Abstract
Double-stranded DNA is a dynamic molecule that adopts different secondary structures. Experimental evidence indicates Z-DNA plays roles in DNA transactions such as transcription, chromatin remodeling and recombination. Furthermore, our computational analysis revealed that sequences with high Z-DNA forming potential at moderate levels of DNA supercoiling are enriched in human promoter regions. However, the actual distribution of Z-DNA segments in genomes of mammalian cells has been elusive due to the unstable nature of Z-DNA and lack of specific probes. Here we present a first human genome map of most stable Z-DNA segments obtained with A549 tumor cells. We used the Z-DNA binding domain, Zα, of the RNA editing enzyme ADAR1 as probe in conjunction with a novel chromatin affinity precipitation strategy. By applying stringent selection criteria, we identified 186 genomic Z-DNA hotspots. Interestingly, 46 hotspots were located in centromeres of 13 human chromosomes. There was a very strong correlation between these hotspots and high densities of single nucleotide polymorphism. Our study indicates that genetic instability and rapid evolution of human centromeres might, at least in part, be driven by Z-DNA segments. Contrary to in silico predictions, however, we found that only two of the 186 hotspots were located in promoter regions.
INTRODUCTION
Left-handed Z-DNA is an alternative secondary structure to the right-handed B-conformer (1). It represents a higher energy state with a short half life, unless stabilized by factors such as negative (−) DNA supercoiling or chemical DNA modification (2,3). Z-DNA occurs preferentially in stretches of alternating purine/pyrimidine residues, where the energetic barrier that accompanies a B-to-Z transition is smallest (2). Potential biological functions of Z-DNA have been investigated for over 30 years, since the first molecular structure was revealed by X-ray crystallography (4). Experimental evidence points to the existence of Z-DNA in living mammalian cells (5) and a functional role in processes such as gene regulation (6,7), nucleosome positioning (8,9), chromatin remodeling (10) and recombination (11,12).
Whole genome mapping of Z-DNA has been limited to in silico predictions (13–15), which showed that high potential Z-DNA forming regions (ZDRs) are located preferentially in close proximity to transcriptional start sites (TSS). This finding together with the fact that translocating RNA polymerases can induce (-) supercoiling in their wake in vivo, raised expectations that segments of Z-DNA exist near TSS in a number of gene regulatory regions during the cell cycle (2,3). However, direct evidence is lacking that this is a widespread phenomenon and, if so, that these ZDRs have a function.
We addressed this topic here and developed first a Z-DNA prediction program to determine genomic ZDRs that undergo structural transitions at different levels of DNA supercoiling. We then used the Zα domain of the human RNA editing enzyme ADAR1 (ZαADAR1) as a Z-DNA-specific probe to obtain direct evidence for the existence of ZDRs in Z-conformation within human cells. ZαADAR1 recognizes Z-DNA through two distinct features: the zigzag phosphate backbone and the syn conformation of one purine residue in a Z-DNA segment. The protein binding site occupies only 6 bp of Z-DNA, yet associates with high affinity to the Z-conformer of many different nucleotide sequences (16,17). Using this versatile probe in conjunction with a dual crosslinking chromatin affinity precipitation (ChAP) strategy, we present here a first map of Z-DNA segments in the human genome and discuss biological implications.
MATERIALS AND METHODS
Construction and purification of probes
Zα (GI:2795789) was amplified by PCR from pET28a-Za77 (17). The product was further amplified by PCR to add sequences encoding tags FLAG and Strep II to the 3′-end. The final product was digested with NdeI and EcoRI, and ligated into pET17b. Mutations were introduced at N173A and Y177A by assembly PCR, and cloned again into pET17b as described above. The purification of probes is described in detail in Supplementary Data.
In vitro binding and crosslinking of Zα to Z-DNA
Genomic DNA fragments containing a d(GT)46 insert were amplified from the promoter region of the mouse mast cell protease 6 gene (18) and inserted into pPGKss-puro vector using EcoRI, which resulted in plasmid pGT-pPGKss-Puro. Total 1.5 μg of supercoiled pGT-pPGKss-puro was mixed with 200 ng of a 360 bp d(GT)46 containing PCR fragment amplified from pGT-pPGKss-puro with primers GT-U 5′CCCTTCTGATGACCACAGGTCAC3′ and GT-L 5′TCCAGACTGCCTTGGGAAAAG3′. The DNA mixture was diluted with 350 μl of HEPES binding buffer (50 mM HEPES pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM DTT) and incubated with 1 μg of purified ZαADAR1, ZαADAR1mut or BSA at room temperature for 15 min. Total 0.5% formaldehyde was added and further incubated for 5 min, followed by an incubation with 125 mM glycine for additional 5 min. Ethanol precipitation was used to purify DNA, which was subsequently cleaved with EcoRI and analyzed by EMSA.
