Abstract
We introduce a technology for the rapid identification and sequencing of conserved DNA elements employing a novel suspension array based on nanoliter (nl)-reactors made from alginate. The reactors have a volume of 35 nl and serve as reaction compartments during monoseptic growth of microbial library clones, colony lysis, thermocycling and screening for sequence motifs via semi-quantitative fluorescence analyses. nl-Reactors were kept in suspension during all high-throughput steps which allowed performing the protocol in a highly space-effective fashion and at negligible expenses of consumables and reagents. As a first application, 11 high-quality microsatellites for polymorphism studies in cassava were isolated and sequenced out of a library of 20 000 clones in 2 days. The technology is widely scalable and we envision that throughputs for nl-reactor based screenings can be increased up to 100 000 and more samples per day thereby efficiently complementing protocols based on established deep-sequencing technologies.
INTRODUCTION
Identification of conserved DNA fragments in poorly characterized DNA samples (e.g. metagenomic or genomic libraries) is a challenge (1,2). We introduce a novel technology based on nl-reactors for screening of large libraries. The technology was adapted to the identification of conserved sequence motifs in the genome of cassava and we argue that any microbial library can be screened in this way. As key steps are based on well-established and robust techniques (e.g. DNA amplification by microbial growth, PCR and Sanger sequencing) the protocol can be easily extended to other applications.
Fishing for conserved DNA motifs
A prerequisite for the successful sequencing of an individual DNA molecule out of a mixture of DNA molecules is the dilution of the DNA molecules down to a level at which single molecules are obtained. Afterwards these single molecules are arrayed in a fashion guaranteeing that no cross-contamination between samples occurs. One of the most-efficient methods for compartmentalization of single DNA molecules is to clone them into replicating elements (e.g. plasmids, fosmids or BACs) (3–5) and to transform microbial hosts with these. By doing so, single DNA molecules are arrayed in a very small volume which in the case of single Escherichia coli cells, for instance, is in the order of a few femtoliters. The DNA is then rapidly amplified upon growth of the clones. However, in order not to mix the clones and therefore also the genetic information during growth, a secondary array for monoseptic (i.e. comprising offspring of one clone only) cell cultivation has to be employed. Such arrays are typically agar plates providing convenient means for monoseptic growth of single cells on solid support (6,7).
However, processing and handling of larger quantities of plates employed for colony expansion and picking is rather laborious and requires a highly automated environment (5). Furthermore, agar plates cannot be used as reaction vessels for in vitro DNA manipulations (e.g. PCR or sequencing). Thus, cells have to be transferred into another array, typically a microtiter plate, thereby slowing down the process and requiring additional equipment. As a result, processing of 100 000 clones per day is probably already close to the practicable limit for most screening applications (8) employing microbial clones. This is one of the reasons why novel technologies (1,9) from within the field of ultra-high-throughput screening (HTS) do not rely on the arraying of single molecules in cells anymore but employ immobilization on surfaces [e.g. microbeads (10,11) or microchips (12)] or in microemulsions (13–17) as main compartmentalization mechanisms.
Still, single molecule arraying in cells has a number of advantages: First, error rates during amplification of the DNA by cell growth are much lower than in vitro (5). Second, the information content per array element, i.e. the length of the screened fragment is much larger than in the competing in vitro systems (3). Third, by the simple application of antibiotic selection pressure, array elements with contents (transformed clones) can be very efficiently separated from empty ones (i.e. clones that do not contain a DNA fragment) (6). Fourth, amplification of certain sequences such as repeating regions requires specific conditions in vitro (9) whereas cells generally provide an appropriate environment for polynucleotide amplification probably with the exception of a few cases where coincidently toxic gene products are synthesized (18). Fifth, cells can also be used for functional screenings, i.e. for screenings where first the phenotype is screened and only clones featuring the desired properties are sequenced (19).
We demonstrate a PCR-based method for HTS of microbial DNA libraries at screening rates of at least 20 000 samples per run. This requires the combination of conventional cloning and bacterial library construction methods with high-throughput technologies for thermocycling and hit-candidate isolation. The core of the technology is the use of hydrogel microcarriers that are produced by laminar jet break-up technology and serve as growth and reaction compartments. The microcarriers have a diameter of ∼400 μm and a volume of ∼35 nl and are subsequently referred to as nl-reactors. The key feature of nl-reactors is their capability to retain high-molecular-weight components such as cells, plasmid constructs or PCR products while at the same time allowing low-molecular-weight components (i.e. nutrients, or mono- and oligonucleotides for PCR) as well as some proteins (in this case DNA polymerase) to continuously access and leave the reactor (see Figure 1). Due to these properties, PCR products are accumulated within the nl-reactors during thermocycling and positive samples can be straightforwardly isolated on the basis of the fluorescence-intensity of nl-reactors after staining of the embedded double-stranded DNA.
