 cells lack EREs and instead have binding sites for Ikaros (IKZF1) and PAX homeobox factors, among others (76), that are also located in the miR-21 promoter. miR-21 is located in the 3′UTR of TMEM49 located at 17q23.1. Using data from Myles Brown's online database of genomic E2-ERα-binding sites in MCF-7 cells from chromatin immunoprecipitation of ERα on-human genome tiled microarray data (ChIP-on-chip) for human chromosome 17 (73) http://research.dfci.harvard.edu/brownlab/datasets/index.php?dir=ER_MCF7_whole_human_genome/, we found that both E2-ERα and RNA polymerase II binding overlap with the 71-bp miR-21 gene (Supplementary Figure 6). AP-1 was shown to activate miR-21 transcription by direct interaction with three binding sites in the miR-21 promoter in response to PMA treatment of HL-60 cells (56). Although both ERα and ERβ interact with AP-1 to regulate gene expression, the direction of regulation (up or down) varies depending on the ligand, cell type, chromatin context and neighboring transcription factor-binding events (77,78). Here we showed that E2 did not alter TMEM49 transcription which supports previous results that TMEM49 and miR-21 are independently regulated (56). Further studies will be required to analyze the precise mechanisms mediating E2 repression of miR-21.
Both E2 and AS-miR-21 induced RASA1 reporter activity; however, the magnitude of luciferase induction was higher with E2 than AS-miR-21. Although normalized relative luciferase between EtOH versus controlAS transfected cells is an unequal comparison, one possible explanation for this difference is that E2 alters the expression of other genes or pathways that selectively impact the RASA1 reporter compared to the other reporters, e.g. TGFB1 and PDCD4, that show similar luciferase activity.
Our data showing the downregulation of miR-21 by E2 correlated with upregulation of PDCD4 RNA and protein (Figure 4B and C) are in agreement with a report that blocking miR-21 using locked nucleic-acid-modified oligonucleotides increased PDCD4 mRNA and protein in MCF-7 cells (79). Furthermore, our results in the transient transfection assays indicate that miR-21 regulates PDCD4 by an MRE in the 3′UTR. The conclusion that E2-increases PDCD4 expression through inhibition of miR-21 expression in MCF-7 cells is further supported by data showing that AS-miR-21 inhibited E2-induced Renilla luciferase activity from the PDCD4 MRE and 3′-UTR in transfected MCF-7 (Figure 2B) and that AS-miR-21 mimics E2-induction of Pdcd4 protein (Figure 5C). Our ERα knockdown experiments indicate that ERα is responsible for the E2-mediated inhibition of miR-21 expression and regulation of PDCD4 as well as other miR-21 target genes. The DPN- induced reduction in PDCD4 mRNA aligns with a report that DPN-activated ERβ inhibits the transcription of PPT-activated ERα target genes in human breast cancer cells (57). The increase seen in Pdcd4 protein after 24 h of DPN treatment may result from a secondary gene effect.
miR-21 functions as an oncogene and modulates tumorigenicity through regulation of Bcl-2 in MCF-7 cells (38). Inhibition of miR-21 expression by AS-miR-21 reduced Bcl-2 protein expression and increased apoptosis in MCF-7 cells in vitro and in tumor xenografts in mice (38). Consistent with these findings, our data demonstrate that both E2 and PPT decrease miR-21 and increase BCL2 mRNA and protein expression in MCF-7 cells. BCL2 expression has long been considered a good prognostic marker in breast cancer (80). DPN increased BCL2 mRNA and protein expression; likely by ERα activation because E2 regulates BCL2 transcription in MCF-7 cells via ERα- Sp1 and AP1 interactions (81), we can not conclude that the increase in BCL2 mRNA is due solely to E2-mediated decreased miR-21. Further studies will be needed to dissect the relative contributions of multiple ERα-mediated pathways controlling BCL2 gene expression.
