Abstract
RECQ5 DNA helicase suppresses homologous recombination (HR) possibly through disruption of RAD51 filaments. Here, we show that RECQ5 is constitutively associated with the MRE11–RAD50–NBS1 (MRN) complex, a primary sensor of DNA double-strand breaks (DSBs) that promotes DSB repair and regulates DNA damage signaling via activation of the ATM kinase. Experiments with purified proteins indicated that RECQ5 interacts with the MRN complex through both MRE11 and NBS1. Functional assays revealed that RECQ5 specifically inhibited the 3′→5′ exonuclease activity of MRE11, while MRN had no effect on the helicase activity of RECQ5. At the cellular level, we observed that the MRN complex was required for the recruitment of RECQ5 to sites of DNA damage. Accumulation of RECQ5 at DSBs was neither dependent on MDC1 that mediates binding of MRN to DSB-flanking chromatin nor on CtIP that acts in conjunction with MRN to promote resection of DSBs for repair by HR. Collectively, these data suggest that the MRN complex recruits RECQ5 to sites of DNA damage to regulate DNA repair.
INTRODUCTION
Proteins belonging to the RecQ DNA helicase family are highly conserved from bacteria to mammals. In human cells, five RecQ homologs have been identified and named RECQ1, BLM, WRN, RECQ4 and RECQ5. Inherited defects in the genes encoding for BLM, WRN and RECQ4 have been found to cause distinct autosomal recessive disorders associated with various forms of genomic instability, premature aging and predisposition to cancer (1). Numerous biochemical and cellular studies have shown that the RecQ homologs in human cells have nonredundant roles in the maintenance of genomic stability (2). Several lines of evidence suggest that BLM suppresses crossovers during homologous recombination (HR) by acting in concert with DNA topoisomerase IIIα to catalyze dissolution of double Holliday junctions (3,4). WRN acts specifically at telomeres to promote lagging-strand replication of G-rich telomeric regions, preventing telomere erosion and subsequent recombination (5,6). RECQ4 accumulates on chromatin during initiation of DNA replication to promote loading of replication factors at unwound origins (7,8).
The role of human RECQ5 protein in the maintenance of genomic stability is not well understood. RECQ5 exists in at least three different isoforms resulting from alternative RNA splicing, but only the largest splice variant, RECQ5β, localizes to the nucleus and possesses DNA helicase activity (9–11). Like other RecQ helicases, RECQ5β promotes branch migration of Holliday junctions and exhibits strand-annealing and strand-exchange activities in vitro (11,12). It can also disrupt RAD51 presynaptic filaments in a reaction dependent on its ATPase activity and the presence of replication protein A (RPA) (13). In proliferating cells, RECQ5β associates with the replication machinery and accumulates at sites of stalled replication forks (12). Hereafter RECQ5β will be referred to as RECQ5.
Mice lacking the Recql5 gene are viable, but are highly prone to various types of cancer (13). Recql5-deficient cells accumulate Rad51 and γ-H2AX foci and are prone to gross chromosomal rearrangements in response to replication stress (13). Moreover, Recql5 deficiency is associated with a significant increase in the frequency of both spontaneous and DNA-damage-induced HR (13,14). These findings establish Recql5/RECQ5 as a tumor suppressor that plays a role in the control of HR, possibly through disruption of inappropriately formed Rad51 filaments (13).
The MRE11–NBS1–RAD50 (MRN) complex exhibits 3′→5′ exonuclease activity on double-stranded (ds) DNA and endonuclease activity on single-stranded (ss) DNA and hairpin DNA structures in a reaction stimulated by ATP (15). The nuclease active site resides in MRE11, while RAD50 contains a bipartite ATP binding domain split by a long repeat region that fold into an anti-parallel coiled-coil domain (16). The coiled-coil domain of RAD50 can dimerize via a hook structure located on the tip of this domain, which gives the MRN complex the capacity to tether DNA ends (16,17). NBS1 does not possess any enzymatic function, but it enhances the endonuclease activity of the MRE11–RAD50 complex (18).
The MRN complex is required for the maintenance of the integrity of DNA replication forks, double-strand break (DSB) repair, G2/M checkpoint activation, telomere length maintenance and meiotic recombination (15,19). Accumulating evidence suggests that it functions as a DSB sensor activating the ATM-dependent signaling pathway, which coordinates cell cycle arrest with DNA repair (20,21). In higher eukaryotes, null mutations in all components of the MRN complex are lethal (19), while hypomorphic mutations in the human MRE11 and NBS1 genes can give rise to Ataxia telangiectasia-like disorder (ATLD) and Nijmegen breakage syndrome (NBS), respectively, which are characterized by neurological abnormalities, radiosensitivity, genomic instability and cancer predisposition (19).
Here we show that RECQ5 and the MRN complex constitutively associate in vivo and colocalize at nuclear foci in response to replication arrest and chromosomal breakage. The interaction between RECQ5 and MRN is direct and can be mediated through both MRE11 and NBS1. We further show that the recruitment of RECQ5 to sites of DNA damage is dependent on MRN. Moreover, biochemical experiments reveal that RECQ5 inhibits the 3′→5′ exonuclease activity associated with the MRE11 protein. Together, these data suggest a functional relationship between RECQ5 and the MRN complex in the cellular response to DNA damage.
MATERIALS AND METHODS
Protein purification
The RECQ5 protein was produced in bacteria as fusion with self-cleaving chitin-binding domain (CBD) and purified as described previously (11). Escherichia coli RecQ was purified as described (22). Baculoviruses expressing (His)6-MRE11, (His)6-NBS1 and RAD50, respectively, were a kind gift from Dr Vilhelm Bohr. The MRN and MR complexes as well as the individual MRN subunits were produced in Sf9 cells infected with appropriate baculoviruses. Cells harvested at 72 h after infection were suspended in buffer A [50 mM sodium phosphate (pH 7.0), 0.3 M NaCl, 0.5% (v/v) Tween 20, 10% (v/v) glycerol, 2 mM β-mercaptoethanol] supplemented with 20 mM imidazole and 2 mM phenylmethylsulfonylfluoride (PMSF) and disrupted by sonication. Cell extract was clarified by centrifugation at 100 000g for 1 h and loaded on a 5-ml HiTrap Ni2+-Sepharose column (GE Healthcare) equilibrated with buffer A containing 20 mM imidazole. Bound proteins were eluted with a linear concentration gradient of imidazole (50 ml; 50–350 mM) in buffer A. Fractions containing MRN proteins were pooled, dialyzed against buffer B [20 mM Tris–HCl (pH 8.0), 100 mM NaCl, 10% (v/v) glycerol and 1 mM DL-Dithiothreitol (DTT)] and loaded onto a 1-ml HiTrap Q Sepharose FF column (GE Healthcare). Bound proteins were eluted with a linear concentration gradient of NaCl (12 ml; 50–500 mM) in buffer B. The RAD50 protein was purified only via HiTrap Q Sepharose FF column as it was expressed without a histidine tag. Purified proteins were stored at −80°C. The concentration of purified proteins was determined by Bradford assay. The values obtained in Bradford assay were divided by predicted molecular mass of appropriate protein to calculate molar concentration. The MRN complex was assumed to have an 2:2:1 (M:R:N) stoichiometry (23).
Antibodies
Rabbit polyclonal antibody against the C-terminal portion of RECQ5 (amino acids 675–991) was affinity purified as described previously (12). Additionally, the following commercially available antibodies were used in this study: mouse monoclonal anti-MRE11 antibody, clone 12D7 (Novus Biologicals), mouse monoclonal anti-RAD50 antibody, clone 13B3 (GeneTex), rabbit polyclonal anti-NBS1 antibody, ab-398 (Abcam), mouse monoclonal anti-NBS1 antibody, clone 1D7 (GeneTex), mouse monoclonal anti-γ-H2AX antibody, clone JBW301 (Upstate Biotechnology), rabbit polyclonal anti-TFIIH p89 antibody (S-19), sc-293 (Santa Cruz Biotechnology), mouse monoclonal anti-RAD51 antibody, ab1837 (Abcam), rabbit polyclonal anti-ATM antibody (Abcam), mouse monoclonal anti-RPA antibody (Ab-3), clone RPA34-20 (Calbiochem), mouse monoclonal anti-Cyclin B1 antibody, clone GNS3 (Upstate) and mouse monoclonal anti-CtIP antibody (24).