Figure 1.
nl-Reactors’ characteristics. nl-Reactors consist of a highly thermostable hydrogel and have a volume of 35 nl (diameter of 400 μm). The porosity of the capsule allows free diffusion of low molecular compounds whereas high-molecular-weight structures cannot penetrate. Embedded E. coli cells can therefore feed on externally provided nutrients and readily expand to colonies that remain immobilized within the nl-reactor. Similarly, larger DNA elements such as plasmids that were liberated from the colonies upon lysis or PCR products formed by thermocycling are also retained, while PCR reagents can continuously access and leave the nl-reactor.
In this article, we report a sequence-based screening of a cassava (Manihot esculenta) shot-gun library in E. coli for identification of microsatellites for gene-marker development (see Figure 2). An equivalent of ∼8 million base pairs was screened within 2 days and it can be foreseen that the method can be readily adapted to uncover the diversity of gene sequences sharing conserved motifs, e.g. within non-sequenced organisms and environmental libraries. The process is easy to scale-up and we envisage that throughput can be easily increased further to 200 million bases or more per day.
Figure 2.
Flow chart—high throughput colony PCR screening in nL-reactors.
MATERIALS AND METHODS
Unless mentioned otherwise, media and media components were obtained from Fluka (Buchs, Switzerland).
Preparation of cell suspension in alginate and production of nl-reactors
Two 40 μl aliquots of electrocompetent E. coli Top10 (F- mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG, Invitrogen, Carlsbad, CA) harboring plasmid pMMBs207–Km14-GFPc (20) with a gfp-mut2 gene constitutively expressed from an unregulated Ptac promoter were transformed with 2 µl of ligation mixture of a cassava library (gDNA digested with Bsp143I (Fermentas International, Burlington, Canada), size selected (350–700 bp) and ligated into a commercially BamHI-digested and dephosphorylated pUC19 vector (Fermentas International, Burlington, Canada). Transformation was carried out with a Genepulser (Biorad, Hercules, CA) at 1.25 kV in a 1 mm cuvette. Cells were incubated directly after the pulse in 1 ml SOC medium (30 min; 37°C) and centrifuged (5 min; 5000 g) in order to remove non-transformed plasmid DNA. The cell-pellet was resuspended in 1 ml SOC medium supplemented with antibiotics (30 mg/l chloramphenicol; 100 mg/l ampicillin), incubated (45 min; 37°C) and centrifuged (5 min; 5000 g). The supernatant was discarded. The cell-pellet was resuspended in 20 ml salt solution (9 g/l NaCl, 1 g/l KCl) containing 25 mg autoclaved graphite powder, added to 20 ml of a sterile-filtered 3% alginate solution in a 50 ml screw-top tube, and mixed by gently inverting the tube manually (5 min; room temperature).
The alginate-cell suspension was immediately processed with a laminar jet break-up encapsulator (Nisco Engineering AG, Zurich) with a nozzle of 150 µm in diameter at a frequency of 1050 Hz. The resulting droplets of ∼500 μm diameter (65 nl) were collected in a continuously stirred beaker (120 ml maximal filling volume; 200 r.p.m.) filled with 100 ml hardening solution (50 mM BaCl2). The nl-reactors were allowed to mature for 30 min which caused the gel-capsules to shrink to diameters of ∼400 µm (35 nL).
Proliferation of E. coli in nl-reactors and determination of colony diameter
The nl-reactors were recovered from the hardening solution by sieving (100 µm Falcon sieve; BD, Franklin Lakes, NJ) and washed three times in 150 ml of growth-medium (4 g/l Bacto yeast extract; 1 g/l Bacto tryptone; 1 g/l glycerol, 1 mM BaCl2; 10 mM Tris–HCl pH 7.0; 30 mg/l chloramphenicol; 100 mg/l ampicillin). Aliquots of ∼3 g wet nl-reactors were added to Petri dishes containing 30 ml of growth medium. The plates were covered with their lids and incubated for 14 h at 30°C. Afterwards ampicillin was added to the medium to 100 mg/l and the plates were incubated for an additional 1.5 h (37°C). nl-Reactors were recovered from the dishes, sieved, washed three times with 50 ml washing solution (10 mM Tris pH 8.0, 0.1 mM BaCl2; pH 8.0) and kept on ice until processing by Complex Object Parametric Analyzer and Sorter (COPAS; Union Biometrica, Holliston, MA). Later the average colony diameter and cell number was determined by analyzing 30 colonies that had been grown within the nl-reactor by fluorescence microscopy under the assumption that a typical E. coli cell volume is 2 fl.