In vitro Chromatin Affinity Precipitation (ChAP)
About 2 × 106 A549 cells were crosslinked with formaldehyde and treated with Triton X-100 as described (10). Cells were washed twice with cold PBS and a buffer containing 50 mM HEPES, pH 8.0, 150 mM NaCl, 1 mM EDTA. Thirty micrograms of purified ZαADAR1, ZαADAR1mut or BSA were diluted into 5 ml of HEPES-binding buffer and added to the cells. After incubation at 4°C for 5 h, cells were washed four times with HEPES-binding buffer at 4°C. For the second crosslinking reaction, 0.5% formaldehyde was added in 10 ml HEPES-binding buffer and incubated for 5 min at room temperature. Crosslinking was terminated as described above, and cells were washed five times with cold PBS and harvested in 4 ml cold ChAP lysis buffer (50 mM HEPES, pH 8.0, 1 mM EDTA, 0.5 mM EGTA, 140 mM NaCl, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, 1 mM PMSF). Nuclei were collected at 1500g for 5 min at 4°C and resuspended in nuclei wash buffer (10 mM Tris–HCl, pH 8.0, 1 mM EDTA, 0.5 mM EGTA, 200 mM NaCl, 1 mM PMSF). Nuclei were harvested again and resuspended in 200 μl of SDS lysis buffer (50 mM Tris–Cl, pH 8.1, 10 mM EDTA, 1% SDS). Chromatin was sonicated to yield DNA fragments of 200–1000 bp using a Vibra Cell Ultrasonic Processor (Sonics and Materials, INC). The lysate was cleared at 16 100g for 10 min at 4°C, and samples were diluted with 1.8 ml of ChAP dilution buffer (16.7 mM Tris–Cl, pH 8.1, 167 mM NaCl, 1.2 mM EDTA, 1.1% Triton X-100, 0.01% SDS). Twenty microliters of BSA-blocked Strep-Tactin beads were added to each sample and incubated at 4°C overnight. Beads were washed 5 times with cold RIPA buffer containing 50 mM Tris–Cl, pH 7.5, 1% NP-40, 1% sodium DOC, 0.1% SDS, 1 mM EDTA, 1 M NaCl, 1.5 M Urea, 0.2 mM PMSF, at 15 min for each wash. This was followed by three washes with cold TBS containing 20 mM Tris–Cl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 10 min for each wash. After elution with buffer E (20 mM Tris–Cl, pH 8.0, 150 mM NaCl, 1mM EDTA, 2.5 mM dethiobiotin), the NaCl concentration was adjusted to 200 mM. Crosslinking was reversed by incubation at 65°C for 8 h, and ChAP DNA samples were recovered by ethanol precipitation in the presence of 20 μg of glycogen and resuspended in 50 μl TE buffer.
Z-DNA library construction
ChAP DNA fragments were blunt-ended by T4 DNA polymerase. Adaptor-L (5′AAACGAATTCGAGGAGATTATGGATCCGAC3′) and adaptor-S (5′pGTCGGATCCATAATCTCCTCGAATTCGT3′) were annealed and ligated to ChAP DNA fragments using T4 DNA ligase (NEB). Fragments were amplified with adaptor-L for 25 cycles using Taq PCR Core Kit (Qiagen) with 1 × Q solution. PCR products were separated on a 1.5% agarose gel in 0.5 × TBE buffer and extracted, followed by digestion with BamHI and separated again on a 1.5% agarose gel. DNA fragments were ligated into the pTZ18R vector cleaved with BamHI and treated with CIP (NEB). DNA was purified by phenol/chloroform extraction and ethanol precipitation, and transformed into 40 μl of Stbl4 competent cells using electroporation (Invitrogen). The resulting DNA library was sequenced at the Beijing Genomics Institute. In total, about 10 500 white colonies were picked and sequenced with primer pTZ-L (5′-GATTACGAATTTAATACGACTCACTA-3′).
ChAP–PCR
Sixteen hotspots consisting of at least four ChAP sequences were subjected for further analysis using ChAP–PCR. PCR primers on the hotspots were designed by Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) or Vector NTI Suite® 8 (Invitrogen). A new batch of ChAP DNA was used as template and each PCR was optimized individually. PCR products were analyzed on agarose gels after EtBr staining. PCR using genomic DNA as template was used as control to check the quality of primers. In order to confirm the specificity of ZαADAR1 binding, seven pairs of primers for hotspot regions or nonhotspot regions were designed, and PCR reactions were performed at 2 mM MgCl2 and Ta = 60°C with 35 cycles. PCR products were quantified with Bio-Rad Quantity One software.
RESULTS
We wrote a computer program dubbed Z-catcher in Perl that identifies ZDRs in entire genomes as a function of σ, the DNA superhelical density of protein-free DNA (http://vhp.ntu.edu.sg/zdna/Z_Catcher.zip). Such an approach provides a useful quantitative value that directly indicates the probability for a particular ZDR to be in the left-handed conformation in the human genome and is, therefore, more informative than previous strategies using a pre-set σ value and the program Z-hunt (14,15). Although σ of overall unconstrained (–) supercoiling in the human genome appears null (19), that of transcription-induced, localized (–) supercoiling reaches high levels, even in the presence of eukaryotic topoisomerases (20,21).
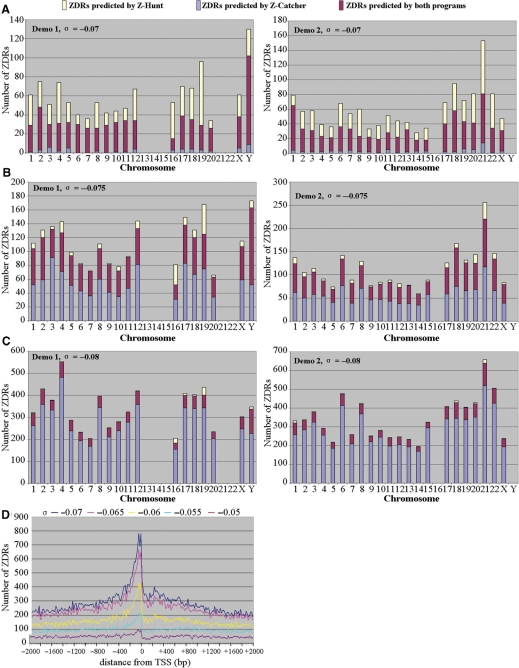
When we subjected the human genome sequence to Z-catcher at σ = –0.07, we found that in agreement with previous in silico analyses using Z-hunt, the occurrence of ZDRs is evenly distributed over the set of human chromosomes (data not shown). In order to directly compare ZDR predictions made by both programs, we used two sets of sequences, labeled Demo 1 and Demo 2, which we randomly selected from the human genome as test sequences. Each set is composed of 24 sequences derived from chromosomal positions 8 400 001 to 8 750 000 in Demo 1, and from positions 42 000 001 to 42 350 000 in Demo 2. Sequences containing undefined bases “N” were excluded, since neither Z-hunt nor Z-catcher can analyze them.