Cell culture and generation of DNA damage
U2OS, HeLa and HEK 293T cells were maintained in Dulbecco modified Eagle's medium (DMEM; OmniLab) supplemented with 10% fetal calf serum (FCS; Life Technologies) and streptomycin/penicillin (100 U/ml). Immortalized ATLD1 cells transduced with retrovirus expressing the wild-type MRE11 cDNA (ATLD1-MRE11) or retrovirus harboring the empty vector were a kind gift from Dr Matthew Weitzman (25) and grown in DMEM supplemented with 20% FCS, streptomycin/penicillin (100 U/ml) and 1 μg/ml puromycin (Sigma-Aldrich). NBS cells stably transfected with vector containing the wild-type NBS1 cDNA (NBS-NBS1) as well as the parental NBS cells (GM7166 VA7) were obtained from Dr Yosef Shiloh and described in (26). Growth medium of complemented NBS cells was supplemented with hygromycin B (100 μg/ml). MDC1+/+ and MDC1−/− mouse embryonic fibroblasts (MEFs) were cultured under the same conditions as U2OS cells (27). Where indicated, cells were treated with 2 mM hydroxyurea (HU) for 16 h or with 20 μM cis-diamminedichloroplatinum (CDDP) for 8 h. Laser microirradiation to generate DNA DSBs in defined nuclear volumes was performed using a MMI CELLCUT system containing a UVA laser of 355 nm (Molecular Machines & Industries). Prior to irradiation, cells were grown for 24 h in the presence of 10 μM bromodeoxyuridine (BrdU). Ionizing radiation (IR) was generated using a Faxitron X-ray system. UV irradiation (at a dose of 40 J/m2) was performed in a UV-Stratalinker 1800 equipped with a 254 nm UV lamp (Stratagene). Immediately after exposure to radiation, cells were placed back to incubator and incubated for various periods of time as indicated. For cell cycle analyses, ethanol-fixed cells were stained with propidium iodide (20 μg/ml; Molecular Probes) and subjected to flow cytometry in a Becton Dickinson cell sorter.
RNA interference
To knock down MRE11 expression in human cells, we employed short-hairpin (sh) RNA technology. The oligonucleotides shRNA-MRE11-top (5′-gatccccacaggagaagagatcaactttcaagagaagttgatctcttctcctgttttttggaaa-3′) and shRNA-MRE11-bottom 5′-agcttttccaaaaaacaggagaagagatcaacttctcttgaaagttgatctcttctcctgtggg-3′) were annealed and ligated into the pSUPER vector (Oligoengine) digested with HindIII and BglII (regions homologous to the MRE11 sequence are underlined in each oligonucleotide). For control RNAi experiments, the oligonucleotides shRNA-C-top (5′-gatccccagacgtgtacacaactagtttcaagagaactagttgtgtacacgtcttttttggaaa-3′) and shRNA-C-bottom (5′-agcttttccaaaaaagacgtgtacacaactagttctcttgaaactagttgtgtacacgtctggg-3′) were annealed and processed as above. These oligonucleotides were designed by scrambling of the shRNA-MRE11 sequence within the MRE11 homology region. The resulting plasmid constructs were further modified by introducing a puromycin resistance marker. To do so, the 1.4 kb BamHI/PvuI fragment of pPUR (Clontech) was ligated into the BamHI and SmaI sites of the pSUPER derivatives. The shRNA plasmids were introduced into U2OS cells by liposomal transfection with Metafectene (Biontex) that was carried out essentially according to the manufacturer instructions. At 24 h posttransfection, cell cultures were supplemented with puromycin (2 μg/ml) to enhance the fraction of shRNA-expressing clones. Cells were usually harvested 3 days after addition of puromycin and analyzed as described below.
The nucleotide sequences of CtIP and control (Ctrl) siRNAs and siRNA transfection method were described previously (24).
Immunofluorescence staining and analyses
Cells grown on cover slips were fixed in methanol for 30 min at −20°C, which was followed by incubation in acetone for 30 s. After blocking in PBS supplemented with 2% FCS (blocking solution), cover slips were incubated overnight at 4°C with appropriate primary antibodies [rabbit polyclonal anti-RECQ5 (1:1000) in combination with either mouse monoclonal anti-RAD50 (1:200) or mouse monoclonal anti-γ-H2AX (1:200); all antibodies were diluted in blocking solution]. After washing with PBS, the cells were incubated with FITC-conjugated sheep anti-rabbit (Sigma; dilution of 1:700) and Texas Red-conjugated donkey anti-mouse (Abcam; dilution of 1:200) secondary antibodies for 1 h at room temperature. The cover slips were then mounted with Vectashield (Vector Labs) and sealed. Images were taken on an Olympus IX81 fluorescence microscope and acquired with a CCD camera (Orca AG, Hamamatsu) using cellR software (Olympus). At least 150 nuclei were analyzed in each experiment.
Immunoprecipitation assay
Cells grown in a 10-cm dish were subjected to trypsinization, harvested by centrifugation and suspended in 0.5 ml of lysis buffer [50 mM Tris–HCl (pH 8.0), 120 mM NaCl, 20 mM NaF, 15 mM sodium pyrophosphate, 0.5 mM sodium orthovanadate and 0.5% (v/v) NP-40] supplemented with a protease inhibitor cocktail (Complete, Mini; Roche). After a 30-min incubation on ice, the lysate was treated with 50 U of RNase-free DNase I (Roche) at 25°C for 30 min, and clarified by centrifugation. Cell extracts (1 mg) were incubated overnight at 4°C with purified rabbit anti-RECQ5 IgGs (1 μg) or with IgGs purified from pre-immune serum (1 μg). Immune complexes were subsequently incubated with protein A/G-agarose beads (20 μl, Santa Cruz Biotechnology) for 1 h at 4°C. After extensive washing with lysis buffer, beads were boiled in SDS–PAGE loading buffer (25 μl) for 10 min to release bound proteins. Immunoprecipitates were analyzed by western blotting. For mass spectrometry (MS) analysis, the immunoprecipitation protocol was scaled up to include 10 mg of total protein. The eluates from protein A/G–agarose beads were concentrated using a speed-vac before loading on a 7.5% (w/v) SDS-polyacrylamide gel.
Liquid chromatography-MS/MS analysis
SDS–PAGE gel was stained with Coomassie Brilliant Blue and sample lanes were cut into 10 slices. Each slice was further diced into smaller cubes (1 mm) and subjected to two cycles of rehydration in 50 mM ammonium bicarbonate (NH4HCO3) and dehydration in 80% (v/v) acetonitrile. Gel pieces were then incubated with 37 mM DTT solution at 50°C for 30 min. After two additional rounds of dehydration in 80% (v/v) acetonitrile, gel pieces were incubated with 20 mM iodoacetamide for 15 min in dark for protein alkylation. After another three rounds of rehydration in 50 mM NH4HCO3 and shrinking in 80% (v/v) acetonitrile, the gel pieces were incubated with freshly diluted trypsin solution (12.5 ng/μl) at 37°C for 4 h and then at 25°C overnight. Digested peptides were finally extracted by 0.1% formic acid (one time) and 80% (v/v) acetonitrile (three times) after which extracts were dried under vacuum. The tryptic peptides were analyzed on a Finnigan LCQ Deca (ThermoFisher, Basel Switzerland). Prior to MS analysis, peptides were separated by an online high-pressure liquid chromatograph (Agilent, Palo Alto, CA) on a C18 reverse phase column using acetonitrile/water system. MS/MS spectra were searched against the human portion (taxonomy ID: 9606) of the UniProt database (http://www.uniprot.org). The peptides identified were interrogated using the Mascot search algorithm (28). Mascot protein score of >45 was considered significant. Individual ions scores >41 indicate identity or extensive homology (P <0.05).
CBD pull-down assay
CBD-tagged RECQ5 was produced in E. coli BL21-CodonPlus(DE3)-RIL cells (Stratagene) as previously described (11). Cells harvested from a 10-ml culture were resuspended in 1 ml of NET-150 buffer [10 mM Tris (pH 8.0), 1 mM EDTA, 150 mM NaCl, 10% (v/v) glycerol and 0.1% (v/v) Triton X-100] supplemented with a protease inhibitor cocktail (Complete, Mini; Roche) and disrupted by sonication followed by centrifugation at 20 000g for 45 min. Clarified cell extract (typically 50 μl) was incubated with 20 μl of chitin beads (NEB) in a total volume of 400 μl of NET-150 buffer for 2 h at 4°C. After extensive washing with NET-150 buffer, beads were incubated with recombinant MRN proteins (1 μg) in 400 μl of NET-150 buffer supplemented with ethidium bromide (50 μg/ml) for 2 h at 4°C. Bound proteins were analyzed by western blotting.
