Abstract
Glis3 is a member of the Krüppel-like family of transcription factors and is highly expressed in islet β cells. Mutations in GLIS3 cause the syndrome of neonatal diabetes and congenital hypothyroidism (NDH). Our aim was to examine the role of Glis3 in β cells, specifically with regard to regulation of insulin gene transcription. We demonstrate that insulin 2 (Ins2) mRNA expression in rat insulinoma 832/13 cells is markedly increased by wild-type Glis3 overexpression, but not by the NDH1 mutant. Furthermore, expression of both Ins1 and Ins2 mRNA is downregulated when Glis3 is knocked down by siRNA. Glis3 binds to the Ins2 promoter in the cell, detected by chromatin immunoprecipitation. Deletion analysis of Ins2 promoter identifies a sequence (5′-GTCCCCTGCTGTGAA-3′) from −255 to −241 as the Glis3 response element and binding occur specifically via the Glis3 zinc finger region as revealed by mobility shift assays. Moreover, Glis3 physically and functionally interacts with Pdx1, MafA and NeuroD1 to modulate Ins2 promoter activity. Glis3 also may indirectly affect insulin promoter activity through upregulation of MafA and downregulation of Nkx6-1. This study uncovers a role of Glis3 for regulation of insulin gene expression and expands our understanding of its role in the β cell.
INTRODUCTION
GLI and GLI-similar (Glis) transcription factors are two closely related subfamilies of Krüppel-like zinc finger (ZF) proteins (1–3). These factors contain five zinc fingers in tandem for DNA binding. The Glis subfamily is comprised of three members Glis1 to 3. Glis proteins contain both repressor and activator domains, suggesting that they may function as positive and negative regulators of gene transcription. During embryonic development, Glis proteins are expressed in a spatially and temporally specific manner, suggesting that they may play key roles in normal organogenesis. Glis1 is involved in the aberrant differentiation observed in psoriatic epidermis (1). Glis2 is highly expressed in the kidney, and loss of Glis2 causes nephronophthisis in humans and mice through increased apoptosis and fibrosis (4,5). Glis2 has also been shown to be involved in the regulation of neuronal differentiation (2). Glis3 is highly expressed in the metanephric mesenchyme during embryonic development and in the uterus, pancreas and kidney of adult mice (3). It was also shown to bind to the promoter of fibroblast growth factor 18 (FGF18) and promote osteoblast differentiation (6).
In vitro assays indicate that all five ZF motifs appear essential for the binding of Glis3 to DNA, and the fourth ZF is also involved in the nuclear localization of Glis3 (3,7). The C-terminal region of the protein contains a strong transactivation domain, and its deletion abolishes transcriptional activity. A recent report identified a consensus binding site for Glis3 to be (G/C)TGGGGGGT(A/C) using a DNA-binding site selection strategy that combines PCR and electrophoretic mobility shift assays (EMSA) (7). However, this sequence was isolated by in vitro experiments and has not been identified in any endogenous Glis3 responsive promoters thus far.
Linkage analysis revealed that mutations in GLIS3 are responsible for the syndrome of neonatal diabetes and congenital hypothyroidism (NDH) (8). Glis3 mRNA is present in human and mouse pancreas from early developmental stages through adulthood, with higher expression in β cells than in other pancreatic islet or exocrine cells (8). However, the functional role of Glis3 in β cells has not been explored. Pancreatic islet development, β-cell maintenance and insulin gene transcription are dependent on multiple islet-enriched transcription factors, including Pdx1 (9–11), MafA (12–14), Pax6 (15,16) and BETA 2/NeuroD1 (17,18). Rat insulin promoter analyses have localized binding of these factors to four promoter regions, C2 (Pax6), A3 (Pdx1), C1 (MafA) and E1 (BETA2/NeuroD1) (19,20). Other factors known to be involved in insulin gene transcription include Nkx6-1 and Pax4, both of which may have a repressive role. Study of these factors has provided insights into the machinery of insulin gene regulation and the pathogenesis of diabetes. As Glis3 is highly expressed in pancreatic islets and mutations in GLIS3 cause neonatal diabetes in humans (8,21), we hypothesized that this factor may regulate insulin gene expression. In this study, we found that Glis3 binds to a cis-acting element (Glis3 response element, Glis3RE) in the rat insulin 2 promoter (RIP) and stimulates its transcription activity. Furthermore, Glis3 interacts with β-cell-specific transcription factors—Pdx1, MafA and NeuroD1—and can interact with these in a synergistic manner to increase insulin promoter activity. In addition, Glis3 also regulates the expression of a number of β cell transcription factors, such as MafA, Nkx6-1 and Pax6. These findings constitute the first evidence that Glis3 regulates insulin gene expression in pancreatic β cells.
MATERIALS AND METHODS
DNA constructs
Using the known coding sequence of mouse Glis3 gene (Genbank accession number NM_175459), we constructed Glis3 cDNA and a Glis3 mutant cDNA (Glis3-NDH1) that corresponds to the sequence in a family with neonatal diabetes and congenital hypothyroidism (NDH1) syndrome (8) by PCR mutagenesis, inserting a C nucleotide at 2930 of wild-type Glis3. The mutant Glis3-NDH1 protein terminates prematurely at aa 844 compared to the full-length protein of 935 aa.
The Glis3 cDNAs as well as control yellow fluorescent protein (YFP) were amplified by using PCR and cloned into the retroviral vector pSRQT (22). Additional constructs were generated for transient transfection by using a human elongation factor 1α (BOS) promoter-driven expression vector pBOS-MCS (23). SRα-Secreted Alkaline Phosphate (SEAP) construct was kindly provided by Dr David Spencer (Department of Immunology, Baylor College of Medicine). The coding sequences of mouse Glis3, Glis3-NDH1, MafA, Pdx1 and NeuroD1 were ligated downstream of a c-myc tag. We constructed a luciferase (Luc) reporter by ligation downstream of the rat insulin 2 promoter (RIP) from −696 to +8 into pGL3-basic (Promega) or pGluc-basic (New England Biolabs) to generate RIP-Luc and RIP-Gluc constructs. Serial deletions of RIP were generated by PCR and subcloned into pGL3-basic. (Glis3RE)5-Luc was constructed by ligation of five copies of complementary oligonucleotides containing the Glis3RE sequence we identified in this article. All expression constructs were confirmed by DNA sequencing.
Cell culture and transfection
Rat 832/13 insulinoma cells (gift of Dr Christopher Newgard, Duke University) were maintained as described (24). We cultured NIH3T3 and BOSC23 cells (retrovirus packaging cell line) in DMEM with 10% FBS. CHO cells were cultured in Ham's F12 supplemented with 10% FBS.
