Abstract
The full understanding of dynamics of cellular processes hinges on the development of efficient and non-invasive labels for intracellular RNA species. Light-up aptamers binding fluorogenic ligands show promise as specific labels for RNA species containing those aptamers. Herein, we took advantage of existing, non-light-up aptamers against small molecules and demonstrated a new class of light-up probes in vitro. We synthesized two conjugates of thiazole orange dye to small molecules (GMP and AMP) and characterized in vitro their interactions with corresponding RNA aptamers. The conjugates preserved specific binding to aptamers while showing several 100-fold increase in fluorescence of the dye (the ‘light-up’ property). In the presence of free small molecules, conjugates can be displaced from aptamers serving also as fluorescent sensors. Our in vitro results provide the proof-of-concept that the small-molecule conjugates with light-up properties can serve as a general approach to label RNA sequences containing aptamers.
INTRODUCTION
The established methods for fluorescent microscopy of intracellular ribonucleic acids (1) rely on either MS2-GFP system (2) or molecular beacons (3). MS2-GFP system suffers from a low signal-to-noise ratio and may be intrusive due to size, while beacons do not readily traverse the cell membranes and are also degraded by nucleases (4).
A different approach, based on small molecules-aptamer pairs with ‘light-up’ properties, has been demonstrated in vitro (4–9), but has yet to be successfully extended to mRNAs in vivo, presumably due to unfavorable characteristics of available fluorophores or insufficient signal-to-noise ratios. Tsien and colleagues were first to report that malachite green in a complex with its RNA aptamer (10) has 2000-fold increase in the quantum yield (5). Sparano and Koide selected the light-up RNA aptamers for quencher modules in the engineered molecular quencher-fluorophore couples (4,6), while Sando's group reported the selection of DNA and RNA aptamers for derivatives of Hoechst 33258 (7,8). Most recently, Armitage and colleagues described the selection of the tightly binding RNA aptamer for an engineered cyanine dye (dimethylindole red) with 60-fold fluorescence enhancement (9).
Instead of isolating a new aptamer, we decided to test whether it is possible to target a non-specific small molecule dye with light-up property, such as thiazole orange (TO) to the existing aptamers. This approach would have an advantage that non-specific light-up probes have well-characterized fluorescent properties (11,12) and have been often already tested in intracellular milieu (13). Our design relied on overcoming the intrinsic lack of specificity of TO through conjugation to small molecules through preferred interactions with a targeted RNA encompassing an aptamer against these small molecules, while still achieving an increase in fluorescence (Scheme 1).
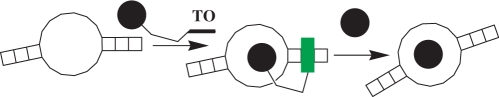
Scheme 1.
Principle behind light-up probes for aptamers and their displacemnt with unconjugated small molecules: An RNA aptamer against a molecule (circle) binds a conjugate between molecule and TO dye, resulting in a fluorescent complex. Presence of free molecule in solution results in displacement of conjugate from the complex and reduction in fluorescence.
Thiazole orange is a weakly fluorescent dye, which becomes brightly fluorescent (quantum yield up to 0.4, excitation and emission close to fluorescein, λex = 480 nm, λem = 530 nm) upon non-specific preferential insertion into double helical nucleic acids (Kd approximately low micromolar, depending on the exact sequence). TO dye has been used in probes targeting nucleic acids, either in conjugates to antisense probes [PNA (14) or DNA (15)] or in conjugates to peptides (16,17) or polyamides (18) for double helical DNA. The common idea behind these approaches was that a sequence-specific targeting module was combined with a non-specific light-up TO dye module, leading to a preferred bivalent binding to a specific sequence coupled with a strong increase in fluorescence.
There are also previous examples of small-molecule-dye conjugates used to study aptamers: for example, Rando's group has previously taken advantage of environment-sensitive properties of pyrene conjugates to study ligand-aptamer interactions. However, in that approach all states had high fluorescence and the λex is in the UV region (19). Interestingly, the same group has also reported paramomycin-TO dyes conjugates as a part of their studies on RNA antagonists, but reported no follow-up observations of light-up properties of these conjugates, albeit fluorescent anisotropy was used for Kd determination (20).