The results of these comparisons show that at σ = −0.07, most ZDRs (91.7%) predicted by Z-catcher were also identified by Z-hunt (Figure 1A). It is also clear that at this σ level, Z-hunt returned more ZDRs than Z-catcher. This changed when we chose σ = –0.075. In this case, Z-catcher returned about 1.5 times more ZDRs than at σ = –0.07. These ZDRs include the majority of those identified by Z-hunt, plus a number of additional ZDRs on each chromosome (Figure 1B). This trend continued when we changed σ to –0.08. Now, the majority of ZDRs returned by Z-catcher is not identified by Z-hunt, while nearly all ZDRs returned by Z-hunt are also predicted by Z-catcher (Figure 1C). Hence, these data show that Z-catcher is a very useful alternative to Z-hunt for in silico ZDR predictions. A detailed description of the former can be found in Supplementary Data.
Figure 1.
Predictions of ZDRs by Z-catcher. (A–C) Comparison of ZDRs identified by Z-catcher and Z-hunt. See text for details. Note that the randomly selected sequences from chromosomes 13, 14, 15, 21 and 22 in Demo 1 contained undefined bases, which prevented in silico analyses. The same applied to sequences from chromosomes 16 and Y in Demo 2. (D) Distribution of ZDRs at various σ levels around TSS in the human genome. All 103 542 representative TSSs of transcription units in the human genome from the Database of Transcriptional Start sites (DBTSS release 5.2.0) were selected. Flanking regions of TSSs 2000 bp upstream and 2000 bp downstream were extracted and subjected to Z-catcher, with σ values ranging from –0.070 to –0.050. The distances between the predicted ZDRs and their corresponding TSSs were calculated and plotted.
Next, we used Z-catcher to perform fine mapping of ZDRs over the entire human genome. The results revealed that they begin to cluster at σ –0.055 between nucleotides + 100 and –600 relative to TSS (Figure 1D). Interestingly, a σ level of –0.06 is in the range of that determined for SV40 DNA purified from nuclei of infected mammalian cells (22). Hence, it seemed indeed likely that some of these ZDRs might adopt a Z-conformation in vivo due to localized (–) DNA supercoiling that may result from nucleosome displacement, transcription, or other changes in chromatin structure, as previously reported for the CSF1 promoter (23).
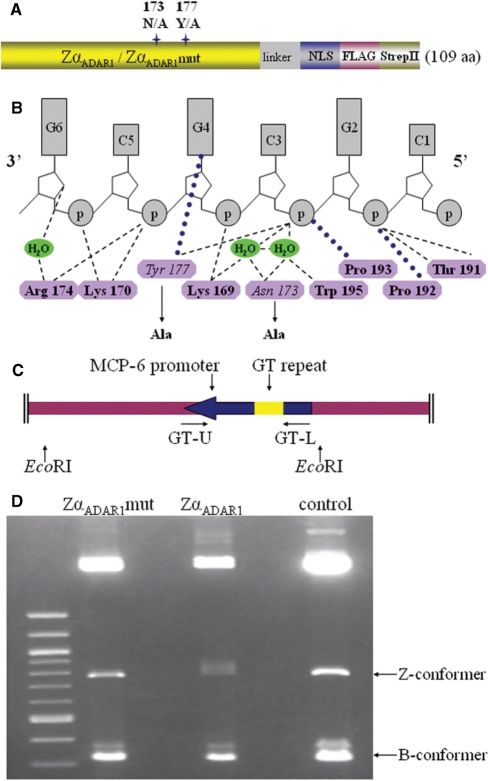
In order to verify that ZDRs predicted by Z-catcher near TSS are in the Z-conformation, we used cell-based assays and ZαADAR1 as the probe (24). We generated a recombinant ZαADAR1 version containing two short tags fused to the C-terminus (Figure 2A). As control probe which cannot recognize Z-DNA, termed ZαADAR1mut, we replaced both tyrosine 177, which specifically interacts with a purine residue in the syn conformation, and asparagine 173, which is engaged in water-mediated contacts with the Z-DNA phosphate backbone, with alanine (Figure 2A and B). Previous data revealed that each of these substitutions alone significantly reduced the binding affinity of ZαADAR1 to Z-DNA (25).
Figure 2.
Construction and characterization of ZαADAR1 probes. (A) Schematic representation of ZαADAR1 and ZαADAR1mut probes. Two tags, FLAG and StrepII, were added to the C-terminus of the probe, plus a nuclear localization signal (NLS). (B) Binding interface between ZαADAR1 and Z-DNA. Nine residues are critical for Z-DNA specific binding of ZαADAR1 (16). (C) Depiction of pGT-pPGKss-puro. The MCP-6 promoter was amplified from the mouse mast cell protease 6 gene and inserted into pPGKss-puro using EcoRI. (D) Specific Z-DNA recognition and crosslinking probed by EMSA. DNA containing the MCP-6 d(GT)46 insert was incubated as supercoiled (Z-conformer) or linear (B-conformer) DNA with ZαADAR1, ZαADAR1mut or BSA control, followed by formaldehyde crosslinking and EMSA. The larger fragments result from EcoRI digestion of the pGT-pPGKss-puro backbone.