We used Lipofectamine 2000 (Invitrogen) for transfection according to the manufacturer's instructions. Retrovirally transduced cells (22) were selected in a medium containing 1.0 µg/ml puromycin and 200 μg/ml hygromycin B for 6–8 days.
For small interfering RNA (siRNA) experiments, all control and Glis3 siRNAs were synthesized by Dharmacon RNAi Technologies (Thermo Fisher Scientific). The rat Glis3 targeting siRNA sequences were: sense, 5′-GCAUCAGUGUACGAUUUUU-3′; antisense, 5′-AAAUCGUACACUGUGAUGCUU-3′. DharmaFECT siRNA transfection reagents were used for delivering siRNA into target cells. The control siRNA is a non-targeting siRNA (Dharmacon RNAi Technologies, D-001810-01-05).
RNA isolation and quantitative polymerase chain reaction (qPCR)
Mouse tissues were dissected, snap frozen in liquid nitrogen and stored at −80°C until use. We isolated pancreatic islets from C57BL/6J mice as described previously (25). We used TRIZOL (Invitrogen, Carlsbad, CA) for RNA extraction and treated the RNA with amplification grade DNase I (Invitrogen) to remove genomic DNA contamination. Reverse transcription was performed with Superscript II reverse transcriptase kit (Invitrogen) using oligo dT.
We performed qPCR using iQ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA) with ROX as the reference dye. The housekeeping gene Cyclophilin A was used as an internal control. Primer sequences are shown in Supplementary Table S1. All primers span exons, except MafA, which has only one exon. Amplification was performed using the Mx3000P Real-time PCR System (Stratagene, La Jolla, CA) with the following parameters: 95°C for 30 s, 55°C for 1 min, 72°C for 30 s for 40 cycles. The fidelity of the PCR products was confirmed by melting curve analysis and gel electrophoresis.
Luciferase assay
For transfection, we seeded cells in 24-well or 48-well culture plates and collected the cultured media (Gluc and SEAP) or harvested the cells (Luc) 48 h later using Lysis Buffer (Promega Corporation, Madison, WI). Luc assays were performed with the Dual-Luciferase Kit (Promega) or Gaussia Luciferase Assay Kit (New England Biolabs). The results were normalized to the activity of the internal control BOS-Renilla Luc or SRα-SEAP Reporter measured with Phospha-Light™ SEAP Reporter Gene Assay System (Applied Biosystems).
Co-immunoprecipitation (Co-IP) assays
Nuclear protein extraction and Co-IP assays were performed using a nuclear complex Co-IP kit (Active Motif, Carlsbad, CA) as described by the manufacturer. We incubated nuclear extract (300 µg) with 3 µg of anti-MafA antibody (Bethyl Laboratories, Inc., Montgomery, TX), anti-Pdx1 antibody (gift of Dr Christopher Wright, Vanderbilt University), anti-NeuroD1 antibody (Santa Cruz Biotechnology, Inc.), or rabbit or goat IgG as control in 500 µl of low salt IP buffer overnight at 4°C. We added Protein-G or A sepharose beads (60 µl, Sigma-Aldrich) to the mixture and incubated for two additional hours with rocking; we then washed the beads six times with the low salt IP buffer. The immunoprecipitated proteins were eluted in 40 µl of 2× SDS sample buffer and boiled for 10 min, prior to analysis by western blotting.
Electrophoretic mobility shift assay (EMSA)
We subcloned the Glis3 zinc finger domain (ZFD, 484–687 aa) into the pGBKT7 vector (Clontech Laboratories, Inc.) and used the TNT Quick Coupled Transcription/Translation Systems (Promega) according to the manufacturer's instructions. This ZFD peptide was used for EMSA using a Panomics ‘gel shift’ kit according to the manufacturer's instructions but with the addition of 100 μM ZnCl2 to the binding buffer. Assays were conducted using a biotin-labeled double-stranded oligonucleotides probe containing the recognition sequence for Glis3 (5′-CTGAGACAATGTCCCCTGCTGTGAACTGGTTCATC-3′). Unlabeled wild-type or mutated Glis3REs double stranded oligonucleotides were added as competitors. Protein–DNA complexes were separated from free probe by 6% non-denaturing polyacrylamide gel electrophoresis using Tris Borate buffer without EDTA.
Chromatin immunoprecipitation (ChIP)
We transfected 832/13 cells (1 × 108) with the c-myc-Glis3 plasmid and, after 48 h, treated them with 1% formaldehyde for 10 min at room temperature. Fixation was stopped with the addition of glycine to a final concentration of 0.125 M and then the cells were treated with lysis buffer (10 mM EDTA, 1% SDS, 1 mM PMSF, 1 µg/ml pepstatin, 1 µg/ml leupeptin, 1 µg/ml aprotinin, 50 mM Tris–HCL, pH 8.1) followed by three 10 s pulses of sonication (40 W). We then incubated the sheared preparations with mouse monoclonal antibody against c-myc overnight at 4°C. Immune complexes were recovered with protein sepharose A/G beads and cross-linking was reversed by heating at 65°C overnight. After removing the protein by treatment with proteinase K, we ethanol precipitated the DNA and resuspended it in 50 µl of dH2O for PCR analysis. Primers spanning the Glis3RE fragment were 5′-AAGGACAAAGAAGGCCTCAC-3′ and 5′-TTAGGGCTGGGGGTTACTGAATC-3′. Primers used as a negative control were 5′-ACCCTGGTCACACAGTAGGC-3′ and 5′-CCTGGTACTTTCTGGGGTC-3′ which span from −5400 to −5150 of the RIP 5′ region.
Statistical analysis
The Student's t-test was used for statistical analysis. Results are presented as the mean ± SD.
RESULTS
Production of mouse Glis3 and Glis3-NDH1 constructs
The mouse Glis3 gene (NM_175459) contains eleven exons, which encode a 935 aa protein of 99.7 kDa (Figure 1A). We isolated a full-length cDNA clone of Glis3 from mouse islet cDNAs. The cDNA encodes a full-length Glis3 (7) protein that is 155 aa longer at the amino-terminus than the previously reported Glis3 variants NΔ155 (7) or Glis3-779 (3), resulting from contributions from an alternate first exon.