Strand exchange assay
Strand exchange assays with RECQ5 were performed essentially as described previously (12). Briefly, reactions were carried at 37°C in buffer containing 20 mM Tris–acetate (pH 7.9), 50 mM KOAc, 10 mM Mg(OAc)2, 1 mM DTT and 50 μg/ml BSA. 1 nM DNA substrates (60-mer/30-mer partial duplex and complementary 60-mer oligonucleotide, of which the former 60-mer oligonucleotide was radiolabeled at 5′ end) were preincubated with 40 nM RECQ5 for 10 min to form a forked duplex. RECQ5-mediated strand exchange between the arms of the fork was initiated by addition of ATP (2 mM). Where required, the annealing mixture was preincubated with 40 nM MRN complex for 5 min prior to addition of ATP. Aliquots (5 μl) from different reaction time points were subjected to electrophoresis in 10% nondenaturing polyacrylamide gel (acrylamide/bis-acrylamide 19:1) run in 1xTBE buffer at 140 V. Gels were dried and subjected to phosphor-imaging analysis using a Typhoon 9400 scanner.
Nuclease assay
The 3′→5′ exonuclease activity of the MR(N) complex was measured using a 5′-tailed oligoduplex prepared by annealing of a 30-mer oligonucleotide (5′-ggagtaaagtactaggtatgtcgacattga-3′) to the 3′-half of a 60-mer oligonucleotide (5′-gaggtcactccagtgaattcgagctcgcagtcaatgtcgacatacctagtactttactcc-3′). The 30-mer oligonucleotide was radiolabeled at the 5′-end prior to annealing. Reactions were conducted at 37°C in buffer containing 30 mM MOPS (pH 7.0), 25 mM KCl, 1 mM DTT, 2 mM MnCl2, 2 mM ATP and 50 μg/ml BSA. Reaction mixtures contained 1 nM DNA substrate, 40 nM MR(N) and varying concentrations of RECQ5 or E. coli RecQ, ranging from 0 to 80 nM. MR(N) and RECQ5 (or RecQ) were preincubated for 5 min on ice before adding to the DNA substrate. Reactions were terminated after 30 min by adding an equal volume of stop solution [80% (v/v) formamide, 10 mM EDTA, 0.1% (w/v) bromophenol blue and 0.1% (w/v) xylene cyanol] and heating at 95°C for 5 min. Reaction aliquots were subjected to electrophoresis in 15% (w/v) polyacrylamide gel containing 8 M urea (acrylamide/bis-acrylamide 19:1) run in 1xTBE buffer at 300 V. Gels were dried and subjected to phosphor-imaging analysis using a Typhoon 9400 scanner. DNA bands in the gel images were quantified using ImageQuant software (Molecular Dynamics).
RESULTS
Physical interaction between RECQ5 and the MRN complex
To identify proteins associated with RECQ5 in vivo, we performed large-scale immunoprecipitation combined with mass spectrometric analysis. Using affinity-purified rabbit polyclonal anti-RECQ5 antibody, we immunoprecipitated RECQ5 from whole cell extract of HEK 293T cells containing 10 mg of protein. IgG purified from preimmune serum was used as the negative control. The immunoprecipitated proteins were separated by SDS–PAGE and visualized by Coomassie Blue staining (Figure S1). The lane containing RECQ5 immunoprecipitate was cut into 10 slices and proteins in each slice were identified by MS as described in Materials and Methods section. This analysis revealed a number of potential RECQ5 interactors involved in DNA metabolism (Figure S1 and Table 1). Our attention was in particular drawn to the MRN complex, a nuclease that plays a pivotal role in the recognition and repair of DSBs (20,21).
Table 1.
A selection of proteins identified by mass spectrometric analysis of the RECQ5 immunoprecipitate from 293T cells
| Protein | Swiss-Prot entry | Sequence coverage (%) | Number of peptides | Mascot protein scorea |
|---|---|---|---|---|
| RECQ5b | O94762 | 18.0 | 57 | 347 |
| MRE11b | P49959 | 13.7 | 43 | 257 |
| RAD50b | Q92878 | 20.1 | 120 | 253 |
| NBS1b | O60934 | 6.1 | 18 | 157 |
| DNA-dependent protein kinase catalytic subunit (DNA-PKcs)b | P78527 | 15.5 | 160 | 1354 |
| RECQ4b | O94761 | 5.2 | 26 | 111 |
| MCM7b | P33993 | 20.4 | 44 | 251 |
| RNA polymerase II largest subunit RPB1b | P24928 | 25.6 | 240 | 1602 |
| RNA polymerase II subunit 2 (RPB2) | P30876 | 12.7 | 43 | 315 |
| Nucleolin | P19338 | 11.4 | 13 | 109 |
| E3 ubiquitin–protein ligase EDD1 (UBR5) | O95071 | 3 | 25 | 370 |
aMascot protein score of >45 was considered significant.
bConfirmed by immunoblotting (IB).
The full list is available upon request.
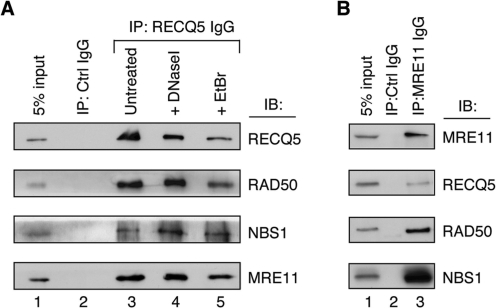
To verify the MS data, the RECQ5 immunoprecipitate from 293T cells was subjected to western blot analysis using antibodies against the individual MRN subunits. We found that this immunoprecipitate contained a large fraction of the endogenous MRN proteins, whereas the control immunoprecipitate obtained with IgG isolated from a pre-immune rabbit serum was devoid of these proteins (Figure 1A, compare lanes 1–3). The amount of MRN detected in RECQ5 immunoprecipitates from cell lysates pre-treated with DNaseI or ethidium bromide was comparable with that detected in RECQ5 immunoprecipitate from nontreated lysate, indicating that the observed association of RECQ5 with MRN proteins is DNA independent (Figure 1A; compare lanes 3, 4 and 5). We also performed a reciprocal co-immunoprecipitation experiment using anti-MRE11 antibody. We found that MRE11 immunoprecipitate from HeLa cells contained a significant amount of RECQ5 (Figure 1B). Collectively, these data indicate that a fraction of the MRN complex stably associates with RECQ5 in vivo.
Figure 1.
RECQ5 associates with the MRN complex in vivo. (A) Co-immunoprecipitation of the MRE11, RAD50 and NBS1 proteins with RECQ5 from total extract of 293T cells (1 mg of protein). Lane 1, 5% of the input material; lane 2, immunoprecipitation using IgG (1 μg) isolated from a preimmune serum; lane 3, immunoprecipitation with affinity-purified rabbit anti-RECQ5 antibody (1 μg); lane 4, the same as lane 3, but the extract was pretreated with DNase I (50 U) for 30 min at 25°C; lane 5, the same as lane 3, but extract was supplemented with ethidium bromide (50 μg/ml). (B) Co-immunoprecipitation of RECQ5 with the MRE11 protein from total extract of HeLa cells. Lane 1, 5% of the input material; lane 2, immunoprecipitation using IgG (1 μg) isolated from a preimmune serum; lane 3, immunoprecipitation using mouse monoclonal anti-MRE11 antibody (1 μg). (A and B) Immunoprecipitated proteins were analyzed by SDS–PAGE followed by IB using antibodies described in Materials and Methods section.
Next, we compared the levels of the MRE11 protein in RECQ5 immunoprecipitates from cells exposed with various genotoxic agents including HU, IR and CDDP. This experiment revealed that the cellular level of the RECQ5–MRN complex is not significantly affected by DNA damage, suggesting that DNA damage does neither induce nor prevent the formation of the RECQ5–MRN complex in vivo (Figure S2A, compare lanes 2–4 with lane 1). We also examined the levels of RECQ5–MRN complex at different stages of the cell cycle. To do so, U2OS cells were synchronized at G1/S transition by HU treatment and then released into drug-free medium. Cells were collected at different time points after the removal of HU and subjected to immunoprecipitation with anti-RECQ5 antibody. In a parallel experiment, cell populations from the individual time points were subjected to FACS analysis to determine the stage of the cell cycle (Figure S2C). We found that the level of the RECQ5–MRN complex remained constant throughout the cell cycle, suggesting a constitutive association (Figure S2B).