COPAS analysis for enrichment of monoclonal microcarriers
Monoseptic nl-reactors harboring one colony only were enriched by COPAS sorting employing the ‘Profiler’ software (21). Pulse shape diagram recording was triggered by the opacity signal [threshold >25 AU (arbitrary units); signal gain factor 1.5; measuring range 0–65500 AU]. nl-Reactor size is expressed as time-of-flight [(ToF); gated range 400–750 ToF, arbitrary units]. Fluorescence signals for colony detection were generally recorded at 510 nm, which is the emission maximum of the employed GFP [ex 488 nm, photon multiplier settings (PMT) 800 V; gain factor 1.0; measuring range 0–65 500 AU; peak-profiling was initiated at >20 000 AU; gated range 60 000–65 500]. The COPAS-device was operated at an average frequency of 30 Hz and coincidence settings for dispensing into microtiter plates were adjusted to ‘pure’-mode which guarantees that nl-reactors are only sorted-out if no other reactor is coincidentally within the same droplet. A detailed description of the cell encapsulation, proliferation and isolation procedure employed for enrichment of monoseptic nl-reactors by COPAS, was published elsewhere (21).
In-bead PCR screening of embedded colonies
A total number of 20 000 enriched monoseptical nl-reactors were suspended in a 50 ml Falcon tube in 10 ml of ddH2O. The cells were lyzed by heat (10 min in a water bath at 96°C). The microcarriers were washed once in 50 ml washing solution and three times in 50 mL ddH2O and directly added to a poly(fluoro acrylate) PCR-bag (Welch Fluorocarbon, Dover, NH; 100 × 100 mm; thickness: 50 μm) containing 10 ml PCR reagent [ddH2O containing: 1 M betaine; 5% DMSO; 1 × PCR buffer; 0.1 mM BaCl2; 0.2 mM of each dNTPs; 0.2 μM primer M13F (–21); 0.2 μM primer GA9; 700 U Taq polymerase; Genscript, Piscataway, NJ]. All gas bubbles were removed, the PCR-bag was air-tightly sealed, and cycled on a custom-built flat-screen thermocycler that consisted of a chamber flanked by aluminum plates which contained channels for water circulation for temperature control (32 cycles; 90 s at 96°C; 180 s at 55°C) exerted by two water-baths (Huber Polystat CC3; set to 96°C and 56°C, respectively). Numerically, controlled thermo-switching between the two water cycles was realized with four valves (m&m international, Bedford, UK) controlling the in- and outlet of the water baths and a customized LabView (National Instruments; Austin; TX) program controlling the relay-switch station USB-Erb24 (Measurement Computing Corp., Norton, MA). After cycling, the beads were recovered from the bag by sieving, washed with 50 ml washing solution and 250 ml ddH2O and suspended in a Petri dish with 20 ml washing solution containing 1× SYBR Green I dye (Invitrogen; Carlsbad; CA) in order to stain PCR-products prior to another COPAS analysis. Microscopic pictures were taken by a Zeiss Axio Star Plus fluorescence microscope (Carl Zeiss AG; Göttingen; Germany) using an excitation filter at 488 nm and an emission long-pass filter >520 nm in combination with phase contrast microscopy.
Screening for PCR positive nl-reactors
All nl-reactors recovered after thermocycling were added to the COPAS sample cup and analyzed. Both, object diameter (based on extinction) as well as fluorescence intensity (510 nm) of each nl-reactor, were measured concomitantly by pulse shape analysis (COPAS profiler software). First, the system was calibrated by analyzing 300 nl-reactors in order to set an adaptive fluorescence intensity threshold just above the main, non-fluorescent, population, which was evaluated to be at 780 AU (at 510 nm). Next, gates for a ToF value of 600–1000 AU were applied for dispensing of nL-reactors into a 96-well microtiter plate. For sampling of the control group, the same ToF gates were applied and 48 putatively PCR-negative nl-reactors featuring signal intensities below the adaptive threshold (600–780 AU) were sorted out. All samples were immediately processed further in order to recover the plasmids (see next section).
Plasmid extraction with cetyl trimethylammonium bromide (CTAB)
For plasmid extraction, we adapted a method developed by Allen et al. (22). An aliquot of 90 μl of freshly prepared and pre-warmed (50°C) CTAB DNA extraction buffer [100 mM Tris–HCl (pH 8.0); 1.4 M NaCl; 2.5% w/v CTAB; 0.5% w/v N-lauryl sarcosine] was added to a well containing a single isolated nl-reactor (one reactor per MT-plate well), plates were air-tightly sealed by 8-cap strips, incubated in a PCR cycler (Mastercycler, Eppendorf, 65°C; 45 min) and every 5 min vigorously agitated by hand for 10 s in order to dissolve the nl-reactor. After centrifugation at 3200 g for 30 s, 90 μl of chloroform/isoamylalcohol (24/1) were added and plates were agitated by hand until turbid emulsions were formed in the wells (∼15 s). Plates were centrifuged (30 min at 3200 g) and an aliquot of 33 μl was withdrawn from the aqueous phase, transferred to another microtiter plate, and DNA was precipitated by ethanol (77 μl per well). After overnight incubation at –80°C, samples were centrifuged (45 min; 3220 g; 0°C) and supernatants were discarded prior to washing (100 μl of 70% EtOH, chilled to –20°C and centrifuged for 30 min; 3220 g; 0°C). Once more, supernatants were discarded and the remaining liquid was allowed to evaporate at room temperature.