PTEN is an important tumor suppressor (82) that has been identified as a breast cancer susceptibility gene (83). miR-21 regulates PTEN in human hepatocellular cancer cells and tumors (35,84) but to our knowledge, no one has examined miR-21 regulation of PTEN in breast cancer. We found that E2, PPT and DPN increased PTEN protein levels without affecting PTEN transcript levels (Figure 4), indicating translational inhibition. Knockdown of ERα by siRNA blocked the E2-mediated downregulation of miR-21 and the E2-induced increase in PTEN, indicating that this effect is mediated via ERα, and commensurate with downregulation of miR-21. With ERβ knockdown, PPT and DPN increased PTEN mRNA; however, because E2, PPT and DPN did not regulate PTEN mRNA in MCF-7 cells, it is likely that this increase is mediated by the loss of the expression of another PTEN transcriptional repressor with ERβ knockdown. Our data contradict a previous report showing no alteration of PTEN expression in MCF-7 cells treated with 100 nM E2 for 24 h (85). This difference may be due to the lower, physiologically relevant E2 concentration and shorter treatment time used here.
In summary, we report for the first time that miR-21 is down-regulated in response to E2 in an ERα-dependent manner and that ERβ regulates basal miR-21 expression. Furthermore, this inhibition correlates with up-regulation of miR-21 targets: PDCD4, PTEN and Bcl-2. The identification of miR-21 as a miRNA regulated by ER may open new avenues for potential therapeutic intervention in breast cancer treatment.
FUNDING
National Institutes of Health R21 CA124811 and an Intramural Research Incentive Grant from the Office of the Senior Vice President for Research [to C.M.K.]. Pre-doctoral fellowship from National Institutes of Environmental Health Sciences T32 ES011564 [to K.A.R.]. Funding for open access charge: National Institutes of Health R21 CA124811 to C.M.K.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Bryan R. Cullen for providing the pri-miR-21 promoter luciferase reporter constructs used in this study. We thank Jeremy S. Harbour and Abirami Krishnasamy for helping with western blots. We thank Drs Barbara J. Clark and Nancy C. Martin for their comments to improve this manuscript. Thanks to Drs Myles Brown and Mathieu Lupien from Harvard University and Dr Ted Kalbflesich (UofL) for their help with the genome analysis for Supplementary Figure 6.
REFERENCES
- 1.Kushner PJ, Agard D, Feng WJ, Lopez G, Schiau A, Uht R, Webb P, Greene G. Oestrogen receptor function at classical and alternative response elements. Novartis Found Symp. 2000;230:20–26. doi: 10.1002/0470870818.ch3. discussion 27–40. [DOI] [PubMed] [Google Scholar]
- 2.Xie W, Duan R, Chen I, Samudio I, Safe S. Transcriptional activation of thymidylate synthase by 17beta-estradiol in MCF-7 human breast cancer cells. Endocrinology. 2000;141:2439–2449. doi: 10.1210/endo.141.7.7538. [DOI] [PubMed] [Google Scholar]
- 3.McKenna NJ, Lanz RB, O'Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 4.Cvoro A, Tzagarakis-Foster C, Tatomer D, Paruthiyil S, Fox MS, Leitman DC. Distinct roles of unliganded and liganded estrogen receptors in transcriptional repression. Mol. Cell. 2006;21:555–564. doi: 10.1016/j.molcel.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 5.Levin ER. Integration of the extranuclear and nuclear actions of estrogen. Mol. Endocrinol. 2005;19:1951–1959. doi: 10.1210/me.2004-0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watson CS, Alyea RA, Jeng YJ, Kochukov MY. Nongenomic actions of low concentration estrogens and xenoestrogens on multiple tissues. Mol. Cell Endocrinol. 2007;274:1–7. doi: 10.1016/j.mce.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher B, Costantino JP, Wickerham DL, Redmond CK, Kavanah M, Cronin WM, Vogel V, Robidoux A, Dimitrov N, Atkins J, et al. Tamoxifen for prevention of breast cancer: report of the national surgical adjuvant breast and bowel project P-1 study. J. Natl Cancer Inst. 1998;90:1371–1388. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- 8.Lewis-Wambi JS, Jordan VC. Treatment of postmenopausal breast cancer with selective estrogen receptor modulators (SERMs) Breast Dis. 2005;24:93–105. doi: 10.3233/bd-2006-24108. [DOI] [PubMed] [Google Scholar]