ZαADAR1 and ZαADAR1mut were expressed in Escherichia coli and purified to homogeneity. We used pull-down assays and confirmed that ZαADAR1 binds specifically to d(GC)16 in the Z-conformation at 150 mM NaCl (Figure S2). Importantly, only ZαADAR1 could be crosslinked with 0.5% formaldehyde to Z-DNA, as demonstrated by EMSA using a genomic target sequence in either the Z- or B-conformation (Figure 2C and D).
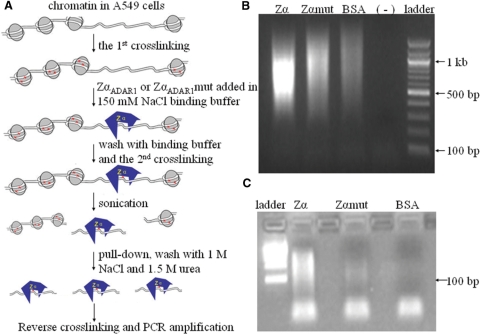
Having established binding specificities and conditions for our probes, we used human A549 lung cancer cells to identify genomic DNA segments in the Z-conformation. We employed a ChAP strategy with dual formaldehyde crosslinking. Cells were first fixed through crosslinking directly on culture plates, incubated with purified probes, which was followed by a second round of crosslinking and ChAP (Figure 3A). After ChAP, DNA fragments were released, linked to adaptors, amplified by PCR, and cloned. Analysis of a sample of the amplified ChAP DNA before cloning revealed that a significant amount of template DNA co-purified only with ZαADAR1 (Figure 3B and C).
Figure 3.
Chromatin Affinity Precipitation (ChAP). (A) Diagram of the experimental strategy. (B) ChAP fragments were amplified by PCR after adaptors ligation. (–) indicates a negative control without added DNA template. (C) PCR products in (B) were digested by BamHI and gel-extracted and analyzed again on an agarose gel.
The resulting genomic library contained about 13 000 clones. After sequencing plus data processing which removed adaptor/vector sequences, 10 005 sequences were subjected to BLASTN (http://blast.ncbi.nlm.nih.gov) or Blat (http://genome.ucsc.edu) for alignment with the human genome build 36.2 (NCBI). We found that 7321 sequences were at least 30 bp long and matched with an identity ≥ 90% and not more than a 1% gap. Total 1715 sequences were mapped to the E. coli genome and 969 were not identified in the NCBI nr (nonredundant) database. Lists of the 10 005 sequences used for the alignment and the 969 unidentified sequences can be found at (http://vhp.ntu.edu.sg/zdna/data.htm).
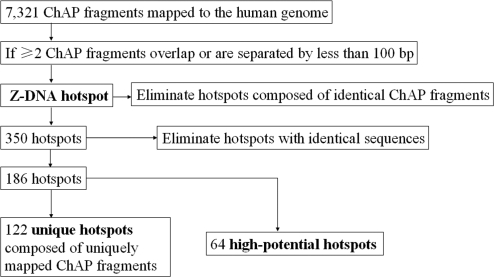
As summarized in a flow chart (Figure 4), we defined a Z-DNA hotspot as a sequence covered by two or more ChAP fragments that either overlapped or are separated by ≤100 bp. This definition was based on the following reasoning: First, genomic fragments containing ZDR(s) in Z-conformation recognized by ZαADAR1 will inevitably exhibit different lengths due to sonication prior to ChAP. This eventually generates a set of overlapping sequences in our library and is, therefore, a strong indicator for specific ZαADAR1 binding. Second, if a genomic region is (–) supercoiled, it is plausible to expect that some ZDRs in Z-conformation might appear in clusters. Therefore, we choose a rather conservative figure of 100 bp as the maximal acceptable distance separating two sequences present in our library. Third, two or more identical ChAP sequences were not considered as they could have been generated as a result of PCR/cloning. It should be highlighted that although we cannot exclude the possibility that sequences which occur only once in our library contain a ZDR(s) in Z-conformation recognizable by our probe, we do not consider them here for further detailed analysis.
Figure 4.
Flow chart for the selection of Z-DNA hotspots based on sequence information. See text for a detailed description.
A total of 350 hotspots fulfilled these criteria. We eliminated next those hotspots with identical sequences which are located at different genomic regions, since this feature made it impossible to determine which one was actually bound by ZαADAR1. The remaining fell into two distinct categories: those that were defined by ChAP fragments which mapped to a unique region in the genome, labeled as ‘unique hotspots’, and so-called ‘high potential hotspots’. The latter were generated by ChAP fragments that individually were found in multiple sequence repeat regions in the genome; however, they clustered according to our hotspot criteria mentioned above only at one unique genomic region (Figure S3). In total, 122 unique and 64 high potential hotspots were finally mapped to human chromosomes (Figure 5; Table S3), and the vast majority (169) was defined by overlapping sequences.
Figure 5.
Z-DNA hotspots mapped to human chromosomes. One hundred and eighty six hotspots were identified and classified as those located in transcribed regions, in centromeres, and unspecified genomic regions. Hotspots were mapped on chromosomes using UCSC Genome Browser, and chromosomes are drawn to scale. Detailed information on all hotspots is listed in Table S3.
The first striking finding was that contrary to expectations based on predictions made by Z-catcher and earlier studies (13–15), only two of our 186 hotspots (1%) were located near TSS (DBTSS 6.01). One was identified in the AK091263 gene on chromosome 6, the other was found inside the human CBX5 gene on chromosome 3 (DBTSS IDs 26805,1 and 28051,3, respectively). It is worth mentioning here that a similar trend was observed for nonhotspot sequences in our library, with 276 of the 6887 (4%) sequences located in TSS.
