Abstract
The molecular basis underlying the aberrant DNA-methylation patterns in human cancer is largely unknown. Altered DNA methyltransferase (DNMT) activity is believed to contribute, as DNMT expression levels increase during tumorigenesis. Here, we present evidence that the expression of DNMT3b is post-transcriptionally regulated by HuR, an RNA-binding protein that stabilizes and/or modulates the translation of target mRNAs. The presence of a putative HuR-recognition motif in the DNMT3b 3′UTR prompted studies to investigate if this transcript associated with HuR. The interaction between HuR and DNMT3b mRNA was studied by immunoprecipitation of endogenous HuR ribonucleoprotein complexes followed by RT–qPCR detection of DNMT3b mRNA, and by in vitro pulldown of biotinylated DNMT3b RNAs followed by western blotting detection of HuR. These studies revealed that binding of HuR stabilized the DNMT3b mRNA and increased DNMT3b expression. Unexpectedly, cisplatin treatment triggered the dissociation of the [HuR-DNMT3b mRNA] complex, in turn promoting DNMT3b mRNA decay, decreasing DNMT3b abundance, and lowering the methylation of repeated sequences and global DNA methylation. In summary, our data identify DNMT3b mRNA as a novel HuR target, present evidence that HuR affects DNMT3b expression levels post-transcriptionally, and reveal the functional consequences of the HuR-regulated DNMT3b upon DNA methylation patterns.
INTRODUCTION
Methylation of DNA at 5-position of cytosine is the predominant epigenetic modification in mammals and regulates critical biological phenomena such as X-chromosome inactivation, genomic imprinting, chromatin structure and regulation of gene expression (1,2) and has been shown to be essential for mammalian development (1). Aberrations in DNA methylation play a causal role in a variety of diseases, including cancer (3). Cancer cells exhibit global DNA hypomethylation and specific promoter hypermethylation of tumor-suppressor genes (4). DNA methylation results from the activity of a family of DNA methyl transferases (DNMTs) that catalyze the addition of a methyl group to cytosine residues at CpG (5). To date, five members of the DNMT family have been described in mammalian cells based on sequence homology within their C-terminal catalytic domain but only three of them have been shown to possess methyltransferase activity. DNMT1 has a preference toward hemimethylated DNA and is responsible for maintaining the methylation patterns following DNA replication (6). The DNMT3 subfamily encodes two functional cytosine methyltransferases, DNMT3a and DNMT3b, which are thought to function as de novo DNA methyltransferases exhibiting equal preference for unmethylated and hemimethylated DNA. DNMT2 and a third homolog of the DNMT3b subfamily, DNMT3L, have failed to show cytosine methyltransferase activity (7). Although Dnmt3a-deficient mice develop to term and appear to be normal at birth, knockout mice deficient in either Dnmt1 or Dnmt3b are embryonic lethal (8). Different studies have shown that DNMTs function in cooperation with each other to facilitate DNA methylation in both human and mouse systems (9,10). For instance, the genetic disruption of the human DNMT3b gene in a colorectal cell line reduced global DNA methylation by less than 3%; however, the genetic disruption of both DNMT1 and DNMT3b nearly eliminated methyltransferase activity and reduced genomic DNA methylation by greater than 95% (10).
All DNMT proteins contain highly conserved COOH-terminal catalytic domains while their NH2-terminal regions are distinct. The NH2-terminal regulatory domain of each DNMT is thought to direct nuclear localization and to mediate interactions with other proteins. Among those DNMTs with proven DNA methyltransferase activity (DNMT1, -3a and -3b), DNMT3b is the only DNMT that is expressed as alternatively spliced variants that affect the integrity of the catalytic domain (11–13). DNMT3b1 and DNMT3b3 are the most highly expressed (14) although only DNMT3b1 and DNMT3b2 have been shown to be catalytically active (15). The role of DNMT3b in DNA methylation is unclear, but it seems to depend on the substrate availability (16,17). The remaining isoforms (DNMT3b4-6) lack an essential conserved motif in the catalytic domain required for enzymatic activity (14,18). On the other hand, mutations in human DNMT3b gene (affecting mainly the catalytic activity) are responsible for the rare autosomal disorder known as ICF syndrome, characterized by variable immunodeficiency, centromeric instability and facial anomalies (19). ICF patients exhibit naturally occurring DNA hypomethylation that affects pericentromeric, and subtelomeric tandem repeats (19). The specific roles of individual DNMT3b splice variants are not fully understood, although DNMT3b isoforms are overexpressed in a variety of human cancers (13). Accordingly, the aberrant methylation of CpG islands in human cancers could be due, in part, to the overexpression of DNMT3b. As the mechanisms underlying the overexpression of DNMT3b in cancer are still unclear, we set out to investigate this topic.
HuR, a protein that binds to target mRNAs and can enhance their stability and modulate their translation, is increasingly recognized as a pivotal regulator of gene expression. Through its post-transcriptional influence on target mRNAs such as those encoding c-fos, c-myc, cyclooxygenase-2, tumor necrosis factor-α, GM-CSF, β-catenin, eotaxin, p27, cyclin A, cyclin B1, cyclin D1, p21, p27, p53 and SIRT1, HuR has been proposed to play major roles in cell proliferation, tumorigenesis, the immune response, senescence and the stress response (20–23). A member of the embryonic abnormal vision family (ELAV), HuR possesses three RNA-recognition motifs through which it binds with high affinity and specificity to target mRNAs bearing AU- and U-rich sequences and modifies their expression by altering their stability, translation, or both (24–26). A signature motif present in HuR target mRNAs was identified, with a specific U-rich primary sequence and a stem-loop secondary structure (27). This HuR motif was successfully used in the prediction of novel HuR target mRNAs such as VDR, HDAC2 and SIRT1 (20,27). The precise mechanisms whereby HuR mediates the stabilization and/or translation of target mRNAs are still poorly understood. However, HuR presence in the cytoplasm appears to be intimately linked to its mRNA-stabilizing function. HuR is predominantly (90%) nuclear in most unstimulated cells, but upon cell stimulation, it can translocate to the cytoplasm where it binds target mRNAs and prevents their decay (26,28,29). Recent studies examining HuR expression in human cancers revealed that the abundance and cytoplasmic localization of HuR protein was significantly greater in malignant tissues than in normal tissues (22,30). Export pathways and signaling cascades that regulate its cytoplasmic abundance have been described (28,31,32). A mechanism through which HuR-binding properties can be modulated has been recently proposed and occurs through HuR phosphorylation by Chk2. HuR phosphorylation at S100 was proposed to reduce, while T118 (and to a lesser extent S88) phosphorylation was proposed to promote HuR binding to target mRNAs (20).
Although the influence of stress model agents (e.g. H2O2 and UVC) on HuR function has been studied extensively, here we sought to test the effect of a chemotherapeutic drug with clinical relevance. Cisplatin is one of the most effective anti-cancer drugs, but the mechanisms underlying its therapeutic action in cancer tissues and its side effects in normal tissues are largely unclear. An en masse search for HuR target mRNAs (27) identified the DNMT3b mRNA as a putative HuR target and computationally detected one hit of the HuR consensus motif in the DNMT3b 3′UTR. Given our long-standing interest in understanding the regulation of DNMTs and DNA methylation (4,33), and the discovery that the DNMT3b mRNA is a predicted target of HuR (27), we set out to investigate if HuR directly regulates the expression of this methyltransferase. HuR was found to associate with the DNMT3b mRNA and to enhance its stability, elevating DNMT3b mRNA steady-state levels. Unexpectedly, cisplatin treatment lowered DNMT3b mRNA and protein expression levels in RKO cells and reduced the stability of the DNMT3b mRNA in a HuR-dependent manner. This effect was caused by the dissociation of HuR from the DNMT3b mRNA following cisplatin treatment, in distinct contrast to the enhanced association of HuR with numerous target mRNAs in response to other stress agents. The dissociation of HuR from the RNP complex was not due to changes in the cytoplasmic abundance of HuR, but instead appeared to be linked to HuR phosphorylation. In turn, modulation of DNMT3b expression by HuR influenced global DNA methylation as well as the methylation of specific DNMT3b target DNA.