- 9.Howell A. New developments in the treatment of postmenopausal breast cancer. Trends Endocrinol. Met. 2005;16:420–428. doi: 10.1016/j.tem.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 10.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 11.Zeng Y. Principles of micro-RNA production and maturation. Oncogene. 2006;25:6156–6162. doi: 10.1038/sj.onc.1209908. [DOI] [PubMed] [Google Scholar]
- 12.Couzin J. Genetics. Erasing microRNAs reveals their powerful punch. Science. 2007;316:530. doi: 10.1126/science.316.5824.530. [DOI] [PubMed] [Google Scholar]
- 13.Cuellar TL, McManus MT. MicroRNAs and endocrine biology. J. Endocrinol. 2005;187:327–332. doi: 10.1677/joe.1.06426. [DOI] [PubMed] [Google Scholar]
- 14.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 15.Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc. Natl Acad. Sci. 2007;104:17719–17724. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volinia S, Calin GA, Liu C-G, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, et al. microRNAs exhibit high frequency genomic alterations in human cancer. PNAS. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 19.Hammond SM. MicroRNAs as oncogenes. Curr. Opin. Genet. Dev. 2005;16:4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 20.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 21.Iorio MV, Ferracin M, Liu C-G, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 22.Jiang J, Lee EJ, Gusev Y, Schmittgen TD. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005;33:5394–5403. doi: 10.1093/nar/gki863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 24.Diederichs S, Haber DA. Sequence variations of microRNAs in human cancer: alterations in predicted secondary structure do not affect processing. Cancer Res. 2006;66:6097–6104. doi: 10.1158/0008-5472.CAN-06-0537. [DOI] [PubMed] [Google Scholar]
- 25.Scott GK, Mattie MD, Berger CE, Benz SC, Benz CC. Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res. 2006;66:1277–1281. doi: 10.1158/0008-5472.CAN-05-3632. [DOI] [PubMed] [Google Scholar]
- 26.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams BD, Furneaux H, White BA. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-{alpha} (ER{alpha}) and represses ER{alpha} messenger RNA and protein expression in breast cancer cell lines. Mol. Endocrinol. 2007;21:1132–1147. doi: 10.1210/me.2007-0022. [DOI] [PubMed] [Google Scholar]
- 28.Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE, Green AR, Ellis IO, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, Ambros VR, Israel MA. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67:2456–2468. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- 30.Giannakakis A, Coukos G, Hatzigeorgiou A, Sandaltzopoulos R, Zhang L. miRNA genetic alterations in human cancers. Exp. Opin. Biol. Ther. 2007;7:1375–1386. doi: 10.1517/14712598.7.9.1375. [DOI] [PubMed] [Google Scholar]
- 31.Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu C-G, Calin GA, Giovannini C, Ferrazzi E, Grazi GL, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 32.Hurteau GJ, Carlson JA, Spivack SD, Brock GJ. Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res. 2007;67:7972–7976. doi: 10.1158/0008-5472.CAN-07-1058. [DOI] [PubMed] [Google Scholar]
- 33.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 34.Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setien F, Casado S, Suarez-Gauthier A, Sanchez-Cespedes M, Gitt A, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 35.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osada H, Takahashi T. MicroRNAs in biological processes and carcinogenesis. Carcinogenesis. 2007;28:2–12. doi: 10.1093/carcin/bgl185. [DOI] [PubMed] [Google Scholar]