We found 66 hotspots in transcribed regions, based on ESTs and mRNAs listed in the UCSC Genome Browser. A closer inspection revealed that the majority (49/74.2%) of these hotspots were located in introns. The remaining fraction was found within exons or splice sites. The level of transcription of these 66 hotspots in A549 cells was checked against the NCBI GEO profile GSM94306 database. Only 31 hotspots matched, with 10 transcribed at high and 21 at low levels.
A previous study found Z-DNA formation in three discrete regions of the human c-myc gene in the vicinity of TSSs. The structural transitions there seemed to be linked to transcription in permeabilized nuclei (26). Since c-myc is transcribed in A549 cells, yet no ZDR of this region is present in our hotspot list, we employed ChAP–PCR and the same primer pairs used earlier (26) to determine whether we can detect an enrichment of these c-myc regions in the ZαADAR1 sample when compared with the control sample obtained with ZαADAR1mut. However, the results showed that no enrichment is seen (data not shown).
We found that 136 hotspots belonged to tandem repeat families; 49 are ALR/alpha satellite sequences and 22 were found in Alu subfamilies. The rest belonged to different repeat classes, including LTR/L1 and MER. Interestingly, 34 hotspots in the ALR/alpha satellite family and 12 non-alpha satellite hotspots mapped to centromeres on 13 different chromosomes (Figure 5). An analysis via the RepeatMasker Web Server (http://www.repeatmasker.org/) identified six non-alpha satellite hotspots as HSATII satellites, one as SST1 satellite, two as BSR/beta satellites and three as nonrepeat sequences. Of the 46 centromeric hotspots, 35 were high potential Z-DNA hotspots, and an analysis of all centromeric hotspots via Z-catcher revealed that 36 were predicted to flip into Z-DNA at a (–) σ level between 0.07 and 0.09, while the remaining 10 required higher levels of torsional strain. However, this was not a particular feature of centromeric hotspots as the remaining 140 also showed a similar dependence for high levels of supercoiling (predicted ZDR sequences within hotspots are listed in Table S4). In addition, the 6887 nonhotspot sequences in our library showed the same tendency to undergo structural transitions at (–) σ levels equal to or below 0.08 (data not shown).
In order to provide further evidence that our map of Z-DNA segments was made up of specific binding sites for ZαADAR1, ChAP–PCR was employed using a different biological sample as starting material. We chose 33 hotspots, each defined by at least four ChAP sequences in our library. We were able to design specific primers for 16 hotspots (Table S5). The results showed that, with two exceptions, ZαADAR1 ChAP samples were markedly enriched with hotspot fragments when compared with ZαADAR1mut or BSA control samples (Figure S4A).
We asked next whether these fragments were also enriched over random fragments in the ZαADAR1 ChAP sample. A second round of ChAP–PCR was therefore performed and normalized to control PCRs using genomic DNA as templates. We designed seven pairs of primers for both hotspot and nonhotspot regions (Table S6). The signal of each PCR was quantified and ratios between ChAP–PCR and control PCRs were calculated and plotted. The results showed that six hotspot PCRs produced stronger signals than nonhotspot PCRs (Figure S4B and C).
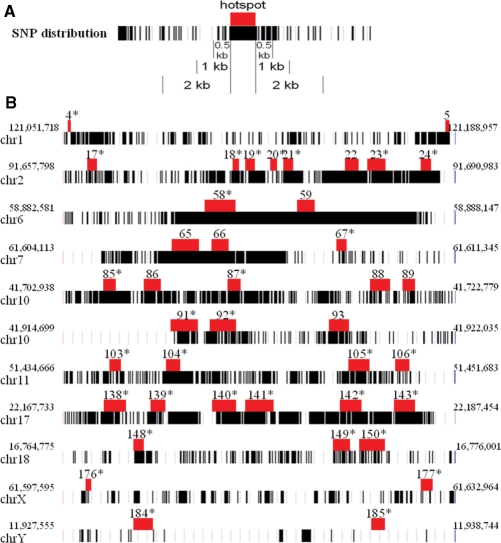
The syn conformation of purines in Z-DNA is more accessible to solvent (27), and the two extruded nucleotides at B–Z junction are potential targets for chemical modification (28). This led us to investigate whether a correlation existed between Z-DNA hotspots and the occurrence of single nucleotide polymorphisms (SNPs) listed in NCBI dbSNP build 128. Both validated RefSNPs and nonvalidated RefSNPs, i.e. all entries in the database, were used for this analysis. RefSNPs in hotspots and in three sections of flanking regions were counted and displayed as SNP density (Figure 6A). Pair-wised Wilcoxon Signed-Rank test was used to compare SNP densities in hotspots and in flanking regions. Interestingly, the 46 hotspots in centromeres correlated with the occurrence of SNPs when validated RefSNPs were considered (Figure 6B, Table 1). This correlation was even more significant (P < 0.001) with nonvalidated RefSNPs. When we considered only the 11 unique hotspots in centromeres with nonvalidated RefSNPs, this correlation was still significant (P < 0.05). However, hotspots found outside centromeres (n = 140) did not reveal a significant enrichment of SNPs (P > 0.1) (Table 1).
Figure 6.
Correlation between Z-DNA hotspots and the occurrence of SNPs in centromeres. (A) Representation of SNP distributions at a hotspot and in flanking regions. Each SNP is represented by a vertical bar. (B) Examples of hotspots and the location of nonvalidated RefSNPs in centromeres. The location of depicted centromere sequences is indicated by the flanking numbers. Hotspots are depicted as red boxes and are drawn to scale. The numbers refer to the hotspot list in Table S3. High potential hotspots were labeled by asterisks.
Table 1.