MATERIALS AND METHODS
Materials and instrumentation
Oligonucleotides were made and DNA/RNAse free HPLC purified by Integrated DNA Technologies, Inc. (Coralville, IA) and were used as received. TO was obtained from Biochemika (distributed by Sigma-Aldrich Co.). Lambda DNA was purchased from Promega (Madison, WI). RNA from baker's yeast, ATP, GTP, CTP, UTP, AMP and GMP were purchased from Sigma-Aldrich Co. (St Louis, MO). 1H NMR spectra were recorded on a Varian NMR 300 (Columbia, MD). Mass spectrometry was performed using a Shimadzu (Piscataway, NJ) LCMS-2010A. High-resolution MS was taken on a JEOL JMS-HX 110 double focusing MS (Tokyo, Japan). For HPLC characterization, we used a Shimadzu (Piscataway, NJ) LC-20AB pump equipped with a SPD-M20A diode array detector. Waters XBRIDGE OST C18 ion-pair, reversed-phase chromatographic column (Milford, MA) was used.
Synthesis of TO-conjugates
TO-hydroxysuccinimidyl ester (TO-OSu)
The TO-carboxylic acid (TO-C10) dye was prepared according to the literature (17). The TO-hydroxysuccinimidyl ester was prepared by stirring TO-carboxylic acid (7.0 mg, 0.0125 mmol), N,N′-dicyclohexylcarbodiimide (DCC) (2.9 mg, 0.0138 mmol) and N-hydroxysuccinimide (HOSu) (1.7 mg, 0.0138 mmol) in 0.5 ml of dichloromethane overnight. The precipitate was filtered, and the solvent evaporated from the filtrate leaving 7.3 mg product (90% crude yield). ESI-MS (positive mode) m/z = 572 (M+).
AMP-NH-linker-NH2
AMP (21 mg, 0.06 mmol) and 2,2′-(ethylenedioxy)bis-ethylamine (0.088 ml, 0.60 mmol) were added to 0.40 ml of dimethyl sulfoxide (DMSO) and stirred. Then, PPh3 (80 mg, 0.30 mmol) and 2,2′-dipyridine disulfide (Aldrithiol-2) (80 mg, 0.36 mmol) were sequentially added. The mixture was stirred at 40°C for 48 h and at this point a clear solution was obtained. The reaction mixture was then added to 20 ml of 2% LiClO4 in acetone, chilled overnight at −20°C, and centrifuged. The precipitate was resuspended in 20 ml of 2% LiClO4 in acetone and centrifuged again. The final precipitate was then extracted twice with 15 ml of methanol. The extracts were combined and solvent evaporated, leaving 22 mg product (75% crude yield). 1H NMR (300 MHz, CD3OD) 8.57 (1 H, s), 8.19 (1 H, s), 6.09 (1 H, d, J = 6.0 Hz), 4.71–4.75 (1 H, dd, J = 5.4 Hz, J = 6.0 Hz), 4.38–4.41 (1 H, dd, J = 3.3 Hz, J = 5.2 Hz), 4.21–4.23 (1 H, m), 4.00–4.03 (2 H, m), 3.37–3.66 (8 H, m), 2.98–3.05 (2 H, dt, J = 6.0 Hz, J = 9.3 Hz) and 2.80 (2 H, t, J = 5.1 Hz). Rf = 0.65, silica, 6:1 MeOH/NH3 aq. ESI-MS (positive mode) m/z = 484 (M+H+), 490 (M+Li+).
TO-AMP
AMP-NH-linker-NH2 (12 mg, 0.025 mmol) in 0.40 ml of dimethylformamide (DMF) and 0.30 ml of water were added to a solution of 0.025 mmol TO-OSu in 0.40 ml of DMF. After 14 h, solvent was evaporated and purification was achieved via preparative reverse phase thin layer chromatography (TLC), Rf = 0.3, MeOH (4 mg, 16% yield). 1H NMR (300 MHz, CD3OD) 8.67 (1 H, d, J = 9.3 Hz), 8.56 (1 H, s), 8.46 (1 H, d, J = 7.2 Hz), 8.13, (1 H, s), 7.91–8.09 (3 H, m), 7.60–7.80 (3 H, m), 7.42–7.49 (2 H, m), 6.93 (1 H, s), 6.09 (1 H, d, J = 5.7 Hz), 4.74 (1 H, t, J = 10.8 Hz), 4.65 (2 H, t, J = 14.7 Hz), 4.41–4.44 (1 H, m), 4.22–4.28 (1 H, m), 4.04 (3 H, s), 4.02–4.06 (2 H, m), 3.47–3.78 (8 H, m), 3.02–3.11 (2 H, dt, J = 6.0 Hz, J = 9.3 Hz), 2.20 (2 H, t, J = 7.2 Hz), 2.00 (2 H, t, J = 6.9 Hz), 1.59 (2 H, m), 1.40–1.35 (2 H, m), 1.20–1.35 (12 H, m) and 0.926 (2 H, t, J = 4.8 Hz). Rf = 0.20, silica, 6:3:1 dioxane/water/NH3 aq. ESI-MS (positive mode) m/z = 956 (M + Na+). HRMS (C46H60N9O9PS): calcd, 934.4051; found, 934.4019.