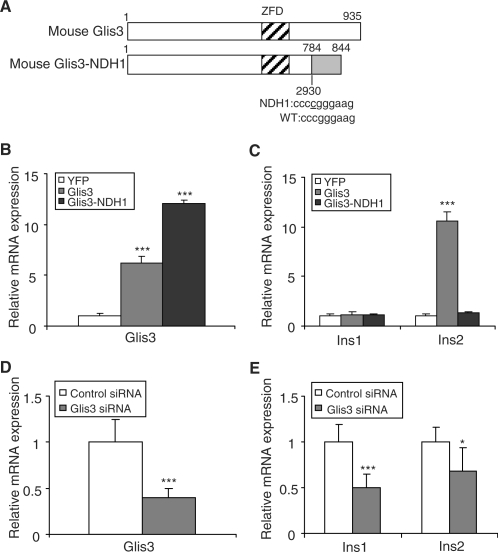
Figure 1.
Glis3 stimulates insulin gene transcription. (A) Schematic representation of the mouse Glis3 and Glis3-NDH1 constructs. (B, C) 832/13 cells were stably infected with mouse Glis3 and Glis3-NDH1 constructs or a YFP control retroviral construct. Mouse/rat Glis3 (B) and rat Ins1 and Ins2 (C) mRNA levels were quantified by qPCR relative to cyclophilin A. The Glis3 and Glis3-NDH1 values are shown relative to YFP-expressing cells and are expressed as the mean ± SD. (D, E) 832/13 cells were transiently transfected with siRNA against rat Glis3 or a control siRNA. Glis3 (D) and rat Ins1 and Ins2 (E) mRNA levels were quantified by qPCR relative to cyclophilin A. The relative Glis3, Ins1 and Ins2 expression levels in Glis3 siRNA transfected cells were compared to those in control siRNA-treated cells and presented as the mean ± SD. Each experiment was performed in triplicate and repeated three times. *P < 0.05; ***P < 0.001.
A cytidine insertion (insC) at the nucleotide 2067 of the human GLIS3 cDNA underlies NDH1 syndrome (8). We generated the homologous mouse 2067-insC mutant, which we call mouse Glis3-NDH1, corresponding to the NDH1 mutation found in human NDH1 syndrome by PCR mutagenesis. The mouse Glis3-NDH1 encodes a 844 aa protein with a calculated MW of 89.8 kDa (Figure 1A).
Glis3 stimulates insulin gene transcription
In mice, we found the level of Glis3 transcripts to be highest in pancreatic islets, followed by kidney (Supplementary Figure S1), in agreement with previous reports (7,8). We examined the effect of stable Glis3 overexpression on insulin gene transcription in insulinoma 832/13 cells. Retrovirally transduced Glis3 and Glis3-NDH1 constructs were strongly expressed and achieved mRNA levels 6–12-fold higher than endogenous Glis3 by qPCR (Figure 1B). Immunoblotting showed that both Glis3 constructs expressed proteins of the expected size in 832/13 cells (data not shown). By qPCR, the level of endogenous rat Ins1 transcript was unchanged but that of Ins2 transcript was increased by more than 10-fold by Glis3 overexpression compared to YFP-expressing control cells (Figure 1C). Expression of Glis3-NDH1 did not affect Ins2 levels compared to the control YFP (Figure 1C), consistent with the absence of a transactivating domain in this truncated factor.
We then knocked down endogenous Glis3 expression with siRNA in 832/13 cells, reducing Glis3 mRNA levels by ∼60% (Figure 1D). Compared to control siRNA treated cells, the expression levels of both Ins1 and Ins2 were significantly decreased when Glis3 was knocked down by siRNA (Figure 1E). A different siRNA against Glis3 resulted in similar results (Supplementary Figure S2). These results strongly suggest that Glis3 is involved in the regulation of insulin gene transcription in β cells.
Glis3 stimulates insulin promoter activity
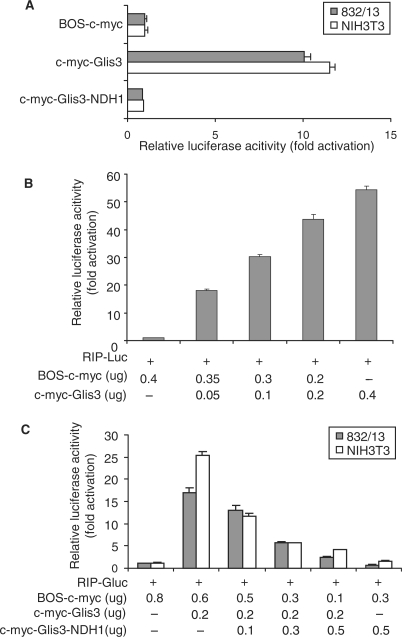
The DNA-binding and transcription activation domains of Glis3 have been examined in non-insulin producing cell lines by Beak et al. (7) and Kim et al. (3). Here, we examined whether Glis3 and Glis3-NDH1 regulate insulin gene promoter activity. Immunoblotting showed that both constructs expressed proteins of the correct sizes in CHO cells (Supplementary Figure S3A) and with similar expression levels in 832/13 cells and NIH3T3 cells (Supplementary Figure S3B). The transcription activation mediated by the different constructs was measured by co-transfection with the RIP-Gaussia luciferase (RIP-Gluc) reporter into 832/13 cells and NIH3T3 cells. In both cell types, Glis3 markedly enhanced RIP-Gluc activity over background (BOS-c-myc). However, the Glis3-NDH1 construct had no effect (Figure 2A). In 832/13 cells, wild-type Glis3 stimulated RIP-Luc activity in a dose-dependent manner (Figure 2B). Moreover, the RIP-Gluc construct showed that c-myc-Glis3-NDH1 inhibited wild-type Glis3 activation of RIP-Gluc in a dose-dependent manner both in a β cell line (832/13) and in a non-β cell line (NIH3T3) (Figure 2C). We speculate that the mechanism of inhibition of RIP by Glis3-NDH1 is due to competition for DNA binding by the mutant, which lacks a transcription activation domain.
Figure 2.
Transcriptional activity of Glis3 and Glis3-NDH1. (A) A β cell line (832/13) and a non-β cell line (NIH3T3) were co-transfected with RIP-Gluc, the internal control SRα-SEAP, and Glis3 or Glis3-NDH1 expression constructs as indicated. Gaussia luciferase assay was performed at 48 h after transfection. The fold change of RIP promoter activity caused by the c-myc-Glis3 constructs were compared to that of the BOS vector expressing the c-myc tag alone. Results are presented as mean ± SD. (B) Dose-dependent induction of RIP-mediated luciferase expression by Glis3. 832/13 cells were co-transfected with RIP-Luc and BOS-c-myc together with increasing amounts of c-myc-Glis3 as indicated. The amount of BOS-c-myc vector was adjusted to maintain the same amount of total DNA in each well. (C) Inhibition of Glis3-induced luciferase expression by the Glis3-NDH1 mutant. 832/13 or NIH3T3 cells were co-transfected with RIP-Gluc, c-myc-Glis3 and BOS-c-myc together with increasing amounts of c-myc-Glis3-NDH1 as indicated.