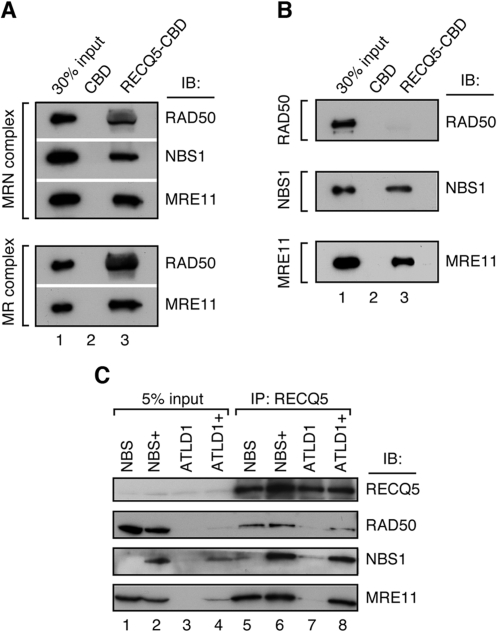
To determine whether RECQ5 and MRN interact directly, we performed affinity pull-down assays with purified recombinant proteins. RECQ5 was expressed in bacteria as a fusion with a CBD tag and bound to chitin beads. The beads were subsequently incubated with purified MRN and MR complexes, respectively, that were produced in insect cells by means of baculovirus system. We found that both complexes were avidly bound to RECQ5 beads, but not to control beads coated with CBD, suggesting that the interaction between RECQ5 and MRN observed in vivo is direct and independent of NBS1 (Figure 2A).
Figure 2.
RECQ5 and the MRN complex interact directly. (A) Binding of purified MRN (‘top panel’) and MR (‘bottom panel’) complexes to chitin beads coated with RECQ5–CBD fusion. (B) Binding of individually purified RAD50 (‘top panel’), NBS1 (‘middle panel’) and MRE11 (‘bottom panel’) proteins to chitin beads coated with RECQ5–CBD fusion. For (A) and (B): lane 1, 30% of input material; lane 2, chitin beads coated with CBD; lane 3, chitin beads coated with RECQ5–CBD fusion protein. Binding reactions were carried out as described in Materials and Methods section. (C) Western blot analysis of RECQ5 immunoprecipitates from total extracts of the following cells: NBS (NBS1 deficiency), NBS1+ (NBS complemented by stable transfection of the NBS1 cDNA), ATLD1 (MRE11 deficiency) and ATLD1+ (ATLD1 complemented by stable transfection of the MRE11 cDNA). Lanes 1–4, 5% of the input material as indicated, lanes 5–8, RECQ5 immunoprecipitates from total extracts (1 mg of protein) of the indicated cells. Blots were probed for the presence of RECQ5, MRE11, RAD50 and NBS1 using antibodies described in Materials and Methods section.
In an attempt to further define the interaction site for RECQ5 on the MRN complex, we tested individually purified MRN subunits for binding to RECQ5–CBD beads. Interestingly, we found that both MRE11 and NBS1, but not RAD50, were bound to RECQ5, suggesting that the interaction between RECQ5 and the MRN complex is mediated not only by MRE11 but also by NBS1 (Figure 2B). To substantiate these findings, we performed immunoprecipitation experiments with total cell extracts from NBS and ATLD1 cells carrying hypomorphic mutations in the NBS1 and MRE11 genes, respectively. We found that the RECQ5 immunoprecipitate from NBS cells contained both MRE11 and RAD50 proteins in similar concentrations as the RECQ5 immunoprecipitate from NBS cells complemented with the wild-type NBS1 cDNA, confirming that RECQ5 binds to the MR complex in a manner independent of NBS1 (Figure 2C, lanes 1, 2, 5 and 6). In ATLD1 cells, the levels of NBS1 and RAD50 proteins were dramatically reduced as previously reported (29). However, a small amount of NBS1 could still be detected in the RECQ5 immunoprecipitate from these cells, indicating that NBS1 can interact with RECQ5 in absence of MRE11 (Figure 2C, lane 7). In MRE11-complemented ATLD1 cells, expression of all three subunits of the MRN complex could again be readily detected by western blot (Figure 2C, lane 4). Accordingly, the RECQ5 immunoprecipitate from total extract of these cells was found to contain substantial amounts of all three MRN proteins (Figure 2C, lane 8).
RECQ5 inhibits the 3′→5′ exonuclease activity of MRN complex
Next, we investigated whether the interaction between RECQ5 and the MRN complex affects the biochemical activities of these proteins. The SDS–PAGE profile of the purified proteins used in this study is shown in Figure S3. First, we tested the effect of MRN on the helicase activity of RECQ5. Previous studies demonstrated that RECQ5 displays a poor 3′→5′ helicase activity on oligonucleotide-based partial duplexes due to a strong strand-annealing activity residing in the C-terminal half of RECQ5 (11). However, RECQ5 could efficiently displace the lagging-strand oligonucleotide from synthetic forked structures with homologous arms where reannealing of this oligonucleotide is prevented by annealing of the parental strands (12). Thus, we evaluated the effect of the MRN complex on this strand-exchange reaction. We found that the rate of RECQ5-mediated strand-exchange was not altered upon addition of the MRN complex, suggesting that the MRN complex does not modulate the helicase activity of RECQ5 (Figure S4).
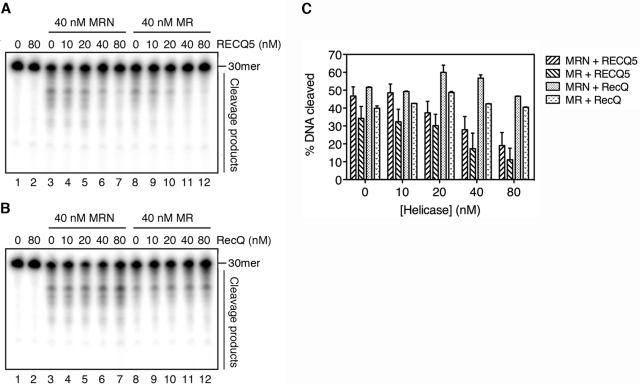
Since the MRN complex has been shown to possess 3′→5′ exonuclease activity (15), we tested the effect of RECQ5 on MRN-mediated exonucleolytic processing of a 30-bp duplex with a 5′-ssDNA tail containing 30 nt. Our initial experiments showed that the MRN complex required manganese and ATP as cofactors to exhibit efficient 3′→5′ exonuclease activity on this DNA substrate (data not shown), which is consistent with previously published data (18,30). Addition of RECQ5 to this reaction resulted in a significant decrease in the exonuclease activity of MRN (Figure 3A, lanes 3–7, and 3C). We did not observe the same effect with the E. coli RecQ helicase, excluding the possibility that the inhibition of the MRE11 nuclease by RECQ5 results from competition between these proteins for the DNA substrate (Figure 3B, lanes 3–7, and 3C).
Figure 3.
RECQ5 inhibits the 3′→5′ exonuclease activity of MR(N). (A) Effect of RECQ5 on the 3′→5′ exonuclease activity of MRN (lanes 3–7) and MR (lanes 8–12). (B) Effect of E. coli RecQ on the 3′→5′ exonuclease activity of MRN (lanes 3–7) and MR (lanes 8–12). In (A) and (B), reaction mixtures were incubated at 37°C for 30 min, and contained 1 nM 5′-tailed oligoduplex substrates (60-mer/30-mer with a 5′-32P label on the shorter strand), 40 nM MRN or MR and various concentrations of RECQ5 or RecQ as indicated. Reaction products were analyzed by denaturing PAGE followed by phosphorimaging as described in Materials and Methods section. (C) Quantification of the gels shown in (A) and (B). Intensity of the DNA substrate band in each lane was measured using ImageQuant software. The obtained values were used to calculate the percentage of cleaved DNA. The values obtained for the reactions carried out in the absence of MR(N) were taken as 100% in these calculations. The data points represent the mean of three independent experiments.