GMP-NH-linker-NH2
GMP (203 mg, 0.5 mmol) and 2,2′-(ethylenedioxy)bis-ethylamine (0.747 ml, 5.0 mmol) were added to 5 ml of DMSO, followed by PPh3 (655 mg, 2.5 mmol) and 2,2′-dipyridine disulfide (Aldrithiol-2) (660 mg, 3.0 mmol). The mixture was stirred at 40°C for 48 h and at this point the clear solution was observed. The product was isolated by precipitation with LiClO4 in acetone (addition of crude reaction to 20 ml of 2% LiClO4 in acetone, overnight incubation at −20°C and centrifugation). The precipitate was resuspended in LiClO4 in acetone (20 ml, 2%) and centrifuged again. The final precipitate was extracted twice with methanol (15 ml). The extracts were combined and solvent evaporated leaving product (70 mg, 60% yield). 1H NMR (300 MHz, CD3OD) 8.00 (1 H, s), 5.81 (1 H, d, J = 6.3 Hz), 4.70–4.75 (1 H, dd, J = 4.8 Hz, J = 10.2), 4.32–4.35 (1 H, m), 4.10–4.16 (1 H, m), 3.92–4.03 (2 H, m), 3.35–3.66 (8 H, m), 2.92–3.22 (2 H, dt, J = 6.6 Hz, J = 9.0 Hz) and 2.74 (2 H, t, J = 6.0 Hz). Rf = 0.8, silica, MeOH/NH3 aq. ESI-MS (positive mode) m/z = 492 (M+).
TO-GMP
A solution of 0.010 mmol TO-OSu in 0.5 ml of dichloromethane was evaporated to a drop of oil in a 1.0 ml Wheaton vial. To this was added GMP-NH-linker-NH2 (26 mg, 0.020 mmol) dissolved in 0.05 ml of DMF and 0.05 ml of 50 mM aq NaHCO3. After 14 h, solvent was evaporated and the residue was purified via preparative reverse phase TLC (3.7 mg, 25% yield). 1H NMR (300 MHz, CD3OD) 8.64 (1 H, d, J = 8.4 Hz), 8.44 (1 H, d, J = 6.9 Hz), 8.04 (1 H, d, J = 8.4 Hz), 7.97 (1 H, d, J = 6.6 Hz), 7.94 (1 H, s), 7.89 (1 H, d, J = 7.8 Hz), 7.57–7.76 (3 H, m), 7.38–7.47 (2 H, m), 6.90 (1 H, s), 5.78 (1 H, d, J = 7.2 Hz), 4.80–4.85 (1 H, m), 4.59 (2 H, t, J = 7.5 Hz), 4.35–4.37 (1 H, m), 4.12–4.18 (1 H, m), 4.01 (3 H, s), 3.98–4.14 (2 H, m), 3.45–3.58 (8 H, m), 2.95–3.10 (2 H, m), 2.14–2.20 (2 H, t, J = 7.5 Hz), 1.96 (2 H, m), 1.56 (2 H, m), 1.25–1.40 (8 H, m), 0.92–0.93 (6 H, m), 0.08–0.15 (2 H, m). R f = 0.45, silica, 11:3:1 Dioxane/Water/NH3 aq. ESI-MS (positive mode) m/z = 950 (M+H+), 972 (M+Na+). HRMS (C46H60N9O10PS): calcd, 950.4000; found, 950.4031. HPLC was used to analyse the purity of conjugates (Supplementary Data).
Fluorescence measurements
Fluorescent spectra were taken on a Perkin-Elmer (San Jose, CA) LS-55 luminescence spectrometer. Experiments were performed at the excitation wavelength of 480 nm and emission scan of 510–595 nm. All measurements were performed in 20 mM Tris–HCl (pH 7.4), 140 mM NaCl, 5 mM KCl and 5 mM MgCl2. The TO-conjugates were dissolved in buffer and 0.2 µm filtered just before measurements. The concentrations of TO-conjugates were calculated based on the absorption at 501 nm (assuming molar extinction coefficients of 63 000 M−1cm−1). Absorbance was determined on an Amersham Biosciences (Piscataway, NJ) Ultrospec 3300 pro UV/visible spectrophotometer. For the studies of interactions with aptamers, TO-conjugates were diluted at the desired concentrations in binding buffer, and series of standard dilutions of aptamers were added and incubated 40 min at room temperature. For displacements by analytes (ATP, GTP, CTP and UTP), series of standard dilutions of analytes were added in the solutions of TO-conjugates (300 nM) and aptamers (600 nM) for 40 min.