Insert amplification by PCR
PCR reagents [25 µl comprising: 1× PCR Buffer, 200 μM of each dNTP, 200 nM of primer M13F(–43) and M13R(+86), 1.75 U Taq DNA polymerase] were added to the dried pellet and the plasmid inserts were amplified by PCR (denaturation at 95°C for 1 min, then 40 cycles of: 95°C for 30 s, 55°C for 30 s, 72°C for 1 min, and final elongation: 72°C for 2 min). After cycling, 5 µl of each sample were loaded on a 1.5% agarose gel in order to determine the number of PCR-amplified fragments per sample, the insert size as well as the approximate PCR product concentration. All samples yielding a single band of at least 100 ng of DNA on the gel (equivalent to 20 ng/μl concentration in the well) were sent out for Sanger sequencing with a M13 reverse primer (GATC Biotech, Konstanz, Germany).
Polymorphism test
Clone-redundancy was tested with the DNA Star Software (DNAstar inc., Madison, WI). Default assembly parameters were used: match size: 12 bp; minimum match percentage: 80%; minimum sequence length 100 bp; maximum added gaps per kb in contig: 70 bp; maximum added gaps per kb in sequence: 70 bp; last group considered: 2; gap penalty: 0.00; gap length penalty: 0.70.
Only sequences with a match to the screening primer of 10 bases or more were used to subsequently design microsatellite markers. Next, PCR primers flanking the CT-rich region were designed by the primer-3 software; http://fokker.wi.mit.edu/primer3/input.htm). Amplicon length polymorphism of microsatellite markers was tested by PCR on seven different cassava cultivars (91/02322; TMS60444; 95/0306; 98/0002; TAI-8; PER-183; COL-1505) using FAM-labeled primers. PCRs were performed in a final volume of 20 µl: 1× PCR Buffer, 200 μM each dNTP, 200 nM of each primer, 5 ng template; 1.75 U Taq DNA polymerase; cycling conditions: initial denaturation: 95°C for 1 min; 35 cycles: 95°C for 30 s, 56°C for 30 s, 72°C for 40 s; final elongation: 72°C for 2 min. Fragments were separated by capillary electrophoresis on an ABI 3700 (Applied Biosystems; Foster City; CA) and analyzed by the GeneMarker (Softgenetics; State College; PA) software. Polymorphic information content (PIC) was calculated according to Anderson et al. (23).
RESULTS
The objective of this study was the detection and sequence determination of specific DNA-motifs from a genomic cassava library using an essentially freely scalable approach employing gel-like, suspended nl-reactors as reaction compartments.
Cell encapsulation and proliferation in nl-reactors
The feasibility of the approach was investigated for a genomic cassava library (average insert length 350–700 bp) in E. coli. The sample throughput was monitored over all process steps (see Table 1). Freshly transformed cells synthesizing a green fluorescent protein (GFP) from Aequorea victoria were embedded into nl-reactors (660 000 reactors total; average degree of occupation with colonies 10.8%) and propagated from single cell status to colonies of ∼50 000 cells. This was confirmed manually by fluorescence microscopy. Next, 20 000 monoseptic nl-reactors were isolated by COPAS-sorting (21). During this step, the number of reactors containing no or more than one colony, and reactor multiplets (24), inevitably formed during nl-reactor production, was reduced to a minimum (Table 1).
Table 1.
Sample flow over all process steps
| Step | Processed nl-reactors | Selected events | Array format |
|---|---|---|---|
| Enrichment | 193 000a | 20 000b | Suspension |
| PCR and hit selection | 20 000 | 66c | Suspension |
| Plasmid isolation | 66 | 52d | 96-well plate |
| Insert sequencing | 52 | 52e | 96-well plate |
| Polymorphic microsatellites | 37f | 11g | 96-well plate |
Overview over sample streams during HTS for novel SSRs (short sequence repeats) in a cassava genomic library.
aThe subset of the 660 000 empty and colonized nl-reactors that was analyzed and sorted in order to isolate 20 000 monoseptic nl-reactors; colony diameter of 32 µm ± 7 µm.
bSorted and monoseptically enriched nl-reactors (still containing ∼1 to 2% polyclonal reactors and roughly 2% multiplets) (21).
cnl-reactors displaying fluorescence intensity >780 AU dispensed into microtiter plate.
dPlasmid extraction confirmed by PCR.
eSanger sequencing.
fSequences matching the screening primer by 10 or more bases (eight sequences were redundant and not used for polymorphism studies).
gBased on polymorphism tests employing seven cassava cultivars (for details see text).
Highly parallelized nl-PCR and PCR-product detection
All 20 000 monoseptic nL-reactors were pooled in one reaction compartment and subjected to 32 rounds of two-step thermocycling with a vector-specific forward primer and a reverse primer with specificity for the desired SSR-sequences, i.e. poly(CT) repeats (see Supplementary Figure 1). After thermocycling, the bead population was stained with the DNA double-strand specific dye SYBR Green I (see Figure 3) and 66 fluorescent reactors were isolated in another COPAS run (Figure 4).