Identification of DNA-binding sequence of Glis3 in the insulin promoter
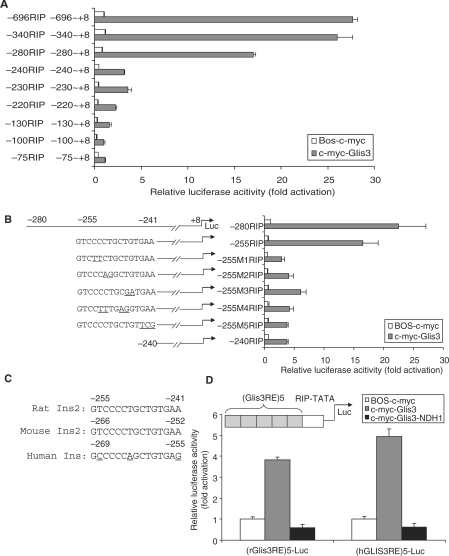
We mapped the region of RIP (−696/+8) which interacts with Glis3 by producing serial 5′ deletion mutants of RIP-Luc constructs. These were co-transfected with c-myc-Glis3 into 832/13 cells. As shown in Figure 3A, deletion of RIP sequences from −696 to −280 produced no major effects on promoter activity. However, further deletion to −240 (−240RIP) decreased Luc reporter activity by over 80% compared to −280RIP. Deleting sequences from −280 to −255 had little additional effect (Figure 3B, first two constructs), suggesting that the critical region was between −255 and −241. To define the cis-acting element further, we generated five mutant constructs (−255 M1 to M5, Figure 3B) within this 15-bp region, all of which (−255M1-M5RIP) were associated with a marked reduction in reporter activity, compared to the −255RIP wild-type construct, suggesting that this entire region was required for Glis3 binding (Figure 3B). Sequence alignment of insulin promoters from mouse, rat and human indicates that this region is highly conserved (Figure 3C), consistent with an important role as the Glis3 response element (Glis3RE). We then ligated five copies of the rat or human Glis3RE sequence to the RIP TATA box [(Glis3RE)5-Luc] and found that both rat and human Glis3RE constructs were activated by c-myc-Glis3, but not c-myc-Glis3-NDH1 (Figure 3D). These results demonstrate that the 15-bp region G(T/C)CCCC(T/A)GCTGTGA(A/G) in the rat ins2 and human insulin promoters functionally interacts with Glis3 and is crucial to promoter activity.
Figure 3.
Identification of Glis3 DNA-binding sequence in RIP region. (A) The rat insulin 5′ flanking serial deletion constructs (0.2 µg) were co-transfected with BOS-c-myc (0.2 µg) or c-myc-Glis3 (0.2 µg). The activity of each construct is expressed relative to the background activity of BOS-c-myc (equal to 1.0). (B) The sequences and activities of the wild-type and mutation constructs (M1–M5) between −255 and −241 of RIP are shown. The mutated nucleotides are underlined. 832/13 cells were co-transfected with c-myc-Glis3 (0.2 µg) and the various RIP mutant constructs (0.2 µg) as indicated. Dual-luciferase assays were performed at 48 h after transfection. The activity of each construct was normalized to that induced by the co-transfection of BOS-c-myc and −280 RIP-Luc. (C) Alignment of the 15-bp sequences (Glis3RE) located between −255 and −241 of the rat Ins2 promoter compared to mouse and human. (D) Schematic view of five tandem copies of the Glis3RE fused upstream of the RIP TATA box to drive expression of luciferase [(Glis3RE)5-Luc]. 832/13 cells were co-transfected with the rat or human (Glis3RE)5-Luc with c-myc-Glis3 or c-myc-Glis3-NDH1 constructs. The activity of each construct was normalized to that obtained with BOS-c-myc. Each experiment was performed in triplicate and repeated three times. Minor variation between experiments in the basal expression with deletion of sequences between −280 and −240, with a decrease of 42.5% in (A) and 32% in (B).
Glis3 binds to the Glis3 response element
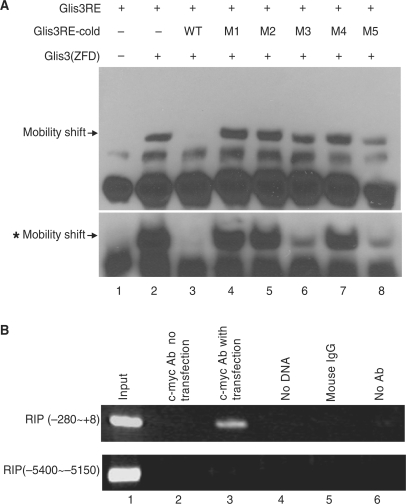
To confirm the physical interaction of Glis3 with the putative rat Glis3RE sequence (GTCCCCTGCTGTGAA), we performed EMSA using the ZF domain Glis3 (ZFD). A biotinylated double-stranded oligonucleotide containing the Glis3RE (−265 to −230) was used as the probe. We found that Glis3 (ZFD) bound to the Glis3RE (−265 to −230) probe and the complex was abolished by a 2-fold molar excess of unlabeled Glis3RE oligonucleotide, whereas 2-fold molar excess of all five mutant Glis3REs corresponding to −255M1 to M5 failed to significantly compete for binding (Figure 4A). In the presence of 20-fold molar excess competitors, M1, M2 and M4 still failed to compete for the binding of Glis3 (ZFD) to wild-type Glis3RE, while M3 and M5 partially competed for the binding (Figure 4A, bottom). Therefore, there is substantial specificity in the binding of Glis3 (ZFD) binds to Glis3RE in vitro.
Figure 4.