Determination of dissociation constants
Dissociation constant K1 (for the interaction between TO-conjugate and aptamer) was determined by measuring the fluorescence intensity (F) as a function of increasing total added aptamer [R]t at a fixed concentration of total TO-conjugate [D]t, and fitting to the following equation (assuming a 1:1 complex, the detailed equation expression in Supplementary Data).
where [R] and [D] were the concentrations of free aptamer and TO-conjugate, respectively. We converted fluorescence unit to µM ([RD] = F/ε, ε = 2941.2 F/µM for TO-AMP, 2384.6 F/µM for TO-GMP) by measuring the fluorescence intensity of 65 nM TO-conjugate in the presence of a saturated aptamer concentration (the conjugates had negligible fluorescence on its own). ε was defined as the conversion coefficient, and [RD] was the concentration of complex of aptamer with TO-conjugate.
In the displacement binding assay, dissociation constant K2 for aptamer with ATP (or GTP) was determined by fluorescence titration of ATP (or GTP) into the complex solution of TO-conjugate and aptamer. The following equation (Supplementary Data) is used for fitting.
[R]t, [D]t were the initial concentrations in the mixing solution of aptamer and TO-conjugate. [A]t was the total concentration of added competitor ATP (or GTP).
RESULTS AND DISCUSSION
The light-up properties of TO-conjugates
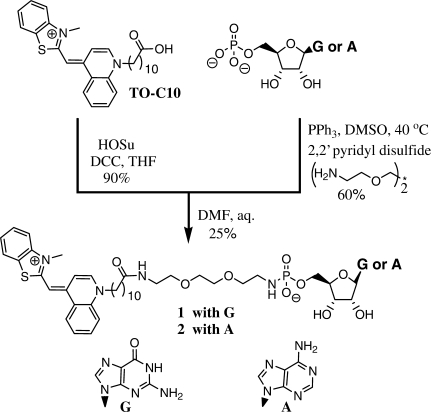
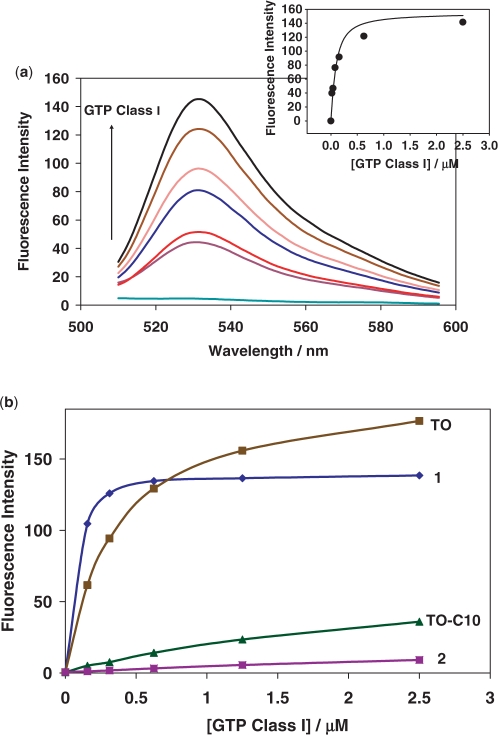
The TO-GMP conjugate 1 was synthesized in two steps (Scheme 2 and Supplementary Data) from the previously reported TO-C10 analog (17). We tested several GTP-binding aptamers (21) reported by Szostak's group with this conjugate (Schemes 2 and 3). The conjugate 1 had negligible fluorescence on its own (lowest spectra in Figure 1a), and showed large (several 100-fold) increase in fluorescence in the presence of various GTP aptamers (Supplementary Data). The strongest fluorescence increase, ∼400-fold, was observed with the aptamer GTP Class I (Scheme 3) and we characterized these interactions more closely (Figure 1a). The apparent Kd of a complex between 1 and this aptamer was approximately 60 nM (Figure 1a, insert), indicating no apparent increase in binding strength over GMP (reported at 23 nM) (21). The substitution of the proposed binding region with the random sequence yielded an aptamer that lost the ability to produce strong fluorescence in the presence of 1 (Supplementary Data).