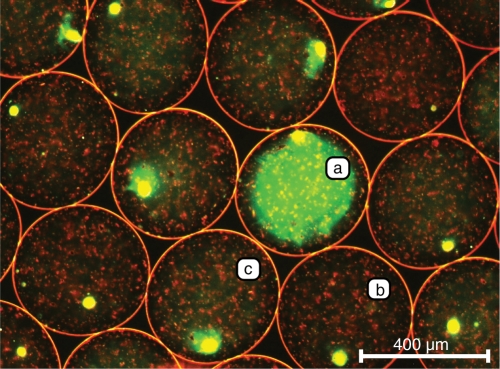
Figure 3.
Monoseptic nl-reactors after two-step PCR and staining of double-stranded DNA with SYBR Green I. Double-stranded DNA entangled in nl-reactors was stained with SYBR Green I dye and fluoresces in a green-yellowish color. (a) nl-reactor filled with fluorescent PCR product. (b) nl-reactor without PCR product, only the SYBR Green I dye binding to the DNA of the lysed colonies fluoresces, though small coronas around the colony debris might develop (c). The small reddish spots within the reactors are graphite particles (see ‘Materials and Methods’ section).
Figure 4.
nl-Reactor analysis by COPAS after thermocycling. nl-Reactor fluorescence (excitation: 488 nm; emission: 510 nm) and size (indexed by ToF) were COPAS-analyzed and 66 reactors above an adaptive fluorescence threshold of 780 AU (dotted line) were isolated. Due to apparatus specific limitations of the COPAS sorter, not all but ∼80% of all reactors above the dotted line could be isolated. In the region between 600 (solid line) and 780 AU (dotted line), 48 nl-reactors were randomly collected and served later as a control. The number of nl-reactors containing inserts that matched to the SSR-specific primer by at least 10 bases (red circles) amounted on 56% above the threshold and on 17% only in the control. Similarly, 23% of all inserts in the putative positive fraction yielded a match of <10 bp (blue triangle) while in the control 56% belonged to that class. nl-Reactors from which plasmid isolation and insert amplification by PCR did not succeed after liquefaction of the gel-carrier are labeled by an ‘X’. Signals beyond TOF = 900 AU stem from nl-reactors with multiple volumes (about 2% of the total) that have not been completely eliminated by COPAS during the first enrichment of monoseptic nl-reactors.
Identification of SSRs of at least 10 bp
After isolation of all 66 putatively positive nl-reactors (fluorescent intensity >780 AU) and dispersion into a micro-titer plate, the plasmid content of the reactors was isolated and presence or absence of the plasmid, presumably harboring the SSR sequence, was verified by amplification of the insert with PCR using two vector-specific primers (see Supplementary Figure 1). Length and quantity of the PCR products was determined and the results indicated that 52 of the 66 reactors (79%) carried a single PCR-product in sufficient quantity (>100 ng) to allow sequencing (Supplementary Figure 1). The sequencing results indicated that 37 plasmids (71%) showed a match of at least 10 bp between sequence and SSR-specific primer, which was defined as a criterion for the presence of a microsatellite. The 15 remaining nl-reactors carried an insert with less than 10 matching bases (4 matches: 6, 5 matches: 1, 7 matches: 3, 8 matches: 3, 9 matches: 2) while of the 37 sequences matching by 10 or more, eight were redundant. In summary, the procedure delivered thus 29 potentially unique and useful SSR markers (Table 2) out of an original library of 20 000 monoseptically enriched nl-reactors. These sequences were then subjected to additional polymorphism studies in seven cassava cultivars.
Table 2.