Glis3 binds to the RIP Glis3 response element (Glis3RE). (A) Synthetic biotinylated RIP sequence from −265 to −230, containing Glis3RE, was used as a probe in EMSA with an in vitro translated Glis3 ZFdomain [Glis3 (ZFD)] peptide. The specificity of the protein–DNA complex (lane 2) was confirmed by competition with a 2-fold molar excess of unlabeled wild-type competitor (lanes 3) but not a 2-fold molar excess of the mutants M1 to M5 (lanes 4–8). The mobility shift bands with asterisk showed the results of similar experiment using 20-fold molar excess wild-type or five mutant competitors. The specific bands are indicated by arrows. (B) The c-myc-Glis3–DNA complex was immunoprecipitated by c-myc antibody from 832/13 cells which had been transfected with c-myc-Glis3. Immunoprecipitated DNA was purified and analyzed by PCR using primers specifically spanning the Glis3RE region (RIP −280 to +8) or region remote from Glis3RE (−5400 to −5150). Lane 1, total input DNA (1:100 dilution); lane 2, DNA immunoprecipitated with c-myc Ab without c-myc-Glis3 transfection; lane 3, DNA immunoprecipitated with c-myc Ab with c-myc-Glis3 transfection; lane 4, no DNA template; lane 5, immunoprecipitation with mouse IgG; lane 6, DNA precipitated in the absence of antibody.
To determine if Glis3 also is bound to the endogenous RIP Glis3RE in the cell, we performed a ChIP assay. 832/13 cells were transfected with c-myc-Glis3 and, 48 h later, c-myc antibody was used to immunoprecipitate the c-myc-Glis3–DNA complexes. Primers corresponding to the RIP fragment spanning −280 to +8 were used for PCR amplification. As shown in Figure 4B, the c-myc antibody was able to immunoprecipitate the RIP region, confirming that Glis3 was bound to the promoter in situ. There was no amplification of DNA for the controls which included absence of DNA template, absence of antibody, use of mouse IgG, or use of c-myc Ab without c-myc-Glis3 transfection. These controls confirmed the specificity of the c-myc-Glis3–DNA interaction. As an additional control, a remote region of the RIP promoter (−5400 to −5150) also was not amplified in the experiment. These results show that Glis3 is capable of binding to the endogenous RIP promoter in 832/13 cells, directly regulating insulin gene transcription.
Glis3 synergistically stimulates insulin transcription activity with Pdx1, MafA and NeuroD1
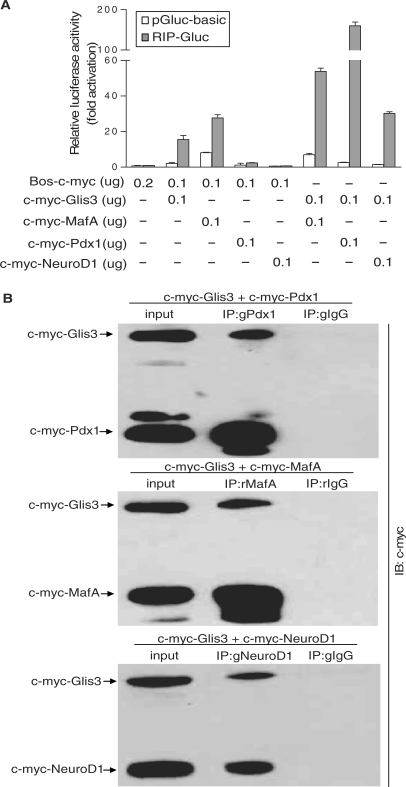
Pdx1, MafA and NeuroD1 are major transcription factors which mediate insulin gene transcription in the β cell and are required for β cell maintenance (12–14). Therefore, we examined the functional interaction of Glis3 with these known insulin gene transactivators. We co-transfected constructs expressing c-myc-Pdx1, c-myc-MafA and c-myc-NeuroD1 with RIP-Gluc into NIH3T3 cells with or without c-myc-Glis3. 832/13 cells were not used for this experiment because we wanted to avoid contributions by the endogenous RIP transactivators. Immunoblotting showed that c-myc-Pdx1, c-myc-MafA and c-myc-NeuroD1 constructs express proteins with correct sizes (Supplementary Figure S2A). RIP-Gluc activity was significantly induced by Glis3, MafA and Pdx1 individually, but not by NeuroD1 (Figure 5A). Co-transfection of c-myc-Glis3 with c-myc-tagged Pdx1, MafA and NeuroD1, revealed a synergistic interaction between Glis3 and all of these factors. These results suggest that Glis3 functionally interacts with other insulin gene transactivators to enhance insulin gene transcription.
Figure 5.
Glis3 synergistically induces RIP-driven transcriptional activity with Pdx1, MafA and NeuroD1. (A) NIH3T3 cells were co-transfected with RIP-Gluc and c-myc-Glis3, c-myc-MafA, c-myc-Pdx1 and c-myc-NeuroD1 individually or in combination with c-myc-Glis3 as labeled. The relative luciferase activity is presented compared to that of Bos-c-myc with pGluc-basic (equal to 1.0). The background luciferase activity of pGluc-basic also is shown. (B) 832/13 cells were transfected with c-myc-Glis3 and c-myc-Pdx1, c-myc-MafA or c-myc-NeuroD1. Forty-eight hours after transfection, nuclear proteins were isolated and immunoprecipitated either with a rabbit anti-mouse MafA, goat anti-mouse Pdx1 or goat anti-mouse NeuroD1 antibody or appropriate control IgGs. The immunoprecipitates were analyzed with western blotting using anti-c-myc antibody. g: goat; r: rabbit.
Glis3 binds to Pdx1, MafA and NeuroD1 in 832/13 cells
Because of the functional interaction of Glis3 with β-cell-specific transcription factors, we examined if Glis3 also physically interacts with them. We expressed c-myc-Glis3 with c-myc-Pdx1, MafA or NeuroD1 in 832/13 cells, and used antibodies to Pdx1, MafA and NeuroD1 to immunoprecipitate the complexes from the nuclear extract. Anti-c-myc antibody was used in western blot analysis to detect Glis3 that had co-precipitated with Pdx1, MafA and NeuroD1 (Figure 5B). The MafA, Pdx1 and NeuroD1 complexes are specific, because pre-immune rabbit or goat IgG did not immunoprecipitate the c-myc-Glis3 protein. We also performed the co-immunoprecipitation experiments in 832/13 cells overexpressing c-myc-Glis3 but without expression of the other factors, in order to examine interactions with endogenous Pdx1, MafA and NeuroD1. Western blot analysis of the immunoprecipitated complexes showed that Glis3 co-precipitated with endogenous Pdx1, MafA, but not NeuroD1 in this case (Supplementary Figure S4). A possible reason for the latter result is that the mouse NeuroD1 antibody could not recognize endogenous rat NeuroD1 protein effectively for immunoprecipitation. Alternatively, the endogenous NeuroD1 may be at a level that is too low for detection.