Scheme 2.
Synthesis of GMP and AMP-TO conjugate (conjugates were made through an identical procedure and with similar yields).
Figure 1.
(a) Fluorescence emission spectra of TO-GMP (65 nM) in the presence of varying concentrations of GTP Class I (0, 0.0195, 0.039, 0.078, 0.1563, 0.625 and 2.5 µM, frssom bottom to top). The insert shows for maximum fluorescence versus aptamer concentration curve fitting (Kd = 60 nM). (b) Dependence of the fluorescence on the concentration of GTP Class I aptamer for: 1; TO; 2 and TO-C10 (all measurements at 50 nM of dyes).
Scheme 3.
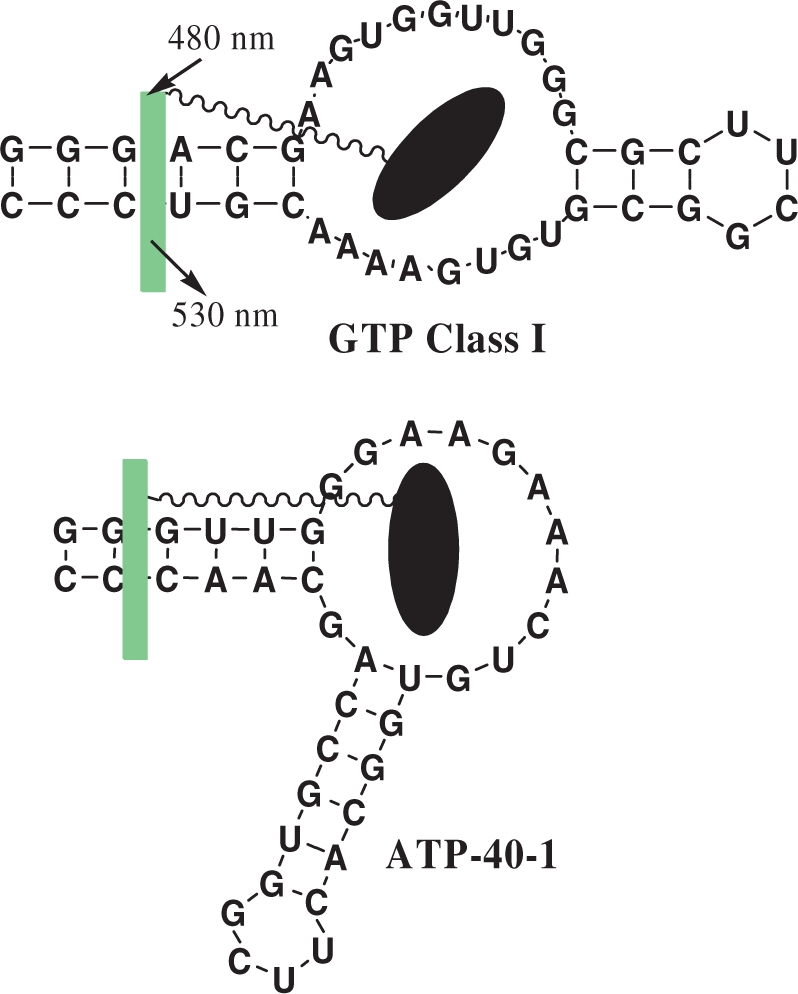
Aptamers tested with their conjugates. Black elipsoid is NMP part of the conjugate, while green fluorescent rectangle is TO-C10 part. In both cases the place of the insertion of TO dyes is shown aribitrarily, although in the case of AMP derivative we have indications based on aptamer modifications (Supplementary Data).
In order to test the generality of the ‘light-up’ approach to label aptamers, we also synthesized TO-AMP conjugate 2 (Scheme 2). The synthetic process and yields closely followed the procedure developed for TO-GMP. The conjugate again had negligible fluorescence on its own, and showed up to 500-fold increase in fluorescence in the presence of the ATP aptamer ATP-40-1 (22) (Supplementary Data). The apparent dissociation constant of a conjugate–aptamer complex was ∼550 nM, indicating a several-fold increase in binding through positive cooperativity between the two binding events [aptamer binds adenosine derivatives in micromolar range (22)]. The aptamer with a single point mutation that abolishes binding [ATP-40-1-G34A, G34 is highly conserved unpaired base (18)] showed no increase in fluorescence beyond what was observed for completely unrelated sequences (Supplementary Data). This result indicates that the binding was small molecule-driven, thus, with a potential for high-selectivity over the unrelated sequences. This conjugate was later used as a fluorescent molecule to study the thermally controlled affinity extraction from aptamer-bound surface on microfluidic device (23). We also synthesized this conjugate with a short linker, and it showed diminished interactions with the aptamer, consistent with bivalent interactions. Finally, we observed diminished interactions when stem without loop was shortened, consistent with intercalation of TO portion in this stem (Supplementary Data).