To investigate whether the inhibition of exonuclease activity of MRN by RECQ5 is dependent on the presence of the NBS1 protein, we evaluated the effect of RECQ5 on the exonuclease activity of the MR complex. The MR complex displayed a similar exonuclease activity as the MRN complex (Figure 3A and B, compare lanes 3 and 8, and C). Most importantly, RECQ5, but not RecQ, was found to inhibit the MR-mediated exonucleolytic processing (Figure 3A and B, lanes 8–12, and C).
Collectively, these data indicate that RECQ5 inhibits the exonuclease activity of MRN most likely as a result of its binding to MRE11.
RECQ5 and the MRN complex colocalize at sites of DNA damage
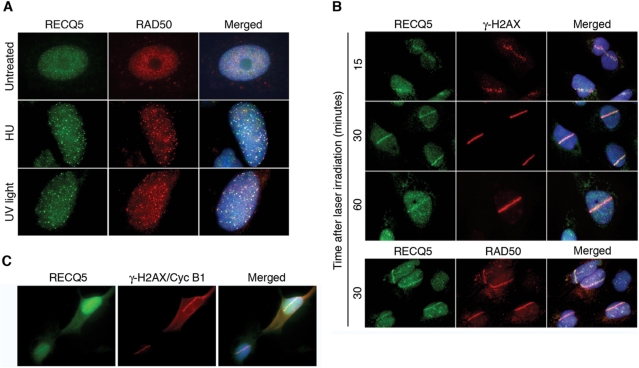
To explore a potential role for RECQ5–MRN interaction in the maintenance of genomic stability, we analyzed the spatial relationship of these proteins in the nucleus of U2OS cells in response to various types of DNA damage including stalled replication forks, DNA adducts and DSBs. After individual DNA-damaging treatments, cell were fixed with methanol, co-immunostained with anti-RECQ5 and anti-RAD50 antibodies and analyzed by fluorescence microscopy. As expected, in majority of unperturbed cells, both RECQ5 and RAD50 were uniformly distributed in the nucleus (Figure 4A, top row). After HU treatment, which results in replication arrest due to depletion of deoxyribonucleotides, both RECQ5 and RAD50 formed bright nuclear foci that extensively colocalized in >70% of cells (Figure 4A, middle row). Moreover, RECQ5 and RAD50 partially colocalized after exposure of cells to UVC light (40 J/m2) that can stall the progression of replication forks through induction of bulky DNA lesions (Figure 4A, bottom row). Thus, it is possible that RECQ5 and the MRN complex cooperate in the processing of stalled replication forks.
Figure 4.
RECQ5 and RAD50 colocalize at sites of DNA damage. (A) Indirect immunofluorescence imaging of RECQ5 and RAD50 in U2OS cells prior to and after DNA replication arrest. Cells were either left untreated or incubated in the presence of HU for 16 h or exposed to UV light at a dose of 40 J/m2 followed by incubation for 6 h. After genotoxic treatments, cells were fixed with methanol and triply stained for RECQ5 (green), RAD50 (red) and DNA (blue) as described in Materials and Methods section. Overlap between the green and red signals in merged images appears yellow. (B) RECQ5 and RAD50 accumulate at laser-induced DNA DSBs. U2OS cells were sensitized with BrdU (10 μM; incubation for 24 h) and subjected to microirradiation with pulsed UVA laser (λ = 355 nm) to generate linear tracks of DSBs. At indicated time points after irradiation, cells were fixed with methanol and co-immunostained either with anti-RECQ5 (green) and anti-γ-H2AX (red) antibodies or with anti-RECQ5 (green) and anti-RAD50 (red) antibodies as indicated. DAPI (blue) was used to stain nuclei. (C) U2OS cells were treated as in (B) and 30 min postirradiation, co-immunostained with antibodies against RECQ5, γ-H2AX (to detect microirradiated tracks) and cyclin B1 (to reveal cells in S/G2). γ-H2AX and cyclin B1 are displayed in the same channel (red). DAPI (blue) was used to stain nuclei.
To generate DSBs, U2OS cells were presensitized by incorporation of BrdU into genomic DNA and then locally exposed to UVA light (355 nm) using micro-laser technology, which results in linear tracks of DSBs across each irradiated nucleus (31). These tracks can be detected by immunofluorescence imaging of phosphorylated histone H2AX (γ-H2AX) that is rapidly generated in the DSB-flanking chromatin by the ATM kinase (32). We found that RECQ5 started to accumulate at DSB tracks as early as 15 min after irradiation, reaching maximal levels at about 30 min after irradiation, and persisted at those sites as long as 1 h after irradiation (Figure 4B, top panel). Double staining for RECQ5 and RAD50 further confirmed these findings (Figure 4B, bottom panel).
Like MRN, RECQ5 was found to accumulate at micro-irradiated areas essentially in all γ-H2AX-positive cells (150 cells evaluated), suggesting that the recruitment of RECQ5 to DSBs is not restricted to any particular phase of the cell cycle. To confirm this assumption, microirradiated cells were doubly stained for RECQ5 and cyclin B1. We found that RECQ5 accumulated in the microirradiated nuclear regions in cyclin B1-positive cells (cells in S and G2) as well as in cells lacking any detectable amount of cyclin B1 (cells in G1) (Figure 4C). These data indicate that RECQ5 accumulates at DSBs throughout the interphase.
The MRN complex is required for the recruitment of RECQ5 to sites of replication arrest
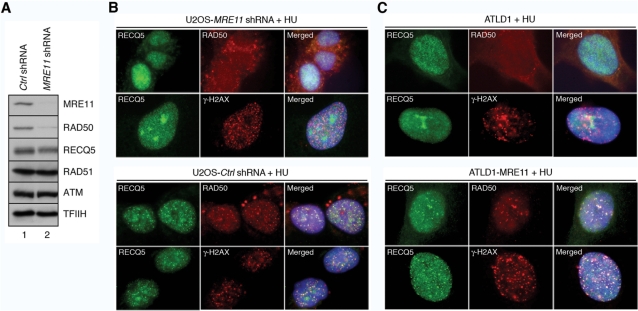
As the MRN complex is well known to function as a DNA damage sensor, we tested the possibility that it mediates the recruitment of RECQ5 to sites of DNA damage. First, we evaluated the effect of MRE11 deficiency on the focal distribution of RECQ5 after HU treatment. To do so, we silenced the expression of the MRE11 gene in U2OS cells using shRNA. We observed a dramatic reduction in the cellular level of MRE11 protein following puromycin selection for cells expressing anti-MRE11 shRNA construct, whereas no reduction in MRE11 levels was observed in cells harboring the control construct expressing a scrambled shRNA (Figure 5A). Depletion of MRE11 by shRNA also dramatically reduced the cellular level of RAD50, which is consistent with low levels of RAD50 found in ATLD1 cells, but it did not affect the level of RECQ5 (Figure 5A, lane 2 and Figure 2C, lane 3). Importantly, we found that shRNA-mediated depletion of MRE11 in U2OS cells completely abolished the formation of RECQ5 foci after HU treatment (Figure 5B, top panel). In contrast, HU-treated U2OS cells expressing the control shRNA contained both RECQ5 and RAD50 foci that showed extensive colocalization (Figure 5B, bottom panel, top row). Essentially the same results were obtained with ATLD1 and ATLD1-MRE11 cells (Figure 5C).
Figure 5.
MRE11 is required for the recruitment of RECQ5 to arrested replication forks. (A) Western blot analysis of extracts of U2OS cells expressing indicated shRNAs. Cells were transfected with appropriate shRNA vector and subjected to puromycin selection for 3 days to enrich the population of transfected cells. Blots were probed for MRE11, RAD50, RECQ5, RAD51, ATM and TFIIH (loading control) using antibodies described in Materials and Methods section. Lane 1, extract from cells expressing control shRNA (Ctrl shRNA); lane 2, extract from cells expressing shRNA targeting the MRE11 transcript (MRE11 shRNA). (B) Effect of shRNA-mediated depletion of MRE11 in U2OS cells on nuclear distribution of RECQ5 in response to HU. Cells were transfected either with the plasmid expressing Ctrl shRNA (‘bottom panel’) or the plasmid expressing MRE11 shRNA (‘top panel’), followed by puromycin selection. Two days after addition of puromycin, HU was added to a final concentration of 2 mM. After 16 h, cells were fixed and co-immunostained either for RECQ5 (green) and RAD50 (red) or for RECQ5 (green) and γ-H2AX (red) as indicated. (C) Nuclear distribution of RECQ5 in ATLD1 (‘top panel’) and ATLD1-MRE11 (‘bottom panel’) cells after replication arrest by HU. Exponentially growing cells were treated with 2 mM HU for 16 h and then fixed and co-immunostained either for RECQ5 (green) and RAD50 (red) or for RECQ5 (green) and γ-H2AX (red) as indicated.