MATERIALS AND METHODS
Cell culture, treatments and siRNA transfections
Human colorectal carcinoma RKO cells were cultured in minimum essential medium (Invitrogen), supplemented with 10% fetal bovine serum and antibiotics. Cells were subjected to Cisplatin (Sigma) treatment at the concentrations and times specified in each experiment. Small interfering RNA (siRNA) targeting HuR was AAGAGGCAATTACCAGTTTCA, DNMT3b siRNA was CAGCTCTTACCTTACCATCGA and the control siRNA was AATTCTCCGAACGTGTCACGT; siRNAs (50 nM, Qiagen) were transfected with Oligofectamine (Invitrogen), and cells were harvested 2–3 days after transfection, as indicated. Lipofectamine 2000 (Invitrogen) was used when plasmids were cotransfected.
Immunoprecipitation of RNP complexes
Immunoprecipitation (IP) of HuR–mRNA complexes from RKO cell lysates was used to assess the association of endogenous HuR with endogenous target mRNAs. The IP assay was performed essentially as described (27,34), except that 100 million cells were used as starting material and the lysate supernatants were precleared for 30 min at 4°C using 15 µg of immunoglobulin G (IgG) (Santa Cruz Biotechnology) and 50 µl of protein A-Sepharose beads (Sigma) that had been previously swollen in NT2 buffer [50 mM Tris (pH 7.4), 150 mM NaCl, 1 mM MgCl2 and 0.05% Nonidet P-40 (NP-40)] supplemented with 5% bovine serum albumin. Beads (100 µl) were incubated (18 h, 4°C) with 30 µg of antibody (either mouse IgG [Santa Cruz Biotechnology] or mouse anti-HuR [Santa Cruz Biotechnology]) and then for 1 h at 4°C with 3 mg of cell lysate. After extensive washes and digestion of proteins in the IP material (34), the RNA was extracted and used to perform reverse transcription (RT) followed by semiquantitative PCR or quantitative PCR (qPCR) to detect the presence of specific target mRNAs using gene-specific primer pairs. GAPDH mRNA was used to normalize the data. All oligonucleotide pairs (5′ and 3′ primers, respectively) used are listed in Supplementary Data.
As with all RNP IP data, we routinely normalized the results and assessed differences in mRNA input by measuring two parameters. First, we measured in parallel the binding of, e.g. DNMT3b mRNA to IgG and to anti-HuR antibodies. Therefore, if the lysate contained different levels of DNMT3b mRNA, these would have been reflected as background association of the mRNA to IgG and beads in the IgG IP samples; the levels of DNMT3b associated with HuR in the HuR IP samples would then reflect the ‘Fold enrichment’ of DNMT3b mRNA in HuR IP relative to IgG IP. Second, we routinely measured (in both HuR IP and IgG IP) the levels of GAPDH mRNA. The GAPDH mRNA is not a target of HuR, but it is an abundant transcript in all RNA preparations and can thus be easily measured as a contaminant in the IP beads, plastic of the microfuge tube, etc. Thus, if there were any minor differences in RNA input, these differences would be measured by quantifying the non-specific presence of GAPDH mRNA in IP materials. Statistical analysis was performed using the Wilcoxon test.
Synthesis of biotinylated transcripts and biotin pull-down analysis
For in vitro synthesis of biotinylated transcripts, reverse-transcribed total RNA was used as the template for PCR amplification using 5′oligonucleotides that contained the T7 RNA polymerase promoter sequence. See Supplementary Data for information on the oligonucleotide pairs used. The PCR-amplified products were resolved on agarose gels and the transcripts were purified and used as templates for the synthesis of the corresponding biotinylated RNAs using T7 RNA polymerase and biotin-CTP (26). Biotin pull-down assays (26) were carried out by incubating 40 µg of cytoplasmic lysate with 0.2 µg of biotinylated transcripts for 30 min at room temperature. Complexes were isolated using streptavidin-conjugated Dynabeads (Dynal), and bound proteins in the pull-down material were analyzed by western blotting using antibodies recognizing HuR (below).
Western blot analysis
Whole-cell lysates were prepared using RIPA buffer, as described (35). For the preparation of cytosolic fractions, cells were scraped in 500 μl of RSB lysis buffer (10 mM Tris–HCl pH 7.5, 10 mM NaCl, 3 mM MgCl2, and inhibitors). After the addition of 1% NP-40, the lysate was incubated on ice for 20 min and centrifuged (2000 r.p.m., 5 min, 4°C), and the supernatant was designated as the soluble cytosolic fraction. The pellet was washed once with RSB/1% NP-40 and twice with RSB before adding NB buffer (10 mM Tris–HCl pH 7.4, 400 mM NaCl, 1 mM EDTA and inhibitors), mixed thoroughly for 15 min at 4°C and centrifuged (11 000 r.p.m., 5 min, 4°C), to obtain the nuclear fraction. Protein lysates were resolved by SDS–PAGE and transferred onto nitrocellulose membranes. Antibodies used to detect HuR, Chk2, phospho-Chk2, β-tubulin and Nucleolin (C23) were from Santa Cruz Biotechnology. A monoclonal antibody-recognizing α-Tubulin was from Sigma. Following secondary antibody incubations, signals were visualized by enhanced chemiluminescence.
mRNA stability
For mRNA half-life assessments, three independent experiments were performed. Actinomycin D (5 µg/ml) was added and total RNA was prepared at the times indicated; mRNA half-lives were calculated after quantifying by RT–qPCR, normalizing to 18S RNA levels (using a 1:20 dilution of the stock sample), plotting on logarithmic scales using GraphPad Prism, and calculating the time period required for a given transcript to undergo a reduction to one-half of its initial abundance (at time zero, before adding actinomycin D) using non-linear regression analysis.
Plasmids and reporter assays
For construction of pMIR-DNMT3b and pMIR-GAPDH, PCR products were prepared with primers spanning the corresponding 3′UTRs of each gene (Supplementary Data) and cloned into the SpeI/HindIII site of plasmid pMIR-report (Ambion). Transient transfection of RKO cultures with pMIR, pMIR-DNMT3b or pMIR-GAPDH was carried out using Lipofectamine 2000 (Invitrogen). Cotransfection of pGL4 plasmid containing Renilla (Promega) served as an internal control. Firefly and Renilla Luciferase activities were measured with the Dual Luciferase Reporter Assay System (Promega), following the manufacturer's instructions. All Firefly luciferase measurements were normalized to Renilla luciferase measurements from the same sample. Statistical analysis was performed using the Wilcoxon test.
Quantification of global 5-methylcytosine content
Briefly, genomic DNA samples were boiled, treated with nuclease P1 (Sigma) for 16 h at 37°C, and with alkaline phosphatase (Sigma) for an additional 2 h at 37°C. After hydrolysis, total cytosine and 5 mC content were measured by HPLC-MS in all samples. The LC-ESI/MS system consisted of an Agilent Series 1100 HPLC system coupled to an Agilent LC/MSD VL mass spectrometer equipped with an electrospray ionization source (Agilent Technology, Palo Alto, CA). Fifty microliters of the hydrolyzed-DNA solution were injected onto an Atlantis dC18 column (2.1 × 150 mm; 5 μm particle size) protected by an Agilent guard column (2.1 × 20 mm; 5 μm particle size) at a constant flow of 0.220 ml min−1. Two buffers, 0.1% formic acid in water (Solvent A) and 0.1% formic acid in 50% water: 50% methanol (Solvent B), were used, with a initial gradient of 5% solvent B, then an increase of solvent B to 50% within 9 min and an isocratic gradient (50% of solvent B) during 25 min. Electrospray source conditions were as described in Friso et al. (36), with minor modifications. A drying gas flow of 10.0 l min−1 was employed, with auxiliary 35 psi gas to assist with nebulization and a drying temperature of 350°C. The mass spectrophotometer was operated at a capillary voltage of 4000 V, and spectra were collected in positive ion mode. Identification of 2′-deoxycytidine (dC) and 5-methyl-2′-deoxycytidine (5 mdC) was obtained by UV detection at A254 and A280. Quantification of global DNA methylation were calculated from integration peak areas of 5 mdC relative to global cytidine (5 mdC + dC). Statistical analysis was performed using an unpaired two-tailed Student t-test with Welch correction.