The selectivity of TO-conjugates
In order to confirm that our targeting with conjugates is indeed selective, we tested the cross-reactivity of both conjugates in the presence of aptamers, and in the presence of unrelated nucleic acids (Supplementary Data). The increase in fluorescence was remarkable only in the presence of targeted aptamers. However, we observed some non-negligible increase in fluorescence with other species as well, indicating that there is residual binding of the TO dye portion to nucleic acids. We tested non-specific responses of λDNA and yeast total RNA with the TO-conjugates. Even 20 times more λDNA or 80 times more yeast total RNA in weight had less 50% response than the target aptamer (Supplementary Data). In order to study the selectivity further, we tested TO dye, its C10-analog, and GMP or AMP conjugate at increasing concentrations of GTP Class I aptamer (Figure 1b). As seen in Figure 1b, original TO-dye (with methyl group in place of C10 spacer), at the same concentrations as conjugate, actually yields a bigger fluorescent signal in the high concentration of GTP Class I aptamer (and all other tested nucleic acids as well, not shown), despite somewhat weaker initial binding. In contrast, under the same conditions the TO-dye analog with a hydrophobic C10 spacer used in 1 and 2 shows very low fluorescent response. Mismatched conjugate 2 is even more selective with only negligible interactions with GTP Class I aptamer.
These observations are consistent with the mechanism of fluorescent increase for our conjugates: The primary binding event of a small molecule to its aptamer increases the effective concentration (24) of TO dye analogs with otherwise lowered affinity to nucleic acids. In retrospective, the success of our design and the high selectivity of fluorescence increase fortuitously relied on the weakened association between nucleic acids and TO by C10 modification. Interestingly, both derivatives of Hoechst 33258 (7,8) and dimethylindole red (9) were engineered to suppress nonspecific DNA or RNA binding, and we seem to have accomplished the same characteristic through a fortuitous accident.
The specific displacement of TO-conjugates by small molecules
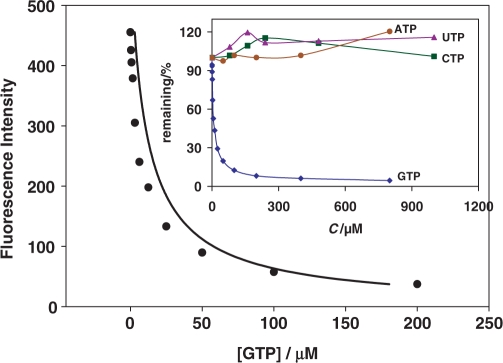
As expected based on equilibrium considerations, conjugate 1 could be displaced from its complex with aptamer by the excess of GTP, but not ATP, CTP or UTP (Figure 2). Importantly, almost complete displacement indicates the binding is directed by the GMP part of the conjugate. Similarly, conjugate 2 could be displaced from its complex after incubation with either ATP or AMP, at almost identical concentrations, consistent with the reported (22) Kd (Supplementary Data). Again, other nucleotides were not capable of displacement, indicating that the selectivity of the original aptamer was conserved in its complex. The complete displacement makes complexes of aptamers and their conjugates interesting as displacement sensors for homogeneous detections, with small molecules as analytes. Although our method is synthetically more challenging than simply using off-the-shelf DNA-binding dyes (25–28), it has an advantage of the increased specificity of interactions with aptamers. Because the binding of light-up probes is directed primarily through small molecule ligands, displacement is almost complete, leading to much better signal than in the previous, non-specific designs (26–28). There are many interesting approaches to aptameric sensors (29,30), but there is only one that uses non-covalently modified RNA aptamer to construct light-up probe for the detection of small molecules and is potentially suitable for expressable intracellular sensors (31). With light-up approach described in this work, we can use TO-conjugates to develop light-up probes for the detection of the other small molecules.
Figure 2.
Fluorescence response of TO-GMP (300 nM) containing GTP Class I (600 nM) in the presence of varying concentrations of GTP (0, 0.39, 0.78, 1.563, 3.125, 6.25, 12.5, 25, 50, 100 and 200 µM). Small insert shows displacement (% remaining versus concentration) in the presence of GTP, and no response to other NTPs.