Further, the MRE11-proficient and MRE11-deficient cells were doubly stained for RECQ5 and γ-H2AX. Histone H2AX is phosphorylated in response to replication arrest in ATR-dependent manner and forms nuclear foci that colocalize with PCNA and hence can serve as markers for stalled replication forks (33,34). As expected, in 70% of MRE11-proficient cells treated with HU, RECQ5 extensively colocalized with γ-H2AX at bright nuclear foci (Figure 5B and C, bottom panels, bottom rows). MRE11-deficient cells also displayed γ-H2AX foci in response to HU treatment (Figure 5B and C, top panels, bottom rows). However, RECQ5 foci were not seen in these cells.
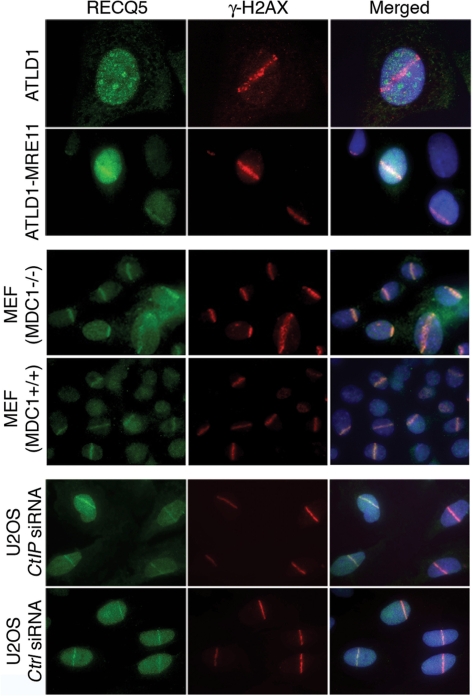
The MRN complex is required for the recruitment of RECQ5 to DSBs
To evaluate whether MRE11 is required for the recruitment of RECQ5 to DSBs, we microirradiated ATLD1 cells with UVA laser and stained them for RECQ5 and γ-H2AX at 30 min after irradiation. We found that none of γ-H2AX-positive ATLD1 cells contained RECQ5 tracks (Figure 6, top panel). In contrast, accumulation of RECQ5 at irradiated areas was seen in ATLD1-McRE11 cells (Figure 6, top panel). Collectively, these data indicate that the recruitment of RECQ5 to DSBs is dependent on a functional MRN complex.
Figure 6.
MRE11, but not MDC1 and CtIP, is required for the recruitment of RECQ5 to laser-induced DSBs. Indicated cell lines were irradiated with UVA laser at 24 h following addition of BrdU (10 μM), fixed at 30 min after the irradiation and co-immunostained to visualize RECQ5 (green) and γ-H2AX (red). DAPI (blue) was used to stain nuclei. For U2OS cell cultures, BrdU was added 2 days after transfection of CtIP or control (Ctrl) siRNA.
The MRN complex has multiple functions in the cellular response to DSBs. Following activation of the ATM kinase by MRN bound to DNA ends, more MRN accumulates in large chromatin domains flanking DSBs, which is likely to enhance the DSB-induced signal by means of a positive feedback loop (32). Recruitment of MRN to DSB-flanking chromatin is critically dependent on MDC1 that forms a complex with MRN in a manner dependent on CK2 phosphorylation and directly interacts with γ-H2AX in the damaged chromatin (35–38). To address whether MDC1 is required for the recruitment of RECQ5 to sites of DSBs, we compared DSB-induced nuclear trafficking of RECQ5 in MDC1+/+ and MDC1−/− MEFs. We found that MDC1 deficiency had no effect on the recruitment of RECQ5 to laser-induced DSB tracks (Figure 6, middle panel). Although this finding does not exclude the possibility that a fraction of the RECQ5 protein associate with DSB-flanking chromatin as a part of the MRN–MDC1 complex, it argues that MRN recruits RECQ5 to broken DNA ends. In both MDC1-proficient and MDC1-deficient MEFs, >97% of irradiated nuclei (out of at least 100 nuclei) contained RECQ5-positive tracks, further supporting the notion that RECQ5 associates with DSBs in a cell-cycle independent manner (data not shown).
MRN, in conjunction with CtIP, promotes resection of DSBs that arise during S/G2 phases of the cell cycle, generating 3′-ssDNA tails for HR repair (24). To address the question whether the recruitment of RECQ5 to DSBs occurs as a consequence of DSB resection, we depleted CtIP from U2OS cell by means of RNA interference (Figure S5A). Cells were fixed at 30 min after laser microirradiation and stained with anti-RECQ5 in combination with either anti-γ-H2AX or anti-RPA antibodies. We found that CtIP depletion had no effect on accumulation of RECQ5 at DSB tracks (Figure 6, bottom panel). In contrast, accumulation of RPA at microirradiated areas was completely abolished upon CtIP depletion (Figure S5B) as previously reported (24). These data indicate that the observed MRE11-dependent accumulation of RECQ5 at DSB sites is not dependent on DSB resection.
DISCUSSION
HR provides the most accurate mechanism for the repair of DSBs and ssDNA gaps which frequently occur in the cell as a result of collision of the DNA replication machinery with DNA lesions and other obstacles or as a result of processing of DNA lesions by DNA repair enzymes (39,40). In mitotic cells, most recombination intermediates are processed through pathways such as synthesis-dependent strand annealing (SDSA) or double-Holliday junction (dHJ) dissolution that do not result in crossovers. These mechanisms are important for avoidance of potentially deleterious chromosomal rearrangements that can arise if the homologous sequence used for HR-mediated repair is located at different chromosomal locus. A key step in HR is the formation of a RAD51-ssDNA filament that mediates the search for sequence homology and associates with the donor sequence forming the so-called D-loop structure (39). Recent studies indicated that the RECQ5 DNA helicase counteracts the formation of crossovers during HR (13,14). Moreover, it has been shown that RECQ5 has the ability to disrupt RAD51 nucleoprotein filaments in vitro (13). However, the importance of this activity in the regulation of HR in vivo still remains to be explored. Furthermore, it is not clear by now how RECQ5 is recruited to the sites of HR.
Here we show that RECQ5 interacts with the MRN complex, a key player in the repair of DNA damage by HR. Our results suggest that this interaction is mediated through MRE11 and NBS1. At the biochemical level, RECQ5 specifically inhibited the 3′→5′ exonuclease activity of the MRN and MR complexes, suggesting that RECQ5 interacts with the nuclease or DNA-binding domains of MRE11 thereby impairing the access of MRE11 to the DNA substrate. At the cellular level, MRE11 was required for the accumulation of RECQ5 at sites of arrested replication forks and sites of DSBs. Interestingly, recruitment of RECQ5 to DSBs was not dependent on MDC1, which mediates chromatin retention of MRN at sites of DNA damage (35,36). Moreover, RECQ5 accumulation at DSBs was not affected upon depletion of CtIP, indicating that it is not dependent on the DNA-end resection of DSBs in S/G2 generating 3′-ssDNA tails to initiate repair by HR (24). Instead, our data argue that RECQ5 migrates to and acts at broken DNA ends as a part of the MRN complex.
In the budding yeast Saccharomyces cerevisiae, the formation of crossovers during HR is counteracted mainly by the Srs2 DNA helicase, which has no obvious sequence homologs in higher eukaryotes (41,42). Like RECQ5, Srs2 has the capacity to disrupt Rad51 filaments formed on ssDNA (43,44). Srs2 also shows preference for unwinding of D-loop structures, and its helicase activity is stimulated by Rad51 filaments on dsDNA, supporting the proposed role for Srs2 in promoting SDSA (45). Strikingly, Srs2 has been shown to physically interact with the DNA-binding domain of Mre11 (46). This, together with our finding of a physical interaction between RECQ5 and the MRN complex in human cells, lends further support to the hypothesis that RECQ5 is a functional ortholog of Srs2 (13). Moreover, our data might explain how these anti-recombinases are delivered to sites of DNA repair. However, the precise role of the MRN complex in the function of these proteins during HR still remains to be addressed.