Analysis of sequence-specific DNA methylation
The methylation status of specific genomic DNA sequences was established by bisulfite genomic sequencing (37). Automatic sequencing of 10 colonies for each sequence was performed to obtain data on the methylation status of every single CpG dinucleotide. Statistical analysis was performed using the McNemar test. Primer sequences are available as Supplementary Data.
RNA extraction, RT and semi-quantitative and qPCR
Total RNA was isolated using phenol/chloroform/isoamyl alcohol, according to manufacturer's protocol (Ambion). Five-microgram aliquots were used to perform RT using random hexamers followed by semiquantitative PCR (semiq-PRC) or qPCR to detect the presence of specific amplicons. For semiq-PCR, PCR conditions were 95°C for 7 min followed by a variable number of cycles (n) of 95°C for 30 s, annealing temperature for 30 s and 72°C for 30 s; a final extension was performed at 72°C for 10 min. The parameter ‘n’ varied depending on the Ct value of the gene (calculated by qPCR); three different n cycles were performed per gene around its Ct value. PCR products were visualized after electrophoresis in 1% agarose gels stained with ethidium bromide. Analysis of real-time qPCR amplification of the cDNA was performed using SYBR Green PCR Master Mix (Applied Biosystems) in a 7900HT fast Real-time PCR System (Applied Biosystems). PCR conditions were 50°C for 2 min, 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Assays were normalized to GAPDH mRNA levels.
RESULTS
HuR associates with DNMT3b mRNA
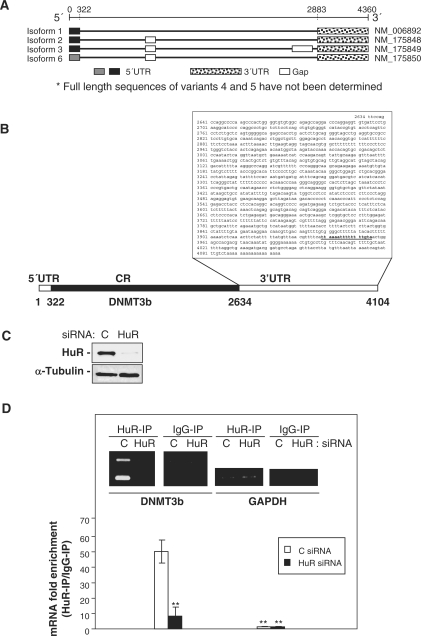
Of the six alternatively spliced transcript variants that have been identified for the human DNMT3b gene, only the full-length sequences of variants 1, 2, 3 and 6 have been determined (Figure 1A). The 3′UTRs of these four DNMT3b transcripts variants are identical, span 1465 nt, and are found to have one computationally predicted hit of an HuR motif in their 3′UTR (27). No HuR hits were found in the mouse DNMT3b gene (27), suggesting that this feature may not be shared among mammalian systems. The identification of the HuR motif in the human DNMT3b predicted that all of the transcript variants could be direct targets of HuR (Figure 1B). To test this possibility, we first assessed whether DNMT3b mRNA associated with HuR by performing immunoprecipitation (IP) assays using anti-HuR antibodies under conditions that preserved the composition of [HuR-mRNAs] RNP complexes. IP was carried out in RKO cells transfected with either control (C) or HuR siRNA, the latter treatment eliciting a significant reduction in HuR levels (5–10% of the HuR levels seen in the control transfection group, Figure 1C). The endogenous association of DNMT3b mRNA with HuR was monitored by isolating RNA from the IP material and subjecting it to RT followed by PCR and quantitative (q) real-time PCR analysis. The DNMT3b primers used for PCR analysis were designed to amplify four different bands; among these, the top band corresponds to variants 1, 2 and 6, and the bottom one to variant 3. Two additional weaker bands can be detected between the top and bottom bands which correspond to variants 4 and 5. For qPCR analysis, specific primers against variant 3 were used. As shown in Figure 1D (graph and inset), the DNMT3b PCR products were dramatically enriched in HuR IP in C siRNA samples compared with control IgG IP samples. For instance, variant 3 was enriched a striking ∼50-fold in the HuR IP as compared with the IgG IP. The absence of enrichment of DNMT3b mRNA in HuR siRNA cells in HuR IP compared with IgG IP supports the specificity of this association. The amplification of a GAPDH PCR product, found in all samples as a low-level contaminating housekeeping transcript, further indicated the specificity of the interaction between HuR and target mRNAs and served to monitor the evenness of sample input.
Figure 1.
HuR associates with the endogenous DNMT3b mRNA. (A) The four alternatively spliced transcript variants for the DNMT3b gene. (B) DNMT3b 3′UTR showing one predicted HuR motif hit. (C) Two days after siRNA transfection, RKO cells were harvested for western blot analysis to monitor the expression of HuR and loading control α-Tubulin. (D) RKO cells were transfected with either control (C) or HuR siRNA and the lysates were used in IP reactions employing anti-HuR antibodies or control IgG1; RNA was subsequently isolated and used in RT reactions (inset). Representative PCR products of all DNMT3b variants visualized in ethidium bromide-stained agarose gels; the levels of GAPDH (a housekeeping mRNA which is not a target of HuR) served to verify equal sample input. (Graph) Fold differences in DNMT3b variant 3 mRNA abundance in HuR IP compared with IgG IP, as measured by RT–qPCR analysis. The means and standard error of the means (SEM) from three independent experiments are represented (**P ≤ 0.01).
[HuR-DNMT3b mRNA] associations were further tested by using biotinylated transcripts spanning the mRNA regions shown (Figure 2, schematic; the star represents the position of the HuR motif within the 3′UTR). Following incubation with RKO cytoplasmic lysates, the interaction between HuR and the different biotinylated transcripts was assessed by biotin pull-down followed by western blot analysis. As shown, HuR only formed complexes with the 3′UTR-DNMT3b (Figure 2). Interestingly, not only did the biotinylated transcript 3′UTR-B (predicted to contain the HuR motif hit) bind to HuR, but the 3′UTR-A transcript (common to all DNMT3b variants) also showed association with HuR, indicating that HuR could interact with multiple regions of the DNMT3b 3′UTR. Supporting this notion is the finding that HuR bound to fragments B1 and B2, which also lack the HuR motif. In the RNP analysis, biotinylated GAPDH 3′UTR (not a target of HuR) served as a negative control transcript while biotinylated p53 3′UTR (a reported HuR target), was used as a positive control. Interestingly, 45% of the cytoplasmic HuR was pulled down with the entire 3′UTR (FL fragment) (Figure 2, graph). We cannot establish whether this association implicated ‘free’ cytoplasmic HuR or HuR that was previously bound to endogenous mRNAs since microccocal nuclease was not added to the samples. Together, these findings indicate that HuR specifically associates with all of the DNMT3b variants, both endogenous and in vitro biotinylated, and that this interaction occurs in at least three different positions of the DNMT3b 3′UTR. From this point forward, unless indicated, the studies have been performed with the DNMT3b variant 3 because it is one of the most abundant splice variants.
Figure 2.
HuR associates in vitro with the DNMT3b mRNA. Schematic representation of the DNMT3b-biotinylated transcripts [CR, CR fragments (A and B), entire 3′UTR (FL), partial 3′UTR (A, B, B1 and B2)] used in biotin pulldown assays. The presence of HuR in the pulldown material was assayed by western blotting; biotinylated GAPDH 3′UTR was included as a negative control. The star represents the position of the HuR motif.