We were not just interested in developing yet another set of sensors for purely analytical purposes. Rather, we were looking for indications that, should we eventually be successful in labeling specific mRNA sequences intracellularly, we could also construct them as expressable intracellular sensors for metabolites. Selectivity of binding (for high signal-to-noise ratio) could be further optimized by using multivalent conjugates (32) combined with multipartite modular aptamers (31). Conjugates of other light-up dyes, including those reported to be specific for their cognate aptamers (5,8,9), can be used as well.
In conclusion, we report an apparently general approach (based on two examples) to target aptamers with tightly-bound light-up probes based on TO-dye conjugates to their small molecules. Aptamers binding to small molecules are usually isolated through in vitro selection and amplification procedure that uses affinity selection for small molecule conjugated to the solid phase of affinity column. Conjugation to the TO dye analog instead of an affinity support, may yield light-up aptamer-conjugate couples in all these cases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation (Biophotonics and ITR); M.N.S is Leukemia and Lymphoma Society Scholar. Funding for open access charge: National Science Foundation-ITR.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Rodriguez AJ, Condeelis J, Singer RH, Dictenberg JB. Imaging mRNA movement from transcription sites to translation sites. Semin. Cell Dev. Biol. 2007;18:202–208. doi: 10.1016/j.semcdb.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertrand EC, Pascal S, Matthias S, Shailesh M, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 3.Bratu DP, Cha BJ, Mhlanga MM, Kramer FR, Tyagi S. Visualizing the distribution and transport of mRNAs in living cells. Proc. Natl Acad. Sci. USA. 2003;100:13308–13313. doi: 10.1073/pnas.2233244100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sparano BA, Koide K. Fluorescent sensors for specific RNA: a general paradigm using chemistry and combinatorial biology. J. Am. Chem. Soc. 2007;129:4785–4794. doi: 10.1021/ja070111z. [DOI] [PubMed] [Google Scholar]
- 5.Babendure JR, Adams SR, Tsien RY. Aptamers switch on fluorescence of triphenylmethane dyes. J. Am. Chem. Soc. 2003;125:14716–14717. doi: 10.1021/ja037994o. [DOI] [PubMed] [Google Scholar]
- 6.Sparano BA, Koide K. A strategy for the development of small-molecule-based sensors that strongly fluoresce when bound to a specific RNA. J. Am. Chem. Soc. 2005;127:14954–14955. doi: 10.1021/ja0530319. [DOI] [PubMed] [Google Scholar]
- 7.Sando S, Narita A, Aoyama Y. Light-up Hoechst-DNA aptamer pair: generation of an aptamer-selective fluorophore from a conventional DNA-staining dye. ChemBioChem. 2007;8:1795–1803. doi: 10.1002/cbic.200700325. [DOI] [PubMed] [Google Scholar]
- 8.Sando S, Narita A, Hayami M, Aoyama Y. Transcription monitoring using fused RNA with a dye-binding light-up aptamer as a tag: a blue fluorescent RNA. Chem. Commun. 2008:3858–3860. doi: 10.1039/b808449a. [DOI] [PubMed] [Google Scholar]
- 9.Constantin TP, Silva GL, Robertson KL, Hamilton TP, Fague K, Waggoner AS, Armitage BA. Synthesis of new fluorogenic cyanine dyes and incorporation into RNA fluoromodules. Org. Lett. 2008;10:1561–1564. doi: 10.1021/ol702920e. [DOI] [PubMed] [Google Scholar]
- 10.Grate D, Wilson C. Laser-mediated, site-specific inactivation of RNA transcripts. Proc. Natl Acad. Sci. USA. 1999;96:6131–6136. doi: 10.1073/pnas.96.11.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee LG, Chen CH, Chiu LA. Thiazole orange: a new dye for reticulocyte analysis. Cytometry. 1986;7:508–517. doi: 10.1002/cyto.990070603. [DOI] [PubMed] [Google Scholar]