Glis3 regulates the expression of β cell enriched transcription factors
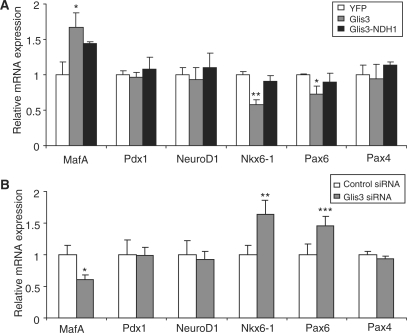
We have shown that Glis3 regulates insulin gene transcription by direct binding to the insulin promoter and also by interacting with insulin gene transcription factors, Pdx1, MafA and NeuroD1. We next examined if Glis3 affected the expression of key β-cell-specific transcription factors. Quantitative PCR showed that the mRNA expression of MafA was significantly upregulated while Nkx6-1 and Pax6 were significantly downregulated in Glis3 stably overexpressing cells as compared to YFP-expressing control cells (Figure 6A); Glis3-NDH1 had no effect on the expression of these genes (Figure 6A). The transcript levels of Pdx1, NeuroD1 and Pax4 were not affected by Glis3 overexpression (Figure 6A). When we knocked down the endogenous Glis3 expression with siRNA in 832/13 cells, MafA expression was significantly decreased whereas Nkx6-1 and Pax6 message levels were significantly increased (Figure 6B), results that were congruent with those of Glis3 overexpression. Because all of these factors directly regulate insulin gene expression (19,20), it appears that Glis3 also has an indirect role in the regulation of the insulin promoter through its effects on the mRNA levels, and possibly protein levels, of these β-cell-specific factors.
Figure 6.
Glis3 regulates islet-enriched transcription factors expression. (A) 832/13 cells were stably infected with Glis3, Glis3-NDH1 or the control YFP retroviral constructs, and MafA, Pdx1, NeuroD1, Nkx6-1, Pax6 and Pax4 mRNA levels were analyzed by qPCR relative to cyclophilin A. The expression values were normalized to those for YFP infected cells and presented as mean ± SD. (B) 832/13 cells were transiently transfected with siRNA against rat Glis3 or control siRNA. MafA, Pdx1, NeuroD1, Nkx6-1, Pax6 and Pax4 mRNA expression were analyzed by qPCR. The expression values were normalized to those for control siRNA treated cells and presented as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001.
DISCUSSION
Glis3 is a member of Krüppel-like ZF proteins and has the potential to function as a transcription activator or repressor (3). It is highly expressed in human pancreatic β cells (8) and mouse islets and rat insulinoma 832/13 cells. The recent discovery that mutations in the GLIS3 gene are responsible for neonatal diabetes and congenital hypothyroidism (NDH) in humans (8) motivated us to focus on the function of this factor in β cells.
In this study, we showed that Glis3 overexpression in rat 832/13 insulinoma cells markedly upregulated Ins2 gene expression. However, both Ins1 and Ins2 were downregulated when endogenous Glis3 was knocked down by siRNA, suggesting that it is involved directly or indirectly in the regulation of both genes. Our data demonstrated that Glis3 is able to directly bind the Glis3RE sequence of the rat Ins2 promoter using ChIP and EMSA, so it likely has a direct effect. The lack of response of Ins1 to Glis3 overexpression may indicate that its expression levels are already near or at maximum in 832/13 cells and an increase in Glis3 levels cannot further enhance Ins1 expression. A similar phenomenon was noted previously for the lack of activation of mouse Ins1, Ins2 and Pdx1 expression by MafB overexpression in βTC3 cells even though it binds to the promoters (26,27). We cannot identify a Glis3RE sequence in the rat Ins1 promoter region. However, Glis3 can increase MafA mRNA expression and as it is known that MafA is capable of activating Ins1 promoter, we speculate that, although the Ins1 promoter is not directly bound by Glis3, Glis3 has effects on other β cell transcription factors which may in turn influence Ins1 expression.
We also showed that Glis3 expression stimulated the rat insulin 2 promoter (RIP-Gluc) in both NIH3T3 cells and 832/13 cells and mapped the regulatory region required for Glis3 activation to a 15-bp sequence (−255 to −241) of the promoter. The site GTCCCCTGCTGTGAA shares 100% homology with a similar region of the mouse Ins2 promoter, and is 80% conserved with a sequence from the human insulin promoter. We showed that Glis3 binds to this sequence and can activate the transcription of both human and rodent (Glis3RE)5-TATA box constructs, confirming its place as a bona fide Glis3 binding and transcription activation site. By ChIP analysis, we confirmed that Glis3 was able to bind to the region of interest in the endogenous Ins2 promoter in 832/13 cells as well.
The sequence we identified differs from the 11 bp consensus identified by Beak et al. (7) (G/C)TGGGGGGGT(A/C) using PCR amplification of random oligonucleotide sequences. Their consensus binds to Glis3 with high affinity and can mediate transcription activation in vitro (7). However, it is an artificial binding sequence that has not been identified as of yet in any 5′ regulatory regions. A computer search also failed to find any matching homologous sequences in the rat Ins2 promoter (−696/+8). The FGF18 promoter contains a Glis3-binding site with the sequence AACCCCCAAA (6). We found a similar sequence AACCCCCAG at −7 to +1 of RIP. However, deletion analysis showed that the proximal promoter region (−75/+1) of RIP is minimally activated by co-transfected Glis3 and this sequence overlaps the transcription initiation site +1, making it unlikely that this region is involved in Glis3-activated gene transcription. The Glis3RE (−269 to −255) identified in this paper partially overlaps with a previously characterized element (GAGACATTTGCCCCCAGCTGT) known as a negative regulatory element (NRE, −279 to −258) (28,29) that lies within the glucose sensing Z element (−292 to −243) of the human insulin promoter (30,31).
The human NDH1 mutation of GLIS3 contains a single base insertion at nucleotide 2067. It encodes a mutant protein that is truncated at its C-terminus at aa 844 due to a translation frameshift (8). The mechanism by which this mutation causes NDH has not been elucidated. Our analysis shows that the analogous mutation of the mouse Glis3 abrogates the transcription activity of the factor, consistent with localization of the activation domain at the C-terminus (7). Furthermore, Glis3-NDH1 acts as a dominant negative factor when expressed together with wild-type Glis3 in insulinoma cells and NIH3T3 cells. The putative mechanism is that Glis3-NDH1 binds to DNA and competes with the wild-type Glis3, but cannot activate transcription. We speculate that the NDH1 mutant protein might profoundly affect wild-type Glis3 function during early pancreas development.