HuR stabilizes the DNMT3b mRNA
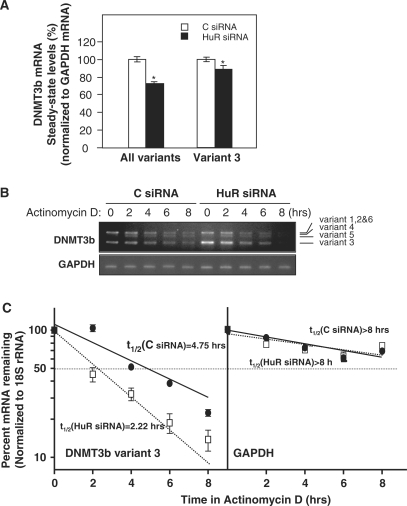
To assess the functional consequences of [HuR-DNMT3b mRNA] interactions, we first measured DNMT3b mRNA levels after silencing HuR. In cells with lowered HuR expression, the steady-state levels of DNMT3b mRNA were reduced by 25% when compared with C siRNA cells; a specific pair of primers were used for RT–qPCR to detect all spliced variants in one single PCR product (Supplementary Data), while a primer pair that specifically detected variant 3 revealed a 12% reduction for this variant (Figure 3A). Since the 3′UTR of the four DNMT3b variants (1, 2, 3 and 6) is identical, these differences might be due to specific sequences in the 5′UTR and CR (Figure 1A). The DNMT3b protein levels could not be studied by western blot or immunofluorescence analyses using any of the commercially available antibodies (data not shown). Contributing to the difficulty in detecting DNMT3b is the fact that DNMT3b protein levels in cancer cell lines is low as compared to the high levels seen in differentiated embryoid bodies (11). To ascertain if the reduction in the DNMT3b mRNA levels was due to changes in mRNA stability, the DNMT3b mRNA stability (t1/2) was analyzed following treatment with actinomycin D to inhibit de novo transcription. The levels of DNMT3b mRNA and the housekeeping control GAPDH mRNA were monitored by RT followed by semiquantitative PCR (using primers that amplify all the DNMT3b variants in different PCR products) and qPCR (using primers that amplify DNMT3b variant 3) (Figure 3B, gel images and graph, respectively). As shown, DNMT3b mRNA was found to be significantly more stable in the control siRNA (C) transfection group, with an estimated t1/2 = 4.75 h for variant 3, while DNMT3b mRNA stability was markedly reduced in the HuR siRNA group, with t1/2 = 2.22 h for variant 3 (Figure 3B). GAPDH mRNA, a very stable mRNA (t1/2 > 8 h) did not show differences between transfection groups, in agreement with the fact that GAPDH mRNA is not a target of HuR target. In summary, HuR specifically enhanced DNMT3b mRNA stability.
Figure 3.
HuR silencing reduces DNMT3b mRNA stability and mRNA levels. (A) Two days after siRNA transfection, RKO cells were harvested, and RNA was analyzed by RT–qPCR (shown as the mean ± SEM from three experiments; *P ≤ 0.05). (B) The DNMT3b mRNA stability after silencing HuR was studied by incubating cells with actinomycin D, extracting total RNA at the times shown, and measuring DNMT3b mRNA levels by RT–PCR. Representative PCR products of all DNMT3b variants visualized in ethidium bromide-stained agarose gels are shown; the levels of GAPDH served to verify equal sample input. (C) DNMT3b variant 3 mRNA levels were measured by RT–qPCR. The data were normalized to 18S (housekeeping) mRNA levels and represented as a percentage of the mRNA levels measured at time 0, before adding actinomycin D, using a semilogarithmic scale. The half-lives (indicated) were calculated as the time required for DNMT3b variant 3 mRNA decrease to 50% of its initial abundance (discontinuous horizontal line). Data represent the mean ± SEM from three independent experiments.
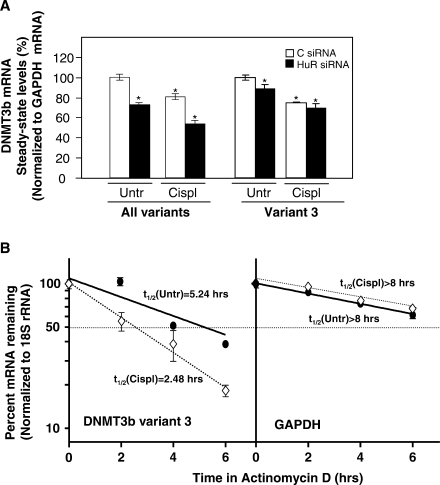
Cisplatin treatment dissociates [HuR-DNMT3b mRNA] complexes, reduces DNMT3b mRNA stability and lowers DNMT3b mRNA abundance
HuR responds rapidly to cellular stress by increasing or decreasing its association to mRNA target genes implicated in cell survival (20,26). Therefore, we extended our studies to examine the functional influence of HuR upon DNMT3b mRNA in response to stress. Exposure of RKO cells to cisplatin (50 µM, 8 h) caused a significant reduction of DNMT3b mRNA levels as compared with untreated cells (Figure 4A). Unexpectedly, the stability of the DNMT3b mRNA was reduced after cisplatin treatment in C siRNA cells (Figure 4B) and HuR siRNA cells (Supplementary Figure 1) as compared to untreated C siRNA cells, suggesting that the lower DNMT3b mRNA abundance (Figure 4A) was due, at least in part, to a cisplatin-triggered decrease in its half-life. The reduction in DNMT3b mRNA levels was further accentuated after treatment with cisplatin, even in cells with silenced HuR, raising the possibility that mechanisms other than dissociation from HuR could also contribute to the loss of DNMT3b mRNA after cisplatin treatment.
Figure 4.
Cisplatin treatment decreases DNMT3b mRNA stability and steady-state levels. (A) Two days after siRNA transfection, cells were treated with 50 μM cisplatin for 8 h, RNA was isolated and RT–qPCR performed (shown as the mean ± SEM from three experiments; *P ≤ 0.05); the data were normalized to GAPDH. (B) The half-lives of DNMT3b variant 3 and housekeeping GAPDH mRNAs in RKO cells transfected with C siRNA (see Supplementary Figure 1 for HuR siRNA cells), either untreated or treated with cisplatin (50 μM), were quantified by using RT–qPCR and calculated as described in the legend of Figure 3B; data represent the mean ± SEM from three independent experiments.
To determine if the reduced DNMT3b mRNA stability after cisplatin treatment was linked to changes in its association with HuR, the levels of these complexes was tested by RNP IP analysis. The amount of DNMT3b mRNA that is constitutively bound to HuR was substantially reduced by the cisplatin treatment, as monitored by RT–qPCR (Figure 5). The reduction in [HuR-DNMT3b mRNA] complexes was time-dependent, decreasing to almost background levels by 2 h after cisplatin treatment, whereas total DNMT3b mRNA levels decreased more slowly, with only a 25–30% decline in variant 3 levels by 8 h after cisplatin treatment, suggesting that the dissociation of this RNP preceded the decay of DNMT3b mRNA (Figures 4A and 5). The association of HuR to other known HuR target transcripts (PTMA, SIRT1, Cyclin D1 and MKP-1 mRNAs) follow distinct patterns and kinetics of binding, indicating that HuR specifically dissociated from DNMT3b mRNA after cisplatin treatment (Figure 5). These observations are consistent with a model whereby cisplatin treatment dissociates DNMT3b mRNA from HuR, in turn triggering DNMT3b mRNA destabilization and reducing DNMT3b mRNA levels.
Figure 5.
Cisplatin treatment decreases [HuR-DNMT3b mRNA] complexes. IP with anti-HuR or IgG antibodies was performed by using lysates that were prepared from either untreated or cisplatin-treated RKO cells (50 μM) at the times indicated; RNA was isolated for RT–qPCR analysis to detect DNMT3b variant 3 mRNA, GAPDH mRNA (a housekeeping control for background binding) and other HuR target mRNAs (shown as the mean ± SEM from three experiments; *P ≤ 0.05; **P ≤ 0.01).