Sequencing results of all clones featuring a primer match of at least 10 bases
| SSR matcha | Integrated signal | Sensor fragmentb | Sequence lengthc | PICd | Subgroups of Cultivarse | |
|---|---|---|---|---|---|---|
| # | [bp] | [AU] | [bp] | [bp] | [–] | [#] |
| 1 | 10 | 811 | 59 | 245 | 0.00 | 7 |
| 2 | 10 | 837 | 269 | 325 | 0.41 | 2; 5 |
| 3 | 10 | 837 | 322 | 338 | 0.00 | 7 |
| 4 | 10 | 978 | 133 | 308 | 0.00 | 7 |
| 5 | 10 | 808/856* | 133 | 174 | 0.00 | 7 |
| 6 | 11 | 789 | 396 | 546 | 0.00 | 7 |
| 7 | 11 | 806 | 59 | 418 | 0.00 | 7 |
| 8 | 11 | 890 | 171 | 504 | 0.00 | 7 |
| 9 | 11 | 940/840/786* | 133 | 236 | 0.00 | 7 |
| 10 | 12 | 802 | 117 | 397 | 0.00 | 7 |
| 11 | 12 | 818 | 200 | 222 | 0.41 | 2; 5 |
| 12 | 12 | 821 | 149 | 610 | 0.00 | 7 |
| 13 | 12 | 825 | 133 | 277 | 0.49 | 3; 4 |
| 14 | 12 | 835 | 124 | 261 | 0.00 | 7 |
| 15 | 12 | 836 | 313 | 384 | 0.00 | 7 |
| 16 | 12 | 863 | 168 | 384 | 0.73 | 3; 1; 1; 1; 1 |
| 17 | 12 | 897 | 149 | 323 | 0.00 | 7 |
| 18 | 12 | 910 | 112 | 261 | 0.00 | 7 |
| 19 | 12 | 882/885/784/861* | 104 | 332 | 0.00 | 7 |
| 20 | 13 | 842 | 118 | 263 | 0.00 | 7 |
| 21 | 13 | 919 | 210 | 280 | 0.78 | 1; 1; 1; 2; 2 |
| 22 | 13 | 1047 | 157 | 416 | 0.41 | 2; 5 |
| 23 | 13 | 909/654/793* | 86 | 315 | 0.00 | 7 |
| 24 | 14 | 815/871/696* | 117 | 348 | 0.41 | 2; 5 |
| 25 | 14 | 823/607* | 117 | 420 | 0.00 | 7 |
| 26 | 15 | 957 | 127 | 237 | 0.65 | 2; 2; 3 |
| 27 | 15 | 1043 | 117 | 462 | 0.41 | 2; 5 |
| 28 | 17 | 908 | 163 | 262 | 0.69 | 1; 1, 2; 3 |
| 29 | 17 | 957 | 157 | 283 | 0.86 | 1; 1; 1; 1; 1; 1; 1; |
aNumber of base pairs matching with the sensing primer.
bLength of the sensor fragment employed for identification of PCR positive clones expressed as the distance of the M13 forward primer to the location at which the screening-primer putatively bound to the insert.
cThe insert was sequenced employing a vector specific M13 reverse primer.
dPIC = polymorphic information content; polymorphisms were tested in seven different cassava cultivars.
eTotal number of cultivars carrying the same microsatellite allele.
*Redundant clones
Bold font: polymorphic microsatellite markers; primers and sequences are listed in Supplementary Data 1.
Within the set of the 48 randomly collected nl-reactors with a fluorescence intensity below the adaptive threshold (i.e. collected from the interval between 600 and 780 AU, Figure 4) that served as a control, plasmid isolation and insert amplification succeeded in 35 cases (73%), and of these only eight clones (23% of 35) had a primer match of 10 or more bases while the remaining 27 clones had a match of nine or less. The results from this control group indicate that tailoring of the adaptive threshold is important in order to obtain a high ratio of nl-reactors featuring a one-to-one ratio between fluorescence intensity and primer match.
Design of microsatellite markers and polymorphism studies
The sequence information obtained from all 29 clones (fluorescence intensity >780 AU) featuring a primer match of 10 or more was utilized for the design of primers for polymorphism studies (Table 2). Generally, a sufficiently large number of bases could be extracted from the data to allow the design of primers likely to target a unique address in the poorly characterized genome of cassava (25).
The polymorphic information content (PIC), a value commonly used in population studies as a measure of genetic marker polymorphism, was then determined in seven cassava cultivars (see Supplementary Figure 1). In total, 11 high-quality microsatellites with a PIC of more than zero had thus been isolated (Table 2). Several microsatellite primer pairs (2, 11, 22, 24 and 27) were polymorphic in only two loci and therefore resulted in a PIC of 0.41. Samples 13, 16, 21, 26 and 28, led to the isolation of markers with an increasing PIC while one marker (sample 29) was found to be indeed polymorph in all cultivars and is therefore rather valuable for population studies.
Thus, 29 individual CT-containing clones had been isolated (Table 2) from a sequence space of at least 7.8 Mbp (see ‘Discussion and conclusions’ section for the rationale). This suggests an SSR frequency of one in 270 kb. This frequency is well within the range of the CT-repeat abundances typically found in higher plants (one in 290 kb) (26). The overall efficiency of the protocol from PCR-product-sensing to polymorphism-isolation is 11 in 66 (17%) which is comparable to a process efficiency of ∼20% generally found in the field of microsatellite marker development (27).
In the sampling window below the adaptive intensity threshold, eight sequences still had a primer match of 10 or more. However, in only four cases, primer match regions were located at a sufficient distance from the multiple cloning site of the vector to enable the straightforward design of primers for polymorphism studies in cassava. Based on the PIC-values, however, none of these microsatellites was of sufficient quality to serve as a marker for future polymorphisms studies in cassava.
Interestingly, the average of the polymorphic information content (PIC) as well as the percentage of polymorphic sequences therefore increased with an increasing number of bases in the insert matching with the screening primer.
DISCUSSION AND CONCLUSIONS
nl-Reactors were employed in a 3D suspension array for the compartmentalized growth of an E. coli library starting from single clones and directly afterwards for a PCR-based colony screening. Large numbers of both, colonized nl-reactors and post-PCR samples were analyzed and sorted by COPAS and the desired fractions of monoseptically inoculated nl-reactors and PCR-positive samples were directly isolated. Furthermore, plasmids identified on the basis of the PCR signal could be easily recovered from the nl-reactors and were subsequently subjected to further characterizations, in this case Sanger-sequencing. The entire procedure did not require more than 48 h with an effective hands-on time of ∼10 h. It required only three microtiter plates, in contrast to current protocols for microsatellite identification which are substantially longer and more expensive (cells are plated on solid support followed by individual PCR screenings or colony hybridizations).