- 37.Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, Wells W, Kauppinen S, Cole CN. Altered microRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–11620. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 38.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 39.Zhou Y, Yau C, Gray JW, Chew K, Dairkee SH, Moore DH, Eppenberger U, Eppenberger-Castori S, Benz CC. Enhanced NF kappa B and AP-1 transcriptional activity associated with antiestrogen resistant breast cancer. BMC Cancer. 2007;7:59. doi: 10.1186/1471-2407-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK, Fedele V, Ginzinger D, Getts R, Haqq C. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol. Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gusev Y, Schmittgen TD, Lerner M, Postier R, Brackett D. Computational analysis of biological functions and pathways collectively targeted by co-expressed microRNAs in cancer. BMC Bioinformatics. 2007;8 (Suppl. 7):S16. doi: 10.1186/1471-2105-8-S7-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hossain A, Kuo MT, Saunders GF. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol. Cell Biol. 2006;26:8191–8201. doi: 10.1128/MCB.00242-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, Li Y. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379. doi: 10.1038/onc.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mattingly KA, Ivanova MM, Riggs KA, Wickramasinghe NS, Barch MJ, Klinge CM. Estradiol stimulates transcription of nuclear respiratory factor-1 and increases mitochondrial biogenesis. Mol. Endocrinol. 2008;22:609–622. doi: 10.1210/me.2007-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneider HP, Jackisch C. Potential benefits of estrogens and progestogens on breast cancer. Int. J. Fertil. Womens Med. 1998;43:278–285. [PubMed] [Google Scholar]
- 47.Deroo BJ, Korach KS. Estrogen receptors and human disease. J. Clin. Invest. 2006;116:561–570. doi: 10.1172/JCI27987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wijayaratne AL, Nagel SC, Paige LA, Christensen DJ, Norris JD, Fowlkes DM, McDonnell DP. Comparative analyses of mechanistic differences among antiestrogens. Endocrinology. 1999;140:5828–5840. doi: 10.1210/endo.140.12.7164. [DOI] [PubMed] [Google Scholar]
- 49.Wijayaratne AL, McDonnell DP. The human estrogen receptor-alpha is a ubiquitinated protein whose stability is affected differentially by agonists, antagonists, and selective estrogen receptor modulators. J. Biol. Chem. 2001;276:35684–35692. doi: 10.1074/jbc.M101097200. [DOI] [PubMed] [Google Scholar]
- 50.Harrington WR, Sheng S, Barnett DH, Petz LN, Katzenellenbogen JA, Katzenellenbogen BS. Activities of estrogen receptor alpha- and beta-selective ligands at diverse estrogen responsive gene sites mediating transactivation or transrepression. Mol. Cell Endocrinol. 2003;206:13–22. doi: 10.1016/s0303-7207(03)00255-7. [DOI] [PubMed] [Google Scholar]
- 51.Gupta M, McDougal A, Safe S. Estrogenic and antiestrogenic activities of 16alpha- and 2-hydroxy metabolites of 17beta-estradiol in MCF-7 and T47D human breast cancer cells. J. Steroid Biochem. Mol. Biol. 1998;67:413–419. doi: 10.1016/s0960-0760(98)00135-6. [DOI] [PubMed] [Google Scholar]
- 52.Power KA, Thompson LU. Ligand-induced regulation of ERalpha and ERbeta is indicative of human breast cancer cell proliferation. Breast Cancer Res. Treat. 2003;81:209–221. doi: 10.1023/A:1026114501364. [DOI] [PubMed] [Google Scholar]
- 53.Hurd C, Dinda S, Khattree N, Moudgil VK. Estrogen-dependent and independent activation of the P1 promoter of the p53 gene in transiently transfected breast cancer cells. Oncogene. 1999;18:1067–1072. doi: 10.1038/sj.onc.1202398. [DOI] [PubMed] [Google Scholar]
- 54.Zampieri L, Bianchi P, Ruff P, Arbuthnot P. Differential modulation by estradiol of P-glycoprotein drug resistance protein expression in cultured MCF7 and T47D breast cancer cells. Anticancer Res. 2002;22:2253–2259. [PubMed] [Google Scholar]