- 12.Nygren J, Svanvik N, Kubista M. The interactions between the fluorescent dye thiazole orange and DNA. Biopolymers. 1998;46:39–51. doi: 10.1002/(SICI)1097-0282(199807)46:1<39::AID-BIP4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 13.Privat E, Melvin T, Asseline U, Vigny P. Oligonucleotide-conjugated thiazole orange probes as “light-up” probes for messenger ribonucleic acid molecules in living cells. Photochem. Photobiol. 2001;74:532–541. doi: 10.1562/0031-8655(2001)074<0532:octopa>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 14.Svanvik N, Westman G, Wang D, Kubista M. Light-up probes: thiazole orange-conjugated peptide nucleic acid for detection of target nucleic acid in homogeneous solution. Anal. Biochem. 2000;281:26–35. doi: 10.1006/abio.2000.4534. [DOI] [PubMed] [Google Scholar]
- 15.Privat E, Asseline U. Synthesis and binding properties of oligo-2′-deoxyribonucleotides covalently linked to a thiazole orange derivative. Bioconjugate Chem. 2001;12:757–769. doi: 10.1021/bc0100051. [DOI] [PubMed] [Google Scholar]
- 16.Thompson M, Woodbury NW. Fluorescent and photochemical properties of a single zinc finger conjugated to a fluorescent DNA-binding probe. Biochem. 2000;39:4327–4338. doi: 10.1021/bi991907g. [DOI] [PubMed] [Google Scholar]
- 17.Carreon JR, Mahon KP, Kelley SO. Thiazole orange-peptide conjugates: sensitivity of DNA binding to chemical structure. Org. Lett. 2004;6:517–519. doi: 10.1021/ol0362818. [DOI] [PubMed] [Google Scholar]
- 18.Fechter EJ, Olenyuk B, Dervan PB. Sequence-specific fluorescence detection of DNA by polyamide-thiazole orange conjugates. J. Am. Chem. Soc. 2005;127:16685–16691. doi: 10.1021/ja054650k. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Rando RR. Specific binding of aminoglycoside antibiotics to RNA. Chem. Biol. 1995;2:281–290. doi: 10.1016/1074-5521(95)90047-0. [DOI] [PubMed] [Google Scholar]
- 20.Tok JBH, Cho J, Rando RR. Aminoglycoside hybrids as potent RNA antagonists. Tetrahedron. 1999;55:5741–5758. [Google Scholar]
- 21.Carothers JM, Oestreich SC, Szostak JW. Aptamers selected for higher-affinity binding are not more specific for the target ligand. J. Am. Chem. Soc. 2006;128:7929–7937. doi: 10.1021/ja060952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sassanfar M, Szostak JW. An RNA motif that binds ATP. Nature. 1993;364:550–553. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen T, Pei R, Stojanovic M, Lin Q. An aptamer-based microfluidic device for thermally controlled affinity extraction. Microfluid Nanofluid. 2008 published online on July 11, 2008. DOI: 10.1007/s10404-008-0322-4. [Google Scholar]
- 24.Mulder A, Huskens J, Reinoudt DN. Multivalency in supramolucular chemistry and nanofabrication. Org. Biomol. Chem. 2004;2:3409–3424. doi: 10.1039/b413971b. [DOI] [PubMed] [Google Scholar]
- 25.Stojanovic MN, Landry DW. Aptamer-based colorimetric probe for cocaine. J. Am. Chem. Soc. 2002;124:9678–9679. doi: 10.1021/ja0259483. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Jiang Y, Zhou C, Fang X. Aptamer-based ATP assay using a luminescent light switching complex. Anal. Chem. 2005;77:3542–3546. doi: 10.1021/ac050165w. [DOI] [PubMed] [Google Scholar]
- 27.Li B, Wei H, Dong S. Sensitive detection of protein by an aptamer-based label-free fluorescing molecular switch. Chem. Commun. 2007:73–75. doi: 10.1039/b612080f. [DOI] [PubMed] [Google Scholar]
- 28.Pei R, Stojanovic MN. Study of thiazole orange in aptamer-based dye-displacement assays. Anal. Bioanal. Chem. 2008;390:1093–1099. doi: 10.1007/s00216-007-1773-2. [DOI] [PubMed] [Google Scholar]
- 29.Collett JR, Cho EJ, Ellington AD. Production and processing of aptamer micorarrays. Methods. 2005;37:4–15. doi: 10.1016/j.ymeth.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 30.Nutiu R, Li Y. Aptamers with fluorescence-signaling properties. Methods. 2005;37:16–25. doi: 10.1016/j.ymeth.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 31.Stojanovic MN, Kolpashchikov DM. Modular aptameric sensors. J. Am. Chem. Soc. 2004;126:9266–9270. doi: 10.1021/ja032013t. [DOI] [PubMed] [Google Scholar]
- 32.Michael K, Wang H, Tor Y. Enhanced RNA binding of dimerized aminoglycosides. Bioorg. Med. Chem. 1999;7:1361–1371. doi: 10.1016/s0968-0896(99)00071-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.