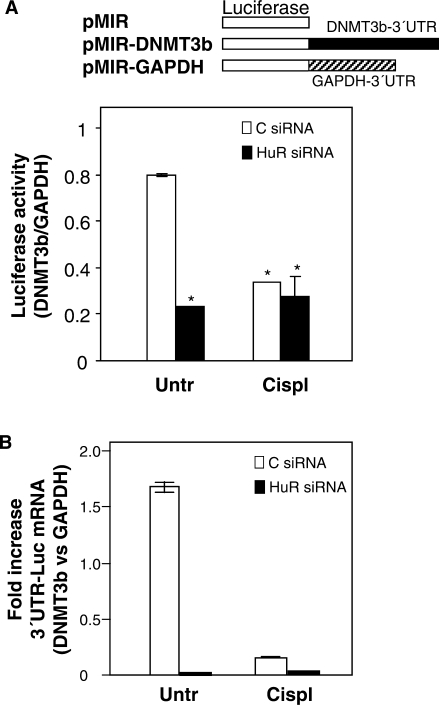
The influence of HuR upon the expression of DNMT3b was further studied through the construction and analysis of luciferase reporter vectors linked to either the full-length DNMT3b 3′UTR or the full-length GAPDH 3′UTR (Figure 6A, schematic). Following transient transfection with the different plasmid constructs, the measurement of luciferase activity showed a marked decrease in the HuR siRNA and cisplatin-treated groups (Figure 6A). Fragments B1, B2 and, to a lesser extent fragment A (Figure 2) recapitulate the effects seen with the full-length DNMT3b 3′UTR (Supplementary Figure 2). Importantly, this reduction was consistent with destabilization of the mRNA, as suggested by the qPCR analysis of the 3′UTR-luc mRNA levels (Figure 6B).
Figure 6.
HuR levels and cisplatin treatment affect the activity of a reporter construct bearing the DNMT3b 3′UTR. Two days after siRNA transfections, the expression vector pMIR-report (firefly luciferase reporter system) containing either DNMT3b 3′UTR (pMIR-3′UTR) or GAPDH 3′UTR (pMIR-GAPDH) were transiently cotransfected into RKO cells along with pGL4-Renilla (used to normalize for transfection efficiency); 24 h later, cells were treated with cisplatin (50 μM, 8 h), and protein and RNA were collected. (A) Protein extracts were used for the detection of firefly and renilla luciferase activities. Graph shows the relative fold increase in firefly luciferase activity seen in the pMIR-DNMT3b transfection group relative to pMIR-GAPDH after normalization to renilla activity. Values represent the means ± SEM from three independent experiments (*P ≤ 0.05). (B) Reverse-transcribed RNA was used for qPCR detection of the reporter DNMT3b 3′UTR-luciferase mRNA relative to the GAPDH 3′UTR-luciferase mRNA. Transfection efficiency was normalized to the amount of pMIR vector present in each sample, which was quantified by qPCR amplification of the CMV promoter region. Data represent the means ± SEM from three independent experiments.
Cisplatin treatment does not change the subcellular localization of HuR
Changes in HuR association to target mRNAs have been largely linked to HuR presence in the cytoplasm. To test if the dissociation of HuR from DNMT3b mRNA after cisplatin treatment was due to changes in HuR cytoplasmic abundance, nuclear and cytoplasmic fractions were collected at different incubation times with cisplatin. Unexpectedly, western blot analysis showed no significant changes in HuR cytoplasmic levels in response to cisplatin (Figure 7). Immunofluorescence staining confirmed the absence of changes (data not shown).
Figure 7.
Cisplatin does not change cytoplasmic HuR levels. Nuclear and cytoplasmic HuR levels were assessed by western blot analysis of RKO cells that were either left untreated or were treated with 50 μM cisplatin for the times shown. α-Tubulin and Nucleolin signals served to show the quality of the preparation of cytoplasmic and nuclear fractions, respectively.
As reported in other cells systems (38), cisplatin treatment triggered the phosphorylation and therefore the activation of the cell cycle checkpoint kinase Chk2 in RKO cells (Supplementary Figure 3A). This finding was particularly important because Chk2 phosphorylates and thereby regulates HuR binding to target mRNAs (20), suggesting that this modification was a potential mechanism responsible for the dissociation of HuR from DNMT3b mRNA after cisplatin treatment. Preliminary data indicate that this is the case as slight shifts in HuR signals were observed by 2D SDS–PAGE after cisplatin treatment (Supplementary Figure 3B).
HuR binding to DNMT3b mRNA influences global DNA methylation as well as the methylation of specific DNMT3b target DNA
We determined whether global DNA methylation was affected by the lowering of DNMT3b levels by HuR. Total cytosine and 5 mC content were measured by high-performance liquid chromatography coupled with mass spectroscopy (HPLC-MS). The percentage of 5 mC relative to the total cytosine content was reduced in both by HuR silencing and by cisplatin treatment (Figure 8A). To study if the decrease in global DNA methylation was due, at least in part, to a direct effect of the DNMT3b activity, we performed bisulfite sequencing on two specific DNA regions that undergo hypomethylation in the ICF syndrome, which is caused by DNMT3b mutations that often affect its methyltransferase activity. Bisulfite sequencing analysis showed a significant decrease in the methylation of the pericentromeric region Sat2 and the subtelomeric D4Z4 tandem repeats in dependence on HuR levels and after cisplatin treatment (Figure 8B).
Figure 8.
HuR silencing and cisplatin treatment affect global DNA methylation and the methylation of specific DNMT3 target DNA sequences. Two days after siRNA transfections, cells were treated with 25 μM cisplatin for 24 h and the DNA was extracted to measure (A) global DNA methylation by HPLC coupled with MS (*P ≤ 0.05; **P ≤ 0.01) and (B) the DNA methylation status of the repetitive sequences D4Z4 and Sat2 by bisulfite sequencing. Black and white squares represent methylated or unmethylated CpGs, respectively (*P ≤ 0.05; **P ≤ 0.01).
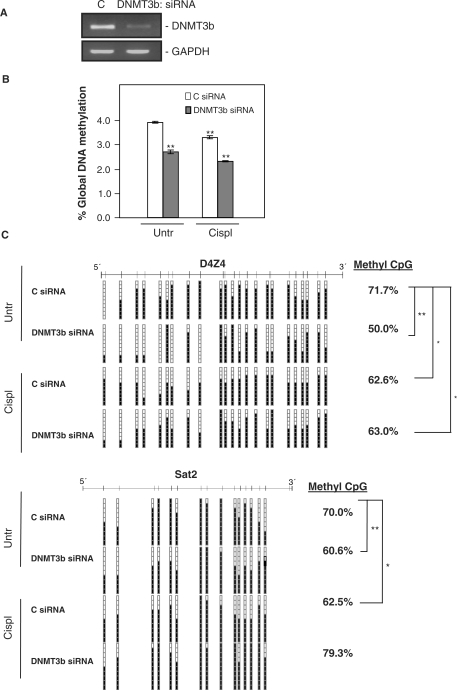
To further strengthen these findings, we extended the analysis of global and regional DNA methylation to cells in which DNMT3b expression was silenced by transfection with a specific siRNA. DNMT3b knockdown reduced DNMT3b mRNA levels down to ∼30% of the levels present in the control transfected cells (Figure 9A). As expected, this intervention caused a reduction in 5 mC content comparable to what was seen after silencing HuR (Figure 9B). Regarding the regional methylation, cells with silenced DNMT3b also showed results comparable to what was seen after HuR silencing (Figure 9C). Taken together, these observations indicate that HuR association with the DNMT3b mRNA influences its expression and activity, which in turn affects global genomic methylation and DNMT3b-specific methylation.
Figure 9.
Effect of DNMT3b silencing on global DNA methylation and methylation of specific DNMT3b target DNA sequences. (A) Two days after transfection with the siRNAs indicated, cells were harvested to check the reduction in DNMT3b mRNA levels by RT–PCR with visualization in agarose gels; GAPDH serves to show the evenness in sample input. Cells that were transfected as explained in (A) were further treated with 25 μM cisplatin for 24 h and the DNA was extracted to measure (B) global DNA methylation by HPLC coupled with MS (*P ≤ 0.05; **P ≤ 0.01) and (C) the DNA-methylation status of the repetitive sequences D4Z4 and Sat2 by bisulfite sequencing. Black and white squares represent methylated or unmethylated CpGs, respectively (*P ≤ 0.05; **P ≤ 0.01).