Interestingly, we have found that RECQ5 accumulates at DSBs throughout the interphase. A recent study demonstrated that RAD51 accumulates at laser-induced damage sites not only in S/G2 phase, where DSBs are repaired by HR, but also in G1 phase where HR repair is suppressed (47). Thus, it is possible that one function of RECQ5 is to prevent RAD51-mediated HR events in G1 cells that could lead to detrimental chromosomal rearrangements because of the lack of an undamaged sister chromatid.
We found that MRE11 was required for the RECQ5 focus formation in response to replication arrest by HU. Given the role for MRN as a DSB sensor, one can assume that these foci represent broken replication forks undergoing HR repair. However, HU-induced DNA DSBs become apparent only after 24 h of continuous HU treatment (48). In contrast, RECQ5 foci could readily be observed at 16 h following addition of HU, suggesting that they rather represent sites of stalled replication forks. Previous studies indicated that the MRN complex is associated with replication forks throughout S-phase via phosphorylation-dependent interaction with RPA (49–51). It was also shown that the MRN complex colocalizes with RPA in response to replication arrest by HU (48,52). Thus, it is possible that the MRN complex tethers RECQ5 to ssDNA regions generated at stalled replication forks to prevent formation of RAD51 filaments and subsequent DNA recombination. Such a ‘fork clearing’ function was proposed for the E. coli UvrD DNA helicase, a bacterial ortholog of Srs2, based on finding that the lethality conferred by uvrD deletion to certain replication mutants is suppressed by inactivation of the genes involved in the RecFOR pathway of HR (53,54). There is also evidence suggesting that Srs2 prevents recombination events at stalled replication forks favoring lesion bypass by translesion polymerases or through template-switching mechanism (55,56).
The BLM helicase also suppresses crossover events during HR as evidenced by elevated rate of sister chromatid exchanges (SCEs) and increased incidence of loss of heterozygocity in BLM-deficient cells relative to normal cells (1). Recent studies demonstrated that, out of human RecQ helicases, only BLM could promote dHJ dissolution, suggesting that BLM is the human counterpart of the yeast Sgs1 helicase (3,4,41). Although there is also some evidence suggesting that BLM functions in the SDSA pathway of HR, genetic studies using mutant mouse cells clearly indicated that BLM and RECQ5 have nonredundant roles in suppression of crossovers, which is similar to the situation of Srs2 and Sgs1 in yeast (14,41,57).
A recent study suggested that the human F-box helicase 1 (FBH1), which is conserved in Schizosaccharomyces pombe, but not in S. cerevisiae, is a functional ortholog of Srs2. It was shown that FBH1 shares homology with Srs2 within the helicase domain and suppresses specific recombination defects of S. cerevisiae srs2 mutants (58). Although it remains to be determined whether FBH1 possesses the ability to displace RAD51 from ssDNA, the existence of both Fbh1 and Srs2 in S. pombe rather suggests that these two helicases use different mechanisms to suppress recombination. This is further supported by the synthetic lethality of mutations in these genes, which is suppressed by deletion of the recombination gene rhp57 (59). Fbh1 is also essential for viability in absence of Rqh1, the sole RecQ homolog of S. pombe (59). Simultaneous inactivation of the FBH1 and BLM homologs in chicken DT40 cells is not lethal, but it results in much higher frequency of SCE relative to SCE levels in the respective single mutants (59). Thus, it seems that BLM, RECQ5 and FBH1 act rather in a complementary fashion during processing of HR intermediates. Since FBH1 helicase can function as the substrate specificity subunit of an SCF ubiquitin ligase complex one can speculate that it regulates recombination by triggering proteolytic degradation of recombination factors (60).
RECQ5 is not the sole human RecQ helicase that interacts with the MRN complex. There is evidence for a functional link between MRN and BLM. Similar to Sgs1, BLM was found to coexist with MRN in a large complex containing proteins involved in the recognition and repair of aberrant DNA structures, although no evidence for a direct BLM–MRN interaction was reported (61). It was also shown that BLM and MRN colocalize at nuclear foci in response to replication stress and that BLM is required for the relocalization of MRN to sites of replication arrest (61,62). Previous studies also demonstrated that the MRN complex physically interacts with WRN, which is required for the resolution of HR intermediates (63–65). Moreover, these studies revealed that the relocalization of WRN to sites of DNA damage is dependent on a functional MRN complex (63,64). However, WRN and RECQ5 differ in the mode of their interaction with MRN. The formation of WRN–MRN complex in vivo is dependent on DNA damage (63,64), whereas, based on our data, RECQ5 appears to constitutively interact with MRN in vivo. The interaction between WRN and the MRN complex is solely mediated by NBS1 (63), while we show that RECQ5 can bind to both MRE11 and NBS1 and that the absence of NBS1 does not affect the association of RECQ5 with the MR complex. At the functional level, MRN dramatically stimulates the helicase activity of WRN (63), whereas it has no effect on the helicase activity of RECQ5. Further studies will definitely be needed to understand the molecular mechanisms underlying the cooperation between RecQ DNA helicases and the MRN complex in the biological processes that enforce genomic stability.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
FUNDING
Swiss National Science Foundation (3100A0-102198, 3100A0-116008); UBS AG, Cancer League of the Canton of Zurich and Czech Science Foundation (GA204/09/0565). Funding for open access charge: Swiss National Science Foundation.
Conflict of interest statement. None declared
Supplementary Material
ACKNOWLEDGEMENTS
We thank Vilhelm Bohr for providing us with baculoviruses expressing MRN proteins, Torsten Kleffmann for help with mass spectrometry analysis, Manuel Stucki for helpful discussions and Christiane König for technical assistance.
REFERENCES
- 1.Hanada K, Hickson ID. Molecular genetics of RecQ helicase disorders. Cell Mol. Life Sci. 2007;64:2306–2322. doi: 10.1007/s00018-007-7121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma S, Doherty KM, Brosh RM., Jr Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem. J. 2006;398:319–337. doi: 10.1042/BJ20060450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 4.Wu L, Chan KL, Ralf C, Bernstein DA, Garcia PL, Bohr VA, Vindigni A, Janscak P, Keck JL, Hickson ID. The HRDC domain of BLM is required for the dissolution of double Holliday junctions. EMBO J. 2005;24:2679–2687. doi: 10.1038/sj.emboj.7600740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 6.Crabbe L, Jauch A, Naeger CM, Holtgreve-Grez H, Karlseder J. Telomere dysfunction as a cause of genomic instability in Werner syndrome. Proc. Natl Acad. Sci. USA. 2007;104:2205–2210. doi: 10.1073/pnas.0609410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, Dunphy WG, Venkitaraman AR. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell. 2005;121:887–898. doi: 10.1016/j.cell.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 8.Matsuno K, Kumano M, Kubota Y, Hashimoto Y, Takisawa H. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase alpha in the initiation of DNA replication. Mol. Cell Biol. 2006;26:4843–4852. doi: 10.1128/MCB.02267-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sekelsky JJ, Brodsky MH, Rubin GM, Hawley RS. Drosophila and human RecQ5 exist in different isoforms generated by alternative splicing. Nucleic Acids Res. 1999;27:3762–3769. doi: 10.1093/nar/27.18.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimamoto A, Nishikawa K, Kitao S, Furuichi Y. Human RecQ5beta, a large isomer of RecQ5 DNA helicase, localizes in the nucleoplasm and interacts with topoisomerases 3alpha and 3beta. Nucleic Acids Res. 2000;28:1647–1655. doi: 10.1093/nar/28.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia PL, Liu Y, Jiricny J, West SC, Janscak P. Human RECQ5β, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004;23:2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanagaraj R, Saydam N, Garcia PL, Zheng L, Janscak P. Human RECQ5β helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 2006;34:5217–5231. doi: 10.1093/nar/gkl677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Y, Lu X, Barnes E, Yan M, Lou H, Luo G. Recql5 and Blm RecQ DNA helicases have nonredundant roles in suppressing crossovers. Mol. Cell Biol. 2005;25:3431–3442. doi: 10.1128/MCB.25.9.3431-3442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D’Amours D, Jackson SP. The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- 16.Assenmacher N, Hopfner KP. MRE11/RAD50/NBS1: complex activities. Chromosoma. 2004;113:157–166. doi: 10.1007/s00412-004-0306-4. [DOI] [PubMed] [Google Scholar]