In addition to direct effects on insulin promoter activity, we showed that Glis3 is capable of interacting with other key β-cell-specific transcription factors. MafA is a crucial factor for the assembly and function of the insulin gene transcription complex. Previous studies have shown that Pdx1, MafA and NeuroD1 synergistically activate the insulin promoter (12–14). Our present results show that Glis3 physically interacts with Pdx1, MafA and NeuroD1 in co-precipitation experiments. In addition, Glis3 shows cooperative interactions with Pdx1, MafA and NeuroD1 for insulin promoter activation. These results lead us to conclude that Glis3 modulates insulin gene expression, both directly through binding to the insulin promoter and indirectly by modulating the activity of other β-cell-enriched transcription factors.
Manipulation of Glis3 levels through stable overexpression or siRNA knock-down also affected the levels of several other Glis3 target genes in β cells, including MafA, Nkx6-1 and Pax6. Therefore, Glis3 also may modulate insulin gene transcription indirectly through changes in the levels of these factors. The mRNA expression of MafA was upregulated in Glis3-overexpressing cells, and downregulated in Glis3-knockdown cells, suggesting that MafA could be a Glis3 target gene. Nkx6-1 is initially expressed broadly in the developing pancreatic bud, but eventually is restricted exclusively to β cells (32). It has been shown that Nkx6-1 can repress insulin promoter activity in β cells (33). Nkx6-1 mRNA expression was significantly downregulated by Glis3 overexpression, and upregulated in cells treated with Glis3 siRNA. Thus, Nkx6-1 could be another mediator of Glis3 action in β cells. Pax6 works through the C2 element of the rat Ins1 promoter (16). Similar to Nkx6-1, Pax6 was decreased in Glis3-overexpressing β cells and increased in Glis3-knockdown cells. We only find that Glis3-NDH1 inhibits wild-type Glis3 when co-expressed during transient transfection. We do not see Glis3-NDH1 inhibition of MafA expression or stimulation of expression of Nkx6-1 and Pax-6 when overexpressed in 832/13 cells. Whether authentic Glis3RE sequences are present in the 5′ regulatory regions of the genes for the affected factors and will require further investigation.
In conclusion, we have demonstrated for the first time that Glis3 regulates insulin gene transcription in a cell line derived from β cells. Furthermore, we have identified a 15-bp sequence of the Ins2 promoter required for Glis3 binding, and this sequence is highly conserved among mouse, rat and human, underscoring its role in insulin gene regulation. Our data indicate that Glis3 can act in concert with other known insulin gene transactivators, such as Pdx1, MafA and NeuroD1. Finally, Glis3 expression also may modulate insulin gene transcription by effects on the levels of other β-cell-enriched transcription factors such as MafA, Nkx6-1 and Pax6.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
US National Institutes of Health (NIH) grant DK-68037 (to L.C.); the Diabetes and Endocrinology Research Center (P30DK079638); the Rutherford Chair from St Luke's Episcopal Hospital; T.T. & W.F. Chao Foundation. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Christopher Newgard for the 832/13 cells, Dr Christopher Wright for goat anti-Pdx1 antibody and Dr David Spencer for SRα-SEAP plasmid. We also thank Lan Li, Weiqin Chen and Naravat Poungvarin, for their contributions to this study.
REFERENCES
- 1.Kim YS, Lewandoski M, Perantoni AO, Kurebayashi S, Nakanishi G, Jetten AM. Identification of Glis1, a novel Gli-related, Kruppel-like zinc finger protein containing transactivation and repressor functions. J. Biol. Chem. 2002;277:30901–30913. doi: 10.1074/jbc.M203563200. [DOI] [PubMed] [Google Scholar]
- 2.Zhang F, Nakanishi G, Kurebayashi S, Yoshino K, Perantoni A, Kim YS, Jetten AM. Characterization of Glis2, a novel gene encoding a Gli-related, Kruppel-like transcription factor with transactivation and repressor functions – roles in kidney development and neurogenesis. J. Biol. Chem. 2002;277:10139–10149. doi: 10.1074/jbc.M108062200. [DOI] [PubMed] [Google Scholar]
- 3.Kim YS, Nakanishi G, Lewandoski M, Jetten AM. GLIS3, a novel member of the GLIS subfamily of Kruppel-like zinc finger proteins with repressor and activation functions. Nucleic Acids Res. 2003;31:5513–5525. doi: 10.1093/nar/gkg776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attanasio M, Uhlenhaut NH, Sousa VH, O’Toole JF, Otto E, Anlag K, Klugmann C, Treier AC, Helou J, Sayer JA, et al. Loss of GLIS2 causes nephronophthisis in humans and mice by increased apoptosis and fibrosis. Nat. Genet. 2007;39:1018–1024. doi: 10.1038/ng2072. [DOI] [PubMed] [Google Scholar]
- 5.Kim YS, Kang HS, Herbert R, Beak JY, Collins JB, Grissom SF, Jetten AM. Kruppel-like zinc finger protein Glis2 is essential for the maintenance of normal renal functions. Mol. Cell Biol. 2008;28:2358–2367. doi: 10.1128/MCB.01722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beak JY, Kang HS, Kim YS, Jetten AM. Kruppel-like zinc finger protein Glis3 promotes osteoblast differentiation by regulating FGF18 expression. J. Bone Mineral Res. 2007;22:1234–1244. doi: 10.1359/jbmr.070503. [DOI] [PubMed] [Google Scholar]
- 7.Beak JY, Kang HS, Kim YS, Jetten AM. Functional analysis of the zinc finger and activation domains of Glis3 and mutant Glis3(NDH1) Nucleic Acids Res. 2008;36:1690–1702. doi: 10.1093/nar/gkn009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Senee V, Chelala C, Duchatelet S, Feng DR, Blanc H, Cossec JC, Charon C, Nicolino M, Boileau P, Cavener DR, et al. Mutations in GLIS3 are responsible for a rare syndrome with neonatal diabetes mellitus and congenital hypothyroidism. Nat. Genet. 2006;38:682–687. doi: 10.1038/ng1802. [DOI] [PubMed] [Google Scholar]