DISCUSSION
HuR dissociation from DNMT3b
The correct regulation of the DNMT3b gene is critical for the maintenance of normal DNA-methylation patterns. Point mutations in human DNMT3b (usually in the catalytic domain) are responsible for the rare autosomal recessive disorder known as ICF (immunodeficiency, centromere instability and facial anomalies) syndrome. ICF patients’ exhibit a loss of methylation at selected centromeric regions and have profound chromosomal structural changes (39). DNMT3b levels are deregulated in cancer and are found to be elevated in a variety of cancer tissues as compared to their normal counterparts (12,14,40–42). Although elevations in DNMT3b levels were long suspected to cause promoter hypermethylation, a common hallmark of cancer, experimental evidence to support this notion was only recently obtained (43). Here, we show that HuR associates with the DNMT3b mRNA and regulates its expression by enhancing DNMT3b mRNA stability. Surprisingly, cisplatin treatment dissociated [HuR-DNMT3b mRNA] RNP complexes, triggering a reduction in DNMT3b mRNA half-life, which in turn decreased DNMT3b expression and activity.
It was interesting to find that the reduced association of HuR and DNMT3b did not arise from changes in HuR cytoplasmic abundance, the main mechanism proposed to regulate HuR function to-date (44). In fact, the extensively documented HuR translocation to the cytoplasm in response to cellular stress has been linked to the increase association of HuR to target mRNAs (22,26,45,46). This was the case for several transcripts (e.g. p21, p53, PTMA and cyclin D1 mRNAs), whose association to HuR increased after irradiation of the cells with UVC (short-wavelength ultraviolet) light (25,26,47) or after cisplatin treatment, as shown in this study (Figure 5). Here, despite extensive analysis of the subcellular localization of HuR after cisplatin treatment, we failed to detect any significant changes in cytoplasmic HuR levels by either western blot analysis (Figure 7) or immunofluorescence (data not shown).
Interestingly, the association of HuR with target mRNAs was recently shown to be modulated by a mechanism involving the phosphorylation of HuR via the kinase Chk2/Cds1 (20), This phosphorylation affects HuR binding in distinct ways depending on which residue is phosphorylated and on the target mRNA. This new mechanism prompted us to analyze the activation of Chk2 after cisplatin treatment. As shown previously (38), cisplatin activated Chk2 (Supplementary Figure 3A), associated with a modest shift in HuR signals by 2D western blot analysis (Supplementary Figure 3B) suggesting that HuR likely becomes phosphorylated in response to cisplatin. This, in turn could lead to the disassembly of HuR from the RNP complex, as shown after activation of Chk2 following exposure to H2O2 (20). Therefore, Chk2 is one potential candidate to explain the loss of HuR binding to DNMT3b mRNA, but this possibility awaits further experimental testing. An additional explanation for the dissociation of [HuR-DNMT3n mRNA] complex could be the induction of conformational changes in the DNMT3b mRNA due to the formation of cisplatin–RNA adducts. The N7 atoms of guanine and adenine are the main binding sites for cisplatin; the resulting adducts could trigger the formation of intra- or interstrand crosslinks, causing local distortions in the nucleic acid structure (48). As the secondary structure of a previously defined HuR signature motif is a stem-loop (27), cisplatin could alter its conformation, thereby inducing the dissociation of HuR from the mRNA. Whether HuR regulates the DNMT3b mRNA stability in response to other stimuli also remains to be tested, but treatment with trichostatin A (TSA, a histone deacetylase inhibitor) lowered DNMT3b mRNA stability (49). Although the authors of this report did not identify the factor(s) implicated in this regulation, it will be interesting to study if challenge with drugs that alter chromatin structure also dissociates HuR from target DNMT3b mRNA.
Coordinate regulation of DNMT3b
The RNA operon theory posits that multiple mRNAs are co-ordinately regulated by RNA-binding proteins and small non-coding RNAs (50). The regulation of DNMT3b mRNA splice variants by HuR supports this concept. HuR binds to the DNMT3b mRNA in at least three different regions of the 3′UTR (Figures 1D and 2, biotinylated fragments A, B1 and B2). The presence of multiple binding sites within the 3′UTR is becoming a common feature among HuR targets mRNA described in the recent years. For instance, Kawai and colleagues found that HuR binds to three different regions of the cytocrome c 3′UTR (51) while the MKP-1 3′UTR showed HuR binding to two non-overlapping segments (24). Why are there multiple HuR-binding sites within a single 3′UTR? Maybe it is to ensure that HuR binds a given mRNA, even under strong pressure from other competing RNA-binding proteins and/or miRNAs with affinities for the same sequences. Supporting this idea is the fact that some of these HuR-binding regions are also bound by other RNA-binding proteins whereas others are exclusively bound by HuR (24,51,52). In the MKP-1 mRNA, a distal biotinylated fragment (C) was bound by HuR, NF90 and TIAR while a middle fragment (B) was only bound by HuR and TIAR (24). Interestingly, we found that AUF1 and to a lesser extent TIA-1 and TIAR, also associated with the DNMT3b mRNA (Supplementary Figure 4). It will be interesting to analyze if they bind to the same regions as HuR. The combination of multiple mRNAs and RNA-binding protein in an RNA operon can help to dynamically determine the abundance and translational status of a given mRNA following biological perturbations. Highlighting this paradigm is the fact that, similar to HuR, AUF1 [which targets for mRNA degradation and has been recently shown to control translation as well (53)] dissociates almost completely from DNMT3b mRNA after cisplatin treatment; by contrast, TIA-1 and TIAR (both translational inhibitors) associate slightly more with the DNMT3b mRNA (Supplementary Figure 4). The changes in binding patterns might be essential in order to meet the requirements of the cell. For instance, the fact that DNMT3b protein is absent in certain stages of germ cell development despite the presence of some of its transcripts (54) suggests the involvement of these translational inhibitors or possibly miRNAs. An additional layer of complexity in this process is potentially provided by miR-29, which binds the DNMT3b3 3′UTR [(55) and data not shown)] and miR-148, which binds the coding regions of these transcripts with the exception of DNMT3b3 mRNA (56).
A scenario wherein multiple trans-acting factors regulate the DNMT3b mRNA could help us to explain the discrepancies that we found in the reduction of DNMT3b mRNA half life (>2-fold) and the changes in steady-state levels (decreased by only 15–25%) when HuR protein levels are reduced. HuR silencing, which clearly reduced DNMT3b mRNA stability (Figure 3C), might permit access to other trans-acting factors (RNA-binding proteins and/or miRNAs) which can further influence the post-transcriptional metabolism of DNMT3b mRNA levels (for example, binding by TIA1 and TIAR might repress DNMT3b translation). Therefore, all the factors that we also found to associate with the DNMT3b 3′UTR (Supplementary Figure 4) should be taken into account, as they will likely influence the final expression levels of DNMT3b. It is also important to note that the mRNA stability is measured after shutting off transcription; thus, we cannot formally exclude the possibilities that DNMT3b levels might be transcriptionally regulated (for instance, transcription could increase to balance the loss of DNMT3b mRNA levels). It also remains possible that the nuclear processing of DNMT3b mRNA and its cytoplasmic export are altered by cisplatin treatment.
All the above considerations could also help to explain the discrepancies we found in the luciferase experiments, as luciferase activity in reporters bearing the DNMT3b 3′UTR were lowered from 0.8 to 0.2 relative units versus mRNA reduction from 1.6 units to <0.1 units when HuR was silenced (Figure 6). It is possible that the aforementioned factors could also be recruited to regulate the endogenous DNMT3b mRNA. In summary, these results indicate that the DNMT3b is tightly regulated through multiple levels of regulation. HuR is a potent regulator of DNMT3b mRNA stability but additional trans-acting factors (RNA-binding proteins and miRNAs) and other levels of regulation (transcription, other mRNA processing) likely help to determine the final levels of the DNMT3b mRNA and protein.