P values of Wilcoxon Signed-Rank tests of SNP densities
| Flanking regions | 0.5 kb | 1 kb | 2 kb |
|---|---|---|---|
| 46 hotspots in centromeres | |||
| All_RefSNPs | <0.001 | <0.001 | <0.001 |
| Validated_RefSNPs | 0.014 | 0.045 | 0.042 |
| 11 unique hotspots in centromeres | |||
| All_RefSNPs | 0.021 | 0.026 | 0.013 |
| Validated_RefSNPs | 0.075 | 0.041 | 0.05 |
| 140 hotspots outside centromeres | |||
| All_RefSNPs | 0.283 | 0.211 | 0.388 |
| Validated_RefSNPs | 0.865 | 0.816 | 0.942 |
| All 186 hotspots | |||
| All_RefSNPs | 0.11 | 0.103 | 0.089 |
| Validated_RefSNPs | 0.238 | 0.325 | 0.219 |
Distributions of RefSNPs were normalized as SNP densities (SNP/bp) for statistical analysis, which showed that only hotspots in centromeres correlated with high densities of SNPs.
DISCUSSION
Our study provided a first Z-DNA map of the human genome generated by a Z-DNA specific, cross-linkable protein probe, termed ZαADAR1. The novel ChAP strategy employed here was based on a previous finding that formaldehyde treatment does not affect binding of ZαADAR1 to Z-DNA (10). It was shown that d(TG) repeats in the CSF1 promoter region were cleaved by ZααFOK in cells fixed with 1% formaldehyde. ZααFOK is a recombinant restriction enzyme created by the fusion of the nuclease domain from FokI endonuclease with two tandem copies of Zα (29).
In our protocol, A549 cells were treated in a similar way and incubated with our protein probes before the second crosslinking, followed by ChAP. Based on the following observations and reasoning, we think that our approach is unlikely to induce B-to-Z structural transitions in appropriate sequence stretches. First, DNA sequences retrieved from ChAP contained very few d(GT)n or d(GC)n repeats. Because d(GT)-containing microsatellites are abundant in the human genome and expected to flip into Z-conformation at moderate levels of (–) supercoiling, it seems unlikely that many of our hotspots were generated by, for example, nucleosome displacement as a result of the chosen experimental strategy. Second, confirmation of our results by ChAP–PCR using a different biological sample indicated that crosslinking of ZαADAR1 occurred at specific genomic DNA segments. Third, the fact that cells were crosslinked before binding of ZαADAR1 made it unlikely that the probe induced a B-to-Z transition, as observed with oligonucleotides in vitro (24). We think that the accompanying introduction of positive supercoiling will impose a very strong energetic barrier in a fixed chromatin background, which prohibits efficient diffusion of supercoils along the chromatin fiber, unless a chromatin domain is already under negative torsional strain that is insufficient to drive a B-to-Z transition. In the latter case, the introduction of positive supercoils due to binding of ZαADAR1 would be cancelled.
Having established a first Z-DNA map of the human genome by applying very stringent selection criteria, a main conclusion is that the in silico predicted enrichment of ZDRs in Z-conformation near TTS could not be verified. In fact, only two hotspots were found near TSS. This finding does not exclude, however, a possibility that some of these ZDRs adopt Z-conformations in other human cells, or undergo structural transitions as a result of different metabolic states of A549 cells. Furthermore, it remains possible that conformation-specific DNA binding proteins mask some Z-DNA sequences in promoter regions. However, the fact that the A549 chromatin is fixed through histone–DNA crosslinking, which also freezes DNA in its topological state, indicates that a pure thermodynamic-driven in silico approach to predict Z-DNA segments in a genome may have severe limitations. The kinetics of Z-DNA formation in regions near TSS may be too slow, perhaps due to efficient diffusion of transcription-induced (–) supercoiling in these regions. Also, the occupancy of ZDRs near TSS by transcription factors could stabilize the B-conformation. Although some ZDRs in promoter regions may actually adopt the Z-conformation in vivo, as recently shown for the CSF1 gene (10), our data indicate that Z-DNA formation is unlikely to be a key component of a widespread mechanism regulating transcription initiation.
We found that 66 hotspots are located in transcribed genomic regions, and 49 of them were in introns. The formation of Z-DNA there might be linked to transcription-induced supercoiling, as suggested previously for the c-myc gene, corticotropin-releasing hormone gene and beta-globin gene (26,30,31). However, we could not confirm Z-DNA formation in the c-myc gene, which might, at least in part, be due to different protocols employed for binding of protein probes to chromatin. Furthermore, the use of a database of transcriptional profiling confirmed that 31 of the 66 hotspots are actually transcribed in A549 cells, with 21 transcribed at low and 10 at high level. No data could be found in the database for the remaining 35 hotspots, which leaves the possibility open that these are also transcribed at some level. We conclude, therefore, that no strong correlation existed between the level of transcription and Z-DNA formation.
A striking result of our study is that 46 (25%) hotspots were located in centromeres of 13 chromosomes. Computational analysis using Z-catcher revealed that all of them will show a structural transition into Z-DNA at σ ≤ –0.09, which is a rather high level of unconstrained (–) supercoiling. It is known that functional eukaryotic centromeres have an irregular nucleosome positioning (32). This may contribute to the generation of high σ levels, perhaps due to active chromatin remodeling. Formation of Z-DNA may in fact stabilize this situation since Z-DNA cannot be incorporated into nucleosomes (8,9). In addition, histone H3 variant CENP-A is found in centromeres and contributes there to a more rigid nucleosome conformation (33). This might also aid in the build-up of torsional strain. It has been reported that scaffold/matrix attachment regions are more abundant in centromeres or neocentromeres than in chromosome arm regions (34). It is very likely that they demarcate regions under high torsional strain and prevent diffusion of negative supercoiling into neighboring chromatin domains. Finally, the prominence of multiple topoisomerase II cleavage sites in human centromeres could imply that these regions have a high level of unconstrained supercoiling (35).