- 9.Ohlsson H, Karlsson K, Edlund T. Ipf1, A homeodomain-containing transactivator of the insulin gene. EMBO J. 1993;12:4251–4259. doi: 10.1002/j.1460-2075.1993.tb06109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peshavaria M, Henderson E, Sharma A, Wright CVE, Stein R. Functional characterization of the transactivation properties of the PDX-1 homeodomain protein. Mol. Cell Biol. 1997;17:3987–3996. doi: 10.1128/mcb.17.7.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohneda K, Mirmira RG, Wang JH, Johnson JD, German MS. The homeodomain of PDX-1 mediates multiple protein-protein interactions in the formation of a transcriptional activation complex on the insulin promoter. Mol. Cell Biol. 2000;20:900–911. doi: 10.1128/mcb.20.3.900-911.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao L, Guo M, Matsuoka TA, Hagman DK, Parazzoli SD, Poitout V, Stein R. The islet beta cell-enriched MafA activator is a key regulator of insulin gene transcription. J. Biol. Chem. 2005;280:11887–11894. doi: 10.1074/jbc.M409475200. [DOI] [PubMed] [Google Scholar]
- 13.Aramata S, Han S, Yasuda K, Kataoka K. Synergistic activation of the insulin gene promoter by the beta-cell enriched transcription factors MafA, Beta2, and Pdx1. Biochim. Biophys. Acta Gene Struct. Expression. 2005;1730:41–46. doi: 10.1016/j.bbaexp.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Docherty HM, Hay CW, Ferguson LA, Barrow J, Durward E, Docherty K. Relative contribution of PDX-1, MafA and E47/beta 2 to the regulation of the human insulin promoter. Biochem. J. 2005;389:813–820. doi: 10.1042/BJ20041891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ritz-Laser B, Estreicher A, Klages N, Saule S, Philippe J. Pax-6 and Cdx-2/3 interact to activate glucagon gene expression on the G(1) control element. J. Biol. Chem. 1999;274:4124–4132. doi: 10.1074/jbc.274.7.4124. [DOI] [PubMed] [Google Scholar]
- 16.Sander M, Neubuser A, Kalamaras J, Ee HC, Martin GR, German MS. Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev. 1997;11:1662–1673. doi: 10.1101/gad.11.13.1662. [DOI] [PubMed] [Google Scholar]
- 17.Naya FJ, Stellrecht CMM, Tsai MJ. Tissue-specific regulation of the insulin gene by A novel basic helix-loop-helix transcription factor. Genes Dev. 1995;9:1009–1019. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- 18.Dumonteil E, Laser B, Constant I, Philippe J. Differential regulation of the glucagon and insulin I gene promoters by the basic helix-loop-helix transcription factors E47 and BETA2. J. Biol. Chem. 1998;273:19945–19954. doi: 10.1074/jbc.273.32.19945. [DOI] [PubMed] [Google Scholar]
- 19.Hay CW, Docherty K. Comparative analysis of insulin gene promoters – implications for diabetes research. Diabetes. 2006;55:3201–3213. doi: 10.2337/db06-0788. [DOI] [PubMed] [Google Scholar]
- 20.Melloul D, Marshak S, Cerasi E. Regulation of insulin gene transcription. Diabetologia. 2002;45:309–326. doi: 10.1007/s00125-001-0728-y. [DOI] [PubMed] [Google Scholar]
- 21.Barbetti F. Diagnosis of neonatal and infancy-onset diabetes. Endocr. Dev. 2007;11:83–93. doi: 10.1159/000111060. [DOI] [PubMed] [Google Scholar]
- 22.Li MV, Chen W, Poungvarin N, Imamura M, Chan L. Glucose-mediated transactivation of carbohydrate response element-binding protein requires cooperative actions from mondo conserved regions and essential trans-acting factor 14-3-3. Mol. Endocrinol. 2008;22:1658–1672. doi: 10.1210/me.2007-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hohmeier HE, Mulder H, Chen GX, Henkel-Rieger R, Prentki M, Newgard CB. Isolation of INS-1-derived cell lines with robust ATP-sensitive K+ channel-dependent and -independent glucose-stimulated insulin secretion. Diabetes. 2000;49:424–430. doi: 10.2337/diabetes.49.3.424. [DOI] [PubMed] [Google Scholar]
- 25.Matsumura K, Chang BHJ, Fujimiya M, Chen W, Kulkarni RN, Eguchi Y, Kimura H, Kojima H, Chan L. Aquaporin 7 is a beta-cell protein and regulator of intraislet glycerol content and glycerol kinase activity, beta-cell mass, and insulin production and secretion. Mol. Cell Biol. 2007;27:6026–6037. doi: 10.1128/MCB.00384-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Artner I, Blanchi B, Raum JC, Guo M, Kaneko T, Cordes S, Sieweke M, Stein R. MafB is required for islet beta cell maturation. Proc. Natl Acad. Sci. USA. 2007;104:3853–3858. doi: 10.1073/pnas.0700013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Artner I, Le Lay J, Hang Y, Elghazi L, Schisler JC, Henderson E, Sosa-Pineda B, Stein R. MafB: an activator of the glucagon gene expressed in developing islet alpha- and beta-cells. Diabetes. 2006;55:297–304. doi: 10.2337/diabetes.55.02.06.db05-0946. [DOI] [PubMed] [Google Scholar]
- 28.Boam DS, Clark AR, Docherty K. Positive and negative regulation of the human insulin gene by multiple trans-acting factors. J. Biol. Chem. 1990;265:8285–8296. [PubMed] [Google Scholar]
- 29.Clark AR, Wilson ME, Leibiger I, Scott V, Docherty K. A silencer and an adjacent positive element interact to modulate the activity of the human insulin promoter. Eur. J. Biochem. 1995;232:627–632. [PubMed] [Google Scholar]
- 30.Pino MF, Ye DZ, Linning KD, Green CD, Wicksteed B, Poitout V, Olson LK. Elevated glucose attenuates human insulin gene promoter activity in INS-1 pancreatic beta-cells via reduced nuclear factor binding to the A5/core and Z element. Mol. Endocrinol. 2005;19:1343–1360. doi: 10.1210/me.2003-0493. [DOI] [PubMed] [Google Scholar]
- 31.Sander M, Griffen SC, Huang J, German MS. A novel glucose-responsive element in the human insulin gene functions uniquely in primary cultured islets. Proc. Natl Acad. Sci. USA. 1998;95:11572–11577. doi: 10.1073/pnas.95.20.11572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sander M, Sussel L, Conners J, Scheel D, Kalamaras J, Dela Cruz F, Schwitzgebel V, Hayes-Jordan A, German M. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of beta-cell formation in the pancreas. Development. 2000;127:5533–5540. doi: 10.1242/dev.127.24.5533. [DOI] [PubMed] [Google Scholar]
- 33.Mirmira RG, Watada H, German MS. Beta-cell differentiation factor Nkx6.1 contains distinct DNA binding interference and transcriptional repression domains. J. Biol. Chem. 2000;275:14743–14751. doi: 10.1074/jbc.275.19.14743. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.