- 55.Grimson A, Farh KK-H, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, Iba H. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J. Mol. Biol. 2008;378:492–504. doi: 10.1016/j.jmb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 57.Sotoca Covaleda AM, van den Berg H, Vervoort J, van der Saag P, Strom A, Gustafsson J-A, Rietjens I, Murk AJ. Influence of cellular ER{alpha}/ER{beta} ratio on the ER{alpha}-agonist induced proliferation of human T47D breast cancer cells. Toxicol. Sci. 2008;105:303–311. doi: 10.1093/toxsci/kfn141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin CY, Strom A, Li Kong S, Kietz S, Thomsen JS, Tee JB, Vega VB, Miller LD, Smeds J, Bergh J, et al. Inhibitory effects of estrogen receptor beta on specific hormone-responsive gene expression and association with disease outcome in primary breast cancer. Breast Cancer Res. 2007;9:R25. doi: 10.1186/bcr1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matthews J, Wihlen B, Tujague M, Wan J, Strom A, Gustafsson J-A. Estrogen receptor (ER) {beta} modulates ER{alpha}-mediated transcriptional activation by altering the recruitment of c-Fos and c-Jun to estrogen-responsive promoters. Mol. Endocrinol. 2006;20:534–543. doi: 10.1210/me.2005-0140. [DOI] [PubMed] [Google Scholar]
- 60.Helguero LA, Faulds MH, Gustafsson JA, Haldosen LA. Estrogen receptors alfa (ERalpha) and beta (ERbeta) differentially regulate proliferation and apoptosis of the normal murine mammary epithelial cell line HC11. Oncogene. 2005;24:6605–6616. doi: 10.1038/sj.onc.1208807. [DOI] [PubMed] [Google Scholar]
- 61.Strom A, Hartman J, Foster JS, Kietz S, Wimalasena J, Gustafsson J-A. Estrogen receptor {beta} inhibits 17{beta}-estradiol-stimulated proliferation of the breast cancer cell line T47D. Proc. Natl Acad. Sci. USA. 2004;101:1566–1571. doi: 10.1073/pnas.0308319100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Behrens D, Gill JH, Fichtner I. Loss of tumourigenicity of stably ER[beta]-transfected MCF-7 breast cancer cells. Mol. Cell Endocrinol. 2007;274:19–29. doi: 10.1016/j.mce.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 63.Pan Q, Luo X, Toloubeydokhti T, Chegini N. The expression profile of micro-RNA in endometrium and endometriosis and the influence of ovarian steroids on their expression. Mol. Hum. Reprod. 2007;13:797–806. doi: 10.1093/molehr/gam063. [DOI] [PubMed] [Google Scholar]
- 64.Hu S-J, Ren G, Liu J-L, Zhao Z-A, Yu Y-S, Su R-W, Ma X-H, Ni H, Lei W, Yang Z-M. MicroRNA expression and regulation in mouse uterus during embryo implantation. J. Biol. Chem. 2008;283:23473–23484. doi: 10.1074/jbc.M800406200. [DOI] [PubMed] [Google Scholar]
- 65.Russo J, Fernandez SV, Russo PA, Fernbaugh R, Sheriff FS, Lareef HM, Garber J, Russo IH. 17-Beta-estradiol induces transformation and tumorigenesis in human breast epithelial cells. FASEB J. 2006;20:1622–1634. doi: 10.1096/fj.05-5399com. [DOI] [PubMed] [Google Scholar]
- 66.Russo J, Tahin Q, Lareef MH, Hu YF, Russo IH. Neoplastic transformation of human breast epithelial cells by estrogens and chemical carcinogens. Environ. Mol. Mutagen. 2002;39:254–263. doi: 10.1002/em.10052. [DOI] [PubMed] [Google Scholar]
- 67.Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, Jacob S, Majumder S. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27(Kip1) J. Biol. Chem. 2008;283:29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frasor J, Chang EC, Komm B, Lin C-Y, Vega VB, Liu ET, Miller LD, Smeds J, Bergh J, Katzenellenbogen BS. Gene expression preferentially regulated by tamoxifen in breast cancer cells and correlations with clinical outcome. Cancer Res. 2006;66:7334–7340. doi: 10.1158/0008-5472.CAN-05-4269. [DOI] [PubMed] [Google Scholar]
- 69.Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, Katzenellenbogen BS. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology. 2003;144:4562–4574. doi: 10.1210/en.2003-0567. [DOI] [PubMed] [Google Scholar]