Influence of cisplatin on HuR function
The finding that cisplatin treatment altered HuR-binding properties has important implications in cancer. Firstly, HuR levels are upregulated in a wide variety of cancers (22,30,57–59). Several studies focusing on cancers of the breast, colon, lung and ovary report a correlation between HuR expression levels and advancing stages of malignancy, supporting a role for HuR in these cancer types (22,57,60,61). HuR is proposed to play an important role in cancer by binding to mRNAs-encoding proteins that participate in malignant transformation (e.g. c-fos, c-myc, COX-2, VEGF and MMP), and modulating their expression via mRNA stabilization and/or altered translation (22,23). Secondly, the cancer-associated overexpression of HuR might contribute to the altered levels of DNMT3b found in many different types of cancer tissues (14,41,42,62). Hypermethylation of the CpG islands in the promoter regions is a common event in many cancers (4). A link between promoter hypermethylation in cancer and DNMT3b was revealed by recent studies which suggested that DNNMT3b was required for tumour development (63) and that the in vivo overexpression of DNMT3b1 caused a loss of imprinting and methylation, leading to the transcriptional silencing of tumour suppressor genes (43). Thirdly, cisplatin is one of the most effective chemotherapeutic agents for cancer treatment. Cisplatin–DNA adducts activate various signal-transduction pathways implicated in DNA–damage recognition and repair, cell-cycle arrest and programmed cell death/apoptosis (48). The finding that cisplatin reduces the binding of HuR to DNMT3b mRNA (and likely other targets) indicate that HuR could be an effector of the changes in gene expression patterns elicited by cisplatin treatment. It will be interesting to evaluate globally the influence of cisplatin on HuR binding to target mRNAs, particularly those which suppress apoptosis such as PTMA (47). The results presented here underscore the importance of HuR as a novel downstream target of cisplatin, pointing to new molecular mechanisms of cisplatin action.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants SAF2007-00027-65134; Consolider CSD2006-49; CANCERDIP FP7-200620; Spanish Ramon & Cajal Programme and the FIS Programme (PI061653) both from the Spanish Ministry of Science and Innovation (to I.L.S.). National Institute on Aging-IRP, National Institutes of Health (to M.G. and K.A.). Funding for open access charge: Fondo de Investigaciones Sanitarias (FIS). Spanish Ministry of Science and Innovation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nick Gilbert for valuable discussions, Francisco Javier Carmona, Vanesa Lafarga, Agustín Fernández, Pilar López, Miguel López, Bianca Barreira, Fernando Setién and Sara Casado for their help.
REFERENCES
- 1.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 2.Robertson KD. DNA methylation and human disease. Nat. Rev. Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 3.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 4.Esteller M. Epigenetics in cancer. N. Engl. J. Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 5.Robertson KD. DNA methylation, methyltransferases, and cancer. Oncogene. 2001;20:3139–3155. doi: 10.1038/sj.onc.1204341. [DOI] [PubMed] [Google Scholar]
- 6.Miremadi A, Oestergaard MZ, Pharoah PD, Caldas C. Cancer genetics of epigenetic genes. Hum. Mol. Genet. 2007;16(Spec No 1):R28–R49. doi: 10.1093/hmg/ddm021. [DOI] [PubMed] [Google Scholar]
- 7.Ferguson-Smith AC, Greally JM. Epigenetics: perceptive enzymes. Nature. 2007;449:148–149. doi: 10.1038/449148a. [DOI] [PubMed] [Google Scholar]
- 8.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 9.Liang G, Chan MF, Tomigahara Y, Tsai YC, Gonzales FA, Li E, Laird PW, Jones PA. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol. Cell Biol. 2002;22:480–491. doi: 10.1128/MCB.22.2.480-491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, Cui H, Feinberg AP, Lengauer C, Kinzler KW, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 11.Chen T, Ueda Y, Xie S, Li E. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J. Biol. Chem. 2002;277:38746–38754. doi: 10.1074/jbc.M205312200. [DOI] [PubMed] [Google Scholar]
- 12.Kanai Y, Hirohashi S. Alterations of DNA methylation associated with abnormalities of DNA methyltransferases in human cancers during transition from a precancerous to a malignant state. Carcinogenesis. 2007;28:2434–2442. doi: 10.1093/carcin/bgm206. [DOI] [PubMed] [Google Scholar]
- 13.Ostler KR, Davis EM, Payne SL, Gosalia BB, Exposito-Cespedes J, Le Beau MM, Godley LA. Cancer cells express aberrant DNMT3B transcripts encoding truncated proteins. Oncogene. 2007;26:5553–5563. doi: 10.1038/sj.onc.1210351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, Jones PA. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 16.Chen ZX, Mann JR, Hsieh CL, Riggs AD, Chedin F. Physical and functional interactions between the human DNMT3L protein and members of the de novo methyltransferase family. J. Cell Biochem. 2005;95:902–917. doi: 10.1002/jcb.20447. [DOI] [PubMed] [Google Scholar]
- 17.Soejima K, Fang W, Rollins BJ. DNA methyltransferase 3b contributes to oncogenic transformation induced by SV40T antigen and activated Ras. Oncogene. 2003;22:4723–4733. doi: 10.1038/sj.onc.1206510. [DOI] [PubMed] [Google Scholar]
- 18.Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell Biol. 2003;23:5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ehrlich M. The ICF syndrome, a DNA methyltransferase 3B deficiency and immunodeficiency disease. Clin. Immunol. 2003;109:17–28. doi: 10.1016/s1521-6616(03)00201-8. [DOI] [PubMed] [Google Scholar]
- 20.Abdelmohsen K, Pullmann R, Jr., Lal A, Kim HH, Galban S, Yang X, Blethrow JD, Walker M, Shubert J, Gillespie DA, et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell. 2007;25:543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 22.Lopez de Silanes I, Fan J, Yang X, Zonderman AB, Potapova O, Pizer ES, Gorospe M. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene. 2003;22:7146–7154. doi: 10.1038/sj.onc.1206862. [DOI] [PubMed] [Google Scholar]
- 23.Lopez de Silanes I, Quesada MP, Esteller M. Aberrant regulation of messenger RNA 3'-untranslated region in human cancer. Cell Oncol. 2007;29:1–17. doi: 10.1155/2007/586139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuwano Y, Kim HH, Abdelmohsen K, Pullmann R, Jr., Martindale JL, Yang X, Gorospe M. MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90. Mol. Cell Biol. 2008;28:4562–4575. doi: 10.1128/MCB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazan-Mamczarz K, Galban S, Lopez de Silanes I, Martindale JL, Atasoy U, Keene JD, Gorospe M. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl Acad. Sci. USA. 2003;100:8354–8359. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang W, Furneaux H, Cheng H, Caldwell MC, Hutter D, Liu Y, Holbrook N, Gorospe M. HuR regulates p21 mRNA stabilization by UV light. Mol. Cell Biol. 2000;20:760–769. doi: 10.1128/mcb.20.3.760-769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl Acad. Sci. USA. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doller A, Akool el S, Huwiler A, Muller R, Radeke HH, Pfeilschifter J, Eberhardt W. Posttranslational modification of the AU-rich element binding protein HuR by protein kinase Cdelta elicits angiotensin II-induced stabilization and nuclear export of cyclooxygenase 2 mRNA. Mol. Cell Biol. 2008;28:2608–2625. doi: 10.1128/MCB.01530-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang W, Fan J, Yang X, Furer-Galban S, Lopez de Silanes I, von Kobbe C, Guo J, Georas SN, Foufelle F, Hardie DG, et al. AMP-activated kinase regulates cytoplasmic HuR. Mol. Cell Biol. 2002;22:3425–3436. doi: 10.1128/MCB.22.10.3425-3436.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brosens LA, Keller JJ, Pohjola L, Haglund C, Morsink FH, Iacobuzio-Donahue C, Goggins M, Giardiello FM, Ristimaki A, Offerhaus GJ. Increased expression of cytoplasmic HuR in familial adenomatous polyposis. Cancer Biol. Ther. 2007;7:424–427. doi: 10.4161/cbt.7.3.5417. [DOI] [PubMed] [Google Scholar]