- 17.de Jager M, van Noort J, van Gent DC, Dekker C, Kanaar R, Wyman C. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell. 2001;8:1129–1135. doi: 10.1016/s1097-2765(01)00381-1. [DOI] [PubMed] [Google Scholar]
- 18.Paull TT, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stracker TH, Theunissen JW, Morales M, Petrini JH. The Mre11 complex and the metabolism of chromosome breaks: the importance of communicating and holding things together. DNA Repair (Amst.) 2004;3:845–854. doi: 10.1016/j.dnarep.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 20.Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–7748. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- 21.Lavin MF, Kozlov S. ATM activation and DNA damage response. Cell Cycle. 2007;6:931–942. doi: 10.4161/cc.6.8.4180. [DOI] [PubMed] [Google Scholar]
- 22.Bernstein DA, Keck JL. Domain mapping of Escherichia coli RecQ defines the roles of conserved N- and C-terminal regions in the RecQ family. Nucleic Acids Res. 2003;31:2778–2785. doi: 10.1093/nar/gkg376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 24.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carson CT, Schwartz RA, Stracker TH, Lilley CE, Lee DV, Weitzman MD. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 2003;22:6610–6620. doi: 10.1093/emboj/cdg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tauchi H, Kobayashi J, Morishima K, Matsuura S, Nakamura A, Shiraishi T, Ito E, Masnada D, Delia D, Komatsu K. The forkhead-associated domain of NBS1 is essential for nuclear foci formation after irradiation but not essential for hRAD50.hMRE11.NBS1 complex DNA repair activity. J. Biol. Chem. 2001;276:12–15. doi: 10.1074/jbc.C000578200. [DOI] [PubMed] [Google Scholar]
- 27.Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, Manis JP, van Deursen J, Nussenzweig A, Paull TT, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 28.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 29.Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 30.Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 31.Lukas C, Bartek J, Lukas J. Imaging of protein movement induced by chromosomal breakage: tiny ‘local’ lesions pose great ‘global’ challenges. Chromosoma. 2005;114:146–154. doi: 10.1007/s00412-005-0011-y. [DOI] [PubMed] [Google Scholar]
- 32.Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward IM, Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 2001;276:47759–47762. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- 34.Limoli CL, Giedzinski E, Bonner WM, Cleaver JE. UV-induced replication arrest in the xeroderma pigmentosum variant leads to DNA double-strand breaks, γ-H2AX formation, and Mre11 relocalization. Proc. Natl Acad. Sci. USA. 2002;99:233–238. doi: 10.1073/pnas.231611798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lukas C, Melander F, Stucki M, Falck J, Bekker-Jensen S, Goldberg M, Lerenthal Y, Jackson SP, Bartek J, Lukas J. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 2004;23:2674–2683. doi: 10.1038/sj.emboj.7600269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 37.Spycher C, Miller ES, Townsend K, Pavic L, Morrice NA, Janscak P, Stewart GS, Stucki M. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. J. Cell Biol. 2008;181:227–240. doi: 10.1083/jcb.200709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melander F, Bekker-Jensen S, Falck J, Bartek J, Mailand N, Lukas J. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J. Cell Biol. 2008;181:213–226. doi: 10.1083/jcb.200708210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krogh BO, Symington LS. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 40.Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robert T, Dervins D, Fabre F, Gangloff S. Mrc1 and Srs2 are major actors in the regulation of spontaneous crossover. EMBO J. 2006;25:2837–2846. doi: 10.1038/sj.emboj.7601158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- 44.Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- 45.Dupaigne P, Le Breton C, Fabre F, Gangloff S, Le Cam E, Veaute X. The Srs2 helicase activity is stimulated by Rad51 filaments on dsDNA: implications for crossover incidence during mitotic recombination. Mol. Cell. 2008;29:243–254. doi: 10.1016/j.molcel.2007.11.033. [DOI] [PubMed] [Google Scholar]
- 46.Chiolo I, Carotenuto W, Maffioletti G, Petrini JH, Foiani M, Liberi G. Srs2 and Sgs1 DNA helicases associate with Mre11 in different subcomplexes following checkpoint activation and CDK1-mediated Srs2 phosphorylation. Mol. Cell Biol. 2005;25:5738–5751. doi: 10.1128/MCB.25.13.5738-5751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim JS, Krasieva TB, Kurumizaka H, Chen DJ, Taylor AM, Yokomori K. Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells. J. Cell Biol. 2005;170:341–347. doi: 10.1083/jcb.200411083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robison JG, Lu L, Dixon K, Bissler JJ. DNA lesion-specific co-localization of the Mre11/Rad50/Nbs1 (MRN) complex and replication protein A (RPA) to repair foci. J. Biol. Chem. 2005;280:12927–12934. doi: 10.1074/jbc.M414391200. [DOI] [PubMed] [Google Scholar]
- 49.Maser RS, Mirzoeva OK, Wells J, Olivares H, Williams BR, Zinkel RA, Farnham PJ, Petrini JH. Mre11 complex and DNA replication: linkage to E2F and sites of DNA synthesis. Mol. Cell Biol. 2001;21:6006–6016. doi: 10.1128/MCB.21.17.6006-6016.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mirzoeva OK, Petrini JH. DNA replication-dependent nuclear dynamics of the Mre11 complex. Mol. Cancer Res. 2003;1:207–218. [PubMed] [Google Scholar]
- 51.Olson E, Nievera CJ, Liu E, Lee AY, Chen L, Wu X. The Mre11 complex mediates the S-phase checkpoint through an interaction with replication protein A. Mol. Cell Biol. 2007;27:6053–6067. doi: 10.1128/MCB.00532-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robison JG, Elliott J, Dixon K, Oakley GG. Replication protein A and the Mre11.Rad50.Nbs1 complex co-localize and interact at sites of stalled replication forks. J. Biol. Chem. 2004;279:34802–34810. doi: 10.1074/jbc.M404750200. [DOI] [PubMed] [Google Scholar]
- 53.Veaute X, Delmas S, Selva M, Jeusset J, Le Cam E, Matic I, Fabre F, Petit MA. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J. 2005;24:180–189. doi: 10.1038/sj.emboj.7600485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Flores MJ, Sanchez N, Michel B. A fork-clearing role for UvrD. Mol. Microbiol. 2005;57:1664–1675. doi: 10.1111/j.1365-2958.2005.04753.x. [DOI] [PubMed] [Google Scholar]
- 55.Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 56.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 57.Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chiolo I, Saponaro M, Baryshnikova A, Kim JH, Seo YS, Liberi G. The human F-Box DNA helicase FBH1 faces Saccharomyces cerevisiae Srs2 and postreplication repair pathway roles. Mol. Cell Biol. 2007;27:7439–7450. doi: 10.1128/MCB.00963-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morishita T, Furukawa F, Sakaguchi C, Toda T, Carr AM, Iwasaki H, Shinagawa H. Role of the Schizosaccharomyces pombe F-box DNA helicase in processing recombination intermediates. Mol. Cell Biol. 2005;25:8074–8083. doi: 10.1128/MCB.25.18.8074-8083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim JH, Kim J, Kim DH, Ryu GH, Bae SH, Seo YS. SCFhFBH1 can act as helicase and E3 ubiquitin ligase. Nucleic Acids Res. 2004;32:2287–2297. doi: 10.1093/nar/gkh534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 62.Franchitto A, Pichierri P. Bloom's syndrome protein is required for correct relocalization of RAD50/MRE11/NBS1 complex after replication fork arrest. J. Cell Biol. 2002;157:19–30. doi: 10.1083/jcb.200110009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cheng WH, von Kobbe C, Opresko PL, Arthur LM, Komatsu K, Seidman MM, Carney JP, Bohr VA. Linkage between Werner syndrome protein and the Mre11 complex via Nbs1. J. Biol. Chem. 2004;279:21169–21176. doi: 10.1074/jbc.M312770200. [DOI] [PubMed] [Google Scholar]
- 64.Franchitto A, Pichierri P. Werner syndrome protein and the MRE11 complex are involved in a common pathway of replication fork recovery. Cell Cycle. 2004;3:1331–1339. doi: 10.4161/cc.3.10.1185. [DOI] [PubMed] [Google Scholar]
- 65.Saintigny Y, Makienko K, Swanson C, Emond MJ, Monnat RJ., Jr Homologous recombination resolution defect in Werner syndrome. Mol. Cell Biol. 2002;22:6971–6978. doi: 10.1128/MCB.22.20.6971-6978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.