The longest distance between the vector-specific primer and an insert-sequence that was complementary to the screening primer was 391 bp (see Table 2). This suggests that during PCR at least 391 bases of each of the 20 000 inserts harbored by the screened nl-reactors were scanned for regions matching the screening primer. Therefore, we reason that at least 7.8 Mbp were sampled during analysis of 20 000 colonies. However, as PCR in solution can be employed for amplification of fragments of several thousand base pairs, we believe that the length of the screened fragments can be improved further.
If compared to the classical approaches with two-dimensional arrays (i.e. microtiter plates or agar plates), the volumes employed here for 3D-arrays for cell expansion (600 ml) and PCR (10 ml), were rather low and could easily be scaled up by a factor of at least 10 without any substantial change of the technology. Furthermore, all samples can be handled in one compartment during each single step in the entire screening protocol, which greatly facilitated the required manipulations such as addition of reagents or incubations. For that reason, no sophisticated robotics had to be employed.
The time-limiting step of the protocol is the standardization of the library after growth by COPAS sorting, which proceeds at a frequency of approximately 30 Hz. From our experience, COPAS devices can be operated for at least seven hours during an 8-h working shift. Still, throughput for this step will remain limited to roughly 750 000 events per shift. All other steps that are carried out in suspensions (nl-reactor production, cell growth, suspension PCR, and isolation of PCR-positives) require approximately another 2 days. Under the conservative assumption that inserts of 2 kb could be screened during PCR and assuming further that roughly 15% of all synthesized nl-reactors contain single colonies (as it was the case in this example), the total number of screened bases could thus easily amount to approx. 200 Mbp per day.
Another obvious measure for further increasing the throughput is of course to replace COPAS-sorting by another technology such as fluorescence-assisted cell sorting (FACS). Sorting particles by FACS will however have to provide a volume in the lower picoliter (pl)-range and such particles can not be reliably produced by the laminar jet technology that was employed for capsule synthesis here. However, we are currently working on the development and standardization of the technologies required for the synthesis of hydrogel pl-reactors.
We argue that the sequence-specific amplification combined with high throughput screening makes this technology suitable for other protocols and generally for all cases where ‘rare’ events are sought. Especially screening of gene homologues or miRNA precursors (28,29) in non-sequenced species and particularly metagenomic approaches, e.g. ‘fishing’ for new enzymes in metagenomic libraries, or localization of insertion sequences in mutagenized genomes (e. g. transposons for random mutagenesis) will be greatly facilitated.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Swiss Commission for Technology and Innovation grants (to M.W., M.H. and R.P.). Funding for open access charge: The Swiss Commission for Technology and Innovation (KTI/CTI).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are indebted to the R'equipe program of the Swiss National Science Foundation for generous support in acquiring the COPAS Plus Biosorter. We also are indebted to H. Hilbi for the gift of plasmid pMMB207–Km14-GFPc, to Bernhard Koller for the support with SSR libraries in preliminary tests, and to Maria Domenica Moccia for the gift of plasmid pCT16 used in optimization experiments. Additionally, we would like to thank the four anonymous reviewers for the valuable input.