- 31.Gallouzi IE, Steitz JA. Delineation of mRNA export pathways by the use of cell-permeable peptides. Science. 2001;294:1895–1901. doi: 10.1126/science.1064693. [DOI] [PubMed] [Google Scholar]
- 32.Kim HH, Abdelmohsen K, Lal A, Pullmann R, Jr., Yang X, Galban S, Srikantan S, Martindale JL, Blethrow J, Shokat KM, et al. Nuclear HuR accumulation through phosphorylation by Cdk1. Genes Dev. 2008;22:1804–1815. doi: 10.1101/gad.1645808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Espada J, Ballestar E, Santoro R, Fraga MF, Villar-Garea A, Nemeth A, Lopez-Serra L, Ropero S, Aranda A, Orozco H, et al. Epigenetic disruption of ribosomal RNA genes and nucleolar architecture in DNA methyltransferase 1 (Dnmt1) deficient cells. Nucleic Acids Res. 2007;35:2191–2198. doi: 10.1093/nar/gkm118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tenenbaum SA, Lager PJ, Carson CC, Keene JD. Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods. 2002;26:191–198. doi: 10.1016/S1046-2023(02)00022-1. [DOI] [PubMed] [Google Scholar]
- 35.Lopez de Silanes I, Galban S, Martindale JL, Yang X, Mazan-Mamczarz K, Indig FE, Falco G, Zhan M, Gorospe M. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol. Cell Biol. 2005;25:9520–9531. doi: 10.1128/MCB.25.21.9520-9531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friso S, Choi SW, Dolnikowski GG, Selhub J. A method to assess genomic DNA methylation using high-performance liquid chromatography/electrospray ionization mass spectrometry. Anal. Chem. 2002;74:4526–4531. doi: 10.1021/ac020050h. [DOI] [PubMed] [Google Scholar]
- 37.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl Acad. Sci. USA. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pabla N, Huang S, Mi QS, Daniel R, Dong Z. ATR-Chk2 signaling in p53 activation and DNA damage response during cisplatin-induced apoptosis. J. Biol. Chem. 2008;283:6572–6583. doi: 10.1074/jbc.M707568200. [DOI] [PubMed] [Google Scholar]
- 39.Ehrlich M, Sanchez C, Shao C, Nishiyama R, Kehrl J, Kuick R, Kubota T, Hanash SM. ICF, an immunodeficiency syndrome: DNA methyltransferase 3B involvement, chromosome anomalies, and gene dysregulation. Autoimmunity. 2008;41:253–271. doi: 10.1080/08916930802024202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin F, Dowdy SC, Xiong Y, Eberhardt NL, Podratz KC, Jiang SW. Up-regulation of DNA methyltransferase 3B expression in endometrial cancers. Gynecol. Oncol. 2005;96:531–538. doi: 10.1016/j.ygyno.2004.10.039. [DOI] [PubMed] [Google Scholar]
- 41.Roll JD, Rivenbark AG, Jones WD, Coleman WB. DNMT3b overexpression contributes to a hypermethylator phenotype in human breast cancer cell lines. Mol. Cancer. 2008;7:15. doi: 10.1186/1476-4598-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roman-Gomez J, Jimenez-Velasco A, Agirre X, Cervantes F, Sanchez J, Garate L, Barrios M, Castillejo JA, Navarro G, Colomer D, et al. Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene. 2005;24:7213–7223. doi: 10.1038/sj.onc.1208866. [DOI] [PubMed] [Google Scholar]
- 43.Linhart HG, Lin H, Yamada Y, Moran E, Steine EJ, Gokhale S, Lo G, Cantu E, Ehrich M, He T, et al. Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes Dev. 2007;21:3110–3122. doi: 10.1101/gad.1594007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keene JD. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc. Natl Acad. Sci. USA. 1999;96:5–7. doi: 10.1073/pnas.96.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Subramaniam D, Ramalingam S, May R, Dieckgraefe BK, Berg DE, Pothoulakis C, Houchen CW, Wang TC, Anant S. Gastrin-mediated interleukin-8 and cyclooxygenase-2 gene expression: differential transcriptional and posttranscriptional mechanisms. Gastroenterology. 2008;134:1070–1082. doi: 10.1053/j.gastro.2008.01.040. [DOI] [PubMed] [Google Scholar]
- 46.Wang JG, Collinge M, Ramgolam V, Ayalon O, Fan XC, Pardi R, Bender JR. LFA-1-dependent HuR nuclear export and cytokine mRNA stabilization in T cell activation. J. Immunol. 2006;176:2105–2113. doi: 10.4049/jimmunol.176.4.2105. [DOI] [PubMed] [Google Scholar]
- 47.Lal A, Kawai T, Yang X, Mazan-Mamczarz K, Gorospe M. Antiapoptotic function of RNA-binding protein HuR effected through prothymosin alpha. EMBO J. 2005;24:1852–1862. doi: 10.1038/sj.emboj.7600661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat. Rev. Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 49.Xiong Y, Dowdy SC, Podratz KC, Jin F, Attewell JR, Eberhardt NL, Jiang SW. Histone deacetylase inhibitors decrease DNA methyltransferase-3B messenger RNA stability and down-regulate de novo DNA methyltransferase activity in human endometrial cells. Cancer Res. 2005;65:2684–2689. doi: 10.1158/0008-5472.CAN-04-2843. [DOI] [PubMed] [Google Scholar]
- 50.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 51.Kawai T, Lal A, Yang X, Galban S, Mazan-Mamczarz K, Gorospe M. Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR. Mol. Cell Biol. 2006;26:3295–3307. doi: 10.1128/MCB.26.8.3295-3307.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liao B, Hu Y, Brewer G. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat. Struct. Mol. Biol. 2007;14:511–518. doi: 10.1038/nsmb1249. [DOI] [PubMed] [Google Scholar]
- 54.Lees-Murdock DJ, Shovlin TC, Gardiner T, De Felici M, Walsh CP. DNA methyltransferase expression in the mouse germ line during periods of de novo methylation. Dev. Dyn. 2005;232:992–1002. doi: 10.1002/dvdy.20288. [DOI] [PubMed] [Google Scholar]
- 55.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl Acad. Sci. USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14:872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blaxall BC, Dwyer-Nield LD, Bauer AK, Bohlmeyer TJ, Malkinson AM, Port JD. Differential expression and localization of the mRNA binding proteins, AU-rich element mRNA binding protein (AUF1) and Hu antigen R (HuR), in neoplastic lung tissue. Mol. Carcinog. 2000;28:76–83. [PubMed] [Google Scholar]
- 58.Erkinheimo TL, Lassus H, Sivula A, Sengupta S, Furneaux H, Hla T, Haglund C, Butzow R, Ristimaki A. Cytoplasmic HuR expression correlates with poor outcome and with cyclooxygenase 2 expression in serous ovarian carcinoma. Cancer Res. 2003;63:7591–7594. [PubMed] [Google Scholar]
- 59.Heinonen M, Bono P, Narko K, Chang SH, Lundin J, Joensuu H, Furneaux H, Hla T, Haglund C, Ristimaki A. Cytoplasmic HuR expression is a prognostic factor in invasive ductal breast carcinoma. Cancer Res. 2005;65:2157–2161. doi: 10.1158/0008-5472.CAN-04-3765. [DOI] [PubMed] [Google Scholar]
- 60.Denkert C, Weichert W, Pest S, Koch I, Licht D, Kobel M, Reles A, Sehouli J, Dietel M, Hauptmann S. Overexpression of the embryonic-lethal abnormal vision-like protein HuR in ovarian carcinoma is a prognostic factor and is associated with increased cyclooxygenase 2 expression. Cancer Res. 2004;64:189–195. doi: 10.1158/0008-5472.can-03-1987. [DOI] [PubMed] [Google Scholar]
- 61.Denkert C, Weichert W, Winzer KJ, Muller BM, Noske A, Niesporek S, Kristiansen G, Guski H, Dietel M, Hauptmann S. Expression of the ELAV-like protein HuR is associated with higher tumor grade and increased cyclooxygenase-2 expression in human breast carcinoma. Clin. Cancer Res. 2004;10:5580–5586. doi: 10.1158/1078-0432.CCR-04-0070. [DOI] [PubMed] [Google Scholar]
- 62.Kanai Y, Ushijima S, Kondo Y, Nakanishi Y, Hirohashi S. DNA methyltransferase expression and DNA methylation of CPG islands and peri-centromeric satellite regions in human colorectal and stomach cancers. Int. J. Cancer. 2001;91:205–212. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1040>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 63.Lin H, Yamada Y, Nguyen S, Linhart H, Jackson-Grusby L, Meissner A, Meletis K, Lo G, Jaenisch R. Suppression of intestinal neoplasia by deletion of Dnmt3b. Mol. Cell Biol. 2006;26:2976–2983. doi: 10.1128/MCB.26.8.2976-2983.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.