Centromeres are spindle attachment sites during mitosis and meiosis, and they mediate proper segregation of chromatids into daughter cells. However, centromeres are extremely diverse in sequence composition, ranging from the 125 bp so-called point sequence in Saccharomyces cerevisiae to highly repetitive satellite DNA in vertebrates. In fact, centromere repeats are the most rapidly evolving sequences in eukaryotic genomes (36), involving sequence expansions and contractions. It has been proposed (36) that these repeat variations might provide a functional advantage to centromeres during female asymmetric meiosis. Based on our results, it is possible that some of these gross deletions plus gene conversions might be triggered by formation of Z-DNA, which was shown to induce genomic recombination (11,37,38).
Alpha-satellites are highly variable in sequence composition (39). Careful examination of SNP distributions in centromeres revealed that they are not evenly spread but clustered in certain segments. Our finding that high SNP densities correlated there strongly with hotspots could indicate that Z-DNA plays a crucial role in the accumulation of SNPs during evolution. Since centromeres are anchor points for the kinetochore during mitosis and meiosis (40), some of their structural features related to function should be conserved. This could lead to Z-DNA formation in neighboring sequences, which might be functionally less constrained and more prone to be mutated and to accumulate SNPs over generations. Z-DNA formation could then serve there as a buffer against very high levels of (–) supercoiling which may build up during mitosis.
Our study provided a strategy to generate a first snapshot map of the most stable Z-DNA segments in the human genome. It will be interesting to compare this map in the future with those obtained during different stages of the cell cycle or with other cell types. Since the formation of Z-DNA can be regarded as a real time indicator for genetic activities in a genome (3), comparative studies could ultimately lead to a better understanding of genome-wide chromatin dynamics and genetic stability.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
ARC grant (12/05) from the Ministry of Education, Singapore, and SEP grant from NTU. Funding for open access charge: SEP grant.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Special thanks go to C.A. Davey, G. Davey and C. Schoenbach for critical comments on the manuscript.
REFERENCES
- 1.Dickerson RE, Drew HR, Conner BN, Wing RM, Fratini AV, Kopka ML. The anatomy of A-, B-, and Z-DNA. Science. 1982;216:475–485. doi: 10.1126/science.7071593. [DOI] [PubMed] [Google Scholar]
- 2.Rich A, Zhang S. Timeline: Z-DNA: the long road to biological function. Nat. Rev. Genet. 2003;4:566–572. doi: 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]
- 3.Droge P. Protein tracking-induced supercoiling of DNA: a tool to regulate DNA transactions in vivo? Bioessays. 1994;16:91–99. doi: 10.1002/bies.950160205. [DOI] [PubMed] [Google Scholar]
- 4.Wang AH, Quigley GJ, Kolpak FJ, Crawford JL, van Boom JH, van der Marel G, Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979;282:680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- 5.Heller DA, Jeng ES, Yeung TK, Martinez BM, Moll AE, Gastala JB, Strano MS. Optical detection of DNA conformational polymorphism on single-walled carbon nanotubes. Science. 2006;311:508–511. doi: 10.1126/science.1120792. [DOI] [PubMed] [Google Scholar]
- 6.Oh DB, Kim YG, Rich A. Z-DNA-binding proteins can act as potent effectors of gene expression in vivo. Proc. Natl Acad. Sci. USA. 2002;99:16666–16671. doi: 10.1073/pnas.262672699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rothenburg S, Koch-Nolte F, Rich A, Haag F. A polymorphic dinucleotide repeat in the rat nucleolin gene forms Z-DNA and inhibits promoter activity. Proc. Natl Acad. Sci. USA. 2001;98:8985–8990. doi: 10.1073/pnas.121176998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong B, Chen S, Kwon JA, Rich A. Characterization of Z-DNA as a nucleosome-boundary element in yeast Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2007;104:2229–2234. doi: 10.1073/pnas.0611447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garner MM, Felsenfeld G. Effect of Z-DNA on nucleosome placement. J. Mol. Biol. 1987;196:581–590. doi: 10.1016/0022-2836(87)90034-9. [DOI] [PubMed] [Google Scholar]
- 10.Liu H, Mulholland N, Fu H, Zhao K. Cooperative activity of BRG1 and Z-DNA formation in chromatin remodeling. Mol. Cell Biol. 2006;26:2550–2559. doi: 10.1128/MCB.26.7.2550-2559.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang G, Vasquez KM. Non-B DNA structure-induced genetic instability. Mutat. Res. 2006;598:103–119. doi: 10.1016/j.mrfmmm.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 12.Wang G, Christensen LA, Vasquez KM. Z-DNA-forming sequences generate large-scale deletions in mammalian cells. Proc. Natl Acad. Sci. USA. 2006;103:2677–2682. doi: 10.1073/pnas.0511084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schroth GP, Chou PJ, Ho PS. Mapping Z-DNA in the human genome. Computer-aided mapping reveals a nonrandom distribution of potential Z-DNA-forming sequences in human genes. J. Biol. Chem. 1992;267:11846–11855. [PubMed] [Google Scholar]
- 14.Champ PC, Maurice S, Vargason JM, Camp T, Ho PS. Distributions of Z-DNA and nuclear factor I in human chromosome 22: a model for coupled transcriptional regulation. Nucleic Acids Res. 2004;32:6501–6510. doi: 10.1093/nar/gkh988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khuu P, Sandor M, DeYoung J, Ho PS. Phylogenomic analysis of the emergence of GC-rich transcription elements. Proc. Natl Acad. Sci. USA. 2007;104:16528–16533. doi: 10.1073/pnas.0707203104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartz T, Rould MA, Lowenhaupt K, Herbert A, Rich A. Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science. 1999;284:1841–1845. doi: 10.1126/science.284.5421.1841. [DOI] [PubMed] [Google Scholar]