REFERENCES
- 1.Hutchison CA., III. DNA sequencing: bench to bedside and beyond. Nucleic Acids Res. 2007;35:6227–6237. doi: 10.1093/nar/gkm688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lorenz P, Eck J. Metagenomics and industrial applications. Nat. Rev. 2005;3:510–516. doi: 10.1038/nrmicro1161. [DOI] [PubMed] [Google Scholar]
- 3.Farrar K, Donnison I. Construction and screening of BAC libraries made from brachypodium genomic DNA. Nat. Protoc. 2007;2:1661–1674. doi: 10.1038/nprot.2007.204. [DOI] [PubMed] [Google Scholar]
- 4.Beja O. To BAC or not to BAC: marine ecogenomics. Curr. Opin. Biotechnol. 2004;15:187–190. doi: 10.1016/j.copbio.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 6.Dahlroth SL, Nordlund P, Cornvik T. Colony filtration blotting for screening soluble expression in Escherichia coli. Nat. Protoc. 2006;1:253–258. doi: 10.1038/nprot.2006.39. [DOI] [PubMed] [Google Scholar]
- 7.Jones P, Watson A, Davies M, Stubbings S. Integration of image analysis and robotics into a fully automated colony picking and plate handling system. Nucleic Acids Res. 1992;20:4599–4606. doi: 10.1093/nar/20.17.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim CG, Fujiyama A, Saitou N. Construction of a gorilla fosmid library and its PCR screening system. Genomics. 2003;82:571–574. doi: 10.1016/s0888-7543(03)00174-5. [DOI] [PubMed] [Google Scholar]
- 9.Shendure J, Ji H. Next-generation DNA sequencing. Nat. Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 10.Freeman A, Cohen-Hadar N, Abramow S, Modai-Hod R, Dror Y, Georgiou G. Screening of large protein libraries by the ‘cell immobilized on adsorbed bead’ approach. Biotechnol. Bioeng. 2004;86:196–200. doi: 10.1002/bit.10883. [DOI] [PubMed] [Google Scholar]
- 11.Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc. Natl Acad. Sci. USA. 2003;100:8817–8822. doi: 10.1073/pnas.1133470100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan JB, Chee MS. Highly parallel genomic assays. Nature Rev. 2006;7:632–644. doi: 10.1038/nrg1901. [DOI] [PubMed] [Google Scholar]
- 13.Lee Y-F, Tawfik DS, Griffiths AD. Investigating the target recognition of DNA cytosine-5 methyltransferase HhaI by library selection using in vitro compartmentalisation. Nucleic Acids Res. 2002;30:4937–4944. doi: 10.1093/nar/gkf617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller OJ, Bernath K, Agresti J, Amitai G, Kelly BT, Mastrobattista E, Taly V, Magdassi S, Tawfik DS, Griffiths AD. Directed evolution by in vitro compartmentalization. Nat. Methods. 2006;3:561–570. doi: 10.1038/nmeth897. [DOI] [PubMed] [Google Scholar]
- 15.Leamon JH, Link DR, Egholm M, Rothberg JM. Overview: methods and applications for droplet compartmentalization of biology. Nat. Methods. 2006;3:541–543. doi: 10.1038/nmeth0706-541. [DOI] [PubMed] [Google Scholar]
- 16.Diehl F, Li M, He Y, Kinzler KW, Vogelstein B, Dressman D. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat. Methods. 2006;3:551–559. doi: 10.1038/nmeth898. [DOI] [PubMed] [Google Scholar]
- 17.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen Z, Dewell SB, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cowan D, Meyer Q, Stafford W, Muyanga S, Cameron R, Wittwer P. Metagenomic gene discovery: past, present, future. Trends Biotechnol. 2005;23:321–329. doi: 10.1016/j.tibtech.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 19.Inglese J, Johnson RL, Simeonov A, Xia M, Zheng W, Austin CP, Auld DS. High-throughput screening assays for the identification of chemical probes. Nat. Chem. Biol. 2007;3:466–479. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- 20.Mampel J, Spirig T, Weber SS, Haagensen JAJ, Molin S, Hilbi H. Planktonic replication is essential for biofilm formation by legionella pneumophila in a complex medium under static and dynamic flow conditions. Appl. Environ. Microbiol. 2006;72:2885–2895. doi: 10.1128/AEM.72.4.2885-2895.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walser M, Leibundgut R, Pellaux R, Panke S, Held M. Isolation of monoclonal microcarriers colonized by fluorescent E. coli. Cytometry Part A. 2008;73A:788–798. doi: 10.1002/cyto.a.20597. [DOI] [PubMed] [Google Scholar]
- 22.Allen GC, Flores-Vergara MA, Krasnyanski S, Kumar S, Thompson WF. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 2006;1:2320–2325. doi: 10.1038/nprot.2006.384. [DOI] [PubMed] [Google Scholar]
- 23.Anderson JA, Churchill GA, Autrique JE, Tanksley SD, Sorrells ME. Optimizing parental selection for genetic linkage maps. Genome. 1993;36:181–186. doi: 10.1139/g93-024. [DOI] [PubMed] [Google Scholar]
- 24.Brandenberger H, Widmer F. Immobilization of highly concentrated cell suspensions using the laminar jet breakup technique. Biotechnol Prog. 1999;15:366–372. doi: 10.1021/bp990033m. [DOI] [PubMed] [Google Scholar]
- 25.Okogbenin E, Marin J, Fregene M. An SSR-based molecular genetic map of cassava. Euphytica. 2006;147:433–440. [Google Scholar]
- 26.Gupta PK, Varshney RK. The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica. 2000;113:163–185. [Google Scholar]
- 27.Squirrell J, Hollingsworth PM, Woodhead M, Russell J, Lowe AJ, Gibby M, Powell W. How much effort is required to isolate nuclear microsatellites from plants? Mol. Ecol. 2003;12:1339–1348. doi: 10.1046/j.1365-294x.2003.01825.x. [DOI] [PubMed] [Google Scholar]
- 28.Terai G, Komori T, Asai K, Kin T. miRRim: a novel system to find conserved miRNAs with high sensitivity and specificity. RNA. 2007;13:2081–2090. doi: 10.1261/rna.655107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohler UWE, Yekta S, Lim LP, Bartel DP, Burge CB. Patterns of flanking sequence conservation and a characteristic upstream motif for microRNA gene identification. RNA. 2004;10:1309–1322. doi: 10.1261/rna.5206304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.