- 70.Frasor J, Stossi F, Danes JM, Komm B, Lyttle CR, Katzenellenbogen BS. Selective estrogen receptor modulators: discrimination of agonistic versus antagonistic activities by gene expression profiling in breast cancer cells. Cancer Res. 2004;64:1522–1533. doi: 10.1158/0008-5472.can-03-3326. [DOI] [PubMed] [Google Scholar]
- 71.Levenson AS, Svoboda KM, Pease KM, Kaiser SA, Chen B, Simons LA, Jovanovic BD, Dyck PA, Jordan VC. Gene expression profiles with activation of the estrogen receptor alpha-selective estrogen receptor modulator complex in breast cancer cells expressing wild-type estrogen receptor. Cancer Res. 2002;62:4419–4426. [PubMed] [Google Scholar]
- 72.Vendrell JA, Bieche I, Desmetz C, Badia E, Tozlu S, Nguyen C, Nicolas JC, Lidereau R, Cohen PA. Molecular changes associated with the agonist activity of hydroxy-tamoxifen and the hyper-response to estradiol in hydroxy-tamoxifen-resistant breast cancer cell lines. Endocr. Relat. Cancer. 2005;12:75–92. doi: 10.1677/erc.1.00899. [DOI] [PubMed] [Google Scholar]
- 73.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 74.Kwon Y-S, Garcia-Bassets I, Hutt KR, Cheng CS, Jin M, Liu D, Benner C, Wang D, Ye Z, Bibikova M, et al. Sensitive ChIP-DSL technology reveals an extensive estrogen receptor {alpha}-binding program on human gene promoters. Proc. Natl Acad. Sci. USA. 2007;104:4852–4857. doi: 10.1073/pnas.0700715104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ludwig LB, Peale FV, Jr, Klinge CM, Bambara RA, Zain S, Hilf R. A microtiter well assay for quantitative measurement of estrogen receptor binding to estrogen-responsive elements. Mol. Endocrinol. 1990;4:1027–1033. doi: 10.1210/mend-4-7-1027. [DOI] [PubMed] [Google Scholar]
- 76.Bourdeau V, Deschenes J, Laperriere D, Aid M, White JH, Mader S. Mechanisms of primary and secondary estrogen target gene regulation in breast cancer cells. Nucleic Acids Res. 2008;36:76–93. doi: 10.1093/nar/gkm945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marino M, Galluzzo P, Ascenzi P. Estrogen signaling multiple pathways to impact gene transcription. Curr. Genomics. 2006;7:497–508. doi: 10.2174/138920206779315737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Safe S, Kim K. Non-classical genomic estrogen receptor (ER)/specificity protein and ER/activating protein-1 signaling pathways. J. Mol. Endocrinol. 2008;41:263–275. doi: 10.1677/JME-08-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J. Biol. Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 80.Schorr K, Li M, Krajewski S, Reed JC, Furth PA. Bcl-2 gene family and related proteins in mammary gland involution and breast cancer. J. Mammary Gland Biol. Neoplasia. 1999;4:153–164. doi: 10.1023/a:1018773123899. [DOI] [PubMed] [Google Scholar]
- 81.Dong L, Wang W, Wang F, Stoner M, Reed JC, Harigai M, Samudio I, Kladde MP, Vyhlidal C, Safe S. Mechanisms of transcriptional activation of bcl-2 gene expression by 17beta-estradiol in breast cancer cells. J. Biol. Chem. 1999;274:32099–32107. doi: 10.1074/jbc.274.45.32099. [DOI] [PubMed] [Google Scholar]
- 82.Li L, Ross AH. Why is PTEN an important tumor suppressor? J. Cell Biochem. 2007;102:1368–1374. doi: 10.1002/jcb.21593. [DOI] [PubMed] [Google Scholar]
- 83.Bradbury AR, Olopade OI. Genetic susceptibility to breast cancer. Rev. Endocr. Metab. Disord. 2007;8:255–267. doi: 10.1007/s11154-007-9038-0. [DOI] [PubMed] [Google Scholar]
- 84.Asangani IA, Rasheed SAK, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 85.Bonofiglio D, Gabriele S, Aquila S, Catalano S, Gentile M, Middea E, Giordano F, Ando S. Estrogen receptor {alpha} binds to peroxisome proliferator-activated receptor response element and negatively interferes with peroxisome proliferator-activated receptor {gamma} signaling in breast cancer cells. Clin. Cancer Res. 2005;11:6139–6147. doi: 10.1158/1078-0432.CCR-04-2453. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.