- 17.Herbert A, Schade M, Lowenhaupt K, Alfken J, Schwartz T, Shlyakhtenko LS, Lyubchenko YL, Rich A. The Zalpha domain from human ADAR1 binds to the Z-DNA conformer of many different sequences. Nucleic Acids Res. 1998;26:3486–3493. doi: 10.1093/nar/26.15.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reynolds DS, Gurley DS, Austen KF, Serafin WE. Cloning of the cDNA and gene of mouse mast cell protease-6. Transcription by progenitor mast cells and mast cells of the connective tissue subclass. J. Biol. Chem. 1991;266:3847–3853. [PubMed] [Google Scholar]
- 19.Sinden RR. DNA Structure and Function. San Diego, CA, USA: Academic Press; 1994. [Google Scholar]
- 20.Wang Z, Droge P. Differential control of transcription-induced and overall DNA supercoiling by eukaryotic topoisomerases in vitro. EMBO J. 1996;15:581–589. [PMC free article] [PubMed] [Google Scholar]
- 21.Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat. Struct. Mol. Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 22.Ambrose C, McLaughlin R, Bina M. The flexibility and topology of simian virus 40 DNA in minichromosomes. Nucleic Acids Res. 1987;15:3703–3721. doi: 10.1093/nar/15.9.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu R, Liu H, Chen X, Kirby M, Brown PO, Zhao K. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell. 2001;106:309–318. doi: 10.1016/s0092-8674(01)00446-9. [DOI] [PubMed] [Google Scholar]
- 24.Herbert A, Alfken J, Kim YG, Mian IS, Nishikura K, Rich A. A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. Proc. Natl Acad. Sci. USA. 1997;94:8421–8426. doi: 10.1073/pnas.94.16.8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schade M, Turner CJ, Lowenhaupt K, Rich A, Herbert A. Structure-function analysis of the Z-DNA-binding domain Zalpha of dsRNA adenosine deaminase type I reveals similarity to the (alpha + beta) family of helix-turn-helix proteins. EMBO J. 1999;18:470–479. doi: 10.1093/emboj/18.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wittig B, Wolfl S, Dorbic T, Vahrson W, Rich A. Transcription of human c-myc in permeabilized nuclei is associated with formation of Z-DNA in three discrete regions of the gene. EMBO J. 1992;11:4653–4663. doi: 10.1002/j.1460-2075.1992.tb05567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zimmerman SB. The three-dimensional structure of DNA. Annu. Rev. Biochem. 1982;51:395–427. doi: 10.1146/annurev.bi.51.070182.002143. [DOI] [PubMed] [Google Scholar]
- 28.Ha SC, Lowenhaupt K, Rich A, Kim YG, Kim KK. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature. 2005;437:1183–1186. doi: 10.1038/nature04088. [DOI] [PubMed] [Google Scholar]
- 29.Kim YG, Kim PS, Herbert A, Rich A. Construction of a Z-DNA-specific restriction endonuclease. Proc. Natl Acad. Sci. USA. 1997;94:12875–12879. doi: 10.1073/pnas.94.24.12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolfl S, Martinez C, Rich A, Majzoub JA. Transcription of the human corticotropin-releasing hormone gene in NPLC cells is correlated with Z-DNA formation. Proc. Natl Acad. Sci. USA. 1996;93:3664–3668. doi: 10.1073/pnas.93.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller V, Takeya M, Brendel S, Wittig B, Rich A. Z-DNA-forming sites within the human beta-globin gene cluster. Proc. Natl Acad. Sci. USA. 1996;93:780–784. doi: 10.1073/pnas.93.2.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marschall LG, Clarke L. A novel cis-acting centromeric DNA element affects S. pombe centromeric chromatin structure at a distance. J. Cell Biol. 1995;128:445–454. doi: 10.1083/jcb.128.4.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Black BE, Brock MA, Bedard S, Woods VL, Jr., Cleveland DW. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc. Natl Acad. Sci. USA. 2007;104:5008–5013. doi: 10.1073/pnas.0700390104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sumer H, Craig JM, Sibson M, Choo KH. A rapid method of genomic array analysis of scaffold/matrix attachment regions (S/MARs) identifies a 2.5-Mb region of enhanced scaffold/matrix attachment at a human neocentromere. Genome Res. 2003;13:1737–1743. doi: 10.1101/gr.1095903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Floridia G, Zatterale A, Zuffardi O, Tyler-Smith C. Mapping of a human centromere onto the DNA by topoisomerase II cleavage. EMBO Rep. 2000;1:489–493. doi: 10.1093/embo-reports/kvd110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 37.Wahls WP, Wallace LJ, Moore PD. The Z-DNA motif d(TG)30 promotes reception of information during gene conversion events while stimulating homologous recombination in human cells in culture. Mol. Cell Biol. 1990;10:785–793. doi: 10.1128/mcb.10.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bacolla A, Jaworski A, Larson JE, Jakupciak JP, Chuzhanova N, Abeysinghe SS, O'Connell CD, Cooper DN, Wells RD. Breakpoints of gross deletions coincide with non-B DNA conformations. Proc. Natl Acad. Sci. USA. 2004;101:14162–14167. doi: 10.1073/pnas.0405974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Charlesworth B, Sniegowski P, Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- 40.Sullivan BA, Blower MD, Karpen GH. Determining centromere identity: cyclical stories and forking paths. Nat. Rev. Genet. 2001;2:584–596. doi: 10.1038/35084512. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.